Abstract
The role of short, large or intermediate normal alleles (ANs) of the ataxin-2 gene in generating expanded alleles (EAs) causing spinocerebellar ataxia type 2 (SCA2) is poorly understood. It has been postulated that SCA2 prevalence is related to the frequency of large ANs. SCA2 shows the highest worldwide prevalence in Cuban population, which is therefore a unique source for studying the relationship between the frequency of large and intermediate alleles and the frequency of SCA2 mutation. Through genetic polymorphism analyses in a comprehensive sample (∼3000 chromosomes), we show that the frequency of large ANs in the ataxin-2 gene is the highest worldwide, although short ANs are also frequent. This highly polymorphic population displayed also high variability in the CAG sequence, featured by loss of the anchor CAA interruption(s). In addition, large ANs showed germinal and somatic instability. Our study also includes related genotypic, genealogical and haplotypic data and provides substantial evidence with regard to the role of large and intermediate alleles in the generation of pathological EAs.
Keywords: SCA2, genetic polymorphisms, ataxin-2, Cuba
Introduction
Spinocerebellar ataxia type 2 (SCA2) is a neurodegenerative disease showing an autosomal dominant inheritance pattern. This disorder is caused by a CAG expansion crossing a certain threshold in the coding region of the ataxin-2 gene.1, 2, 3 SCA2 patients exhibit a progressive cerebellar syndrome characterized by ataxic gait, cerebellar dysarthria, dysmetria and dysdiadochokinesia. Normal SCA2 gene/ataxin-2 CAG tracts range from 13 to 31 repeats, the most frequent being that of 22 trinucleotides.4, 5 Ataxic phenotype occurs when the repeat is larger than 34 CAG.6 Triplet repeats between 32–34 fall in the gray zone for penetrance, whereas 37–75 CAG repeats are fully penetrant.6 Only few patients having 32 and 33 CAG repeats have been reported so far, with very late onset – between 50 and 60 years of age.4, 7, 8 Extremely large expansions of 109, 200 and 500 CAG in infants have also been observed,9, 10, 11 but are rarer.
The CAG sequence in the SCA2 locus is cryptic in nature, showing CAA interruptions. Normal SCA2 alleles contain CAA interruptions, whereas the vast majority of expanded alleles are uninterrupted.1, 2, 3 The most common configuration is (CAG)8CAA(CAG)4CAA(CAG)8. It is claimed that the CAA interruptions confer genomic stability to the CAG tracts.12, 13 The absence of interruptions, on the other hand, predisposes the CAG tracts to undergo expansion and eventually to reach the pathological threshold in analogy to the absence of interruptions in the CGG and CAG tracts in fragile X syndrome and SCA1, respectively.12, 13, 14, 15, 16 In a first effort to decipher which factors underlie the prevalence of SCAs, it was found that a high frequency of large normal alleles (ANs) was closely linked to a high prevalence of SCA2 in Caucasians, whereas low large ANs results in a very low prevalence of SCA2 in the Japanese population.17 Later, some authors suggested that it is biased to only use CAG length as a marker for determining the propensity of certain alleles to be predisposed or prone to undergo expansion reaching the pathological range.18 This argument is also supported by the fact that in East India, where SCA2 prevalence is high,19, 20, 21 large ANs are spared and clustered to limited ethnic groups.22 Therefore, the role of large ANs as a possible source of SCA2 expansion and the mechanism by which this might happen is poorly understood. Here, we performed a comprehensive study to determine the frequencies of ANs in the Cuban population and their relationship with the high prevalence of SCA2. We analyzed CAG sizes, sequences, as well as genealogical and microsatellite haplotype data to gain insights into the mechanism underlying the prevalence of SCA2 in Cuba. The main questions were as follows: (1) What is the relative frequency of ANs? (2) Are the frequency of large ANs and the prevalence of SCA2 associated? In answering these questions, we compared our data with other studies performed in large populations. We found a strong relationship between the SCA2 prevalence and the relative frequency of large ANs.
Methods
Design of the study
The study was performed in three phases: (1) a nation-wide screening for SCA, conducted during 2003–2007; (2) a survey for neurological consultations to SCA2 relatives identified in the former phase; and (3) nation-wide recruitment of the general non-SCA2 population (not related healthy subjects), in 2009. The study was conducted by the National Center for the Research and Rehabilitation of the Hereditary Ataxias (CIRAH) in the city of Holguín, which is the main referral center for these conditions and a neurological center of excellence in the country.22 The Ethics Committee of the National Centre for the Research and Rehabilitation of the Hereditary Ataxias approved the research protocol and all studied subjects signed an informed consent form after being explained the purpose and methods of the research.
CAG length estimation by fragment analysis
Peripheral blood leukocytes were extracted using EDTA as anticoagulant, and genomic DNA was isolated using standard methods. Molecular diagnosis was performed to estimate the number of CAG repeats at the SCA2 locus according to the methods described elsewhere.1, 2 CAG length was determined by using Alfexpress II sequencing system and the PCR fragments run with ReproGel high resolution (GE Healthcare, Buckinghamshire, UK). All repeat sizes were compared with the standards, before which repeat size was confirmed by DNA sequence analysis. Also, internal (100 and 300 bp) and external (50–500-step, 50 bp) Alfexpress ladders were used to extrapolate the fragment size. Traces were analyzed using the software Allelelink according to the manufacturer's specifications. In all, 200 ng of genomic DNA from peripheral blood was used in each PCR aimed at somatic mosaicism determination, which was defined by Matsuura et al.23
CAG substructure deciphering
In total, 81 normal chromosomes with 13–31 CAG repeats representing the majority of allelic classes were sequenced. Reactions were performed by using either DAN1–DAN2 or SCA2A–SCA2B (Tib-Mol-Biol, Berlin, Germany) and amplified fragments were excised from the gel using GFX band extraction kit (GE Healthcare). Each allele was sequenced directly in both directions, forward and reverse, using the Thermosequenase Cy5 Dye Terminator Sequencing kit (GE Healthcare) and run as explained above. Sequence data were managed by using Alf win Sequence analyzer 2.10.
