Abstract
Human alterations to the environment can exert strong evolutionary pressures, yet contemporary adaptation to human-mediated stressors is rarely documented in wildlife populations. A common-garden experimental design was coupled with comparative transcriptomics to discover evolved mechanisms enabling three populations of killifish resident in urban estuaries to survive normally lethal pollution exposure during development, and to test whether mechanisms are unique or common across populations. We show that killifish populations from these polluted sites have independently converged on a common adaptive mechanism, despite variation in contaminant profiles among sites. These populations are united by a similarly profound desensitization of aryl-hydrocarbon receptor-mediated transcriptional activation, which is associated with extreme tolerance to the lethal effects of toxic dioxin-like pollutants. The rapid, repeated, heritable and convergent nature of evolved tolerance suggests that ancestral killifish populations harboured genotypes that enabled adaptation to twentieth-century industrial pollutants.
Keywords: comparative transcriptomics, adaptation, Fundulus heteroclitus, ecological genomics
1. Introduction
Modern humans have acted as agents of abrupt environmental change [1,2] usually resulting in extirpation of wildlife populations. Rarely, strong evolutionary pressures can drive adaptation of species that are targets for eradication, such as agricultural pests [3] and pathogenic bacteria [4]. Yet, contemporary adaptation to unintentional human-mediated stressors is rarely documented in non-targeted wildlife populations, especially vertebrate species. Evidence of successful contemporary adaptation is apparent in large, persistent populations of killifish (Fundulus heteroclitus) residing in densely populated US Atlantic coast urban estuaries. While these estuaries are bedded with sediments contaminated with unique mixtures of lethal pollutants [5], resident killifish populations are tolerant to a widely distributed class of persistent, bioaccumulative and toxic pollutants. These ‘dioxin-like pollutants’, which include 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD, dioxin) and some planar polychlorinated biphenyl (PCB) congeners whose toxicity is known to be largely mediated through the aryl-hydrocarbon receptor (AHR), are particularly toxic to the early development of fishes [6].
Despite differences in the magnitude and pattern of contamination at each polluted site, resident populations share some similarities: pollution tolerance has evolved quickly (based on the history of sediment contamination at each site), populations exhibit similar levels of protection from extreme exposures to dioxin-like compounds, and protection is accompanied by poor induction of mixed function oxygenase activity, a distal target of the ligand-activated AHR pathway [5]. Yet, population genetic studies [7,8] and data presented here indicate that tolerant populations share more recent ancestry with nearby sensitive populations than with other tolerant populations, suggesting that dioxin-tolerant phenotypes have evolved repeatedly in wild killifish populations. For example, F. heteroclitus show a general pattern of isolation by distance [7,9], and neutrally evolving markers show that geographically proximate populations share greatest similarity independent of pollution exposure history [8]. We used a common-garden comparative transcriptomics approach to discover the mechanistic basis of tolerant phenotypes, and to test whether the same mechanisms are responsible for converged tolerant phenotypes in these parallel-evolved killifish populations. Our experimental framework was designed to: (i) distinguish population variation that was habitat induced from variation that is heritable, (ii) distinguish population variation that is neutral from variation, that is adaptive, and (iii) distinguish population-specific adaptive mechanisms from common and converged adaptive mechanisms. Our data confirm and extend results from a previous comparison of a single pair of tolerant and sensitive populations [10].
2. Material and methods
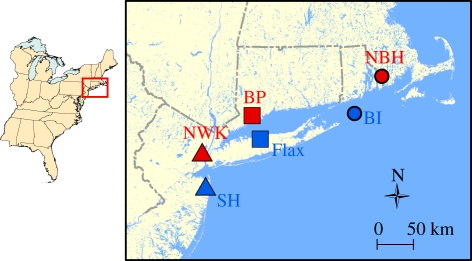
Mature fish were collected from six sites including three highly polluted estuaries (New Bedford Harbor, MA, USA; Bridgeport, CT, USA; and Newark Bay, NJ, USA), and nearby comparatively clean reference sites (Block Island, RI, USA; Flax Pond, NY, USA; and Sandy Hook, NJ, USA; figure 1). Coordinates (latitude/longitude) for source populations can be found in Nacci et al. [5]. Polluted sites are characterized by high sediment concentrations of PCBs [5], but also include dioxins and polycyclic aromatic hydrocarbons, which all act partly or fully through the AHR, and other mechanistically unrelated pollutants such as metals [5]. Adult fish collected from contaminated sites were held in flow-through aquaria in the laboratory for 6–24 months before spawning, which permits substantial depuration of contaminants. Offspring of this spawning (F1) were raised in the laboratory for 2–3 years before spawning, minimizing any potential for contaminant transfer from the field-derived generation. Offspring from the spawning of F1 individuals (F2) were used for the experiments reported here. That is, fish from polluted sites were raised for two generations (F1 and F2) in a common clean environment with reference site fish to isolate the heritable component of population variation in pollution tolerance.
