Abstract
Seasonal migration in birds is known to be highly labile and subject to rapid change in response to selection, such that researchers have hypothesized that phylogenetic relationships should neither predict nor constrain the migratory behaviour of a species. Many theories on the evolution of bird migration assume a framework that extant migratory species have evolved repeatedly and relatively recently from sedentary tropical or subtropical ancestors. We performed ancestral state reconstructions of migratory behaviour using a comprehensive, well-supported phylogeny of the Parulidae (the ‘wood-warblers’), a large family of Neotropical and Nearctic migratory and sedentary songbirds, and examined the rates of gain and loss of migration throughout the Parulidae. Counter to traditional hypotheses, our results suggest that the ancestral wood-warbler was migratory and that losses of migration have been at least as prevalent as gains throughout the history of Parulidae. Therefore, extant sedentary tropical radiations in the Parulidae represent losses of latitudinal migration and colonization of the tropics from temperate regions. We also tested for phylogenetic signal in migratory behaviour, and our results indicate that although migratory behaviour is variable within some wood-warbler species and clades, phylogeny significantly predicts the migratory distance of species in the Parulidae.
Keywords: evolution of migration, ancestral state reconstruction, phylogenetic signal, Parulidae, seasonal migration
1. Introduction
Our understanding of the evolution of seasonal migration in birds has been revolutionized by experimental demonstrations that many aspects of this behaviour are heritable and subject to rapid change in response to selection [1–3]. This information, coupled with the hypothesis that migratory genes are universal and ancestral in birds [2–4], has led to the conclusion that transitions from sedentary to migratory behaviour, or vice versa, require only selection acting on a pre-existing genetic programme, rather than the repeated evolution of an innovation [2,4–8]. Although the deep ancestry of migration confounds our ability to discern the ultimate origins of migratory behaviour in birds, the apparently dynamic changes in migration throughout extant avian lineages allow comparative investigation of the phylogenetic patterns of ancestry, homoplasy and retention of migration [6,9,10]. Such comparisons can help explain how and why migratory behaviour has changed through evolutionary time and across speciation events.
Despite the apparent rapidity with which migration can evolve, the degree of homoplasy in migratory behaviour has not been fully tested. Helbig [4] hypothesized that migratory behaviour changes so rapidly that it should show little or no phylogenetic signal even within genera; in other words, phylogenetic relationships should neither predict nor constrain whether a species is migratory [7,8]. Helbig [4] further argued that the increased natal dispersal often associated with seasonal migration [11–14] leads to homogenizing gene flow, which should constrain migratory, taxa from speciating on the breeding grounds. The combination of multiple origins of migration within lineages and failure of migratory species to speciate in situ led Helbig [4] to hypothesize that migrants should be most closely related to sedentary or less migratory species, as opposed to other migratory species. Most studies that have examined migration in a phylogenetic context have found evidence for multiple origins of migratory behaviour within the focal lineage [9,15–21] (but see [22,23]), but no study has yet formally tested for phylogenetic signal in migratory behaviour. Examination of the phylogenetic signal of migration within bird lineages will help clarify the degree to which extant migratory birds have speciated in situ, versus having arisen repeatedly from sedentary ancestors.
Regardless of the amount of change in migratory behaviour among lineages, we cannot assume that the dominant direction of change within these lineages has been from sedentary to migratory behaviour. Nevertheless, discussions or models of how migratory behaviour evolves within some lineages but not in others often presuppose that the ancestral state of the migratory population in question was sedentary, and then consider the ecological, behavioural or genetic conditions under which migration could evolve from a hypothetical sedentary population (e.g. [24–26]). For example, both Berthold [27] and Rappole [28] noted that temperate, migratory species usually have close relatives in the tropics that are sedentary or partially migratory, and they treated this as evidence that sedentary behaviour is ancestral in those lineages. They interpreted this pattern as supporting a traditional hypothesis that migration in birds evolves out of the tropics through the extension of ancestral tropical or subtropical breeding ranges into temperate regions (the ‘southern home theory’), as opposed to the extension of an ancestral non-breeding range from temperate to tropical regions to avoid deteriorating seasonal conditions (the ‘northern home theory’; reviewed in [8,25,28,29]).
However, the region where migration evolves within a lineage may be different from the region where a lineage has its phylogenetic origins [6,8,29]. Furthermore, having relatives that are tropical sedentary species does not imply that this is the ancestral condition; within a given lineage, migration may be ancestral, and extant sedentary species may represent a derived condition through losses of migratory behaviour. Therefore, the ancestral state of migration must be analysed in a phylogenetic framework [6,9]. Previous studies that have reconstructed the ancestral state of migration in genera or families in an explicit phylogenetic context have found either equivocal results or results in favour of a sedentary ancestor [9,15–23]. Thus, current phylogenetic evidence supports the idea that extant migratory species are often derived from their sedentary tropical or subtropical relatives [25,27,28,30], and hence that migration among extant birds has evolved relatively recently compared with the total age of the lineages from which migratory species are derived.
Phylogenetic patterns of migration therefore inform our understanding of the age of migratory behaviours and pathways, as well as the prevalence of losses versus gains of migration through time [30,31]. In this paper, we examine phylogenetic patterns of migratory behaviour in the Parulidae, using a comprehensive, well-supported molecular phylogeny [32]. The Parulidae (New World warblers, or ‘wood-warblers’) are restricted to the Western Hemisphere but span a wide spectrum of migratory behaviours, from mostly sedentary tropical genera to some of the most celebrated champions of long-distance migration [33,34]. As a whole, they are one of the most prevalent, familiar and well-studied components of the Nearctic–Neotropical migratory avifauna, and as individual species they have served as models in a wide variety of studies of avian ecology and evolution [35–40]. Here, we ask whether migratory or sedentary behaviour is ancestral in the Parulidae, whether migration tends to be lost or gained across the phylogeny and whether migratory behaviour shows a lack of phylogenetic signal consistent with Helbig's hypotheses that migratory species should represent multiple independent origins of migration from sedentary lineages rather than speciation of migratory species in situ.
