Abstract
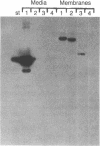
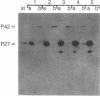
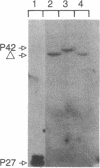
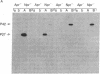
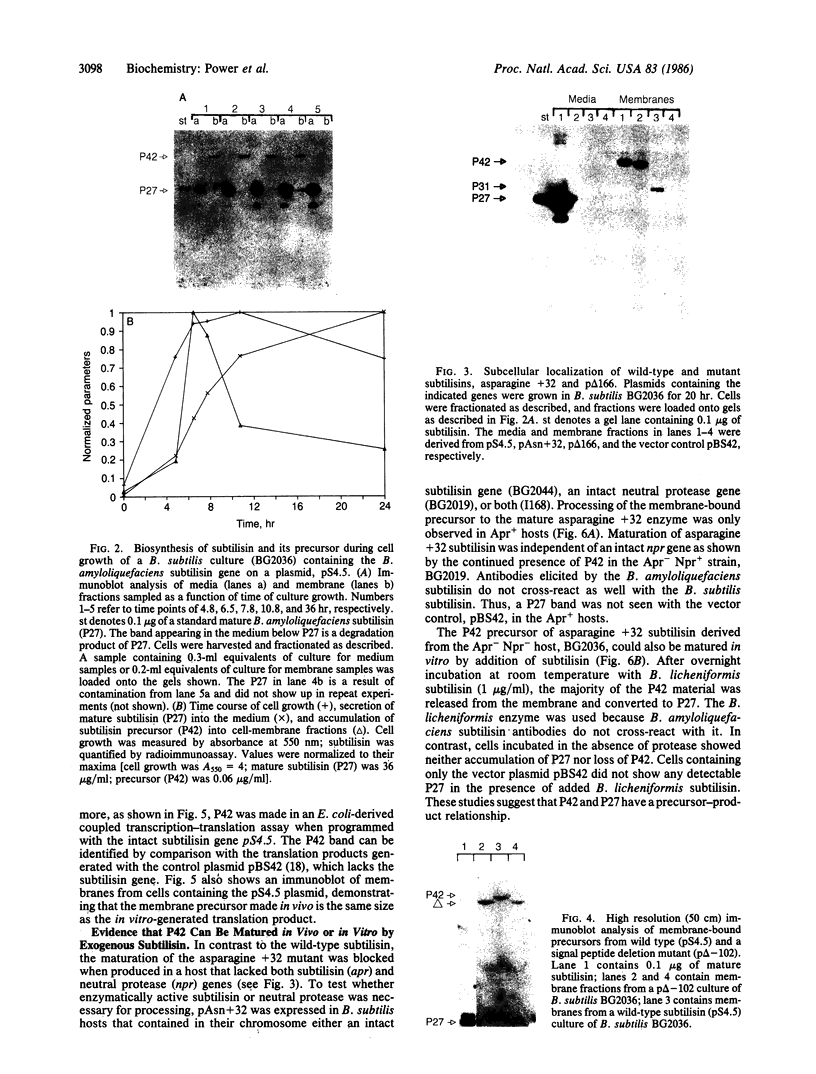
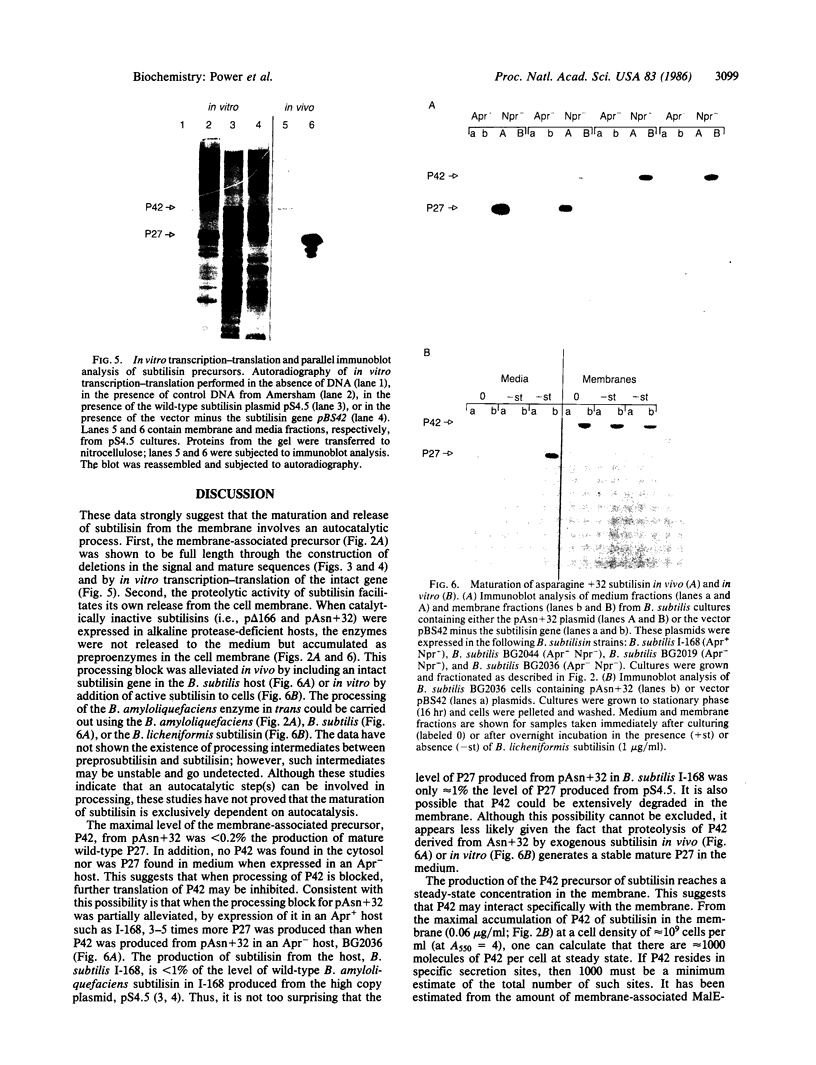
The sequence of the cloned Bacillus amyloliquefaciens subtilisin gene suggested that this secreted serine protease is produced as a larger precursor, designated preprosubtilisin [Wells, J. A., Ferrari, E., Henner, D. J., Estell, D. A. & Chen, E. Y. (1983) Nucleic Acids Res. 11, 7911-7925]. Biochemical evidence presented here shows that a subtilisin precursor is produced in Bacillus subtilis hosts. The precursor is first localized in the cell membrane, reaching a steady-state level of approximately equal to 1000 sites per cell. Mutations in the subtilisin gene that alter a catalytically critical residue (i.e., aspartate +32----asparagine), or delete the carboxyl-terminal portion of the enzyme that contains catalytically critical residues, block the maturation of this precursor. This block occurs when these mutant genes are expressed in B. subtilis hosts where the chromosomal subtilisin gene has been deleted. When the mutant B. amyloliquefaciens subtilisins are expressed in B. subtilis hosts that contain an intact chromosomal subtilisin gene, the mutant precursors are processed to a mature form and released to the medium. Such processing, in trans, of the precursor is also demonstrated in vitro by addition of active subtilisin. Thus, the release of subtilisin from the cell membrane is dependent on an autoproteolytic process that appears to be novel among secreted proteins.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adelman J. P., Hayflick J. S., Vasser M., Seeburg P. H. In vitro deletional mutagenesis for bacterial production of the 20,000-dalton form of human pituitary growth hormone. DNA. 1983;2(3):183–193. doi: 10.1089/dna.1983.2.183. [DOI] [PubMed] [Google Scholar]
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band L., Henner D. J. Bacillus subtilis requires a "stringent" Shine-Dalgarno region for gene expression. DNA. 1984;3(1):17–21. doi: 10.1089/dna.1.1984.3.17. [DOI] [PubMed] [Google Scholar]
- Benson S. A., Hall M. N., Silhavy T. J. Genetic analysis of protein export in Escherichia coli K12. Annu Rev Biochem. 1985;54:101–134. doi: 10.1146/annurev.bi.54.070185.000533. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crea R., Horn T. Synthesis of oligonucleotides on cellulose by a phosphotriester method. Nucleic Acids Res. 1980 May 24;8(10):2331–2348. doi: 10.1093/nar/8.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenth J., Hol W. G., Jansonius J. N., Koekoek R. Subtilisin Novo. The three-dimensional structure and its comparison with subtilisin BPN'. Eur J Biochem. 1972 Mar 27;26(2):177–181. doi: 10.1111/j.1432-1033.1972.tb01754.x. [DOI] [PubMed] [Google Scholar]