Data management and statistical analyses
Availability of a large number of SCA2 and non-SCA2 carrier chromosomes (n=2695) and the general population (GP) (n=80) allowed us to determine the distribution of the number of CAG repeats in ANs at the SCA2 locus. Non-SCA2 carrier chromosomes were defined as the new mutation (NM) group given its familial relationship with SCA2 families, because all of them segregated or were related to SCA2 families. CAG distributions were determined by pooling chromosomes. Genotypes only included healthy individuals. Means, variances, ranges and skewness were determined for the distributions of ANs at the SCA2 locus in the individuals. To perform statistical analyses of the differences in the frequencies of large ANs between Cuban and other populations, we defined large ANs as in Takano et al.17 Thus, for the purposes of this study, all alleles smaller than 22 repeats were defined as short alleles and those longer than 22 CAG repeats were considered as large alleles. Differences in the relative frequencies of the large ANs were analyzed by means of the χ2-test with Yates's correction when necessary. Associations were also assessed using the Fisher's exact test. Data were arranged in excel sheets, managed and analyzed as counts and frequencies. The normality of each distribution was assessed by the Kolmogorov–Smirnov test (K–S). All statistical analyses were performed using the STATISTICA data analysis software system version 6 (StatSoft Inc., 2003, Tulsa, OK, USA). The null hypotheses were rejected at P≤0.05.
Results
Normal genetic polymorphism according to CAG repeat length (13–31 CAG)
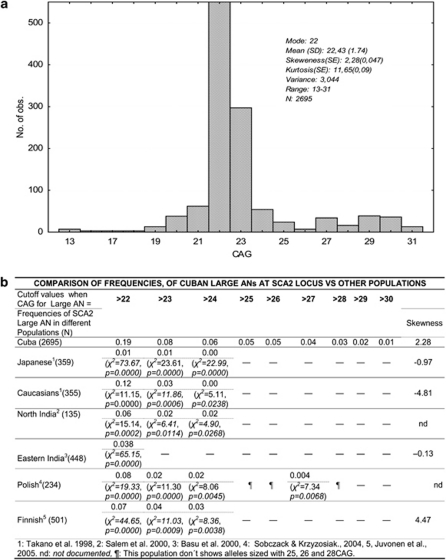
The normal CAG repeat size from 2695 (K–S, P<0.01) chromosomes of the Cuban NM population shows a modal distribution (Figure 1a) (mode and median=22 CAG), with 22 CAG being the most frequent allele (n=2046/2695, 75.92%). This frequency is the lowest reported as compared with other populations: East Indian,24 91% (χ2=51.53, d.f.=1, P=0.000); Polish,18 91.2% (χ2=27.85, d.f.=1, P=0.0000); and Finnish,25 92.81% (χ2=71.62, d.f.=1, P=0.0000). CAG mean was 22.43±0.033; while in the populations previously reported, it was as follows: Japanese,17 21.86; Caucasians,17 22.24; East Indian,24 21.95; Finnish,25 22.2; Canadian,26 22.2; four human populations mixed,27 22.15; and Czech, 22.3.28 The range of the CAG is distributed continuously from 13 to 31 CAG and encompasses almost all the expected allelic classes in this numeric series (17 observed/18 expected) – with a kurtosis of 11.66 and a variance of 3.04 vs 1.21 for other populations worldwide.27 The allele with 13 CAG repeats is exclusively found in the Cuban population and that with 26 CAG in both Cuban and Czech populations.28 The distribution is non-symmetrical (skewness 2.28), with a clear tendency toward the large CAG range. The mode at 22 CAG splits the distribution into short ANs sized from 13 to 21 CAG and large ANs sized from 23 to 31 CAG, showing 7 and 9 allelic classes, respectively (χ2=0.03, d.f.=1, P=0.86) (Figure 1a). Short AN frequencies are as follows: 21 (2.30%), 20 (1.41%), 19 (0.48%), while in the large AN zone those sized with 23 (11.02%), 24 (2.00%), 29 (1.44%), 30 (1.33%), 27 (1.26%) and 25 (0.89%) CAG units are represented more (Figure 1a). Alleles falling in the positive tail (ie, large ANs) are highly over-represented compared with those included in the negative tail (ie, short ANs) (χ2=204.71, d.f.=1, P=0.0000).
Figure 1.
Unexpanded genotype frequency distribution at the ataxin-2 gene in 2695 NM Cuban chromosomes and frequency of large ANs in Cuba vs different populations. (a) CAG distribution at the SCA2 locus of the ANs in the Cuban population related to SCA2 families (NM). The distribution is skewed toward large ANs. The shortest alleles found are those sized 13 and 14 CAG and the largest are 30 and 31 CAG. Alleles with >23 CAG repeats are over-represented over all large and short ANs. (b) Comparison of the CAG size frequency of large ANs in Cuba vs other populations. Allele frequencies in Cuba were grouped by CAG size; frequencies of large normal allele (>22 CAG) and other alleles (≤22 CAG) were compared with the frequency of large ANs in other populations by χ2 or Fisher's exact test. Frequencies were tabulated in a 2 × 2 contingency table with 2 d.f. for comparison. Because the frequency of alleles sized 22 CAG may be higher with respect to both groups, short (<22 CAG) and large alleles (>22 CAG), we also applied a component analysis by χ2 and Fisher's exact test, excluding such alleles, and included in the table only alleles either >22 CAG or <22 CAG. In the table each line shows the frequency of large ANs in each population and the resulting comparison of the frequency of each allele when grouped according to CAG cutoff (ie, >22 CAG, >23 CAG, >24 CAG, and so on). Frequencies of alleles in each region were taken from the literature as shown in the table and the Cuban frequency was determined in the current work.