Figure 1.
Atlantic Coast of the USA. Tolerant killifish populations (red) are from New Bedford Harbor (NBH) MA, USA; Bridgeport (BP) CT, USA; and Newark (NWK) NJ, USA. Reference killifish populations (blue) are from Block Island (BI) RI, USA; Flax Pond (Flax) NY, USA; and Sandy Hook (SH), NJ, USA. Precise locations (coordinates) can be found in Nacci et al. [5]. Symbols (circles, squares, triangles) unite geographically paired populations, whereas colour (red or blue) unite populations with similar pollution exposure histories.
Embryos were exposed at day 1 post-fertilization for 7 days during development to the model pollutant PCB-126 (3,3′,4,4′,5-pentachlorobiphenyl). A subset of embryos were snap-frozen in liquid nitrogen on day 10 (post-organogenesis, stage 34 [11]) of development for transcriptome profiling (n = 5 per treatment) after each embryo was assessed for developmental abnormalities, where an abnormality rating was assigned to each embryo on a scale ranging from zero (no abnormalities) to four (severe abnormalities) as described in Whitehead et al. [10]. Remaining embryos were allowed to hatch and survivorship was calculated out to 7 days post-hatch [5]. Chemical exposures included a vehicle (acetone) control (0 ng l−1 PCB-126) and PCB-126 nominal exposure concentrations of 2, 20 and 200 ng l−1 for reference populations and 200, 2000 and 20 000 ng l−1 for tolerant populations.
Whole embryos were homogenized and total RNA extracted in Trizol reagent (Invitrogen), antisense-RNA (aRNA) prepared using the MessageAmp II aRNA amplification kit (Ambion), purified aRNA coupled with Alexa fluor dyes (Alexa fluor 555 and 647; Molecular Probes, Inc.), then competitively hybridized to custom microarrays (Agilent eArray Design ID 027999) in a loop design (three loops, one for each geographical pair of populations) where hybridized pairs were balanced across treatments. Separate hybridizations were performed for each of five replicate embryos (biological replicates) per treatment, including a dye swap. Data were Lowess normalized then mixed-model normalized using Jmp Genomics (SAS Inc.). Log2-transformed normalized data were fit to a mixed linear model, which specified main effects of ‘population’ and ‘dose’ including an interaction term (to identify population-dependent dose–responses) using Jmp Genomics (SAS Inc.). Replicate individuals (n = 5) within treatments were treated as random effects. Two ANOVAs were performed. The first ANOVA compared transcriptional responses with PCB exposure among populations in response to a common PCB dose (200 ng l−1). A second ANOVA compared transcriptional responses with an ‘effects-matched’ range of doses, where the dose range for reference (2, 20 and 200 ng l−1) and tolerant (200, 2000 and 20 000 ng l−1) populations captured the nominal no observable phenotypic effect dose and lowest observable effect dose for each population (figure 2). Statistical significance was determined at p < 0.01. Principal component analysis (PCA) and classification analysis were performed using Mev [12]. For PCA, treatment averages were clustered using the median centring mode and 10 neighbours for k-nearest neighbour imputation. Classification analysis was by partial least squares [13] and used polychotomous discriminant analysis option as the classification algorithm.
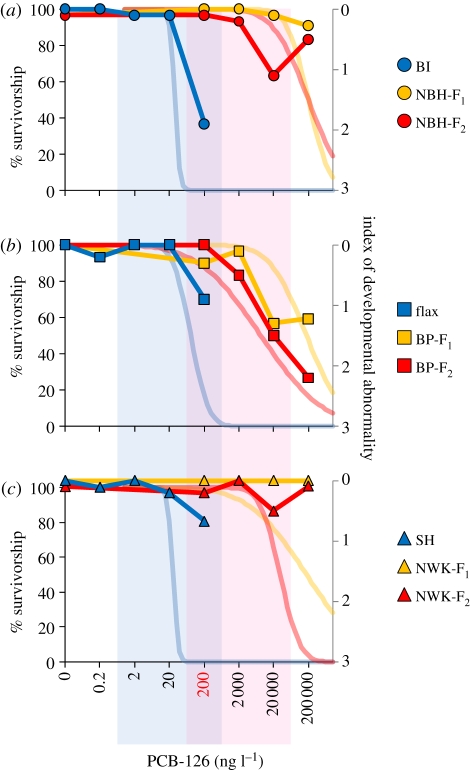
Figure 2.