- Inouye M., Halegoua S. Secretion and membrane localization of proteins in Escherichia coli. CRC Crit Rev Biochem. 1980;7(4):339–371. doi: 10.3109/10409238009105465. [DOI] [PubMed] [Google Scholar]
- Ito K., Bassford P. J., Jr, Beckwith J. Protein localization in E. coli: is there a common step in the secretion of periplasmic and outer-membrane proteins? Cell. 1981 Jun;24(3):707–717. doi: 10.1016/0092-8674(81)90097-0. [DOI] [PubMed] [Google Scholar]
- Julius D., Brake A., Blair L., Kunisawa R., Thorner J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984 Jul;37(3):1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- Julius D., Schekman R., Thorner J. Glycosylation and processing of prepro-alpha-factor through the yeast secretory pathway. Cell. 1984 Feb;36(2):309–318. doi: 10.1016/0092-8674(84)90224-1. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973 Nov 15;80(4):575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Markland F. S., Smith E. L. Subtilisin BPN. VII. Isolation of cyanogen bromide peptides and the complete amino acid sequence. J Biol Chem. 1967 Nov 25;242(22):5198–5211. [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Michaelis S., Beckwith J. Mechanism of incorporation of cell envelope proteins in Escherichia coli. Annu Rev Microbiol. 1982;36:435–465. doi: 10.1146/annurev.mi.36.100182.002251. [DOI] [PubMed] [Google Scholar]
- Millet J. Characterization of proteinases excreted by Bacillus subtilis Marburg strain during sporulation. J Appl Bacteriol. 1970 Mar;33(1):207–219. doi: 10.1111/j.1365-2672.1970.tb05245.x. [DOI] [PubMed] [Google Scholar]
- Norris K., Norris F., Christiansen L., Fiil N. Efficient site-directed mutagenesis by simultaneous use of two primers. Nucleic Acids Res. 1983 Aug 11;11(15):5103–5112. doi: 10.1093/nar/11.15.5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn M. J., Wu H. C. Proteins of the outer membrane of gram-negative bacteria. Annu Rev Microbiol. 1980;34:369–422. doi: 10.1146/annurev.mi.34.100180.002101. [DOI] [PubMed] [Google Scholar]