The exclusive distribution of Cuban large ANs prompted us to compare the frequencies of such alleles with other populations. It is noteworthy that the frequency of large ANs (>22 CAG) was significantly higher in Cubans than in Japanese17 (χ2=73.67, d.f.=1, P=0.0000), Caucasians17 (χ2=11.15, d.f.=1, P=0.0000), North Indians21 (χ2=15.14, d.f.=1, P=0.0002), East Indians24 (χ2=65.15, d.f.=1, P=0.0000) and Polish18 (χ2=19.33, d.f.=1, P=0.0000) (Figure 1b). Extending our cutoff values to >23 CAG or >24 CAG disclosed even more significant differences between our and the aforementioned populations. Curiously, there were populations lacking alleles larger than 25 CAG triplets (Japanese, Caucasians, Indians, except Czechs) and others showing gaps (Polish) in the large AN zone, contrasting with Cubans, who showed a continuous distribution of such alleles.
Intermediate alleles (32 and 33 CAG)
We extended our analysis to include those alleles sized 32 and 33 CAG (intermediate alleles (IAs)), thus enlarging our sample to 2722 chromosomes. Cuban chromosomes carrying 32 and 33 CAG repeats were as frequent in our population (0.55% and 0.44%, respectively) as those with 28 (0.58%) and 31 (0.47%) CAG repeats. The paired comparison between IAs and the frequency of short ANs disclosed highly significant differences in the frequencies of IAs vs those short ANs with sizes ranging from 13 to 18 CAG (IAs: 0.99% vs short ANs: 0.58%, χ2=85.80, d.f.=1, P=0.0000, Fisher's exact test P=0.0000). No differences were observed when the rest of the short ANs with 18, 19 and 21 CAG repeats were added (data not shown). In all, 25 genotypes with IAs were found. All these alleles were found in SCA2 families, that is, the NM population. The genotypic frequencies were 22/32 (n=11/25, 44%); 22/33 (n=7/25, 28%); 23/32 (n=3/25, 12%); 30/32 (n=1/25, 4%); 23/33 (n=1/25, 4%); 33/41 (n=1/25, 4%); and 33/43 (n=1/25, 4%). Only two IAs were associated with age at disease onset at 34 and 45 years, with genotypes of 22/32 CAG and 23/32 CAG, respectively (penetrance 8%) (Table 1).
Table 1. Genotypic frequency of IAs.
| Genotype | Freq (%) | Cases sick (age at onset years) |
|---|---|---|
| 22/32 | 14 | 1 (34 years) |
| 22/33 | 28 | — |
| 23/32 | 12 | 1 (45 years) |
| 30/32 | 4 | — |
| 23/33 | 4 | — |
| 33/41 | 4 | — |
| 33/43 | 4 | — |
Normal genotypic distribution
The genotypic distribution of 836 healthy subjects from the NM group revealed that the most frequent combination was 22/22 CAG (60.88%), followed by 22/23 (13.75%), 23/23 (2.99%), 22/29 (2.15%), 22/30 (2.15%), 22/27(1.91%), 21/21(1.55%) and 20/20 (0.47%). The distribution was skewed toward the positive tail of the distribution, with 28 vs 12 genotypic classes in the negative tail (comparison of number of allelic classes χ2=0.36, d.f.=1, P=0.55) (Supplementary Table S1 and Figure S1). Excluding the central genotype 22/22, we divided the distribution into short (with at least one short AN) and large genotypes (with at least one large AN). Both of these categories were significantly different with regard to the presence of the second allele (ie, short ANs in large genotypes and vice versa) in each group (χ2=288.46, d.f.=1, P=0.0000, Fisher's exact test P=0.0000). Moreover, large genotypes were more enriched, with 22 CAG alleles as compared with the short genotypes (200/572 (35%) vs 20/114 (18%), respectively, χ2=13.24, d.f.=1, P=0.0003, Fisher's exact test P=0.0000) (Table 2). Therefore, the large chromosomes associate preferentially with 22 CAG and with large than with short ANs. We focused on the comparison excluding the 22/23 genotype that accounts for the 42.59% of the large genotypes, but the association yet remained highly significant (155 vs 57, χ2=7.28, d.f.=1, P=0.007, Fisher's exact test P=0.0078).
Table 2. Comparisons of the frequencies of short ANs, large ANs and 22 CAG normal alleles in the short, normal or large genotypes.
| Counts (frequency of alleles) | ||||
|---|---|---|---|---|
| Genotypes (n) | Short ANs | Large ANs | 22 CAG | Total (2n) |
| Short heterozygotes (57) | 78 (0.68)a | 16 (0.14) | 20 (0.17) | 114 |
| Large heterozygotes(286) | 16 (0.02) | 356 (0.62)b | 200 (0.34)c | 572 |
χ2=288.46, d.f.=1, P=0.0000 (short ANs in short heterozygote group vs short ANs in large heterozygote group).
χ2=288.46, d.f.=1, P=0.0000 (large ANs in large heterozygote group vs large ANs in short heterozygote group).
χ2=13.24, d.f.=1, P=0.0003 (22 CAG ANs in each large or short heterozygote groups).
