Abstract
We outline a descriptive framework of how candidate alleles of the immune system associate with infectious diseases in natural populations of animals. Three kinds of alleles can be separated when both prevalence of infection and infection intensity are measured—qualitative disease resistance, quantitative disease resistance and susceptibility alleles. Our descriptive framework demonstrates why alleles for quantitative resistance and susceptibility cannot be separated based on prevalence data alone, but are distinguishable on infection intensity. We then present a case study to evaluate a previous finding of a positive association between prevalence of a severe avian malaria infection (GRW2, Plasmodium ashfordi) and a major histocompatibility complex (MHC) class I allele (B4b) in great reed warblers Acrocephalus arundinaceus. Using the same dataset, we find that individuals with allele B4b have lower GRW2 infection intensities than individuals without this allele. Therefore, allele B4b provides quantitative resistance rather than increasing susceptibility to infection. This implies that birds carrying B4b can mount an immune response that suppresses the acute-phase GRW2 infection, while birds without this allele cannot and may die. We argue that it is important to determine whether MHC alleles related to infections are advantageous (quantitative and qualitative resistance) or disadvantageous (susceptibility) to obtain a more complete picture of pathogen-mediated balancing selection.
Keywords: qualitative resistance, quantitative resistance, susceptibility allele, major histocompatibility complex (MHC) class I, avian malaria
1. Introduction
Why is it so that some host individuals within a population become very ill from a specific pathogen while others do not? Parasite species and parasite genetics, as well as host genetics and condition, are all important explanatory parameters for how hosts respond to infections [1–3]. These factors, as well as the environmental context (e.g. the temporal and spatial distribution of the parasite and the host), determine whether a host in a natural population becomes exposed and infected by a parasite or not, i.e. these factors determine parasite prevalence [4,5]. The parasite infection intensity, on the other hand, is a measure of the severity of the infection in hosts that have contracted the infection [6,7]. Hosts that completely prevent any establishment of a specific parasite have a qualitative form of disease resistance, while hosts that only limit the deleterious effects of a specific parasite have a quantitative form of resistance [8]. Most studies on disease resistance to date in natural populations have correlated candidate immune system alleles (‘immuno-allele’) with prevalence of infection, and not with infection intensity [9–11]. Some studies have found negative associations between prevalence of infection and an immuno-allele, interpreted as an advantageous (resistance) allele, while others have found positive associations, interpreted as a disadvantageous (susceptibility) allele. However, when studying naturally infected wild animals, an allele that is positively correlated with prevalence of infection does not have to be disadvantageous. To explain this more clearly, we present a descriptive framework on how immuno-alleles may associate with infectious diseases in study systems that cannot be sampled repeatedly (before, during and after selection from an infection), which is often the case in natural populations. We outline three scenarios where both prevalence of infection and infection intensity are included (figure 1).
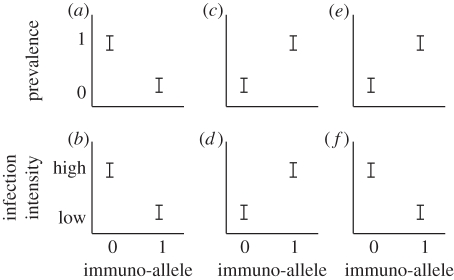
Figure 1.
Hypothetical associations in natural populations between prevalence of infection and infection intensity, respectively, and three different types of immuno-alleles: (a,b) a qualitative disease-resistance allele, (c,d) a susceptibility allele and (e,f) a quantitative disease-resistance allele. A qualitative disease-resistance allele is negatively associated with both (a) prevalence of infection and (b) infection intensity. A susceptibility allele is positively associated with both (c) prevalence of infection and (d) infection intensity. A quantitative disease-resistance allele is positively associated with (e) prevalence of infection but (f) negatively associated with infection intensity.
(i) In the first scenario, we assume that the immuno-allele either provides an individual with full protection against the infection (qualitative resistance), or that it rapidly clears the infection (quantitative resistance). In studies of natural populations, we will rarely be able to separate between these two scenarios, so in the present study we call them jointly qualitative resistance alleles. Qualitative resistance results in a negative association between the observed prevalence of infection and the immuno-allele (figure 1a) and a negative association between infection intensity and the immuno-allele (figure 1b). A disease-resistance allele that gives complete resistance against the infection will be advantageous for the bearer in populations where the parasite is present. (ii) An immuno-allele that makes an individual more prone to become infected is a disease-susceptibility allele. A susceptibility allele will be associated with both higher prevalence (figure 1c) and higher intensity of infection (figure 1d), and it will be of a disadvantage to the bearer in populations where that parasite is present. However, the susceptibility allele may be advantageous when that parasite is absent owing to costs of carrying a disease-resistance allele. Balanced polymorphisms between resistance and susceptibility alleles have been reported frequently, e.g. in studies of wild plants [7,12]. (iii) An immuno-allele that does not give complete resistance against the parasite, but instead suppresses the development of the infection, yields association patterns resembling both a susceptibility (for prevalence; figure 1e) and a qualitative resistance allele (for infection intensity; figure 1f). This kind of immuno-allele is also a disease-resistance allele, although it does not give complete resistance to the parasite but rather keeps the infection intensity low (quantitative resistance). A quantitative disease-resistance allele prevents a mortality-causing infection from becoming too severe, which makes infected individuals with the allele more likely to survive until the time of sampling than individuals without the allele. A subset of the individuals that do not carry the quantitative resistance allele has already died from the infection (black hole in the sampling [13]) which results in a positive association between prevalence of infection and the immuno-allele (figure 1e). From the human literature, it is known that, e.g. HIV infections do not develop into AIDS when the carrier has a resistance haplotype that suppresses the infection [14], and malaria (Plasmodium falciparum) does not develop into severe cerebral malaria when the carrier has a resistance haplotype that suppresses the infection [15]. Animals in natural populations cannot be sampled for prevalence of infection continuously and it is therefore difficult to separate susceptibility (disadvantageous) and quantitative resistance (advantageous) alleles on prevalence data alone. However, we argue that these two, fundamentally different, types of infection-associated alleles may be distinguishable based on their associations with infection intensity. It is therefore crucial to include data on both prevalence and infection intensity when evaluating the true effect of immuno-alleles in natural populations. Furthermore, it is critical to analyse the severity of infections, as well as the timing of sampling in relation to the main selective event.
