Abstract
Aim: Recent advances in quantitative methods and sensitive imaging techniques of trace elements provide opportunities to uncover and explain their biological roles. In particular, the distribution of selenium in tissues and cells under both physiological and pathological conditions remains unknown. In this work, we applied high-resolution synchrotron X-ray fluorescence microscopy (XFM) to map selenium distribution in mouse liver and kidney. Results: Liver showed a uniform selenium distribution that was dependent on selenocysteine tRNA[Ser]Sec and dietary selenium. In contrast, kidney selenium had both uniformly distributed and highly localized components, the latter visualized as thin circular structures surrounding proximal tubules. Other parts of the kidney, such as glomeruli and distal tubules, only manifested the uniformly distributed selenium pattern that co-localized with sulfur. We found that proximal tubule selenium localized to the basement membrane. It was preserved in Selenoprotein P knockout mice, but was completely eliminated in glutathione peroxidase 3 (GPx3) knockout mice, indicating that this selenium represented GPx3. We further imaged kidneys of another model organism, the naked mole rat, which showed a diminished uniformly distributed selenium pool, but preserved the circular proximal tubule signal. Innovation: We applied XFM to image selenium in mammalian tissues and identified a highly localized pool of this trace element at the basement membrane of kidneys that was associated with GPx3. Conclusion: XFM allowed us to define and explain the tissue topography of selenium in mammalian kidneys at submicron resolution. Antioxid. Redox Signal. 16, 185–192.
Introduction
Biological trace elements occur in very small amounts in organisms but often have essential functions. The metabolism of trace elements is tightly regulated to avoid toxicity (owing to their high reactivity) while maintaining the concentrations required for metabolic processes. Several regulatory mechanisms have evolved to control transport, trafficking, and utilization of trace elements. For example, most elements have specific importers and exporters; there are also designated proteins, metallothioneins, capable of binding excess amounts of trace elements (copper, zinc) through their cysteine residues (9). One important biological trace element is selenium (Se). Although this element is toxic at higher concentrations, it is an essential dietary trace element in humans and other mammals (24). In mammals, Se occurs primarily in a protein-based form (selenocysteine and selenomethionine residues) and, at lower levels, in the form of low molecular weight compounds (selenite, selenide, monomethylselenol, dimethylselenide, trimethylselenonium, L-selenomethionine, and Se-methyl-L-selenocysteine) (22).
The incorporation of Se into proteins in mammals is a complex process that involves several levels of regulation (2). Selenocysteine (Sec) is encoded by a UGA codon, and its insertion requires the presence of a specific stem–loop structure, designated the SECIS element, in the 3′-UTR (31). The human selenoproteome is encoded by 25 selenoprotein genes, and there are 24 such genes in rodents (17). Selenoproteins are involved in central cellular processes, such as regulation of redox homeostasis, biosynthesis of thyroid hormones, glucose metabolism, and male reproduction (3). Se deficiency has been associated with cancer, arthritis, immunological defects, aging, and infertility (11, 24). In addition, an endemic cardiomyopathy, Keshan disease, occurs in certain Se-deficient areas of China and can be treated by oral Se supplementation (33). Se metabolic disorders also include Kashin-Beck disease, myxedematous cretinism, and male infertility (27). It is thought that dietary Se plays an important role in cancer prevention; however, controversial results were obtained in a human clinical trial that used selenomethionine as a source of Se to prevent prostate cancer. This trial found no effect of Se supplementation on prostate cancer incidence (19); however, these results in part can be explained by the high Se baseline of the participants and the chemical form of Se used in the trial (12).
Innovation.
In this work, high resolution synchrotron X-ray fluorescence microscopy was used, for the first time, to image Se in mammalian liver and kidney. Both tissues contained uniformly distributed Se, but kidney also showed highly localized circular structures of Se surrounding proximal tubules. We found that this signal represents GPx3, which was secreted from these tubules and remained bound to the basement membrane. It represented approximately 20% of the Se pool in mouse kidney, and an even a higher fraction of this element in the kidney of the naked mole rat. We demonstrate that XFM is a useful tool for studying Se trafficking and distribution in mammals at tissue and cellular levels. Advances in XFM should further help to increase sensitivity and resolution of this method and make it amenable for detection of lower amounts of Se and its distribution in cellular compartments.