Large ANs in the GP
By active screening, we randomly selected 80 chromosomes in the GP (K–S, P<0.01) (Supplementary Figure S2). The analysis revealed the same combination, with at least 25% (n=20/80) of large ANGP's. Short ANGP's were only sized with 16, 20 and 21 repeats (n=30/80, 28.75%), but were over-represented compared with short ANNM's (n=129/2695 (4.78%), χ2=153.93, d.f.=1, P=0.0000). The paired comparison between the frequencies of short ANGP and large ANGP revealed only marginal differences (n=30/80 (28.75%) vs n=20/80 (25%), χ2=2.91, d.f.=1, P=0.08, Fisher's exact test P=0.06). Variance of the CAG repeats belonging to the GP distribution was 5.31, with a positive kurtosis of 4.44±0.532. Interestingly, we found an expanded allele with 35 CAG repeats without direct evidence of any other pathogenic CAG expansion in the family; this allele was not previously registered during the time of our national screening.5 The overall distribution in GP samples was skewed, although less (1.66±0.26 vs 2.29±0.047) than in the NM samples. Large ANGP's appeared much more frequently than large ANNM's, but did not yield significant differences (n=20/80, 25.00% vs n=536/2695, 19.29% χ2=1.27, d.f.=1, P=0.2604).
CAG>CAA substructure of Cuban ATXN2 alleles with 13–33 CAG repeats
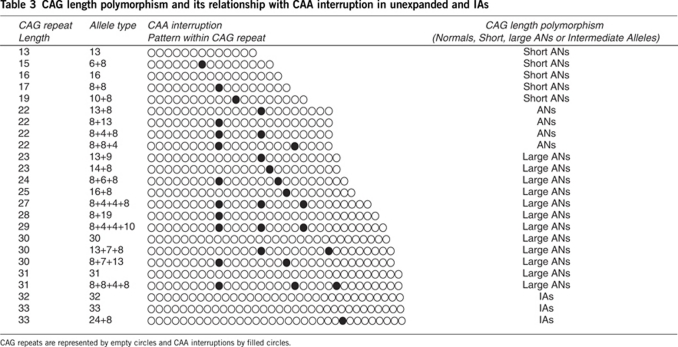
To characterize the allelic diversity of the CAG tract substructure, 81 normal chromosomes were analyzed, encompassing the whole spectrum of Cuban normal CAG alleles (range 13–31 CAG). Of the 36 allelic worldwide classes, 24 were found in the Cuban population (Table 3). Other populations and ethnic groups are much less diverse, for example, Indian29 (14), Yoruba in Ibadan, Nigeria (YRI)30 (11), Polish18(11) and French2 (7) (Supplementary Table S2).
Table 3. CAG length polymorphism and its relationship with CAA interruption in unexpanded and IAs.
Nine new alleles with regard to the CAG/CAA configuration were exclusively linked to the Cuban population and were sized 13, 16, 22, 24, 28, 30 and 31 CAG, respectively: (CAG)13; (CAG)16; (CAG)8CAA(CAG)8CAA(CAG)4; (CAG)8CAA(CAG)6CAA(CAG)8; (CAG)8CAA(CAG)19; CAG30; (CAG)8CAA(CAG)7CAA(CAG)13; (CAG)31; and (CAG)8CAA(CAG)8CAA(CAG)4(CAG)8 (Table 3 and Supplementary Table S2).
Among the short ANs, those sized with 15, 17 and 19 trinucleotides had one CAA interruption, whereas alleles with 13 and 16 units did not contain such interruptions. Four CAA interruption patterns, (CAG)8CAA(CAG)4CAA(CAG)8 or 8+4+8, (CAG)8CAA(CAG)8CAA(CAG)4 or 8+8+4, (CAG)13CAA(CAG)8 or 13+8 and inverted 8+13, were found in alleles with 22 repeats (Table 4). The large AN zone was more enriched in allelic variants with regard to the configuration of the internal CAA interruption 10/20 classes (50%). Alleles with 27, 29 and 31 CAG repeats had three CAA interruptions, alleles with 24 and 30 CAG repeats contained two CAA interruptions, those with 23, 25 and 28 CAG repeats had one CAA interruption, and those with 30 and 31 CAG repeats were uninterrupted (Table 4).
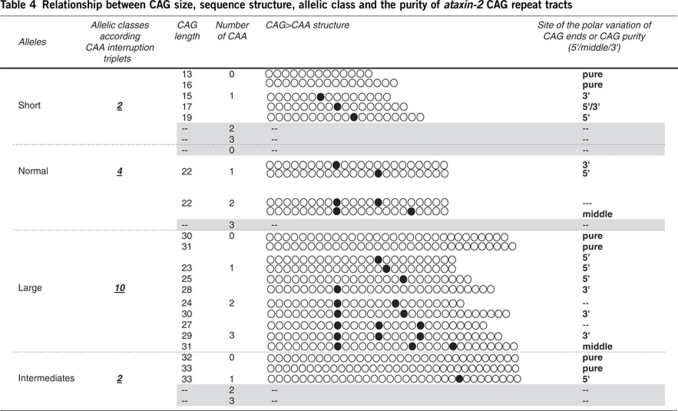
Table 4. Relationship between CAG size, sequence structure, allelic class and the purity of ataxin-2 CAG repeat tracts.