The major histocompatibility complex (MHC) genes are the most variable genes known in vertebrates and they encode cell-surface proteins that are vital in the acquired immune system [16]. It is generally agreed that this high genetic variation is mainly maintained by some kind of parasite-mediated balancing selection [17]. MHC-dependent disease resistance and susceptibility to infection have been reported in all kinds of vertebrates (from fish to mammals) and for several sorts of infections (protozoa, viruses, bacteria and helminths) [9–11,15,18]. However, MHC genes do not only give resistance to infections but they are also frequently associated with autoimmunity in humans and mice [19,20], although less is known about autoimmunity in wild birds. Interestingly, the MHC haplotype that gives quantitative resistance to HIV (prevents the development into AIDS) is also associated with an increased risk for autoimmunity [14].
MHC genes are probably the best candidate genes to study genetically determined disease resistance in wild animals [9]. We think that MHC genes in birds and avian malaria constitute a highly suitable parasite–host system for investigating qualitative resistance, quantitative resistance and susceptibility to infection in natural populations (figure 1). Positive associations between certain MHC alleles and prevalence of avian malaria infections have been reported in wild populations of house sparrows Passer domesticus and great reed warblers Acrocephalus arundinaceus [21–24]. However, nothing is known about infection intensities in these studies, so we do not know if the positive associations between prevalence of infection and MHC alleles are owing to susceptibility or quantitative resistance alleles (figure 1). Loiseau et al. [23] and Bonneaud et al. [21] interpreted the positive associations between the MHC alleles and prevalence of infection in house sparrows as evidence for susceptibility alleles, while Westerdahl et al. [24] argued that the positive association found in great reed warblers was owing to quantitative resistance.
Avian malaria can be a severe disease in many bird species and it constitutes a very species-rich group of parasites where local host populations often can be infected by multiple species [25]. It is therefore possible to measure selection from several parasites within a single host species [26,27]. Birds that become infected with a novel malaria parasite experience an initial acute phase response to the infection, with typically a high percentage of infected red blood cells, whereafter those that survive continue to carry the malaria as a chronic infection (chronic phase) [28,29]. This means that the chronic phase infection can be measured over long time periods. The acute phase infection intensity is an important factor for determining the risk of mortality in human and rodent malaria infections [30], and it has also been found that a high infection intensity of acute phase malaria may also cause mortality in birds [29,31].
We have previously investigated the prevalence of avian malaria infections, Plasmodium sp., in a natural population of great reed warblers and here the birds are infected with 10 different Plasmodium lineages, two of which are rather common: Plasmodium relictum (GRW4) occurs in 16 per cent and Plasmodium ashfordi (GRW2) in 6 per cent of the breeding individuals [32]. Both GRW4 and GRW2 are transmitted at the great reed warbler's wintering sites in Africa south of the Sahara [27]. From experimental studies, where great reed warblers were inoculated with GRW2 or GRW4 infections, it has been shown that the acute infection intensity of GRW2 is an order of magnitude higher than that of GRW4 [29], suggesting that GRW2 infections would result in a higher mortality rate than GRW4 infections. GRW2 is thus potentially a severe infection for great reed warblers while GRW4 is more benign.
The great reed warbler has at least eight MHC class I (MHC-I) genes and the expressed MHC-I alleles show evidence of positive selection on the DNA sequence level and of temporal selection between breeding cohorts in our study population [33–35]. In particular, one MHC-I allele (B4b) varies significantly in frequency between breeding cohorts and this particular allele is also positively correlated with the prevalence of GRW2 malaria infection [24]. The main aim of the present study is to determine if the previously identified candidate MHC-I allele (B4b; [24]) is a susceptibility allele or a quantitative resistance allele for avian malaria (figure 1c–f). We investigate to what extent MHC-I explains the infection intensities of avian malaria in a natural bird population and test for associations between MHC-I alleles and avian malaria infection intensities of the severe P. ashfordi (GRW2) infection and the more benign P. relictum (GRW4) infection.
2. Methods
(a). Great reed warbler study population
The present study is based on life-history data collected on great reed warblers breeding at lake Kvismaren (59°10′ N, 15°25′ E), south Central Sweden, between 1987 and 1996. Daily visits were performed throughout the breeding season (May to July) and the majority of the breeding males and females (>98%) were ringed with one aluminium ring and a unique combination of two to three colour rings. Detailed descriptions on the fieldwork and measures of life history can be found in, e.g. Bensch and co-workers [36,37]. We collected a blood sample of 20–100 µl from each captured bird, which was stored in SET-buffer at −20°C until DNA extraction.