2. Material and methods
We performed comparative phylogenetic analyses using a comprehensive, highly resolved molecular phylogeny that is based on 10 442 nucleotides of mitochondrial and nuclear DNA [32]. The phylogeny includes 108 out of 110 possible species of Parulidae, including recently extinct taxa, and there is strong support that these wood-warbler taxa comprise a monophyletic group [32]. We used the fully resolved tree generated with maximum likelihood from Lovette et al. [32]. The topology contains many clades that do not correspond to traditionally recognized Parulidae genera [41], but for clarity we use the revised generic nomenclature proposed by Lovette et al. [32]. To account for phylogenetic uncertainty, we verified results from the maximum-likelihood topology by repeating analyses across a posterior distribution of 1000 trees generated with Bayesian methods by Lovette et al. [32] (electronic supplementary material). Prior to analysis, all outgroups used for phylogeny construction by Lovette et al. were pruned from the trees and all topologies were made ultrametric with branch lengths proportional to time using penalized likelihood [42] (electronic supplementary material).
(a). Ancestral state reconstructions
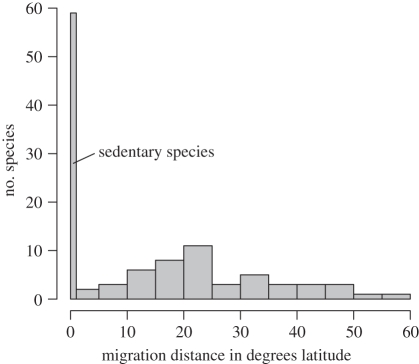
We gathered literature data on migratory behaviour and migration distance for each species [33,34,43–47]. We coded a discrete binary character with states migratory (49 species) and non-migratory (59 species) and a continuous character of migration distance, each distance corresponding to degrees of latitude between the midpoints of each species's breeding and wintering ranges (figure 1). For the binary character, we defined migratory species as those in which at least some individuals migrate seasonally between different latitudinal geographical breeding and wintering ranges. This includes eight species that are either partial migrants [27], in which not all individuals across a broad range or population migrate (e.g. Geothlypis trichas), or species that are fully migratory over most of their range but also have isolated sedentary populations (e.g. Setophaga dominica). In general, migratory behaviour is strongly correlated with breeding in the temperate zone in Parulidae [34], and the migratory character state does not include tropical species that may undertake short-distance altitudinal migration, nor two species that have limited seasonal movements at the fringes of their ranges (Setophaga pitiayumi and Basileuterus lachrymosa). We also reconstructed ancestral states of a binary character for which the eight species whose migratory status is variable were coded as sedentary, as opposed to migratory, to test the robustness of our conclusions to potential ambiguities in character-state coding.
Figure 1.
Histogram of migratory distances of all 108 Parulidae species included in the analysis. Distances correspond to the degrees of latitude between each species's breeding range midpoint and non-breeding range midpoint.
We first performed analyses on the fully resolved maximum-likelihood topology. We used Mesquite 2.71 [48] to perform exploratory ancestral state reconstruction in both maximum-parsimony and maximum-likelihood frameworks. Final maximum-likelihood analyses on this topology were performed in the R packages Ape [49] and Diversitree [50,51]; results from maximum parsimony were consistent with those from maximum likelihood and are not shown. For maximum-likelihood analysis, we performed likelihood ratio tests to determine the best-fitting model of character evolution based on likelihoods generated in the program Bayes Traits Multistate [52–54]. A two-rate (Mk2) model of character evolution was not significantly better than a one-rate (Mk1) model for the binary migration character (LR = 0.530, p > 0.05, df = 1). Therefore, we used the simpler, one-rate model to reconstruct ancestral states [53–55]; results from exploratory analyses using Mk2 are consistent with the results presented here using Mk1 (electronic supplementary material, table S1).
If migratory or sedentary behaviour has influenced the diversification rates of parulid species, then ancestral state reconstructions from Mk1 or Mk2 models can misrepresent character history [51,56–58]. Migratory behaviour has been suggested to either dampen [4,11] or promote [14,59] speciation. Therefore, we used the binary-state speciation and extinction (BiSSE) model [51] as implemented in Diversitree to test the influence of the binary migration characters on speciation and extinction rates, as well as the inferred character history of migration under models that incorporate character-dependent diversification rates. Using the BiSSE model, we also estimated relative rates of losses versus gains of migratory behaviour with maximum-likelihood and Markov chain Monte Carlo methods (details in the electronic supplementary material). Ancestral state reconstructions under the full BiSSE model and a variety of constrained models were consistent with ancestral state reconstructions under the Mk1 model (electronic supplementary material, figures S4 and S5 and table S1), so we performed further analyses (below) under the simpler Mk1 model.
For all maximum-likelihood ancestral state reconstructions, we used branch lengths proportional to time as generated by penalized likelihood in r8s [60]. Some authors [53,61] recommend transforming branch lengths to increase the fit of the data to the maximum-likelihood model for ancestral state reconstructions. We used Bayes Traits to estimate the maximum-likelihood value of the branch length scaling parameter kappa for the binary migration character under the Mk1 model [53,54]. Likelihood ratio tests indicated that the maximum-likelihood value of kappa provided a significantly better fit to the data than a model without branch length scaling (LRT = 11.45, p < 0.001, d.f. = 1). However, the maximum-likelihood value of kappa was estimated to be zero. Using a kappa scaling factor of zero is equivalent to converting all branch lengths to one, which implies that change occurs rapidly at the time of speciation rather than gradually throughout time with more change accumulating along longer branches [62]. It is plausible that changes in migration could occur rapidly during speciation events, as well as gradually in proportion to the lengths of the branches. Therefore, we performed maximum-likelihood ancestral state reconstructions of the binary character using branch lengths proportional to time and compared the results with reconstructions in which the branch lengths were converted to one.
We verified the conclusions of our ancestral state reconstructions on the maximum-likelihood topology by performing Bayesian analysis across the posterior set of 1000 trees in Bayes Traits. This analysis used a Bayesian reversible-jump Markov chain Monte Carlo (RJMCMC) method to estimate transition rate parameters, ancestral states and likelihoods across the tree set [52]. Using this method, we reconstructed ancestral states of all nodes found in the maximum-likelihood topology across the set of Bayesian trees. Additionally, we examined support for the migratory versus sedentary state at each of the basal nodes in the tree using Bayes factor tests in conjunction with the RJMCMC analysis (full details in the electronic supplementary material).