According to the previous criteria stated by Choudhry et al,29 the polar variation of the internal CAG tracts was analyzed. Five alleles showed a continuous pure CAG configuration (alleles: 13, 16, 30, 31 and 33, range 13–33 CAG). In the remaining 16 alleles, the 5′ and 3′ tracts were much more variable (range 6–16 and 8–19, respectively) than the middle tract (range 4–8). Large ANs accounted for the vast majority of the length variability at each end (5′ tract: range 8–16 CAG; 3′ tract: range 8–19 CAG; and middle tract: range 4–8 CAG). On the contrary, short ANs displayed variability only at the 3′ tract (range 6–10 CAG). IAs (32–33 CAG alleles) showed CAG/CAA polymorphism as well. Three of these alleles had uninterrupted CAG tracts. CAA interruptions were frequent in 33 CAG alleles (14 chromosomes), all having the (CAG)24CAA(CAG)8 structure (Table 4).
CAG>CAA, short tandem repeat and CAG length polymorphism analysis in large ANs in SCA2 families
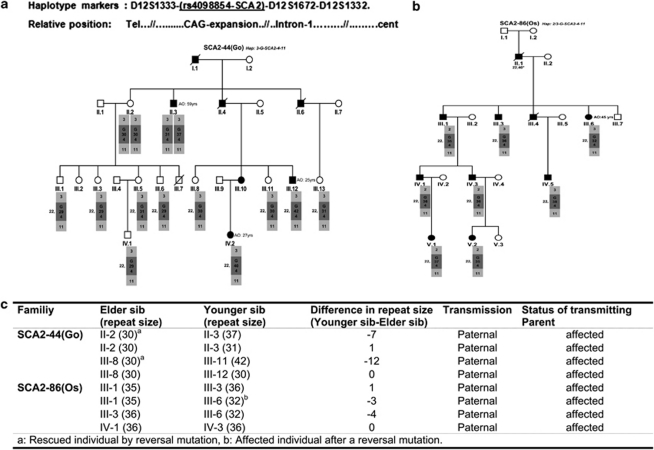
A combined analysis of four genetic polymorphisms, short tandem repeat (STR) haplotype, CAG length, the CAG → CAA change (SNP database, SNP ID No. rs4098854) and sequence architecture was carried out in two SCA2 Cuban families with expanded and large ANs (Figures 2a and b). Marked association between the disease locus and the haplotypic run 3-G-4-11 at D12S1332–(A/G)–D12S1672–D12S1333 loci (allele 3, 202 bp; allele 4, 287 bp; allele 11, 235 bp) was observed. In the case of family 86, the allele 3 at D12S1332 in II-1 expanded and became allele 2 (204 bp). It was then passed with the rest of the truncated haplotype 4-11 to the successive generations IV and V. Homozygotes for the whole haplotype block was a feature in the family SCA2-44 (Go) (individuals II-2 and II-3) (Figure 2a). In the first branch of this pedigree, discrete vertical, sib–ships and horizontal, sib–sib CAG instabilities (±1 CAG) in individuals III-1, III-3, III-5, III-6 and IV-1 were observed (Figures 2a and c). These CAG instabilities reached the peak normality, 31 CAG, in the son (III-5) and grandson (IV-1) of II-2. Furthermore, reverse mutations or contractions to 30 repeats resulted in non-ataxic phenotype in individuals II-2, III-8, III-12 and III-13 of the SCA2-44 (Go) family. Under a similar back mutation, however, the ataxic phenotype was retained with onset at 45 years in the female III-6, belonging to family SCA2-86 (Os) (Figure 2b). All alleles were sequenced and no interruptions were detected in the CAG tracts.
Figure 2.
Haplotype of STR microsatellites around the SCA2 CAG in two pedigrees and analysis of sib–sib instability. STR haplotype segregating into two proven SCA2 families with large ANs. Un/expanded alleles and the STR microsatellite haplotypes are indicated in the boxes (gray). Genotype of the individual II-1 was taken from Santos et al.4 and is marked with asterisk. Age at onset in individuals II-3, III-12, IV-2 and III-6, of pedigrees 44 and 86, respectively, are denoted. (a and b) Families SCA2-44 (Go) and SCA2-86 (Os) of the Cuban kindred. Note that in the first branch of family 44, there are slight instabilities in descendants of the homozygous female II-2 with the haplotype 3-4-11. (c) Analysis of CAG instabilities in both pedigrees in sib–sib pairs. CAG contractions are prominent, leading to back or reversal mutations that rescued 2/5 individuals of the SCA2 phenotype. The contraction in the female III-6 was able to retain the SCA2 phenotype with a mild ataxic course. Note: The normal alleles of the STR have been omitted, and only disease alleles are shown.
Large ANs are somatically unstable
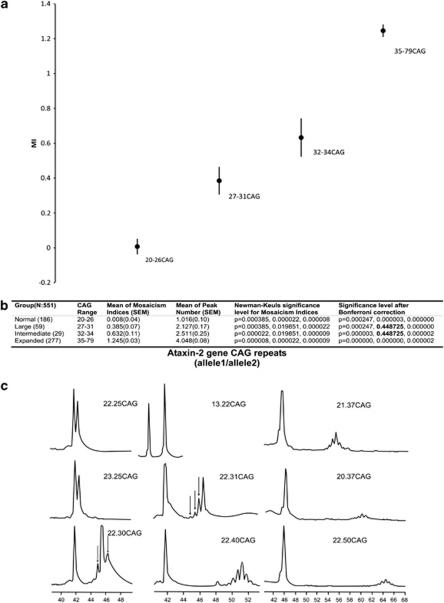
To provide insights into the behavior of large ANs, we analyzed somatic mosaicism in 245 alleles, CAG range: 20–31. Large ANs displayed different morphologies as compared with 22 CAG alleles (Supplementary Figure S3c). Through associative analysis using CAG cutoff ranges and stability criteria according to the fragment morphology, we clearly discriminated two CAG ranges (20–26 CAG and 27–31 CAG) with different stabilities (χ2=159.80, Fisher's exact test P=0.0000, OR: infinitum and ROC curve sensitivity: 100%, specificity: 91%) (Supplementary Figures S3a and b). Furthermore, the comparison disclosed highly significant differences and strong effect of the CAG length in the somatic mosaicism, assessed either as mosaicism index (MI) or as peak numbers (F(1,243)=131.57, P=0.00, ANOVA followed by Bonferroni post hoc test, 20–26 vs 27–31 CAG range, MI±SEM: 0.00±0.016, 0.385±0.028, respectively, P=0.000) (Supplementary Figures S3a and b). According to our current results with regard to the differences between somatic mosaicism and the known phenotypic range of SCA2, we extended the previous comparison to a cohort of 551 alleles with the following ranges: normal (20–26), large (27–31), intermediate (32–34) and expanded (35–79). In addition to the differences in normal CAG repeat ranges, the levels of somatic mosaicism of 32–34 CAG alleles were lower than those observed in the full penetrant expanded alleles (35–79 CAG) (MI±SEM: 0.632±0.11 and 2.51±0.25, respectively, P=0.000) (Figures 3a–c). These results remained significant after adjustment for multiple comparisons. However, the levels of somatic mosaicism for the 27–31 CAG and 32–34 CAG repeat ranges were not different (MI±SEM: 0.385±0.07 and 0.632±0.11, respectively, P=0.448725) (Figure 3b).