(b). Malaria screening protocol: parasite quantification
DNA was extracted using standard phenol/chloroform methods [38] and diluted to a concentration of 1 ng µl−1 for quantification of P. ashfordi (GRW2) and P. relictum (GRW4) infection intensity. For measuring infection intensity, we used real-time qPCR with lineage-specific primers (GRW2/8F, GRW2/9R, GRW4/11F, GRW4/11R) to amplify a portion (101 bp) of the cytochrome b gene [29]. It has previously been confirmed that this assay gives very good agreement with parasitaemia estimated by traditional microscopic methods [29]. To get accurate measurements of host DNA in q-PCR, a second reaction was carried out with specific primers [39] SFSR3Fb (5′-ACTAGCCCTTTCAGCGTCATGT-3′ and SFSR3Rb 5′-CATGCTCGGGAACCAAAGG-3′) that amplify a single copy nuclear sequence of a host DNA region (114 bp), which is ultra-conserved across vertebrates [40]. Each reaction of 25 µl included 5 µl (1 ng µl−1) DNA, 12.5 µl Supermix (Platinum SYBR Green qPCR Supermix-UDG, Invitrogen), 0.1 µl ROX, 0.5 µl (GRW2) and 1 µl (GRW4/SFSR3) of each primer, 0.5 µl MgCl2 only for GRW4 and ddH2O. Thermal cycling conditions started with an initial incubation at 50°C for 2 min and 95°C for 2 min, followed by 42 cycles at 95°C for 15 s, 55°C for 30 s (57°C for SFSR3) and at 72°C for 30 s (Mx3000 real-time PCR instrument, Stratagene). Each DNA sample was run in duplicate and scored as average values. On each 96-plate we ran a dilution series (five times dilution) of one sample with known infection level (as determined by microscopy), which was used to calculate the standard curve and the relative infection levels. For quantification of host DNA, we ran standard curves by diluting great reed warbler DNA with ddH2O in five step—five time dilutions (5, 1, 0.2, 0.04 and 0.008 ng ml−1). We discarded and re-ran reactions that produced standard curves that were steeper than −3.8, as this is indicative of inefficient amplifications and errors in the qPCR estimation. We recalculated the relative infection level after adjusting for the total host DNA content in each reaction.
(c). Major histocompatibility complex-I screening protocol
We used sequence-specific amplification of exon 3 to get a measure of functional MHC-I variation (exon 3 encodes an important part of the peptide-binding region of MHC-I). Several transcribed loci were amplified simultaneously using two different primer combinations: HN36–GC46 and HN38–GC46 [35]. These PCR products were 260 bp (primers not included; the entire exon is 274 bp and were separated by the denaturing gradient gel electrophoresis (DGGE) method [35]. The DGGE method separates PCR fragments based on base pair composition [41]. Our sequence-specific primer protocol amplifies 2–12 exon 3 sequences per individual and an individual with few exon 3 sequences is more homozygous than an individual with many alleles, the latter being more heterozygous [35]. We call these exon 3 sequences ‘MHC alleles’ although we are aware that they stem from several loci [35]. The rationale behind this is that in passerine birds, MHC alleles cannot be separated by locus, and furthermore because identical alleles can occur in several loci [13,34].
(d). Statistics
We analysed the relationship between number of MHC alleles as well as presence/absence of the MHC-I allele B4b on the intensity of GRW2 and GRW4 malaria infections among recruited infected great reed warblers (i.e. birds aged more than 1 year and that have returned to lake Kvismaren after wintering in Africa) using a univariate general linear model (GLM). Non-significant factors (p > 0.10) were excluded from the model (number of MHC-I alleles at p = 0.204 and (number of MHC-I alleles)2 at p = 0.443). There were seven double-infected individuals out of 95 (88 single-infected with GRW2 or GRW4 and seven infected with both GRW2 and GRW4) and these were included both as GRW2 and GRW4 infection intensities (n = 102). The significant results from the GLM remained the same when GRW4 infection intensities of the double-infected individuals were excluded from the GLM analysis (data not shown). Neither the infection intensity for GRW2 and GRW4, respectively, differed between single- and double-infected individuals (t-tests—GRW2: t22 = 0.011, p = 0.99; GRW4 t76 = 0.62, p = 0.53) nor did the number of MHC alleles (GRW2: U-test22, p = 0.601; GRW4: U-test76, p = 0.330).
Theory predicts that individuals with the broadest MHC repertoire, i.e. the combination of peptides that could be bound by an individual's MHC molecules, should have an acquired immunity that is able to respond to the widest range of antigens. It is to date not clear if individuals with the largest number of alleles or an optimal number of alleles have the broadest repertoire, so we tested both the total number of alleles and the optimal number of alleles (the χ2 function) against GRW2 and GRW4 infection intensities [42]. We also compared the number of alleles between individuals infected with GRW2 and GRW4, respectively, using a U-test. Here, all GRW4 double-infected individuals were excluded (and hence n = 95) because each individual should contribute only once to the total and the optimal number of alleles. The infection intensities were log-transformed to become normally distributed. All tests are two-tailed and the values are given as mean ± s.e. All analyses were conducted using the statistical package PASW v. 18 (SPSS Inc., Chicago, IL, USA).
3. Results
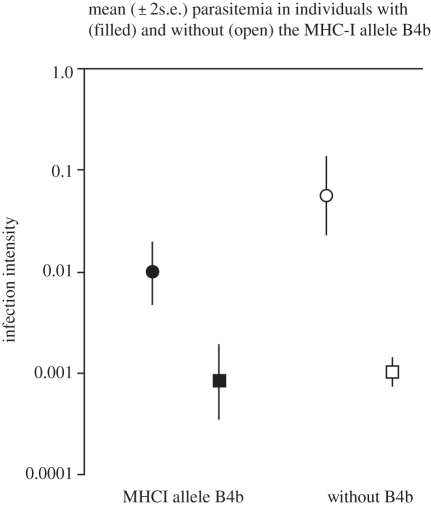
All individuals included in the present study were infected with avian malaria, either the parasite P. ashfordi (GRW2) and/or P. relictum (GRW4). The infection intensity (log-transformed) was significantly higher for GRW2 than for GRW4 (GRW2: −1.49 ± 0.16; GRW4: −3.01 ± 0.07; GLM (malaria species) F1,98 = 61.62, p < 0.001; figure 2). Individuals that carried the candidate MHC-I allele B4b [24] had a significantly lower GRW2 infection intensity than birds not carrying this allele (with B4b = −2.02 ± 0.16, without B4 = −1.27 ± 0.19; GLM (MHC-I allele B4b) F1,98 = 5.53, p = 0.021; figure 2). However, for infection intensity of GRW4 there was no difference in infection intensity whether an individual carried the B4b allele or not (with B4b = −3.08 ± 0.18, without B4 = −2.99 ± 0.08; t76 = 0.46, p = 0.65; figure 2). There was a tendency for an interaction between the two malaria parasite species (GRW2 and GRW4) and the MHC-I allele B4b, with MHC-I allele B4b lowering the GRW2 but not the GRW4 infection intensity (GLM (malaria species × MHC-I allele B4b) F1,98 = 3.49, p = 0.065). The previously identified candidate allele B4b, known to be positively correlated with prevalence of GRW2 infection, is associated with lower GRW2 infection intensity and is therefore an immuno-allele that seems to provide quantitative resistance (cf. figure 1c–f).