Several valuable knockout mouse models have been developed to study Se metabolism, such as liver-specific Sec tRNA[Ser]Sec knockout mice, which lack expression of all selenoproteins specifically in hepatocytes. These mice are viable and show dramatically decreased Se levels in plasma (8). The use of this model revealed an essential role of selenoproteins in liver as well as compensatory mechanisms that protect against selenoprotein deficiency. Glutathione peroxidase 3 (GPx3, also known as plasma glutathione peroxidase) is an extracellular selenoprotein that belongs to the glutathione peroxidase family. In mammals, it is mainly expressed in proximal convoluted tubule cells in the kidney (28). After secretion, it circulates in plasma, but a recent study suggested that a significant portion of GPx3 remains bound to the basement membranes in kidneys (20). The exact function of GPx3 in plasma or at the basement membrane is not known. Decreased expression of this protein may lead to arterial ischemic stroke and cerebral venous thrombosis (29). GPx3 knockout mice are viable, but show increased platelet-dependent thrombus due to the decreased antioxidant capacity in plasma (14). As a member of the glutathione peroxidase family, GPx3 can function as an antioxidant enzyme; however, it is unclear what the endogenous source of reducing equivalents is that supports its peroxidase activity.
Another plasma selenoprotein, Selenoprotein P (SelP), contains 10 Sec residues and is a major plasma selenoprotein. It is primarily expressed in liver and serves as a Se delivery protein (4). It has also been shown that SelP delivers Se to the brain (13). SelP knockout mice are infertile due to impaired spermatogenesis and show severe neurological defects. Specific re-expression of SelP in the liver restores Se transport and rescues phenotypes associated with SelP deficiency (25). Using X-ray fluorescence microscopy (XFM), we recently showed that SelP provides testes with Se where it is used for synthesis of the mitochondrial form of glutathione peroxidase 4 (16, 20).
Naked mole rats are another valuable rodent model to study the role of Se in biology and medicine. These mouse-sized eusocial animals live in subterranean colonies in eastern Africa. The lifespan of naked mole rats exceeds 30 years and they are not known to develop cancer (5, 18, 26). We recently found that naked mole rats are characterized by overall lower levels of Se (15). Mapping Se in tissues may provide new and important insights on the role of Se in cancer prevention and aging.
Se distribution and relocalization during physiological and pathological processes in tissues and cells have not been characterized. Se content in mammalian tissues is quite low (compared to other elements, such as zinc and iron). Therefore, accurate and sensitive detection of cellular Se is challenging. In this study, we utilized high resolution synchrotron XFM to map the content and distribution of Se in liver and kidney. In addition to imaging, we analyzed Se levels by inductively-coupled plasma mass spectrometry (ICP-MS). These analyses were coupled with imaging Se in the kidney of knockout models of two major plasma selenoproteins, SelP and GPx3. Finally, we analyzed Se in the kidney of mice and naked mole rats.
Results
Uniform distribution of Se in mouse liver
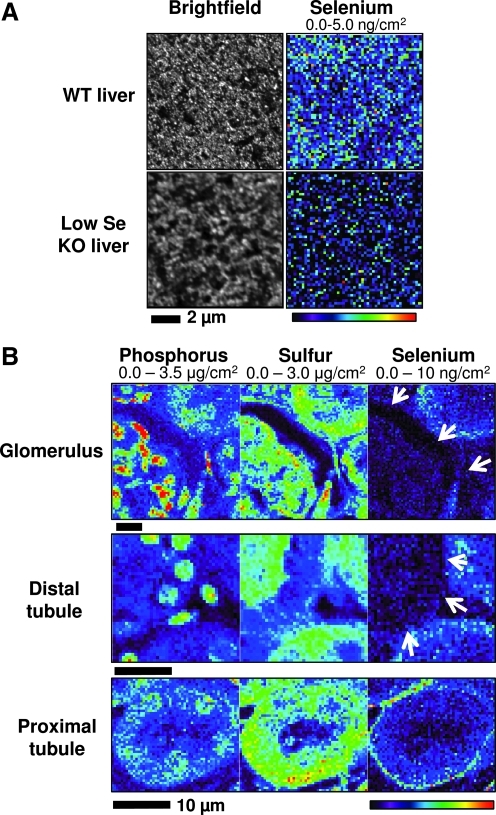
We recently reported the first use of XFM to image Se distribution in mouse testes and sperm (16); however, the distribution of Se in other mammalian organs remains unknown. In the current work, we applied synchrotron XFM to analyze the spatial distribution of Se in mouse tissues, focusing on liver and kidney, which are organs with higher levels of Se compared to most other organs. Analysis of paraffin-embedded liver sections of a wild-type mouse maintained on a regular diet (0.4 ppm Se) revealed a Se signal in liver (Fig. 1A). The assignment of the Se signal was verified by recording single point spectra. Se maps of multiple independent liver samples showed a uniform distribution of this element in hepatocytes. In contrast, Se could not be unambiguously detected in the livers of mice characterized by liver-specific knockout of Sec tRNA[Ser]Sec (8) maintained on a Se-deficient diet (Fig. 1A). This diet has a four-fold lower Se level than regular chow and is often used to achieve decreased expression of all selenoproteins in mouse tissues. Although some background signal was recorded for knockout samples, integral spectra showed that it was considerably lower than the signal in wild-type tissue (Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/ars).