Figure 3.
Multiple comparisons of the somatic mosaicism indices harboring 20–79 CAG repeats in the ataxin-2 gene. (a) One-way ANOVA comparison for somatic mosaicism in a cohort of 551 un/expanded alleles. Each range showed differences either in the somatic mosaicism indices or in peak number. (b) Post hoc Newman–Keuls multiple comparisons of the mosaicism indices in the studied sample. Only alleles sized between 27–31 and 32–34 CAG were not different after corrections. Data are presented as mean and standard error of mean (SEM). (c) Somatic mosaicism of unexpanded and expanded pathological (causing SCA2) ATXN2 alleles; arrowheads point to the somatic mosaicism in large ANs.
Discussion
We found that large alleles and IAs are very frequent in the Cuban population. This may be relevant for the understanding of SCA2 origin in Cuba, because though the prevalence of SCA2 in Cuba is the highest worldwide,5 the basis for this31 is poorly understood yet. The frequency of large ANs in Cuba is the highest compared with other populations having a high prevalence of SCA220, 21 and even other populations with a high frequency of such alleles.18 This highly significant frequency of large ANs and other alleles distinct from 22 CAG suggests that the SCA2 locus in Cuba is highly polymorphic. The similar distribution in the sample of non-related chromosomes in the general population, collected for our study, further supports the relative abundance of large ANs in Cuba. Taken together, the genetic variance in the CAG>CAA sequence was also the highest, showing novel alleles situated at the peak of normality (29, 30, 31 CAG; Table 3), completely lacking stability-mediating CAA interruptions (Figure 3). Collectively, the data may indicate a causal relationship between the overall abundance of large ANs with CAG purity and the frequency of expanded SCA2 alleles. It is well known that Cuba has the highest frequency of SCA2 mutation worldwide,5 a fact that supports our hypothesis. It is conceivable that alleles with high normal CAG provide the source of de novo mutations that further contributes to the high SCA2 prevalence in the Cuban population.
Our study also revealed that large ANs were more frequent than short ANs. Previously, it has been stated that longer alleles in the normal range present a higher probability of pathogenic expansion than do shorter alleles.32, 33 Therefore, this makes more likely the contribution of the former group to the onset of de novo mutations. In this state, IAs with 32–35 CAG repeats that may have evolved from large ANs with 23–31 CAG repeats are intermediates, which in successive generations would give rise to full penetrant alleles (37–79 CAG) associated with SCA2, ALS,34 FTDP-U,34 or pure parkinsonism.
Alleles with 27–31 CAG were somatically unstable, similar to expanded alleles (Figures 3a and b), providing significant evidence for the increased instability of large ANs, and pointing to 27 CAG as the threshold for IAs. The high odds ratio and the specificity seen in the 27–31 CAG alleles suggest that instabilities would be associated to 27–31 CAG alleles and are more likely to occur in these rather than in the shorter ones (20–26 CAG alleles). The proposed 27 CAG as the lower threshold for intermediate or indeterminate penetrance alleles is in agreement with the situation in SCA7, for which de novo mutations have been proven, and the lower threshold for mitotic and meiotic instabilities has been set at 27 CAG.35 In addition, polyglutamine stretches between 27 and 33 CAG repeats in ataxin-2 are associated with an increased risk for ALS,34 suggesting a different molecular nature and behavior of ≥27 CAG alleles, which is similar to that of 32–34 CAG alleles (Figure 3b). The association of IAs with other phenotypes alongside SCA2, for example, ALS, FTDP-U and PD in Cuba, is currently being studied because it cannot be neglected in our population (Laffita-Mesa et al, in preparation).
Given the fact that somatic mosaicism could be responsible for the juvenile onset and different SCA2 course and phenotypes (unpublished data), and that mosaicism has been found in SCA2 brain and cerebellum,23 our finding of large ANs being somatically unstable is important for considering these alleles as risk alleles for intergenerational instabilities. Moreover, they are likely to contribute to other neurodegenerative pathologies (ie, ALS and FTDP-U).34 The lack of differences between this group and the 32–33 CAG places these alleles in the category of pathological CAG expansions, which is supported by recent findings.34 Somatic instability in blood might be used as a diagnostic and prognostic measurement in these pathologies given that ataxin-2 is ubiquitously expressed, although it eminently affects CNS.