Figure 2.
Plasmodium ashfordi GRW2 (circles) and Plasmodium relictum GRW4 (squares) infection intensity in great reed warblers that carry (filled) or do not carry (open) the MHC-I allele B4b (GLM: malaria species, F1,98 = 61.62, p < 0.001; MHC-I allele B4b, F1,98 = 5.53, p = 0.021; malaria species × MHC-I allele B4b, F1,98 = 3.49, p = 0.065).
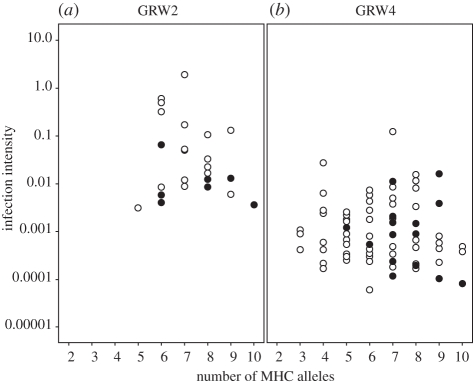
No association was found between the total number of MHC alleles and the ability to suppress the GRW2 or GRW4 infection intensities. Likewise, there was no support for an optimal number of MHC alleles in suppressing the GRW2 or GRW4 infection intensities (excluded factors GLM (number of MHC-I alleles)2 at p = 0.443, number of MHC-I alleles at p = 0.204; figure 3). However, individuals with a GRW2 infection had a larger number of MHC alleles than individuals with a GRW4 infection (U-test100, p = 0.047; GRW2 = 7.25 ± 0.26; GRW4 = 6.45 ± 0.20; figure 3).
Figure 3.
(a) Plasmodium ashfordi GRW2 infection intensity in great reed warblers with 5–10 MHC alleles (mean value: 7.2 alleles). (b) Plasmodium relictum GRW4 infection intensity in great reed warblers with 3–10 MHC alleles (mean value: 6.4 alleles). Individuals that carry the MHC-I allele B4b are denoted with filled circles and individuals that do not carry B4b are denoted with open circles.
4. Discussion
Our framework on how immuno-alleles associate with infectious diseases in natural populations demonstrated that it is essential to interpret both prevalence of infection and infection intensity in natural study systems (figure 1). A disadvantageous susceptibility allele and an advantageous quantitative resistance allele cannot be separated based on prevalence data alone, although these alleles are clearly distinguishable using infection intensity data. In the present study, we wanted to determine if the previously found candidate MHC-I allele B4b, identified based on prevalence of infection [24], was an avian malaria susceptibility or quantitative resistance allele. We used qPCR to estimate infection intensities of one benign P. relictum (GRW4) and one severe P. ashfordi (GRW2) lineage of avian malaria and found that birds carrying the MHC-I allele B4b had significantly lower GRW2 infection intensity than individuals without this allele (figure 2), bearing in mind that our previous study showed a higher prevalence of GRW2 in individuals having allele B4b than in those lacking the allele. Our data are in accordance with the quantitative resistance allele scenario (suppression of infection), because we found a positive association between prevalence of infection and the MHC-I allele B4b (figure 1e) and a negative association between infection intensity and the MHC-I allele B4b (figure 1f). These results imply that great reed warblers carrying the MHC-I allele B4b are more likely to survive an (acute phase) GRW2 P. ashfordi malaria infection than individuals without this allele, consistent with patterns found in both human and mice malaria where there is a strong positive correlation between infection intensity and mortality [30]. In contrast, we found no association between the B4b allele and the more benign GRW4 P. relictum infection (figure 2). Our present result on the GRW2 infection is in agreement with the classical finding by Hill et al. [15] in humans where children with certain MHC haplotypes did not develop severe malaria, although all children in the study suffered from the human P. falciparum malaria infection. The parallel scenario for great read warblers would be that GRW2 naive individuals carrying the MHC-I allele B4b do not develop severe GRW2 infection, while great reed warblers without the B4b allele do.
Several studies from natural populations have found associations between prevalence of infection and certain MHC alleles or MHC heterozygosity [22,43–45], but only a handful of studies have yet investigated associations between infection intensities and certain MHC alleles/MHC heterozygosity in natural populations [46–49]. Kloch et al. [47] recently showed that an MHC class IIB allele (Mygl-DRB*028) was associated with lower infection intensities of the nematode Aspiculuris tetraptera across three subpopulations of bank voles Myodes glareolus. These negative associations between infection intensity and an MHC allele indicate qualitative or quantitative resistance (these cannot be separated here as we do not know the prevalence of infection; figure 1b,f). In striped mice Rhabdomys pumilio and yellow-necked mice Apodemus flavicollis, there were positive associations between both prevalence of nematodes and specific MHC class IIB alleles and nematode intensity and specific MHC class IIB alleles, hence suggesting susceptibility alleles (figure 1c,d) [46,49]. Finally, our present study on great reed warblers implied an allele for quantitative resistance (figure 1e,f). The two studies on susceptibility alleles in rodents (mentioned above) report positive associations between MHC class IIB alleles and prevalence of infection and infection intensity, respectively, of relatively benign nematode infections, while we report associations between an MHC-I allele and prevalence and infection intensity of a severe avian malaria infection in great reed warblers. Different types of disease patterns (qualitative and quantitative resistance, and susceptibility, figure 1) may be typical for different levels of infection severity. The selective event during severe infections could result in mortality of individuals that are less able to suppress the acute infection, e.g. individuals without an MHC resistance allele that suppress infection intensity. In more benign infections, the selective event is less critical and individuals without a resistance allele will most often survive the infection. It is reasonable that severe infections need to be suppressed to a larger extent than benign infections. However, complete absence of benign helminth infections could actually be a more severe problem for an individual than having the infection, because individuals who manage to completely clear helminth infections may become very sick owing to the pathology of the actual immune response [50].