Survivorship and developmental effects upon exposure to increasing log10 doses of PCB-126 (ng l−1). Light blue, red and yellow curves represent embryonic and larval survivorship out to 7 days post-hatch (primary y-axis) modelled from Nacci et al. [5]. Bold blue, red and yellow points and curves represent average severity of developmental abnormality (secondary y-axis) as a function of PCB-126 dose. Each panel includes data from geographically paired populations (symbols and abbreviations as in figure 1). Colours unite populations with similar pollution exposure histories: blue is for reference populations, whereas yellow and red are for tolerant population F1 and F2 embryos, respectively. Vertical light blue and light pink panels highlight ‘effects-matched’ dose ranges used for comparative transcriptomics profiling for reference and tolerant populations, respectively, where the 200 ng l−1 dose (red) is common to all populations. (a) Dark blue circles, BI; yellow circles, NBH-F1; red circles, NBH-F2. (b) Dark blue squares, flax; yellow squares, BP-F1; red squares, BP-F2. (c) Blue triangles, SH; yellow triangles, NWK-F1; red triangles, NWK-F2.
3. Results and discussion
Since early life stages are particularly sensitive to the toxicity of dioxin-like compounds, F1 and F2 embryos were challenged with up to six log doses of 3′3′4′4′5 penta-chlorinated biphenyl (PCB-126), a prototypical dioxin-like compound. In replicate studies [5], lethal responses were assessed (median lethal concentrations, LC50s) at 7 days post-hatching, and were between two and three orders of magnitude greater for F1 embryos from polluted sites compared with those from reference sites (figure 2). That PCB tolerance was similar in F1 and F2 embryos from polluted sites confirmed that tolerance is heritable, and therefore genetically based in all tolerant populations.
To offer insight into the functional basis of divergence in tolerance between populations, we sampled embryos from tolerant and reference populations at pre-lethal times (day 10 of development, post-organogenesis [11]) and at sub-lethal exposure concentrations. Since dioxin-like compounds are particularly and characteristically disruptive to cardiovascular system development in fishes and mammals [14,15], microscopic observation of transparent embryos permitted quantitative assessment of tolerance prior to RNA isolation, scoring for the presence and severity of developmental abnormalities. Parallel with differences in lethality, cardiovascular system developmental abnormalities emerged at PCB doses between two to three orders of magnitude greater for embryos from polluted sites compared with those from reference sites (figure 2). Transcriptional responses to PCB exposure were compared among populations in response to a common PCB dose (200 ng l−1) and to an ‘effects-matched’ range of doses which captured the nominal no observable phenotypic effect dose and lowest observable effect dose for each population (figure 2).
Since evolutionary divergence in gene expression is governed by both neutral and adaptive processes [16], we designed our population contrasts to specifically isolate the component of population variation that rejects neutral patterns of population divergence in favour of patterns of divergence that are putatively adaptive. Populations were compared by geographical region (comparisons among geographical pairs) and by pollution history (fish from highly polluted habitats versus fish from clean habitats). Geographical pairs of populations share more recent ancestry, so if trait divergence is governed by historical demographic processes, then population pairs should share greatest similarity. We hypothesized that population divergence in expression for genes that are not transcriptionally responsive to the environment is most probably governed by neutral evolutionary processes, thereby reflecting the historical demography of populations. Since chemical pollution primarily accounts for habitat variation among the native sites of our experimental populations, pollutant exposure is the environmental variable that we manipulate. Conversely, genes that are responsive to the environment are more likely to be the targets of selective processes. For example, genes that were transcriptionally responsive to osmotic stress were more likely to show adaptive patterns of divergence among fish populations distributed across an environmental salinity gradient than genes that were not transcriptionally responsive [17]. Average expression levels for genes that were not PCB-responsive (2208 genes) clustered populations by geographical neighbour, rather than by tolerance phenotype (figure 3a). Indeed, less than 1 per cent of the non-responsive genes clustered populations by tolerance phenotype using classification analysis. Similarly, population genetic studies have tended to show that tolerant populations share more recent ancestry with nearby sensitive populations than with other tolerant populations [7,8]. These results are consistent with the hypothesis that local pollution tolerance has evolved multiple times independently, especially since natural selection can act efficiently within F. heteroclitus by virtue of large local population size and low migration rates [7,18].
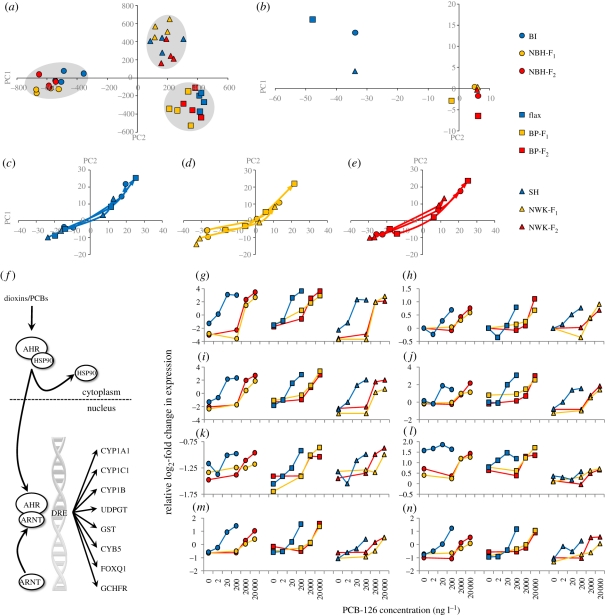
Figure 3.