(b). Phylogenetic signal
A phylogenetic signal in the binary migration character is evident from simple examination of the topology (figure 2): the most prominent pattern is a large clade of mostly migratory species (genus Setophaga), sister to a clade of mostly non-migratory species. Of greater interest is the phylogenetic signal in migratory distance at multiple levels within the tree, as migratory distance is thought to be highly labile [4], and therefore examination of phylogenetic signal in migratory distance is a more conservative test than one that only examines the presence of migratory behaviour. We tested for phylogenetic signal in migration distance in each of three maximum-likelihood ultrametric phylogenies: (i) the whole family phylogeny; (ii) a phylogeny in which the largest clade of non-migratory species (Myiothlypis + Basileuterus + Cardellina + Myioborus; node 6, figure 2) was removed to eliminate the bias of many closely related taxa with a migratory distance of zero, thereby making support for phylogenetic signal in migratory distance more conservative; and (iii) a phylogeny consisting only of the monophyletic Setophaga (node 7, figure 2) to test for signal in migration distance at the generic level in a genus of mostly migratory species that has been of particular historical interest to biologists [35,37]. For each topology, we estimated the phylogenetic signal of the continuous distance character from squared tree length, implemented using weighted squared-change parsimony in Mesquite [48], and from phylogenetic independent contrasts (PICs) implemented in the R package Picante [63]. Both methods estimate the fit of the character-state data to a Brownian motion model of evolution given the topology. Departures from Brownian motion imply either greater or lesser phylogenetic signal in the character-state data than expected if the character states were to evolve along the branches under Brownian motion [61]. Squared tree length minimizes the sum of squared character-state differences between internal nodes and nodes and tips [64,65], and can be thought of as equivalent to the number of character steps for a discrete character [66], whereas the PIC method estimates the variance of PICs for a character. We estimated the significance of both measures of phylogenetic signal with randomization tests by comparing the observed values (squared tree length or PIC variance) of migration distance with a null distribution from 1000 characters with randomly permuted tip data. If the observed value is less than 95 per cent of the values in the null distribution, the character can be considered to be a good fit to the topology under Brownian motion and therefore to have phylogenetic signal [61,65]. Finally, we tested for phylogenetic signal using Blomberg et al.'s K statistic [61], implemented in Picante, which standardizes departures in character evolution from Brownian motion across phylogenies. The K statistic was calculated on the maximum-likelihood topology and across the set of 1000 Bayesian trees to account for phylogenetic uncertainty.
Figure 2.
Ancestral state reconstructions of the binary migration character across the maximum-likelihood topology using the Mk1 model and branch lengths proportional to time. Black circles at the tips indicate migratory species, and white circles indicate sedentary species. Letters next to the tips indicate geographical breeding ranges (N, North America; M, Mexico and/or Central America; C, Caribbean islands; S, South America; W, widespread across all regions). Pie charts at the nodes represent proportional maximum-likelihood support for the migratory (black) and sedentary (white) character states from the ancestral state reconstruction. Asterisks next to species names indicate species that have variable migratory strategies and were coded as sedentery in exploratory analyses (electronic supplementary material).
3. Results
(a). Ancestral state reconstructions
We performed ancestral state reconstructions using a variety of models and parameters, and results consistently supported a migratory ancestral parulid. The basal three nodes were reconstructed with strong support for the migratory state in the Mk1 analysis (nodes 1–3, figure 2) as well as in all other analyses, including the BiSSE models, analyses with branch lengths converted to one and analyses with the eight species with variable migratory behaviour coded as sedentary instead of migratory (electronic supplementary material, table S1 and figures S1–S5). The reconstruction of an ancestral parulid as migratory was also robust to analyses across the set of 1000 trees from the Bayesian posterior distribution, using both maximum-likelihood and RJMCMC analyses in Bayes Traits (electronic supplementary material, table S1 and figures S6 and S7). Bayes factor tests of the likelihood of the migratory state at the basal nodes from the RJMCMC Bayesian sampling indicate strong or very strong support for the migratory state at nodes 1–3 and positive support for the migratory state at node 4 (electronic supplementary material, table S1).
Migration has been lost and regained numerous times during the radiation of the wood-warblers. Estimates of the node or lineage where migration was initially lost in Parulidae are variable depending on the model of ancestral state reconstruction and the topology used, as support for the migratory versus sedentary states at nodes 4 and 5 varies between methods (figure 1; electronic supplementary material, table S1 and figures S6 and S7). Similarly, estimates of the precise number of state changes in either direction are variable across models, as inferring these changes requires assigning majority likelihood support to internal nodes throughout the tree that have weak or variable support for either character state depending on the model. A more informative method to test the directionality of state change is to examine the rates of change from the migratory to the sedentary state [57], which we did using maximum-likelihood and MCMC methods in Diversitree (electronic supplementary material).
Maximum-likelihood estimates of character-state transition rates in Diversitree found a higher rate of loss of migration than gain in the full BiSSE model and all constrained models with unequal transition rates (electronic supplementary material). However, as with the tests of Mk2 versus Mk1 models, likelihood ratio tests of the BiSSE models indicated that the models with unequal transition rates (i.e. more parameters) were not significantly better than the models with equal transition rates. MCMC analysis found a higher mean rate of loss of migration than gain in all models with unequal transition rates (electronic supplementary material, table S2), but broad overlap in the posterior probability distributions of the two character-state transitions; the rate of loss of migration was higher than the rate of gain, with posterior probability of 0.648–0.783 depending on the model (electronic supplementary material, table S3). However, when the eight species with variable migratory status were coded as sedentary instead of migratory, the ratio of the rate of loss to the rate of gain of migration was higher, and posterior distributions of the transition rates had less overlap; the rate of loss of migration was higher than the rate of gain, with posterior probability of 0.771–0.977 depending on the model (electronic supplementary material, table S3). These analyses suggest that the rate of loss of migration in Parulidae has been higher than the rate of gain in Parulidae, though the significance of this conclusion depends on the character-state coding. Maximum-likelihood and MCMC analyses in Diversitree of speciation and extinction rates were not significantly different for the migratory and sedentary states under either character-state coding, though speciation rates tended towards being somewhat higher for the sedentary state, with posterior probability of 0.578–0.752 (electronic supplementary material, table S3).
(b). Phylogenetic signal
The squared tree length and PIC variance of migration distance on the phylogeny were significantly lower than the values for the null model, demonstrating significant phylogenetic signal for migratory distance (table 1). This result applies to the entire topology, as well as to a topology in which the largest clade of non-migratory species (node 6, figure 2) was removed. Therefore, even without the bias of a large clade of warbler species in which nearly all members have migration distances of zero, migratory distance shows significant phylogenetic signal in the Parulidae phylogeny. A separate analysis for the genus Setophaga (node 7, figure 2), whose members are mostly migratory but vary widely in migration distance, also revealed significant phylogenetic signal for migration distance. Blomberg's K statistic was less than 1.0 for all topologies (table 1), indicating that phylogenetic signal in migration distance is lower than would be expected if the character was evolving under perfect Brownian motion. However, the K values of 0.896 for the whole maximum-likelihood topology and 0.880 for the Setophaga clade, as well as a mean K of 0.793 across the set of 1000 Bayesian trees, are close to 1.0. For a behavioural character as labile as migratory tendency, these values indicate phylogenetic signal in the data [61].