Loiseau et al. [23] found positive and negative associations between identical MHC alleles and prevalence of one avian malaria infection (P. relictum, cytochrome b lineages SGS1 and GRW11) across 13 populations of house sparrows. This result was interpreted as host–parasite antagonistic coevolution, a potential mechanism for spatial diversifying evolution on MHC. It would be very interesting to know if the house sparrow MHC alleles that are both positively and negatively related to avian malaria infections are (i) both susceptibility and qualitative resistance alleles or (ii) both quantitative and qualitative resistance alleles (figure 1). From the plant literature, it is well established that resistance alleles may be costly and are selected against when the infection is rare, resulting in a balanced polymorphism between resistance and susceptibility alleles [7]. Naturally, we do not have the resolution in the bird MHC–avian malaria study system that is found in the plant resistance/susceptibility system [51]. In order to interpret positive and negative associations between MHC alleles and prevalence of avian malaria infections correctly, we must know the fitness consequences of each malaria strain. If it is not possible to interpret each strain's fitness consequence, we suggest that it is partly possible to overcome this problem by measuring infection intensity rather than only the prevalence of infection. Spurgin & Richardson [52] recently reviewed mechanisms behind ‘pathogen-mediated selection’ (heterozygote advantage, negative frequency-dependent selection and diversifying selection in a variable environment) and suggested ways to distinguish these mechanisms in natural populations. We would like to add one additional parameter to ‘pathogen-mediated selection’. To elucidate what maintains MHC polymorphism in natural populations, we should not only focus on the mechanism, but also seek to understand when candidate MHC alleles are advantageous (i.e. quantitative and qualitative resistance alleles) and when they are disadvantageous (i.e. susceptibility alleles) to be better able to interpret patterns of balancing selection.
Nowak et al. [53] proposed that individuals with an optimal MHC repertoire have an adaptive immune system that could respond to a wider range of antigens when compared with individuals with a narrower or wider MHC repertoire. An optimal MHC repertoire was recently evaluated and elaborated on by Woelfing et al. [54]. However, it is to date not completely clear whether it is individuals with an optimal number of MHC alleles or individuals with the highest number of MHC alleles that have the broadest MHC repertoire, i.e. ability to bind most antigens. Yet, an important factor for how successful MHC molecules will bind and recognize peptides from pathogens is the binding properties and the stability of each MHC molecule per se [14,55]. Several studies of natural populations have found that an optimal number of MHC alleles are most advantageous for avoiding infections [45,47,56,57]. In the great reed warbler, we found neither any association between MHC diversity (presented as the number of MHC alleles), nor support for an optimal number of MHC alleles in controlling the intensity of either GRW2 or GRW4 infections. One explanation for the absence of such associations could be that we studied the birds in spring and summer at their European breeding sites, which is after the main selective event of the acute-phase GRW2 infection had occurred. We were therefore able to screen only those individuals who had survived the acute phase of the GRW2 infection. Interestingly, among these survivors, the GRW2-infected individuals had more MHC alleles than individuals carrying the less pathogenic GRW4 infection (figure 3). We interpret this result as being caused by selection against individuals with a low number of MHC alleles during the acute phase of GRW2, but not GRW4, infections in Africa. Individuals with the GRW4 infection have the same number of MHC alleles (6.45 ± 0.20) as individuals who were screened as non-infected with avian malaria in a previous study (6.36 ± 0.10; [24]). Individuals with many MHC alleles are more likely to carry rare advantageous alleles that can bind antigens from the GRW2 parasite and individuals with few MHC alleles would therefore more often be selected against.
Our interpretation that individuals with the MHC-I allele B4b can suppress GRW2 malaria infections is based on infection intensity data collected during the chronic phase of the malaria infection. However, from a survival perspective, it is likely that it is suppression during the acute phase that is most critical [30]. Recent results showed that the infection intensity of the chronic GRW2 infection could be predicted from infection intensity of the acute infection (M. Asghar, H. Westerdahl, P. Zethindjiev, D. Hasselquist and S. Bensch, unpublished data). This association between infection intensities during the acute and chronic phases is a very important finding lending strong support to the hypothesis that the chronic infections that we measured in great reed warblers at Lake Kvismaren are correlated with the preceding acute phase of infection that occurred in tropical Africa. If the acute and chronic phase infections were not correlated, a quantitative measure of infection intensity would not have been more informative than a simple qualitative estimate.
Wild birds that have an acute phase malaria infection are very unlikely to be caught in mist nets, because severely infected birds mainly sit still to recover from the infection or to avoid predators [25,58]. So, even in study populations where birds are monitored at sites and times of the year where there is transmission of malaria parasites, it is still difficult to observe and catch ill birds. We know from captive malaria-naive birds that were experimentally infected with Plasmodium that this infection can reach high intensities during the acute phase [28,29]. It is therefore likely that great reed warblers that get severe avian malaria in the wild in Africa have very high infection intensities. These birds will only rarely be recruited into the breeding population in Europe and constitute an event where we very rarely will be able to collect data on chronic phase infection.
In conclusion, we measured both prevalence of infection and infection intensity to determine whether our candidate MHC-I allele B4b was a qualitative disease resistance or a susceptibility allele. Our data show that great reed warblers carrying the candidate MHC-I allele B4b manage to suppress the acute GRW2 avian malaria infection more successfully than individuals without this allele. The B4b allele provides quantitative resistance to the malaria GRW2 infection. We suggest that individuals who are able to suppress the infection survive the acute phase infection more frequently, and this is in line with our finding of a positive association between prevalence of infection and the candidate MHC allele B4b. Our study also highlights a general problem when studying the prevalence of infection and disease resistance in natural populations, and we claim that candidate alleles that provide quantitative disease resistance, qualitative disease resistance and susceptibility to disease, can only be separated adequately based on data on both prevalence of infection and infection intensity.