FIG. 1.
XFM of Se in mouse liver and kidney. (A) XFM scans of livers from wild-type and liver-specific Sec tRNA[Ser]Sec knockout mice. Paraffin embedded sections (5 μm) from livers of C57BL/6J mice (WT liver) on a normal diet and the corresponding mice characterized by liver-specific Sec tRNA[Ser]Sec knockout maintained on a Se-deficient diet (low Se KO liver) were mounted on silicon nitride windows, and both light microscope and XFM images were obtained. Se maps are shown in the figure. Maximum and minimum threshold values are given above each image. The scan was obtained by using 13 keV incident energy with a dwell time of 4 sec per pixel and 0.4 μm steps through the sample. (B) XFM of Se in different regions of mouse kidney. Paraffin sections of mouse kidney were mounted on silicon nitride windows and analyzed using XFM. Se distribution was analyzed in glomeruli, proximal tubules, and distal tubules. Arrows indicate the areas of low Se signal. Phosphorus and sulfur maps were used to visualize the morphology of the tissue sections. The data were obtained using 13 keV incident energy with a dwell time of 1.8 sec per pixel and 1 μm steps through the samples. Maximum and minimum threshold values are given above each image. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Highly localized Se in the basement membrane of proximal tubules in mouse kidney
We further examined the Se distribution in mouse kidney. In contrast to liver, Se was heterogeneously distributed in the kidney. The XFM map indicated two Se pools, one of which was distributed across various structures, while the other was represented by a highly localized signal (Fig. 1B). Glomeruli are responsible for blood filtration and therefore may be expected to show elevated Se levels. However, no increased Se was detected in glomeruli, where this element was uniformly distributed. The localized Se distribution was observed in the form of thin circular structures 15–25 μm in diameter. These structures specifically encircled renal proximal tubules and were identified as the basement membrane. Distal tubules, like other parts of the kidney, showed a uniform Se distribution (Fig. 1B). The specific Se distribution to the basement membrane of proximal tubules was repeatedly observed in the analyzed samples.
In addition to Se, we imaged other elements. Chemically, Se is very similar to sulfur, but unlike Se, sulfur was uniformly distributed throughout mouse kidney (Fig. 1B). Phosphorus was associated with cell nuclei, consistent with its high occurrence in DNA. None of the other elements detected under XFM analysis showed circular structures co-localizing with Se maps.
Se imaging in the kidney of the naked mole rat
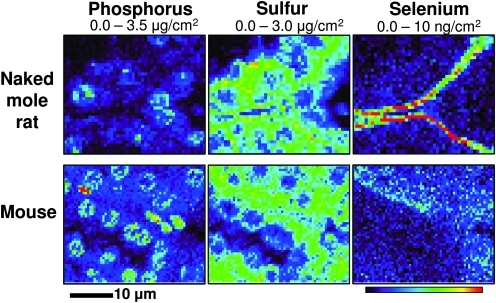
We further examined the Se distribution in the kidney of the naked mole rat (H. glaber) to test whether the highly localized Se signal in the proximal tubule was specific to mice or if it also occurred in other mammals. We recently found that naked mole rats are characterized by overall lower levels of Se due to low expression of GPx1 (15). Paraffin-embedded kidney sections from a 6-year-old naked mole rat were imaged for Se content and distribution using XFM. While the uniformly distributed Se was decreased in the naked mole rat (Table 1), the highly localized basement membrane Se was significantly higher than that in the mouse (Fig. 2). To further characterize Se levels in naked mole rat kidneys, we analyzed trace elements in tissue lysates using ICP-MS. When compared to the mouse, kidney lysates of the naked mole rat had lower Se levels (Table 1).