Table 1.
Results for the tests of phylogenetic signal using squared tree length and phylogenetic independent contrasts. Node numbers refer to figure 2.
| squared tree length |
phylogenetic independent contrasts |
||||||
|---|---|---|---|---|---|---|---|
| tree | actual length | null model mean length | p | actual variance | null model mean variance | p | K |
| full phylogeny | 41 767.51 | 173 964.16 | p < 0.001 | 395.04 | 1646.92 | p = 0.001 | 0.896 |
| clade at node 5 removed | 36 021.01 | 104 873.97 | p < 0.001 | 563.648 | 1621.327 | p = 0.001 | 0.59 |
| Setophaga (node 7) only | 20 421.67 | 49 009.34 | p < 0.001 | 638.177 | 1555.911 | p = 0.002 | 0.88 |
| 1000 Bayesian trees | — | — | — | — | — | — | 0.793 (mean) |
4. Discussion
This study is the first to provide evidence for ancestral migratory behaviour in a diverse mixed radiation of migratory and sedentary bird species. Sedentary wood-warblers, including several Caribbean island endemics and a large radiation of Central and South American species, probably represent repeated losses of migratory behaviour and subsequent speciation. This pattern provides the first evidence at the family level that the diversity of migratory strategies observed within bird lineages—even those with large radiations of sedentary tropical species—may be driven as much or more by losses of migration as they are by gains.
These findings contradict the assumption that temperate migratory species of parulid warblers are derived from their subtropical or tropical sedentary relatives [27,28]. The assumption of sedentary ancestry has provided a framework for mechanistic theories on the evolution of migration among lineages of Nearctic–Neotropical migrants (e.g. [25,28]). It is important to note, however, that our results neither support nor contradict the unifying prediction of ‘southern ancestral home’ theories, which states that migration tends to evolve through a shift of the breeding range out of ancestral tropical or subtropical regions, rather than extensions of an ancestral temperate non-breeding range to tropical regions [8,16,67]. Our results imply only that the ancestral parulid was probably a Nearctic–Neotropical migrant, and that extant sedentary taxa represent a derived condition within the family. Analysis deeper in the nine-primaried oscine phylogeny (the older and larger songbird clade that includes the Parulidae) is required to determine whether the tropical ranges of extant sedentary warblers represent a reversal to an older Neotropical ancestry, versus the alternative scenario that nine-primaried oscines have deep phylogenetic origins in North America [68]. Hence, where and when a transition to migratory behaviour may have taken place in the ancestral lineages that predate the Parulidae requires further analysis deeper in the avian tree of life.
This study also demonstrates that, within the Parulidae, migratory species are generally more closely related to other migratory species than to non-migrants, whereas Helbig [4] predicted that migratory behaviour is so labile that migratory species should be most closely related to non-migratory or less migratory species (phylogenetic ‘antisignal’ [61]). This hypothesis has in general been supported by interspecific phylogenetic studies [9,17,18] as well as intraspecific population-level studies [69–71]. Although the variability of migratory behaviour within some wood-warbler species and clades suggests migration may be labile in Parulidae [34,72–76], our tests of phylogenetic signal nevertheless indicate that the migratory distance of a wood-warbler species is significantly predicted by its phylogenetic relationships.
We do not interpret this phylogenetic signal to be representative of phylogenetic constraint, per se, on the migratory behaviour of parulids [61,77]. Rather, the close ties between migratory behaviour and breeding latitude in Parulidae suggest that breeding latitude, not phylogeny, most directly influences the migratory behaviour of these populations [7,8]. Although there is growing evidence that seasonal movements among tropical species are more widespread than traditionally thought [78–80], and several species of tropical parulids are known to engage in altitudinal or localized seasonal movements (C. Merkord 2010, personal communication) [43,44], it remains true that breeding latitude is an important predictor of the pronounced seasonal latitudinal migrations in wood-warblers and many other taxa: every parulid species that breeds north of the Mexican plateau or southern Florida exhibits seasonal migratory behaviour, whereas all parulid species breeding south of this latitudinal boundary are predominately sedentary [34]. However, the degree to which phylogeny may influence the latitudinal ranges of parulids—and thus influence their migratory behaviour—by way of ecological requirements, colonization propensity or dispersal ability remains uncertain, as do the potential phylogenetic constraints on the ability of migratory species to evolve adaptations to survive harsh, resource-depleted non-breeding climates and become year-round residents in temperate regions.
The patterns of migratory gains and losses in the wood-warblers are further relevant to testing the potential causal relationships between migration and phylogeny: it has been proposed that migratory behaviour may dampen [4,11] or promote [59] speciation rates, both of which would influence patterns of phylogenetic signal in migration. Although it is widely accepted that migratory species experience homogenizing gene flow that reduces rates of intraspecific differentiation [14,81,82], the relationship between migration and the behaviours that influence gene flow, such as natal philopatry and colonization ability, is poorly understood [13,83–85]. Additionally, there is compelling evidence that differences in migratory behaviour may promote reproductive isolation between populations [70,86–89]. The general clustering of wood-warbler species into clades with similar migratory behaviour, and our results that migratory behaviour has a significant phylogenetic signal in Parulidae, suggest that speciation has occurred among migratory wood-warbler species throughout the history of Parulidae. However, the variability of migratory distance and behaviour within these clades and within some species suggests that there is no simple relationship between speciation and migratory gains or losses in this group. Furthermore, our analysis of character-dependent diversification rates in Diversitree did not find a significant relationship between migratory behaviour and speciation, extinction or net diversification rates. However, breeding latitude and migratory behaviour are tightly correlated in Parulidae, and recent evidence suggests that breeding latitude may influence speciation rate in New World birds [90,91]. Therefore, we interpret this latter result cautiously, and recommend that future analyses of character-dependent diversification rates for migratory behaviour should ideally incorporate other clades that contain both sedentary temperate species and migratory tropical or subtropical species in order to reduce the potential confounding influence of breeding latitude.