Acknowledgements
The present study was financed by grants from the Swedish Research Council (VR, 621-2006-4551 to H.W.), (VR, 621-2007-5193 to S.B.) and (VR, 621-2007-5385 to D.H.), from the Swedish Research Council Formas (217-2006-375 to D.H.), from Lunds Djurskyddsfond to H.W. and D.H., and from Higher Education Commission of Pakistan (HEC) to M.A. Comments from and discussions with Barbara Tschirren, Lars Råberg and Kristin Scherman greatly improved the manuscript. This is report number 162 from the Kvismare Bird Observatory.
References
- 1.De Roode J. C., Gold L. R., Altizer S. 2007. Virulence determinants in a natural butterfly-parasite system. Parasitology 134, 657–668 10.1017/S0031182006002009 (doi:10.1017/S0031182006002009) [DOI] [PubMed] [Google Scholar]
- 2.Greenwood B. M., Marsh K., Snow R. 1991. Why do some African children develop severe malaria? Parasitol. Today 7, 277–281 10.1016/0169-4758(91)90096-7 (doi:10.1016/0169-4758(91)90096-7) [DOI] [PubMed] [Google Scholar]
- 3.Miller L. H., Baruch D. I., Marsh K., Doumbo O. K. 2002. The pathogenic basis of malaria. Nature 415, 673–679 10.1038/415673a (doi:10.1038/415673a) [DOI] [PubMed] [Google Scholar]
- 4.Bensch S., Akesson S. 2003. Temporal and spatial variation of haemoprotozoans in Scandinavian willow warblers. J. Parasitol. 89, 388–391 10.1645/0022-3395(2003)089[0388:TASVOH]2.0.CO;2 (doi:10.1645/0022-3395(2003)089[0388:TASVOH]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 5.Yohannes E., Hansson B., Lee R. W., Waldenström J., Westerdahl H., Åkesson M., Hasselquist D., Bensch S. 2008. Isotope signatures in winter moulted feathers predict malaria prevalence in a breeding avian host. Oecologia 158, 299–306 10.1007/s00442-008-1138-3 (doi:10.1007/s00442-008-1138-3) [DOI] [PubMed] [Google Scholar]
- 6.Beldomenico P. M., Begon M. 2010. Disease spread, susceptibility and infection intensity: vicious circles? Trends Ecol. Evol. 25, 21–27 10.1016/j.tree.2009.06.015 (doi:10.1016/j.tree.2009.06.015) [DOI] [PubMed] [Google Scholar]
- 7.Bergelson J., Purrington C. B. 1996. Surveying patterns in the cost of resistance in plants. Am. Nat. 148, 536–558 10.1086/285938 (doi:10.1086/285938) [DOI] [Google Scholar]
- 8.Gandon S., Michalakis Y. 2000. Evolution of parasite virulence against qualitative or quantitative form of resistance. Proc. R. Soc. Lond. B 267, 985–990 10.1098/rspb.2000.1100 (doi:10.1098/rspb.2000.1100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernatchez L., Landry C. 2003. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J. Evol. Biol. 16, 363–377 10.1046/j.1420-9101.2003.00531.x (doi:10.1046/j.1420-9101.2003.00531.x) [DOI] [PubMed] [Google Scholar]
- 10.Piertney S. B., Oliver M. K. 2006. The evolutionary ecology of the major histocompatibility complex. Heredity 96, 7–21 10.1038/sj.hdy.6800724 (doi:10.1038/sj.hdy.6800724) [DOI] [PubMed] [Google Scholar]
- 11.Sommer S. 2005. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front. Zool. 2, 1–18 10.1186/1742-9994-2-1 (doi:10.1186/1742-9994-2-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purrington C. B., Bergelson J. 1999. Exploring the physiological basis of costs of herbicide resistance in Arabidopsis thaliana. Am. Nat. 154, 82–91 10.1086/303285 (doi:10.1086/303285) [DOI] [PubMed] [Google Scholar]
- 13.Westerdahl H. 2007. Passerine MHC: genetic variation and disease resistance in the wild. J. Ornithol. 148, 469–477 10.1007/s10336-007-0230-5 (doi:10.1007/s10336-007-0230-5) [DOI] [Google Scholar]
- 14.Kosmrlj A., et al. 2010. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature 465, 350–354 10.1038/nature08997 (doi:10.1038/nature08997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill A. V. S., et al. 1991. Common West African HlA antigens are associated with protection from severe malaria. Nature 352, 595–600 10.1038/352595a0 (doi:10.1038/352595a0) [DOI] [PubMed] [Google Scholar]
- 16.Klein J. 1986. Natural history of the major histocompatibility complex, 1st edn New York, NY: John Wiley and Sons [Google Scholar]
- 17.Hedrick P. W. 2002. Pathogen resistance and genetic variation at MHC loci. Evolution 56, 1902–1908 10.1111/j.0014-3820.tb00116.x (doi:10.1111/j.0014-3820.tb00116.x) [DOI] [PubMed] [Google Scholar]
- 18.Worley K., Collet J., Spurgin L. G., Cornwallis C., Pizzari T., Richardson D. S. 2010. MHC heterozygosity and survival in red junglefowl. Mol. Ecol. 19, 3064–3075 10.1111/j.1365-294X.2010.04724.x (doi:10.1111/j.1365-294X.2010.04724.x) [DOI] [PubMed] [Google Scholar]
- 19.Fujisawa T., et al. 2006. MHC-linked susceptibility to type 1 diabetes in the NOD mouse: further localization of Idd16 by subcongenic analysis. Ann. NY Acad. Sci. 1079, 118–121 10.1196/annals.1375.017 (doi:10.1196/annals.1375.017) [DOI] [PubMed] [Google Scholar]