(a–e) Principal components (PC) analyses of gene expression differences. Blue circles, BI; yellow circles, NBH-F1; red circles, NBH-F2; blue squares, Flax; yellow squares, BP-F1; red squares BP-F2; blue triangles, SH; yellow triangles, NWK-F1; red triangles, NWK-F2. (a) Genes that are population-variable in expression only (not dose–responsive) cluster geographical pairs of populations, irrespective of exposure history. (b) Response trajectories for genes that show population-specific dose–response (AHR signalling targets) at the common dose (200 ng l−1 minus control) show that F1 and F2 embryos from tolerant populations converge on the same response. (c–e) Population-specific dose–response genes show identical response trajectories through PC space (PC1 and PC2) at ‘effects-matched’ doses (see text and figure 2), which are offset by two orders of magnitude between (c) reference, (d) tolerant F1 and (e) tolerant F2 embryos, where the base of each line represents the control dose and the line connects through the lowest to middle to highest dose (at arrow head) for each population. (f) Canonical dioxin/PCB-induced aryl-hydrocarbon-mediated signalling pathway and gene battery turned on in our data. (g–n) Each panel shows population-specific dose–response of expression for AHR gene battery members showing divergence between tolerant and sensitive populations but convergence among tolerant populations. (g) CYP1A1; (h) GST; (i) CYP1C1; (j) CYB5; (k) CYP1B; (l) FOXQ1; (m) UDPGT; (n) GCHFR. Tolerant populations show a similar dose–response to PCB-126 as sensitive populations, but at concentrations two orders of magnitude higher. Common symbols (circles, squares or triangles) unite geographically paired populations as in panels (a–e), and blue represents sensitive populations, yellow represents tolerant populations F1 and red tolerant populations F2. Dependent variable is PCB-126 concentration (ng l−1) and independent variable is relative log2 fold change in gene expression level.
In contrast to environmentally unresponsive genes, genes that show population-dependent dose–response in expression are most likely to reflect evolved functional differences between populations. Comparing patterns of population-specific divergence allowed us to test whether derived responses were unique to each tolerant population or common across all tolerant populations. We compared gene expression response with the common 200 ng l−1 dose (relative to control) among populations, and 14 genes showed expression that was dose–responsive (p < 0.01, for main effect) and population-dependent (p < 0.01, for interaction). Population comparisons showed that dose–response trajectories for these 14 genes were the same for F1 and F2 embryos from all three tolerant populations, but distinct from expression response trajectories for sensitive reference populations (figure 3b). This pattern does not support unique evolved responses for each tolerant population, but rather is consistent with the hypothesis that tolerant populations converged on a heritable, common, non-neutral and putatively adaptive functional response to a shared evolutionary challenge. Importantly, constitutive differences in gene expression in a static common-garden do not reveal mechanisms of adaptive divergence in killifish populations [19] probably as a result of developmental canalization, whereas challenge with a model pollutant is necessary to reveal adaptive population divergence in the genomic response function (figure 3).
The cluster of tolerant population-specific dose–responsive genes consists mainly of genes that are transcriptionally induced by the AHR-mediated signalling pathway, which activates a canonical battery of genes [20] (figure 3f). This gene battery is strongly induced at 2–20 ng l−1 PCB-126 in fish populations derived from clean environments, whereas F1 and F2 embryos derived from polluted habitats are refractory to AHR gene battery induction (figure 3 g–n). The AHR gene battery is ultimately induced in tolerant populations at 2000 ng l−1, thus requiring doses 100 to 1000 times higher than in sensitive reference populations (figure 3g–n). Induction of this gene battery is highly correlated with the emergence of toxic effects in all populations, which is consistent with demonstration that PCB toxicity is largely mediated through AHR activation (e.g. [21]). Indeed, at effects-matched doses, the trajectories of expression response of the group of 14 tolerant population-specific dose–responsive genes, which includes the AHR gene battery, are highly correlated (figure 3c–e; average correlation for across all population pairs is 0.98 for PC1 and 0.97 for PC2).