- Philipp M., Bender M. L. Kinetics of subtilisin and thiolsubtilisin. Mol Cell Biochem. 1983;51(1):5–32. doi: 10.1007/BF00215583. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Stahl M. L., Ferrari E. Replacement of the Bacillus subtilis subtilisin structural gene with an In vitro-derived deletion mutation. J Bacteriol. 1984 May;158(2):411–418. doi: 10.1128/jb.158.2.411-418.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanov V. M., Strongin A. Y., Izotova L. S., Abramov Z. T., Lyublinskaya L. A., Ermakova L. M., Baratova L. A., Belyanova L. P. Intracellular serine protease from Bacillus subtilis. Structural comparison with extracellular serine proteases-subtilisins. Biochem Biophys Res Commun. 1977 Jul 11;77(1):298–305. doi: 10.1016/s0006-291x(77)80196-4. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasantha N., Thompson L. D., Rhodes C., Banner C., Nagle J., Filpula D. Genes for alkaline protease and neutral protease from Bacillus amyloliquefaciens contain a large open reading frame between the regions coding for signal sequence and mature protein. J Bacteriol. 1984 Sep;159(3):811–819. doi: 10.1128/jb.159.3.811-819.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. A., Ferrari E., Henner D. J., Estell D. A., Chen E. Y. Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtilisin in Bacillus subtilis. Nucleic Acids Res. 1983 Nov 25;11(22):7911–7925. doi: 10.1093/nar/11.22.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. A., Vasser M., Powers D. B. Cassette mutagenesis: an efficient method for generation of multiple mutations at defined sites. Gene. 1985;34(2-3):315–323. doi: 10.1016/0378-1119(85)90140-4. [DOI] [PubMed] [Google Scholar]
- Wong S. L., Price C. W., Goldfarb D. S., Doi R. H. The subtilisin E gene of Bacillus subtilis is transcribed from a sigma 37 promoter in vivo. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1184–1188. doi: 10.1073/pnas.81.4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. I., Gitschier J., Lasky L. A., Lawn R. M. Base composition-independent hybridization in tetramethylammonium chloride: a method for oligonucleotide screening of highly complex gene libraries. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1585–1588. doi: 10.1073/pnas.82.6.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C. S., Alden R. A., Kraut J. Structure of subtilisin BPN' at 2.5 angström resolution. Nature. 1969 Jan 18;221(5177):235–242. doi: 10.1038/221235a0. [DOI] [PubMed] [Google Scholar]
- Yang M. Y., Ferrari E., Henner D. J. Cloning of the neutral protease gene of Bacillus subtilis and the use of the cloned gene to create an in vitro-derived deletion mutation. J Bacteriol. 1984 Oct;160(1):15–21. doi: 10.1128/jb.160.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]