The distorted assortment shown in Table 2 may reflect some selective advantage. Significant distortion either of homozygous or of heterozygous Ataxin-2-deficient mice with regard to wild type might be reminiscent of the current distortion.36 Short ANs might be variants with partial loss of function and large ANs variants with partial gain of function. The former group may be negatively selected. As it has been shown previously, levels of Ataxin-2 protein have a very important relationship with its biological function.37, 38 The partial gain of function may confer better functionality (translatability and/or protein stability), leading to an increase in Ataxin-2 levels and a positive selection in the population, a notion that is in agreement with the current selection of pre-expansions in the ATXN2 locus.30
In conclusion, we have shown that the highest worldwide concentration of large ANs underlies the highest worldwide prevalence and incidence rates of SCA2. We have found that Cuban population is highly polymorphic at the SCA2 locus. Precisely, these polymorphisms are central for the generation of NMs. These polymorphisms deviate from the focal length (22 CAG) and harbor short and large CAG repeats with polar loss of the CAA interruptions, which are essential for genomic stability. The genomic instability of larger-than-27 CAG alleles supports the proposal that expansions arise from those alleles showing augmented CAG, with either 5′ or 3′ CAA loss, under a predisposed haplotype. The weight of the evidence seems to involve a premutation with a predisposed haplotype as an early step in the foundational event of SCA2 in Cuba; it is more likely than the introduction of the mutation properly. It seems that the enlargement of the CAG is linked to the conservation of certain adaptive functions, with a further selection of the more advantageous variants.
Acknowledgments
We are indebted to our Cuban SCA2 patients and their relatives. We are grateful to Dr Peter O Bauer for his critical review and correction of the English. This work was supported in part by Third World Academy of Sciences -TWAS- in the framework of the Research Grant Project 06-329 RG/BIO/LA UNESCO FR: 3240157855 all to JMLM and a grant of the Cuban Ministry of Health and the Science and Technology Ministry.
Author Contributions
Study concept and design: Laffita-Mesa. Acquisition of data: Laffita-Mesa, Santos-Falcón, González-Zaldívar, Almaguer-Mederos, Almaguer-Gotay, Vázquez-Mojena, Rodríguez-Labrada. Analysis and interpretation of data: Laffita-Mesa, Velázquez-Pérez, Rodríguez-Labrada, Cruz-Mariño. Drafting of the manuscript: Laffita-Mesa. Critical revision of the manuscript for important intellectual content: Vázquez-Mojena, Rodríguez-Labrada, Almaguer-Gotay. Statistical analysis: Laffita-Mesa. Obtained funding: Laffita-Mesa and Velázquez-Pérez. Study supervision: Laffita-Mesa.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Pulst SM, Nechiporuk A, Nechiporuk T, et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- Imbert G, Saudou F, Yvert G, et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat Genet. 1996;14:285–291. doi: 10.1038/ng1196-285. [DOI] [PubMed] [Google Scholar]
- Sanpei K, Takano H, Igarashi S, et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat Genet. 1996;14:277–284. doi: 10.1038/ng1196-277. [DOI] [PubMed] [Google Scholar]
- Santos N, Aguiar J, Fernandez J, et al. Molecular diagnosis of a sample of the Cuban population with spinocerebellar ataxia type 2. Biotecnol Aplic. 1999;16:219–221. [Google Scholar]
- Velázquez Pérez L, Sánchez Cruz G, Santos Falcón N, et al. Molecular epidemiology of spinocerebellar ataxias in Cuba: Insights into SCA2 founder effect in Holguin. Neurosci Lett. 2009;454:157–160. doi: 10.1016/j.neulet.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Cancel G, Durr A, Didierjean O, et al. Molecular and clinical correlations in spinocerebellar ataxia 2: a study of 32 families. Hum Mol Genet. 1997;6:709–715. doi: 10.1093/hmg/6.5.709. [DOI] [PubMed] [Google Scholar]
- Futamura N, Matsumura R, Fujimoto Y, et al. CAG repeat expansions in patients with sporadic cerebellar ataxia. Acta Neurol Scand. 1998;98:55–59. doi: 10.1111/j.1600-0404.1998.tb07378.x. [DOI] [PubMed] [Google Scholar]
- Fernandez M, McClain ME, Martinez RA, et al. Late-onset SCA2: 33 CAG repeats are sufficient to cause disease. Neurology. 2000;55:569–572. doi: 10.1212/wnl.55.4.569. [DOI] [PubMed] [Google Scholar]
- Spears MD, Melton S, Mao Q, et al. Ataxia and progressive encephalopathy in a 4-year-old girl. Lab Med. 2010;41:5–9. [Google Scholar]
- Mao R, Aylsworth AS, Potter N, et al. Childhood-onset ataxia: testing for large CAG-repeats in SCA2 and SCA7. Am J Med Genet. 2002;110:338–345. doi: 10.1002/ajmg.10467. [DOI] [PubMed] [Google Scholar]
- Babovic-Vuksanovic D, Snow K, Patterson MC, et al. Spinocerebellar ataxia type 2 (SCA 2) in an infant with extreme CAG repeat expansion. Am J Med Genet. 1998;79:383–387. [PubMed] [Google Scholar]
- Pearson CE, Eichler EE, Lorenzetti D, et al. Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry. 1998;37:2701–2708. doi: 10.1021/bi972546c. [DOI] [PubMed] [Google Scholar]
- Chung MY, Ranum LP, Duvick LA, et al. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat Genet. 1993;5:254–258. doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]