As more comprehensive species-level phylogenies are published, ancestral state reconstructions of migratory behaviour in other taxa will increase our understanding of general patterns of change in migratory behaviour across the avian tree of life, and in aggregate will shed light on the deeper history of migration in birds. Detailed population genetic studies can similarly inform our understanding of changes in migratory behaviour. For example, Milá et al. [71] found evidence that migration in chipping sparrows Spizella passerina was driven by range expansions from sedentary populations in Mexico following glacial episodes; conversely, Kondo et al. [92] concluded that the short-distance, subtropical species Icterus abeillei diverged from the highly migratory Icterus galbula following a reduction of migratory distance. All of these approaches will help integrate our increasing knowledge of how and why migratory systems change in ecological time with our inferences of broader patterns of phylogenetic change through evolutionary time.
Acknowledgements
We are grateful to John Bates, Kim Bostwick, Deren Eaton, Shannon Hackett, Boris Igić, Steffi Kautz, Corrie Moreau, Trevor Price, Dan Rabosky, Andy Raduski, Rick Ree, Ben Rubin, Aaron Savit, Jason Weckstein, Jason Weir and an anonymous reviewer for comments on earlier versions of the manuscript and/or help with methodology. We thank Chris Merkord for sharing his knowledge of altitudinal migration in Parulidae. This research was supported by a Rawlings Cornell Presidential Research Scholarship and the Cornell College Scholars programme.
References
- 1.Berthold P., Helbig A., Mohr G., Querner U. 1992. Rapid microevolution of migratory behavior in a wild bird species. Nature 360, 668–670 10.1038/360668a0 (doi:10.1038/360668a0) [DOI] [Google Scholar]
- 2.Berthold P. 1999. A comprehensive theory for the evolution, control and adaptability of avian migration. Ostrich 70, 1–11 10.1080/00306525.1999.9639744 (doi:10.1080/00306525.1999.9639744) [DOI] [Google Scholar]
- 3.Pulido F. 2007. The genetics and evolution of avian migration. Bioscience 57, 165–174 10.1641/B570211 (doi:10.1641/B570211) [DOI] [Google Scholar]
- 4.Helbig A. 2003. Evolution of bird migration: a phylogenetic and biogeographic perspective. In Avian migration (eds Berthold P., Gwinner E., Sonnenschein E.), pp. 81–95 Berlin, Germany: Spring-Verlag [Google Scholar]
- 5.Pulido F., Berthold P., van Noordwijk A. J. 1996. Frequency of migrants and migratory activity are genetically correlated in a bird population: evolutionary implications. Proc. Natl Acad. Sci. USA 93, 14 642–14 647 10.1073/pnas.93.25.14642 (doi:10.1073/pnas.93.25.14642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zink R. 2002. Towards a framework for understanding the evolution of avian migration. J. Avian Biol. 33, 433–436 10.1034/j.1600-048X.2002.03081.x (doi:10.1034/j.1600-048X.2002.03081.x) [DOI] [Google Scholar]
- 7.Alerstam T., Hedenström A., Åkesson S. 2003. Long-distance migration: evolution and determinants. Oikos 103, 247–260 10.1034/j.1600-0706.2003.12559.x (doi:10.1034/j.1600-0706.2003.12559.x) [DOI] [Google Scholar]
- 8.Salewski V., Bruderer B. 2007. The evolution of bird migration: a synthesis. Naturwissenschaften 94, 268–279 10.1007/s00114-006-0186-y (doi:10.1007/s00114-006-0186-y) [DOI] [PubMed] [Google Scholar]
- 9.Outlaw D., Voelker G., Mila B., Girman D. 2003. Evolution of long-distance migration in and historical biogeography of Catharus thrushes: a molecular phylogenetic approach. Auk 120, 299–310 10.1642/0004-8038(2003)120[0299:EOLMIA]2.0.CO;2 (doi:10.1642/0004-8038(2003)120[0299:EOLMIA]2.0.CO;2) [DOI] [Google Scholar]
- 10.Piersma T., Pérez-Tris J., Mouritsen H., Bauchinger U., Bairlein F. 2005. Is there a ‘migratory syndrome’ common to all migrant birds? Ann. NY Acad. Sci. 1046, 282–293 10.1196/annals.1343.026 (doi:10.1196/annals.1343.026) [DOI] [PubMed] [Google Scholar]
- 11.Keast A. 1958. The relationship between seasonal movements and the development of geographic variation in the Australian Chats (Epthianura Gould and Ashbyia North (Passeres: Muscicapidae, Malurinae)). Austr. J. Zool. 6, 53–68 10.1071/ZO9580053 (doi:10.1071/ZO9580053) [DOI] [Google Scholar]
- 12.Paradis E., Baillie S. R., Sutherland W. J., Gregory R. D. 1998. Patterns of natal and breeding dispersal in birds. J. Anim. Ecol. 67, 518–536 10.1046/j.1365-2656.1998.00215.x (doi:10.1046/j.1365-2656.1998.00215.x) [DOI] [Google Scholar]
- 13.Winkler D. 2005. How do migration and dispersal interact? In Birds of two worlds: the ecology and evolution of migration (eds Greenberg R., Marra P.), pp. 401–413 Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 14.Winker K. 2010. On the origin of species through heteropatric differentiation: a review and a model of speciation in migratory animals. Ornithol. Monogr. 69, 1–30 10.1525/om.2010.69.1.1 (doi:10.1525/om.2010.69.1.1) [DOI] [Google Scholar]
- 15.Chesser R. 2000. Evolution in the high Andes: the phylogenetics of Muscisaxicola ground-tyrants. Mol. Phylogenet. Evol. 15, 369–380 10.1006/mpev.1999.0774 (doi:10.1006/mpev.1999.0774) [DOI] [PubMed] [Google Scholar]
- 16.Joseph L., Lessa E., Christidis L. 1999. Phylogeny and biogeography in the evolution of migration: shorebirds of the Charadrius complex. J. Biogeogr. 26, 329–342 10.1046/j.1365-2699.1999.00269.x (doi:10.1046/j.1365-2699.1999.00269.x) [DOI] [Google Scholar]