- 20.Zenewicz L. A., Abraham C., Flavell R. A., Cho J. H. 2010. Unraveling the genetics of autoimmunity. Cell 140, 791–797 10.1016/j.cell.2010.03.003 (doi:10.1016/j.cell.2010.03.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonneaud C., Perez-Tris J., Federici P., Chastel O., Sorci G. 2006. Major histocompatibility alleles associated with local resistance to malaria in a passerine. Evolution 60, 383–389 10.1111/j.0014-3820.2006.tb01114.x (doi:10.1111/j.0014-3820.2006.tb01114.x) [DOI] [PubMed] [Google Scholar]
- 22.Loiseau C., Zoorob R., Garnier S., Birard J., Federici P., Julliard R., Sorci G. 2008. Antagonistic effects of a MHC class I allele on malaria-infected house sparrows. Ecol. Lett. 11, 258–265 10.1111/j.1461-0248.2007.01141.x (doi:10.1111/j.1461-0248.2007.01141.x) [DOI] [PubMed] [Google Scholar]
- 23.Loiseau C., Zoorob R., Robert A., Chastel O., Julliard R., Sorci G. 2011. Plasmodium relictum infection and MHC diversity in the house sparrow (Passer domesticus). Proc. R. Soc. B 278, 1264–1272 10.1098/rspb.2010.1968 (doi:10.1098/rspb.2010.1968) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westerdahl H., Waldenstrom J., Hansson B., Hasselquist D., von Schantz T., Bensch S. 2005. Associations between malaria and MHC genes in a migratory songbird. Proc. R. Soc. B 272, 1511–1518 10.1098/rspb.2005.3113 (doi:10.1098/rspb.2005.3113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valkiunas G. 2005. Avian malaria parasites and other haemosporidia. Boca Raton, FL: CRC Press [Google Scholar]
- 26.Hellgren O., Krizanauskiene A., Valkiunas G., Bensch S. 2007. Diversity and phylogeny of mitochondrial cytochrome B lineages from six morphospecies of avian Haemoproteus (Haemosporida: Haemoproteidae). J. Parasitol. 93, 889–896 10.1645/GE-1051R1.1 (doi:10.1645/GE-1051R1.1) [DOI] [PubMed] [Google Scholar]
- 27.Waldenstrom J., Bensch S., Kiboi S., Hasselquist D., Ottosson U. 2002. Cross-species infection of blood parasites between resident and migratory songbirds in Africa. Mol. Ecol. 11, 1545–1554 10.1046/j.1365-294X.2002.01523.x (doi:10.1046/j.1365-294X.2002.01523.x) [DOI] [PubMed] [Google Scholar]
- 28.Palinauskas V., Valkiunas G., Bolshakov C. V., Bensch S. 2008. Plasmodium relictum (lineage P-SGS1): effects on experimentally infected passerine birds. Exp. Parasitol. 120, 372–380 10.1016/j.exppara.2008.09.001 (doi:10.1016/j.exppara.2008.09.001) [DOI] [PubMed] [Google Scholar]
- 29.Zehtindjiev P., Ilieva M., Westerdahl H., Hansson B., Valkiunas G., Bensch S. 2008. Dynamics of parasitemia of malaria parasites in a naturally and experimentally infected migratory songbird, the great reed warbler Acrocephalus arundinaceus. Exp. Parasitol. 119, 99–110 10.1016/j.exppara.2007.12.018 (doi:10.1016/j.exppara.2007.12.018) [DOI] [PubMed] [Google Scholar]
- 30.Mackinnon M. J., Read A. F. 2003. The effects of host immunity on virulence–transmissibility relationships in the rodent malaria parasite Plasmodium chabaudi. Parasitology 126, 103–112 10.1017/S003118200200272X (doi:10.1017/S003118200200272X) [DOI] [PubMed] [Google Scholar]
- 31.Atkinson C. T., van Riper C. 1991. Pathogenicity and epizootiology of avian haematozoa: Plasmodium, Leucocytozoon and Haemoproteus. In Bird–parasite interactions (eds Loye J. E., Zuk M.), pp. 19–48 New York, NY: Oxford University Press [Google Scholar]
- 32.Bensch S., Waldenstrom J., Jonzen N., Westerdahl H., Hansson B., Sejberg D., Hasselquist D. 2007. Temporal dynamics and diversity of avian malaria parasites in a single host species. J. Anim. Ecol. 76, 112–122 10.1111/j.1365-2656.2006.01176.x (doi:10.1111/j.1365-2656.2006.01176.x) [DOI] [PubMed] [Google Scholar]
- 33.Westerdahl H., Hansson B., Bensch S., Hasselquist D. 2004a. Between-year variation of MHC allele frequencies in great reed warblers: selection or drift? J. Evol. Biol. 17, 485–492 10.1111/j.1420-9101.2004.00711.x (doi:10.1111/j.1420-9101.2004.00711.x) [DOI] [PubMed] [Google Scholar]
- 34.Westerdahl H., Wittzell H., von Schantz T. 1999. Polymorphism and transcription of MHC class I genes in a passerine bird, the great reed warbler. Immunogenetics 49, 158–170 10.1007/s002510050477 (doi:10.1007/s002510050477) [DOI] [PubMed] [Google Scholar]
- 35.Westerdahl H., Wittzell H., von Schantz T., Bensch S. 2004. MHC class I typing in a songbird with numerous loci and high polymorphism using motif-specific PCR and DGGE. Heredity 92, 534–542 10.1038/sj.hdy.6800450 (doi:10.1038/sj.hdy.6800450) [DOI] [PubMed] [Google Scholar]
- 36.Bensch S., Hasselquist D. 1991. Territory infidelity in the polygynous great reed warbler Acrocephalus arundinaceus: the effect of variation in territory attractiveness. Anim. Ecol. 60, 857–871 10.2307/5418 (doi:10.2307/5418) [DOI] [Google Scholar]
- 37.Bensch S., Hasselquist D., von Schantz T. 1994. Genetic similarity between parents predicts hatching failure: non-incestuous inbreeding in the great reed warbler? Evolution 48, 317–326 10.2307/2410095 (doi:10.2307/2410095) [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- 39.Asghar M., Hasselquist D., Bensch S. 2011. Are chronic haemosporidian infections costly in wild birds? J. Avian Biol. 42 10.1111/j.1600-0587.2011.05281.x (doi:10.1111/j.1600-0587.2011.05281.x) [DOI] [Google Scholar]