At effects-matched doses, which are offset by two orders of magnitude between tolerant and reference populations, many genes (706, p < 0.01) show a common transcriptional response across all populations (see the electronic supplementary material), including the AHR gene battery. This group of AHR transcriptional targets includes phase I and II metabolism genes (cytochrome P450s, UDP-glucuronosyltransferase, glutathione S-transferase, cytochrome b5), forkhead box protein Q1, which is associated with dioxin-induced craniofacial malformation in developing fish [22], and GTP cyclohydrolase feedback regulatory protein, which is associated with nitric oxide synthase uncoupling leading to increased vascular superoxide and cardiovascular disease observed with dioxin exposure [23] (figure 3g–n). Thus, the threshold for PCB activation of AHR signalling has shifted two to three orders of magnitude higher in tolerant compared with reference populations. Consistent with this shift, PCB body burdens carried by fish resident in the NBH polluted site are between two to three orders of magnitude higher than in fish from clean sites [5]. The fitness advantage and ecological relevance of this tolerance are clear, since this shift in threshold activation of AHR appears scaled appropriately for the level of chemical risk posed by native polluted habitats.
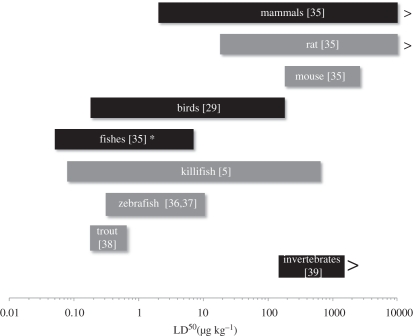
AHR signalling is normally activated by dioxin-like compounds and largely mediates their toxicity [21,24–27]. Given the nodal position of AHR in a regulatory network and the necessity and sufficiency of AHR activation for dioxin toxicity, AHR activation offers a large mutational target of the sort that tends to be preferred targets for repeated evolution [28]. Molecular variants in the AHR gene are associated with dioxin tolerance in birds [29], mammals [30] and fishes [31]. Rats that carry a mutation in the AHR transactivation domain are relatively dioxin-resistant, though only a subset of the AHR transcriptional targets is refractory to dioxin induction [32]. By contrast, our data show that resistant killifish populations display global blockade of AHR-mediated transcriptional activation upon low- to intermediate-level PCB exposures, resembling the transcriptional response to dioxins in AHR-knockout mice [33]. Unlike other animal models, including another coastal fish [31], currently identified AHR variants do not appear to account for derived tolerance in killifish [34], yet the dioxin-tolerance range spanned by populations within F. heteroclitus is unprecedented, and to our knowledge far exceeds that of any other species even including all species of fishes (figure 4).
Figure 4.
Ranges of within and among-species variation in dioxin sensitivity (LD50, ug kg−1). Black bars indicate sensitivity ranges for broad taxonomic groups. Grey bars indicate sensitivity ranges among populations or strains within species. Asterisk indicates not including killifish.
While exploiting variation within traditional laboratory models is useful for studying mechanisms of dioxin toxicity, intraspecific variation in killifish reveals a conserved mechanism of dioxin tolerance derived by natural selection in the wild. Insecticide resistance offers comparable examples of repeated adaptation to anthropogenic poisons, where the selection pressure was extreme, the mutational target was large and population sizes were large [3]. Where dioxin tolerance in killifish diverges from examples of derived insecticide resistance is that insecticides were specifically designed to poison and kill target organisms by interfering with specific molecular targets, whereas toxicity to wild fish from dioxin pollution was not intentional. Evolutionary mutants are emerging as powerful models for biomedical research, because the types of mutations favoured by natural evolutionary forces may more realistically represent models of genomic variation contributing to human disease, compared with traditional mutant models created in the laboratory [40]. Since exposure to dioxin-like chemicals is an important contemporary risk to human health [41], the nature of variation in sensitivity to these chemicals that we detect in killifish could be useful for identifying human individuals or populations particularly at risk from exposure.
Human-induced environmental changes offer exceptional opportunities to study evolutionary phenomena in real time. That profound tolerance to environmental pollution evolved rapidly and repeatedly by the same mechanism and with no apparent erosion of population genetic diversity [42] is extraordinary, especially since even large populations are unlikely to survive rates of environmental change that exceed 1–2 per cent of a phenotypic standard deviation per generation [43]. This suggests that tolerance-enabling alleles are not rare in F. heteroclitus populations [44], and that these alleles were repeatedly swept to fixation in habitats receiving chemical contaminants. Large population size to harbour high allelic diversity may be necessary to enable persistence of wild populations given the rapid pace of some human-induced environmental change.
Acknowledgements
All work was conducted according to the US EPA's Atlantic Ecology Division Animal Care and Maintenance Practices and Policies (2004), which ensure the ethical treatment of all animals used in the conduct of this research.
A.W. and D.N. designed experiments. D.N. and D.C. maintained fish colonies, conducted embryo PCB-exposure experiments, and collected and analysed survivorship data. W.P. collected transcriptomics data. A.W. analysed and interpreted transcriptomics data. A.W. and D.N. wrote the manuscript. This is contribution number AED-11-014 of the US Environmental Protection Agency, Office of Research and Development, National Health and Environmental Effects Research Laboratory, Atlantic Ecology Division, which partially supported this research. This manuscript has been reviewed and approved for publication by the US EPA. Approval does not signify that the contents necessarily reflect the views and policies of the US EPA. Mention of trade names, products or services does not convey and should not be interpreted as conveying, official US EPA approval, endorsement or recommendation. This research was partially supported by funding from the National Science Foundation (grants EF-0723771 and BES-0652006 to A.W.).