- 17.Kondo B., Omland K. 2007. Ancestral state reconstructions of migration: multistate analysis reveals rapid changes in New World orioles (Icterus spp.). Auk 124, 410–419 10.1642/0004-8038(2007)124[410:ASROMM]2.0.CO;2 (doi:10.1642/0004-8038(2007)124[410:ASROMM]2.0.CO;2) [DOI] [Google Scholar]
- 18.Outlaw D., Voelker G. 2006. Phylogenetic tests of hypotheses for the evolution of avian migration: a case study using the Motacillidae. Auk 123, 455–466 10.1642/0004-8038(2006)123[455:PTOHFT]2.0.CO;2 (doi:10.1642/0004-8038(2006)123[455:PTOHFT]2.0.CO;2) [DOI] [Google Scholar]
- 19.Winker K., Pruett C. 2006. Seasonal migration, speciation, and morphological convergence in the genus Catharus (Turdidae). Auk 123, 1052–1068 10.1642/0004-8038(2006)123[1052:SMSAMC]2.0.CO;2 (doi:10.1642/0004-8038(2006)123[1052:SMSAMC]2.0.CO;2) [DOI] [Google Scholar]
- 20.Rheindt F., Christidis L., Norman J. 2008. Habitat shifts in the evolutionary history of a Neotropical flycatcher lineage from forest and open landscapes. BMC Evol. Biol. 8, 193. 10.1186/1471-2148-8-193 (doi:10.1186/1471-2148-8-193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cicero C., Johnson N. 1998. Molecular phylogeny and ecological diversification in a clade of New World songbirds (genus Vireo). Mol. Ecol. 7, 1359–1370 10.1186/1471-2148-8-193 (doi:10.1186/1471-2148-8-193) [DOI] [PubMed] [Google Scholar]
- 22.Burns K. 1998. Molecular phylogenetics of the genus Piranga: implications for biogeography and the evolution of morphology and behavior. Auk 115, 621–634 [Google Scholar]
- 23.Cicero C., Johnson N. 2002. Phylogeny and character evolution in the Empidonax group of tyrant flycatchers (Aves: Tyrannidae): a test of W. E. Lanyon's hypothesis using mtDNA sequences. Mol. Phylogenet. Evol. 22, 289–302 10.1006/mpev.2001.1054 (doi:10.1006/mpev.2001.1054) [DOI] [PubMed] [Google Scholar]
- 24.Cox G. 1968. The role of competition in the evolution of migration. Evolution 22, 180–192 10.2307/2406662 (doi:10.2307/2406662) [DOI] [PubMed] [Google Scholar]
- 25.Cox G. 1985. The evolution of avian migration systems between the temperate and tropical regions of the New World. Am. Nat. 126, 451–474 10.1086/284432 (doi:10.1086/284432) [DOI] [Google Scholar]
- 26.Levey D., Stiles F. 1992. Evolutionary precursors of long-distance migration: resource availability and movement patterns in neotropical landbirds. Am. Nat. 140, 447–476 10.1086/285421 (doi:10.1086/285421) [DOI] [Google Scholar]
- 27.Berthold P. 2001. Bird migration: a general survey, 2nd edn. New York, NY: Oxford University Press [Google Scholar]
- 28.Rappole J. 1995. The ecology of migrant birds: a neotropical perspective. Washington, DC: Smithsonian Institution Press [Google Scholar]
- 29.Bell C. 2005. The origin and development of bird migration: comments on Rappole and Jones, and an alternative evolutionary model. Ardea 93, 115–123 [Google Scholar]
- 30.Joseph L. 2005. Molecular approaches to the evolution and ecology of migration. In Birds of two worlds: the ecology and evolution of bird migration (eds Greenberg R., Marra P.), pp. 18–26 Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 31.Bruderer B., Salewski V. 2008. Evolution of bird migration in a biogeographical context. J. Biogeogr. 35, 1951–1959 10.1111/j.1365-2699.2008.01992.x (doi:10.1111/j.1365-2699.2008.01992.x) [DOI] [Google Scholar]
- 32.Lovette I., et al. 2010. A comprehensive multilocus phylogeny for the wood-warblers and a revised classification of the Parulidae (Aves). Mol. Phylogenet. Evol. 57, 1–18 10.1016/j.ympev.2010.07.018 (doi:10.1016/j.ympev.2010.07.018) [DOI] [PubMed] [Google Scholar]
- 33.Dunn J., Garrett K. 1997. A field guide to warblers of North America. Boston, MA: Houghton-Mifflin [Google Scholar]
- 34.Curson J., Quinn D., Beadle D. 1994. Warblers of the Americas. Boston, MA: Houghton-Mifflin [Google Scholar]
- 35.Macarthur R. 1958. Population ecology of some warblers of northeastern coniferous forests. Ecology 39, 599–619 10.2307/1931600 (doi:10.2307/1931600) [DOI] [Google Scholar]
- 36.Marra P., Hobson K., Holmes R. 1998. Linking winter and summer events in a migratory bird by using stable-carbon isotopes. Science 282, 1884–1886 10.1126/science.282.5395.1884 (doi:10.1126/science.282.5395.1884) [DOI] [PubMed] [Google Scholar]
- 37.Lovette I., Bermingham E. 1999. Explosive speciation in the New World Dendroica warblers. Proc. R. Soc. Lond. B 266, 1629–1636 10.1098/rspb.1999.0825 (doi:10.1098/rspb.1999.0825) [DOI] [Google Scholar]
- 38.Rubenstein D., Chamberlain C., Holmes R., Ayres M., Waldbauer J., Graves G., Tuross N. 2002. Linking breeding and wintering ranges of a migratory songbird using stable isotopes. Science 295, 1062–1065 10.1126/science.1067124 (doi:10.1126/science.1067124) [DOI] [PubMed] [Google Scholar]
- 39.Lovette I., Hochachka W. 2006. Simultaneous effects of phylogenetic niche conservatism and competition on avian community structure. Ecology 87, S14–S28 10.1890/0012-9658(2006)87[14:SEOPNC]2.0.CO;2 (doi:10.1890/0012-9658(2006)87[14:SEOPNC]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 40.Rabosky D., Lovette I. 2008. Density-dependent diversification in North American wood warblers. Proc. R. Soc. B 275, 2363–2371 10.1098/rspb.2008.0630 (doi:10.1098/rspb.2008.0630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.American Ornithologists' Union 1998. Checklist of North American birds, 7th edn. Washington, DC: American Ornithologists' Union [Google Scholar]
- 42.Sanderson M. 2002. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 19, 101–109 [DOI] [PubMed] [Google Scholar]