- Kunst CB, Leeflang EP, Iber JC, et al. The effect of FMR1 CGG repeat interruptions on mutation frequency as measured by sperm typing. J Med Genet. 1997;34:627–631. doi: 10.1136/jmg.34.8.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Holden JJ, Popovich BW, et al. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet. 1994;8:88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- Zhong N, Ju W, Pietrofesa J, et al. Fragile X ‘gray zone' alleles: AGG patterns expansion risks and associated haplotypes. Am J Med Genet. 1996;64:261–265. doi: 10.1002/(SICI)1096-8628(19960809)64:2<261::AID-AJMG5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Takano H, Cancel G, Ikeuchi T, et al. Close associations between prevalences of dominantly inherited spinocerebellar ataxias with CAG-repeat expansions and frequencies of large normal CAG alleles in Japanese and Caucasian populations. Am J Hum Genet. 1998;63:1060–1066. doi: 10.1086/302067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak K, Krzyzosiak WJ. Patterns of CAG repeat interruptions in SCA1 and SCA2 genes in relation to repeat instability. Hum Mutat. 2004;24:236–247. doi: 10.1002/humu.20075. [DOI] [PubMed] [Google Scholar]
- Wadia NH, Swami RK. A new form of heredo-familial spinocerebellar degeneration with slow eye movements (nine families) Brain. 1971;94:359–374. doi: 10.1093/brain/94.2.359. [DOI] [PubMed] [Google Scholar]
- Sinha KK, Worth PF, Jha DK, et al. Autosomal dominant cerebellar ataxia: SCA2 is the most frequent mutation in eastern India. J Neurol Neurosurg Psychiatry. 2004;75:448–452. doi: 10.1136/jnnp.2002.004895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem Q, Choudhry S, Mukerji M, et al. Molecular analysis of autosomal dominant hereditary ataxias in the Indian population: high frequency of SCA2 and evidence for a common founder mutation. Hum Genet. 2000;106:179–187. doi: 10.1007/s004390051026. [DOI] [PubMed] [Google Scholar]
- Becker M. For neurologists in Cuba, hope is not embargoed. Lancet Neurol. 2009;8:1088–1089. doi: 10.1016/S1474-4422(09)70314-X. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Sasaki H, Yabe I, et al. Mosaicism of unstable CAG repeats in the brain of spinocerebellar ataxia type 2. J Neurol. 1999;246:835–839. doi: 10.1007/s004150050464. [DOI] [PubMed] [Google Scholar]
- Basu P, Chattopadhyay B, Gangopadhaya PK, et al. Analysis of CAG repeats in SCA1, SCA2, SCA3, SCA6, SCA7 and DRPLA loci in spinocerebellar ataxia patients and distribution of CAG repeats at the SCA1, SCA2 and SCA6 loci in nine ethnic populations of eastern India. Hum Genet. 2000;106:597–604. doi: 10.1007/s004390000320. [DOI] [PubMed] [Google Scholar]
- Juvonen V, Hietala M, Kairisto V, et al. The occurrence of dominant spinocerebellar ataxias among 251 Finnish ataxia patients and the role of predisposing large normal alleles in a genetically isolated population. Acta Neurol Scand. 2005;111:154–162. doi: 10.1111/j.1600-0404.2005.00349.x. [DOI] [PubMed] [Google Scholar]
- Butland SL, Devon RS, Huang Y, et al. CAG-encoded polyglutamine length polymorphism in the human genome. BMC Genom. 2007;8:126. doi: 10.1186/1471-2164-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres AM, Lao O, Soldevila M, Calafell F, Bertranpetit J. Dynamics of CAG repeat loci revealed by the analysis of their variability. Hum Mutat. 2003;21:61–70. doi: 10.1002/humu.10151. [DOI] [PubMed] [Google Scholar]
- Bauer PO, Kotliarova SE, Matoska V. Fluorescent multiplex PCR – fast method for autosomal dominant spinocerebellar ataxias screening. Russ J Genet. 2005;41:675–682. [PubMed] [Google Scholar]
- Choudhry S, Mukerji M, Srivastava AK, et al. CAG repeat instability at SCA2 locus: anchoring CAA interruptions and linked single nucleotide polymorphisms. Hum Mol Genet. 2001;10:2437–2446. doi: 10.1093/hmg/10.21.2437. [DOI] [PubMed] [Google Scholar]
- Yu F, Sabeti PC, Hardenbol P, et al. Positive selection of a pre-expansion CAG repeat of the human SCA2 gene. PLoS Genet. 2005;1:e41. doi: 10.1371/journal.pgen.0010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A, Magarino C, Gispert S, et al. Genetic mapping of the spinocerebellar ataxia 2 (SCA2) locus on chromosome 12q23–q24.1. Genomics. 1995;25:433–435. doi: 10.1016/0888-7543(95)80043-l. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Amos W, Leggo J, et al. Mutational bias provides a model for the evolution of Huntington's disease and predicts a general increase in disease prevalence. Nat Genet. 1994;7:525–530. doi: 10.1038/ng0894-525. [DOI] [PubMed] [Google Scholar]
- Cleary JD, Pearson CE. The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenet Genome Res. 2003;100:25–55. doi: 10.1159/000072837. [DOI] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nat Genet. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanin G, Giunti P, Belal GD, et al. De novo expansion of intermediate alleles in spinocerebellar ataxia 7. Hum Mol Genet. 1998;7:1809–1813. doi: 10.1093/hmg/7.11.1809. [DOI] [PubMed] [Google Scholar]
- Kiehl TR, Nechiporuk A, Figueroa KP, et al. Generation and characterization of Sca2 (ataxin-2) knockout mice. Biochem Biophys Res Commun. 2006;339:17–24. doi: 10.1016/j.bbrc.2005.10.186. [DOI] [PubMed] [Google Scholar]
- Satterfield TF, Pallanck LJ. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum Mol Genet. 2006;15:2523–2532. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, et al. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.