References
- 1.Palumbi S. R. 2001. Humans as the world's greatest evolutionary force. Science 293, 1786–1790 10.1126/science.293.5536.1786 (doi:10.1126/science.293.5536.1786) [DOI] [PubMed] [Google Scholar]
- 2.Smith T. B., Bernatchez L. 2008. Evolutionary change in human-altered environments. Mol. Ecol. 17, 1–8 10.1111/j.1365-294X.2007.03607.x (doi:10.1111/j.1365-294X.2007.03607.x) [DOI] [PubMed] [Google Scholar]
- 3.Ffrench-Constant R. H., Daborn P. J., Le Goff G. 2004. The genetics and genomics of insecticide resistance. Trends Genet. 20, 163–170 10.1016/j.tig.2004.01.003 (doi:10.1016/j.tig.2004.01.003) [DOI] [PubMed] [Google Scholar]
- 4.Walsh C. 2000. Molecular mechanisms that confer antibacterial drug resistance. Nature 406, 775–781 10.1038/35021219 (doi:10.1038/35021219) [DOI] [PubMed] [Google Scholar]
- 5.Nacci D., Champlin D., Jayaraman S. 2010. Adaptation of the estuarine fish Fundulus heteroclitus (Atlantic killifish) to polychlorinated biphenyls (PCBs). Estuaries Coasts 33, 853–864 10.1007/s12237-009-9257-6 (doi:10.1007/s12237-009-9257-6) [DOI] [Google Scholar]
- 6.Tillitt D. E., Cook P. M., Giesy J. P., Heideman W., Peterson R. E. 2008. Reproductive impairment of great lakes lake trout by dioxin-like chemicals. In The toxicology of fishes (eds Di Giulio R. T., Hinton D. E.), pp. 819–876 Boca Raton, FL: Taylor and Francis [Google Scholar]
- 7.Duvernell D. D., Lindmeier J. B., Faust K. E., Whitehead A. 2008. Relative influences of historical and contemporary forces shaping the distribution of genetic variation in the Atlantic killifish, Fundulus heteroclitus. Mol. Ecol. 17, 1344–1360 10.1111/j.1365-294X.2007.03648.x (doi:10.1111/j.1365-294X.2007.03648.x) [DOI] [PubMed] [Google Scholar]
- 8.Williams L., Oleksiak M. 2008. Signatures of selection in natural populations adapted to chronic pollution. BMC Evol. Biol. 8, 282. 10.1186/1471-2148-8-282 (doi:10.1186/1471-2148-8-282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams S. M., Lindmeier J. B., Duvernell D. D. 2006. Microsatellite analysis of the phylogeography, Pleistocene history and secondary contact hypotheses for the killifish, Fundulus heteroclitus. Mol. Ecol. 15, 1109–1123 10.1111/j.1365-294X.2006.02859.x (doi:10.1111/j.1365-294X.2006.02859.x) [DOI] [PubMed] [Google Scholar]
- 10.Whitehead A., Triant D. A., Champlin D., Nacci D. 2010. Comparative transcriptomics implicates mechanisms of evolved pollution tolerance in a killifish population. Mol. Ecol. 19, 5186–5203 10.1111/j.1365-294X.2010.04829.x (doi:10.1111/j.1365-294X.2010.04829.x) [DOI] [PubMed] [Google Scholar]
- 11.Armstrong P. B., Child J. S. 1965. Stages in normal development of Fundulus heteroclitus. Biol. Bull. 128, 143–168 10.2307/1539545 (doi:10.2307/1539545) [DOI] [Google Scholar]
- 12.Saeed A. I., et al. 2006. TM4 Microarray Software Suite. Methods Enzymol. 411, 134–193 10.1016/S0076-6879(06)11009-5 (doi:10.1016/S0076-6879(06)11009-5) [DOI] [PubMed] [Google Scholar]
- 13.Nguyen D. V., Rocke D. M. 2002. Tumor classification by partial least squares using microarray gene expression data. Bioinformatics 18, 39–50 10.1093/bioinformatics/18.1.39 (doi:10.1093/bioinformatics/18.1.39) [DOI] [PubMed] [Google Scholar]
- 14.Antkiewicz D. S., Burns C. G., Carney S. A., Peterson R. E., Heideman W. 2005. Heart malformation is an early response to TCDD in embryonic zebrafish. Toxicol. Sci. 84, 368–377 (doi:10.1093/toxsci/kfi073) [DOI] [PubMed] [Google Scholar]
- 15.Thackaberry E. A., Nunez B. A., Ivnitski-Steele I. D., Friggins M., Walker M. K. 2005. Effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin on murine heart development: alteration in fetal and postnatal cardiac growth, and postnatal cardiac chronotropy. Toxicol. Sci. 88, 242–249 10.1093/toxsci/kfi302 (doi:10.1093/toxsci/kfi302) [DOI] [PubMed] [Google Scholar]
- 16.Whitehead A., Crawford D. L. 2006. Variation within and among species in gene expression: raw material for evolution. Mol. Ecol. 15, 1197–1211 10.1111/j.1365-294X.2006.02868.x (doi:10.1111/j.1365-294X.2006.02868.x) [DOI] [PubMed] [Google Scholar]