- 43.Howell S., Webb S. 1995. A guide to the birds of Mexico and northern Central America. Oxford, UK: Oxford University Press [Google Scholar]
- 44.Ridgely R. S., Tudor G. 1994. The birds of South America, vol. 1: The oscine passerines. Austin, TX: University of Texas Press [Google Scholar]
- 45.Stiles F. G., Skutch A. F. 1989. A guide to the birds of Costa Rica. Ithaca, NY: Comstock [Google Scholar]
- 46.Ridgely R. S., Greenfield P. J. 2001. The birds of Ecuador, vol. 1: status, distribution and taxonomy. Ithaca, NY: Cornell University Press [Google Scholar]
- 47.Poole A. (ed.) The birds of North America online. Ithaca, NY: Cornell Laboratory of Ornithology; See http://bna.birds.cornell.edu.bnaproxy.birds.cornell.edu/BNA/ [Google Scholar]
- 48.Maddison W., Maddison D. 2009. Mesquite: a modular system for evolutionary analysis, v. 2.71. See http://mesquiteproject.org
- 49.Paradis E., Claude J., Strimmerk K. 2004. APE: analysis of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 10.1093/bioinformatics/btg412 (doi:10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 50.FitzJohn R. G. 2010. Diversitree: comparative phylogenetic tests of diversification. R package, v. 0.6-5. (See http://www.zoology.ubc.ca/prog/diversitree)
- 51.Maddison W., Midford P., Otto S. 2007. Estimating a binary character's effect on speciation and extinction. Syst. Biol. 56, 701–710 10.1080/10635150701607033 (doi:10.1080/10635150701607033) [DOI] [PubMed] [Google Scholar]
- 52.Pagel M., Meade A., Barker D. 2004. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53, 673–684 10.1080/10635150490522232 (doi:10.1080/10635150490522232) [DOI] [PubMed] [Google Scholar]
- 53.Pagel M. 1994. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc. R. Soc. Lond. B 255, 37–45 10.1098/rspb.1994.0006 (doi:10.1098/rspb.1994.0006) [DOI] [Google Scholar]
- 54.Pagel M. 1999. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst. Biol. 48, 612–622 10.1080/106351599260184 (doi:10.1080/106351599260184) [DOI] [Google Scholar]
- 55.Mooers A., Schluter D. 1999. Reconstructing ancestor states with maximum likelihood: support for one- and two-rate models. Syst. Biol. 48, 623–633 10.1080/106351599260193 (doi:10.1080/106351599260193) [DOI] [Google Scholar]
- 56.Goldberg E., Kohn J., Lande R., Roberston K., Smith S., Igić B. 2010. Species selection maintains self-incompatibility. Science 330, 493–495 10.1126/science.1194513 (doi:10.1126/science.1194513) [DOI] [PubMed] [Google Scholar]
- 57.Goldberg E., Igić B. 2008. On phylogenetic tests of irreversible evolution. Evolution 62, 2727–2741 10.1111/j.1558-5646.2008.00505.x (doi:10.1111/j.1558-5646.2008.00505.x) [DOI] [PubMed] [Google Scholar]
- 58.FitzJohn R., Maddison W., Otto S. 2009. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 58, 595–611 10.1093/sysbio/syp067 (doi:10.1093/sysbio/syp067) [DOI] [PubMed] [Google Scholar]
- 59.Winker K. 2000. Migration and speciation. Nature 404, 36. 10.1038/35003651 (doi:10.1038/35003651) [DOI] [PubMed] [Google Scholar]
- 60.Sanderson M. 2003. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19, 301–302 10.1093/bioinformatics/19.2.301 (doi:10.1093/bioinformatics/19.2.301) [DOI] [PubMed] [Google Scholar]
- 61.Blomberg S., Garland T., Ives A. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745 10.1111/j.0014-3820.2003.tb00285.x (doi:10.1111/j.0014-3820.2003.tb00285.x) [DOI] [PubMed] [Google Scholar]
- 62.Pagel M. 1997. Inferring evolutionary processes from phylogenies. Zool. Scripta 26, 331–348 10.1111/j.1463-6409.1997.tb00423.x (doi:10.1111/j.1463-6409.1997.tb00423.x) [DOI] [Google Scholar]
- 63.Kembel S. W., Cowan P. D., Helmus M. R., Cornwell W. K., Morlon H., Ackerly D. D., Blomberg S. P., Webb C. O. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464 10.1093/bioinformatics/btq166 (doi:10.1093/bioinformatics/btq166) [DOI] [PubMed] [Google Scholar]
- 64.Maddison W. P. 1991. Squared-change parsimony reconstructions of ancestral states for continuous-valued characters in a phylogenetic tree. Syst. Zool. 40, 304–314 10.2307/2992324 (doi:10.2307/2992324) [DOI] [Google Scholar]
- 65.Cubo J., Ponton F., Laurin M., de Margerie E., Castanet J. 2005. Phylogenetic signal in bone microstructure of sauropsids. Syst. Biol. 54, 562–574 10.1080/10635150591003461 (doi:10.1080/10635150591003461) [DOI] [PubMed] [Google Scholar]
- 66.Maddison W. P., Slatkin M. 1991. Null models for the number of evolutionary steps in a character on a phylogenetic tree. Evolution 45, 1184–1197 10.2307/2409726 (doi:10.2307/2409726) [DOI] [PubMed] [Google Scholar]
- 67.Gauthreaux S. A. 1982. The ecology and evolution of avian migration systems. In Avian biology, vol. 6 (eds Farner D. S., King J. R., Parkes K. C.), pp. 93–168 New York, NY: Academic Press [Google Scholar]
- 68.Barker F. K., Cibois A., Schikler P., Feinstein J., Cracraft J. 2004. Phylogeny and diversification of the largest avian radiation. Proc. Natl Acad. Sci. USA 101, 11 040–11 045 10.1073/pnas.0401892101 (doi:10.1073/pnas.0401892101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joseph L., Wilke T., Alpers D. 2003. Independent evolution of migration on the South American landscape in a long-distance temperate-tropical migratory bird, Swainson's flycatcher (Myiarchus swainsoni). J. Biogeogr. 30, 925–937 10.1046/j.1365-2699.2003.00841.x (doi:10.1046/j.1365-2699.2003.00841.x) [DOI] [Google Scholar]
- 70.Pérez-Tris J., Bensch S., Carbonell R., Helbig A. J., Tellería J. L. 2004. Historical diversification of migration patterns in a passerine bird. Evolution 58, 1819–1832 10.1554/03-731 (doi:10.1554/03-731) [DOI] [PubMed] [Google Scholar]