- 40.Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W. J., Mattick J. S., Haussler D. 2004. Ultraconserved elements in the human genome. Science 304, 1321–1325 10.1126/science.1098119 (doi:10.1126/science.1098119) [DOI] [PubMed] [Google Scholar]
- 41.Myers R. M., Maniatis T., Lerman L. S. 1987. Detection and localization of single base changes by denaturing gradient gel-electrophoresis. Methods Enzymol. 155, 501–527 10.1016/0076-6879(87)55033-9 (doi:10.1016/0076-6879(87)55033-9) [DOI] [PubMed] [Google Scholar]
- 42.Nowak M. A., Tarczyhornoch K., Austyn J. M. 1992. The optimal number of major histocompatibility complex-molecules in an individual. Proc. Natl Acad. Sci. USA 89, 10 896–10 899 10.1073/pnas.89.22.10896 (doi:10.1073/pnas.89.22.10896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langefors A., Lohm J., Grahn M., Andersen O., von Schantz T. 2001. Association between major histocompatibility complex class IIB alleles and resistance to Aeromonas salmonicida in Atlantic salmon. Proc. R. Soc. Lond. B 268, 479–485 10.1098/rspb.2000.1378 (doi:10.1098/rspb.2000.1378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madsen T., Ujvari B. 2006. MHC class I variation associates with parasite resistance and longevity in tropical pythons. J. Evol. Biol. 19, 1973–1978 10.1111/j.1420-9101.2006.01158.x (doi:10.1111/j.1420-9101.2006.01158.x) [DOI] [PubMed] [Google Scholar]
- 45.Wegner K. M., Kalbe M., Kurtz J., Reusch T. B. H., Milinski M. 2003. Parasite selection for immunogenetic optimality. Science 301, 1343. 10.1126/science.1088293 (doi:10.1126/science.1088293) [DOI] [PubMed] [Google Scholar]
- 46.Froeschke G., Sommer S. 2005. MHC class II DRB variability and parasite load in the striped mouse (Rhabdomys pumilio) in the southern Kalahari. Mol. Biol. Evol. 22, 1254–1259 10.1093/molbev/msi112 (doi:10.1093/molbev/msi112) [DOI] [PubMed] [Google Scholar]
- 47.Kloch A., Babik W., Bajer A., Sinski E., Radwan J. 2010. Effects of an MHC-DRB genotype and allele number on the load of gut parasites in the bank vole Myodes glareolus. Mol. Ecol. 19, 255–265 10.1111/j.1365-294X.2009.04476.x (doi:10.1111/j.1365-294X.2009.04476.x) [DOI] [PubMed] [Google Scholar]
- 48.Meyer-Lucht Y., Sommer S. 2005. MHC diversity and the association to nematode parasitism in the yellow-necked mouse (Apodemus flavicollis). Mol. Ecol. 14, 2233–2243 10.1111/j.1365-294X.2005.02557.x (doi:10.1111/j.1365-294X.2005.02557.x) [DOI] [PubMed] [Google Scholar]
- 49.Meyer-Lucht Y., Sommer S. 2009. Number of MHC alleles is related to parasite loads in natural populations of yellow necked mice, Apodemus flavicollis. Evol. Ecol. Res. 11, 1085–1097 [Google Scholar]
- 50.Rook G. A. W. 2009. Review series on helminths, immune modulation and the hygiene hypothesis: the broader implications of the hygiene hypothesis. Immunology 126, 3–11 10.1111/j.1365-2567.2008.03007.x (doi:10.1111/j.1365-2567.2008.03007.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Todesco M., et al. 2010. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465, 632–636 10.1038/nature09083 (doi:10.1038/nature09083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spurgin L. G., Richardson D. S. 2010. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. R. Soc. B 277, 979–988 10.1098/rspb.2009.2084 (doi:10.1098/rspb.2009.2084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nowak M. A., Tarczyhornoch K., Austyn J. M. 1992. The optimal number of major histocompatibility complex-molecules in an individual. Proc. Natl Acad. Sci. USA 89, 10 896–10 899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woelfing B., Traulsen A., Milinski M., Boehm T. 2009. Does intra-individual major histocompatibility complex diversity keep a golden mean? Phil. Trans. R. Soc. B 364, 117–128 10.1098/rstb.2008.0174 (doi:10.1098/rstb.2008.0174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roder G., Geironson L., Darabi A., Harndahl M., Schafer-Nielsen C., Skjodt K., Buus S., Paulsson K. 2009. The outermost N-terminal region of tapasin facilitates folding of major histocompatibility complex class I. Eur. J. Immunol. 39, 2682–2694 10.1002/eji.200939364 (doi:10.1002/eji.200939364) [DOI] [PubMed] [Google Scholar]
- 56.Harf R., Sommer S. 2005. Association between major histocompatibility complex class II DRB alleles and parasite load in the hairy-footed gerbil, Gerbillurus paeba, in the southern Kalahari. Mol. Ecol. 14, 85–91 10.1111/j.1365-294X.2004.02402.x (doi:10.1111/j.1365-294X.2004.02402.x) [DOI] [PubMed] [Google Scholar]
- 57.Madsen T., Ujvari B. 2006. MHC class I variation associates with parasite resistance and longevity in tropical pythons. J. Evol. Biol. 19, 1973–1978 10.1111/j.1420-9101.2006.01158.x (doi:10.1111/j.1420-9101.2006.01158.x) [DOI] [PubMed] [Google Scholar]
- 58.Moller A. P., Nielsen J. T. 2007. Malaria and risk of predation: a comparative study of birds. Ecology 88, 871–881 10.1890/06-0747 (doi:10.1890/06-0747) [DOI] [PubMed] [Google Scholar]