- 17.Whitehead A., Roach J. L., Zhang S., Galvez F. 2011. Genomic mechanisms of evolved physiological plasticity in killifish distributed along an environmental salinity gradient. Proc. Natl Acad. Sci. USA 108, 6193–6198 10.1073/pnas.1017542108 (doi:10.1073/pnas.1017542108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sweeney J., Deegan L., Garritt R. 1998. Population size and site fidelity of Fundulus heteroclitus in a macrotidal saltmarsh creek. Biol. Bull. 195, 238–239 10.2307/1542858 (doi:10.2307/1542858) [DOI] [PubMed] [Google Scholar]
- 19.Bozinovic G., Oleksiak M. F. 2010. Embryonic gene expression among pollutant resistant and sensitive Fundulus heteroclitus populations. Aquat. Toxicol. 98, 221–229 10.1016/j.aquatox.2010.02.022 (doi:10.1016/j.aquatox.2010.02.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hankinson O. 1995. The aryl-hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 35, 307–340 10.1146/annurev.pa.35.040195.001515 (doi:10.1146/annurev.pa.35.040195.001515) [DOI] [PubMed] [Google Scholar]
- 21.Jonsson M. E., Jenny M. J., Woodin B. R., Hahn M. E., Stegeman J. J. 2007. Role of AHR2 in the expression of novel cytochrome p450 1 family genes, cell cycle genes, and morphological defects in developing zebra fish exposed to 3,3 ′,4,4 ′,5-pentachlorobiphenyl or 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 100, 180–193 10.1093/toxsci/kfm207 (doi:10.1093/toxsci/kfm207) [DOI] [PubMed] [Google Scholar]
- 22.Planchart A., Mattingly C. J. 2010. 2,3,7,8-Tetrachlorodibenzo-p-dioxin upregulates FoxQ1b in zebrafish jaw primordium. Chem. Res. Toxicol. 23, 480–487 10.1021/Tx9003165 (doi:10.1021/Tx9003165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kopf P. G., Huwe J. K., Walker M. K. 2008. Hypertension, cardiac hypertrophy, and impaired vascular relaxation induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin are associated with increased superoxide. Cardiovasc. Toxicol. 8, 181–193 10.1007/s12012-008-9027-x (doi:10.1007/s12012-008-9027-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antkiewicz D. S., Peterson R. E., Heideman W. 2006. Blocking expression of AHR2 and ARNT1 in zebrafish larvae protects against cardiac toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 94, 175–182 10.1093/toxsci/kfl093 (doi:10.1093/toxsci/kfl093) [DOI] [PubMed] [Google Scholar]
- 25.Bunger M. K., Glover E., Moran S. M., Walisser J. A., Lahvis G. P., Hsu E. L., Bradfield C. A. 2008. Abnormal liver development and resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in mice carrying a mutation in the DNA binding domain of the aryl hydrocarbon receptor. Toxicol. Sci. 106, 83–92 10.1093/toxsci/kfn149 (doi:10.1093/toxsci/kfn149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carney S. A., Peterson R. E., Heideman W. 2004. 2,3,7,8-tetrachlorodibenzo-p-dioxin activation of the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator pathway causes developmental toxicity through a CYP1A-independent mechanism in zebrafish. Mol. Pharmacol. 66, 512–521 [DOI] [PubMed] [Google Scholar]
- 27.Prasch A. L., Teraoka H., Carney S. A., Dong W., Hiraga T., Stegeman J. J., Heideman W., Peterson R. E. 2003. Aryl hydrocarbon receptor 2 mediates 2,3,7,8-tetrachlorodibenzo-p-dioxin developmental toxicity in zebrafish. Toxicol. Sci. 76, 138–150 10.1093/toxsci/kfg202 (doi:10.1093/toxsci/kfg202) [DOI] [PubMed] [Google Scholar]
- 28.Gompel N., Prud'homme B. 2009. The causes of repeated genetic evolution. Dev. Biol. 332, 36–47 10.1016/j.ydbio.2009.04.040 (doi:10.1016/j.ydbio.2009.04.040) [DOI] [PubMed] [Google Scholar]
- 29.Head J. A., Hahn M. E., Kennedy S. W. 2008. Key amino acids in the aryl hydrocarbon receptor predict dioxin sensitivity in avian species. Environ. Sci. Technol. 42, 7535–7541 10.1021/Es801082a (doi:10.1021/Es801082a) [DOI] [PubMed] [Google Scholar]