- 71.Milá B., Smith T. B., Wayne R. K. 2006. Postglacial population expansion drives the evolution of long-distance migration in a songbird. Evolution 60, 2403–2409 10.1111/j.0014-3820.2006.tb01875.x (doi:10.1111/j.0014-3820.2006.tb01875.x) [DOI] [PubMed] [Google Scholar]
- 72.Buerkle C. 1999. The historical pattern of gene flow among migratory and nonmigratory populations of prairie warblers (Aves: Parulinae). Evolution 53, 1915–1924 10.2307/2640450 (doi:10.2307/2640450) [DOI] [PubMed] [Google Scholar]
- 73.Milá B., Smith T. B., Wayne R. K. 2007. Speciation and rapid phenotypic differentiation in the yellow-rumped warbler Dendroica coronata complex. Mol. Ecol. 16, 159–173 10.1111/j.1365-294X.2006.03119.x (doi:10.1111/j.1365-294X.2006.03119.x) [DOI] [PubMed] [Google Scholar]
- 74.Milá B., Girman D. J., Kimura M., Smith T. B. 2000. Genetic evidence for the effect of a postglacial population expansion on the phylogeography of a North American songbird. Proc. R. Soc. Lond. B 267, 1033–1040 10.1098/rspb.2000.1107 (doi:10.1098/rspb.2000.1107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boulet M., Gibbs H. L. 2006. Lineage origin and expansion of a Neotropical migrant songbird after recent glaciation events. Mol. Ecol. 15, 2505–2525 10.1111/j.1365-294X.2006.02956.x (doi:10.1111/j.1365-294X.2006.02956.x) [DOI] [PubMed] [Google Scholar]
- 76.Davis L., Roalson E., Cornell K., McClanahan K., Webster M. 2006. Genetic divergence and migration patterns in a North American passerine bird: implications for evolution and conservation. Mol. Ecol. 15, 2141–2152 10.1111/j.1365-294X.2006.02914.x (doi:10.1111/j.1365-294X.2006.02914.x) [DOI] [PubMed] [Google Scholar]
- 77.Blomberg S., Garland T. 2002. Tempo and mode in evolution: phylogenetic inertia, adaptation and comparative methods. J. Evol. Biol. 15, 899–910 10.1046/j.1420-9101.2002.00472.x (doi:10.1046/j.1420-9101.2002.00472.x) [DOI] [Google Scholar]
- 78.Jahn A., Levey D., Smith K. 2004. Reflections across hemispheres: a system-wide approach to New World bird migration. Auk 121, 1005–1013 10.1642/0004-8038(2004)121[1005:RAHASA]2.0.CO;2 (doi:10.1642/0004-8038(2004)121[1005:RAHASA]2.0.CO;2) [DOI] [Google Scholar]
- 79.Winker K., Escalante P., Rappole J., Ramos M., Oehlenschlager R., Warner D. 1997. Periodic migration and lowland forest refugia in a ‘sedentary’ neotropical bird, Wetmore's Bush-Tanager. Conserv. Biol. 11, 692–697 10.1046/j.1523-1739.1997.95450.x (doi:10.1046/j.1523-1739.1997.95450.x) [DOI] [Google Scholar]
- 80.Helm B., Gwinner E. 2006. Migratory restlessness in an equatorial nonmigratory bird. PLOS Biol. 4, 611–614 10.1371/journal.pbio.0040110 (doi:10.1371/journal.pbio.0040110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barrowclough G. 1980. Gene flow, effective population sizes, and genetic variance components in birds. Evolution 34, 789–798 10.2307/2408033 (doi:10.2307/2408033) [DOI] [PubMed] [Google Scholar]
- 82.Zink R. 1996. Comparative phylogeography in North American birds. Evolution 50, 308–317 10.2307/2410802 (doi:10.2307/2410802) [DOI] [PubMed] [Google Scholar]
- 83.Bensch S. 1999. Is the range size of migratory birds constrained by their migratory program? J. Biogeogr. 26, 1225–1235 10.1046/j.1365-2699.1999.00360.x (doi:10.1046/j.1365-2699.1999.00360.x) [DOI] [Google Scholar]
- 84.Böhning-Gaese K., González-Guzmán L. I., Brown J. H. 1998. Constraints on dispersal and the evolution of the avifauna of the Northern Hemisphere. Evol. Ecol. 12, 767–783 10.1023/A:1006538414645 (doi:10.1023/A:1006538414645) [DOI] [Google Scholar]
- 85.Thorup K. 2006. Does the migration programme constrain the dispersal and range sizes of migratory birds? J. Biogeogr. 33, 1166–1171 10.1111/j.1365-2699.2006.01487.x (doi:10.1111/j.1365-2699.2006.01487.x) [DOI] [Google Scholar]
- 86.Ruegg K. 2008. Genetic, morphological and ecological characterization of a hybrid zone that spans a migratory divide. Evolution 62, 452–466 10.1111/j.1558-5646.2007.00263.x (doi:10.1111/j.1558-5646.2007.00263.x) [DOI] [PubMed] [Google Scholar]
- 87.Irwin D. E. 2009. Speciation: new migratory direction provides route towards divergence. Curr. Biol. 19, R1111–R1113 10.1016/j.cub.2009.11.011 (doi:10.1016/j.cub.2009.11.011) [DOI] [PubMed] [Google Scholar]
- 88.Irwin D., Irwin J. 2005. Siberian migratory divides. In Birds of two worlds: the ecology and evolution of bird migration (eds Greenberg R., Marra P.), pp. 27–40 Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 89.Bearhop S., Fiedler W., Furness R., Votier S., Waldron S., Newton J., Bowen G., Berthold P., Farnsworth K. 2005. Assortative mating as a mechanism for rapid evolution of a migratory divide. Science 310, 502–504 10.1126/science.1115661 (doi:10.1126/science.1115661) [DOI] [PubMed] [Google Scholar]
- 90.Weir J., Schluter D. 2007. The latitudinal gradient in recent speciation and extinction rates of birds and mammals. Science 315, 502–504 10.1126/science.1135590 (doi:10.1126/science.1135590) [DOI] [PubMed] [Google Scholar]
- 91.Weir J., Price T. 2011. Limits to speciation inferred from times to secondary sympatry and ages of hybridizing species along a latitudinal gradient. Am. Nat. 177, 462–469 10.1086/658910 (doi:10.1086/658910) [DOI] [PubMed] [Google Scholar]
- 92.Kondo B., Peters J., Rosensteel B., Omland K. 2008. Coalescent analysis of multiple loci support a new route to speciation in birds. Evolution 62, 1182–1191 10.1111/j.1558-5646.2008.00345.x (doi:10.1111/j.1558-5646.2008.00345.x) [DOI] [PubMed] [Google Scholar]