- 30.Pohjanvirta R., et al. 1999. Physicochemical differences in the AH receptors of the most TCDD-susceptible and the most TCDD-resistant rat strains. Toxicol. Appl. Pharmacol. 155, 82–95 10.1006/taap.1998.8565 (doi:10.1006/taap.1998.8565) [DOI] [PubMed] [Google Scholar]
- 31.Wirgin I., Roy N. K., Loftus M., Chambers R. C., Franks D. G., Hahn M. E. 2011. Mechanistic basis of resistance to PCBs in Atlantic Tomcod from the Hudson river. Science. 331, 1322–1325(doi:10.1126/science.1197296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moffat I., Boutros P., Chen H., Okey A., Pohjanvirta R. 2010. Aryl hydrocarbon receptor (AHR)-regulated transcriptomic changes in rats sensitive or resistant to major dioxin toxicities. BMC Genom. 11, 263. 10.1186/1471-2164-11-263 (doi:10.1186/1471-2164-11-263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tijet N., Boutros P. C., Moffat I. D., Okey A. B., Tuomisto J., Pohjanvirta R. 2006. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol. Pharmacol. 69, 140–153 10.1124/mol.105.018705 (doi:10.1124/mol.105.018705) [DOI] [PubMed] [Google Scholar]
- 34.Hahn M. E., Karchner S. I., Franks D. G., Merson R. R. 2004. Aryl hydrocarbon receptor polymorphisms and dioxin resistance in Atlantic killifish (Fundulus heteroclitus). Pharmacogenetics 14, 131–143 10.1097/00008571-200402000-00007 (doi:10.1097/00008571-200402000-00007) [DOI] [PubMed] [Google Scholar]
- 35.Korkalainen M. 2005. Structure and expression of principal proteins involved in dioxin signal transduction and potentially in dioxin sensitivity. In Publications of the National Public Health Insitute, p. 79 Finland: National Public Health Insitute [Google Scholar]
- 36.Elonen G. E., Spehar R. L., Holcombe G. W., Johnson R. D., Fernandez J. D., Erickson R. J., Tietge J. E., Cook P. M. 1998. Comparative toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin to seven freshwater fish species during early life-stage development. Environ. Toxicol. Chem. 17, 472–483 10.1002/etc.5620170319 (doi:10.1002/etc.5620170319) [DOI] [Google Scholar]
- 37.Waits E. R., Nebert D. W. In press Genetic architecture of susceptibility to PCB126-induced developmental cardiotoxicity in zebrafish. Toxicol. Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carvalho P. S. M., Noltie D. B., Tillitt D. E. 2004. Intra-strain dioxin sensitivity and morphometric effects in swim-up rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 137, 133–142 10.1016/j.cca.2003.12.005 (doi:10.1016/j.cca.2003.12.005) [DOI] [PubMed] [Google Scholar]
- 39.West C. W., Ankley G. T., Nichols J. W., Elonen G. E., Nessa D. E. 1997. Toxicity and bioaccumulation of 2,3,7,8-tetrachlorodibenzo-p-dioxin in long-term tests with the freshwater benthic invertebrates Chironomus tentans and Lumbriculus variegatus. Environ. Toxicol. Chem. 16, 1287–1294 [Google Scholar]
- 40.Albertson R. C., Cresko W., Detrich H. W., Postlethwait J. H. 2009. Evolutionary mutant models for human disease. Trends Genet. TIG 25, 74–81 10.1016/j.tig.2008.11.006 (doi:10.1016/j.tig.2008.11.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.EPA US 2004. Dioxin Reassessment. National Academy of Sciences (NAS) Review Draft 2004. U.S. Environmental Protection Agency, Washington, D.C., EPA/600/P-00/001Cb
- 42.McMillan A. M., Bagley M. J., Jackson S. A., Nacci D. E. 2006. Genetic diversity and structure of an estuarine fish (Fundulus heteroclitus) indigenous to sites associated with a highly contaminated urban harbor. Ecotoxicology 15, 539–548 10.1007/s10646-006-0090-4 (doi:10.1007/s10646-006-0090-4) [DOI] [PubMed] [Google Scholar]
- 43.Lynch M. 1996. A quantitative-genetic perspective on conservation issues. In Conservation genetics: case histories from nature (eds Avise J. C., Hamrick J. L.), pp. 471–501 New York, NY: Chapman & Hall [Google Scholar]
- 44.Colosimo P. F., et al. 2005. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307, 1928–1933 10.1126/science.1107239 (doi:10.1126/science.1107239) [DOI] [PubMed] [Google Scholar]