Table 1.
Quantification of Se in Mouse and Naked Mole Rat Kidney Using XFM and ICP-MS
| Method | Tissue | Naked Mole Rat | Mouse |
|---|---|---|---|
| XFM ng/mm3 tissue | Kidney, basement membrane* | 10.1±0.51 | 6.94±0.83 |
| Kidney, proximal tubule | 1.72±0.49 | 2.91±1.28 | |
| ICP-MS fg/μg total protein | Kidney* | 27.1±1.92 | 77.6±9.47 |
Data represents means±SD, n=3 for XFM and n=5 for ICP-MS measurements.
Values (naked mole rat vs. mouse) are statistically significant (P<0.05).
FIG. 2.
Distribution of elements in mouse and naked mole rat kidneys. Paraffin-embedded kidney sections (5 μm) from mice and naked mole rats were mounted on silicon nitride windows and elemental distribution of phosphorus, sulfur, and Se was mapped using synchrotron XFM. Phosphorus and sulfur maps were used to visualize the morphology of the tissue sections. Images were obtained with 1.5 sec dwell time and 1 μm steps at the incident energy 13 keV. Maximum and minimum threshold values are given above each image. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Se in the basement membrane of proximal tubules represents GPx3
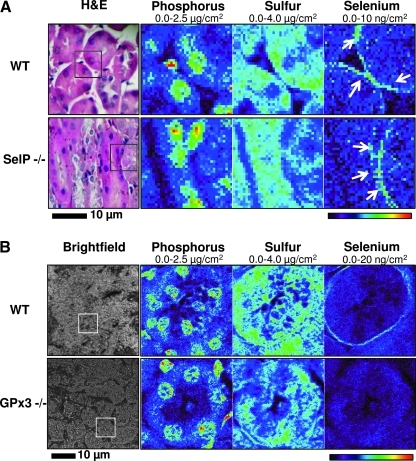
To characterize the source of Se in the basement membranes of proximal tubules in kidneys, we utilized knockout mouse models. We analyzed samples from SelP and GPx3 knockout mice to test if the localized Se signal is associated with the expression of these selenoproteins. As mentioned above, SelP is the most abundant plasma selenoprotein in mammals; it is synthesized primarily in the liver and delivers Se to other organs. Using XFM, we first imaged Se distribution in SelP knockout and corresponding wild type mice and found that the basement membrane Se was preserved in SelP knockout mice (Fig. 3A). Further XFM analyses did not show significant changes in the Se content of proximal tubules in SelP-/- and control mice (Table 2).
FIG. 3.
Elemental distribution in kidneys of wild-type and selenoprotein knockout mice. (A) Elemental distribution in kidneys of wild-type and SelP knockout mice. Mouse tissues were fixed and paraffin embedded. Five-micron sections were mounted on the silicon nitride windows. Se, P, and S distributions of the boxed region (H&E image) were analyzed using XFM. Following elemental mapping, the morphology of the tissues was confirmed using hematoxylin/eosin staining (H&E). Se was primarily localized to the basement membrane of proximal tubules (arrows). This distribution was not altered in the SelP knockout. (B) Elemental distribution in kidneys of control and GPx3 knockout mice. Mouse tissues were fixed and paraffin embedded. Five-micron sections were mounted on the silicon nitride windows. P, S, and Se inside the boxed region were imaged using XFM. Phosphorus and sulfur images are used to show tissue morphology. Images were obtained with 1.5 sec dwell time and 1 μm steps at the incident energy 13 keV. Maximum and minimum threshold values are given above each image. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Table 2.
Quantification of Se in Kidney of WT, GPx3 Knockout and SelP Knockout Mice
| |
Kidney Selenium, ng/mm3 |
|
|---|---|---|
| Mouse genotype | Basement membrane | Proximal tubule |
| WT control SelP +/+ | 7.2±1.1 | 3.5±0.1* |
| SelP −/− | 6.9±0.4 | 3.1±0.1* |
| WT control GPx3 +/+ | 11.5±0.4* | 3.7±1.0 |
| GPx3 −/− | 3.3±0.1* | 3.1±0.1 |
Data represents means±SD. Mean content was calculated using ROI analysis (n=3).
Values (control vs. knokcout) are statistically significant (P<0.05).
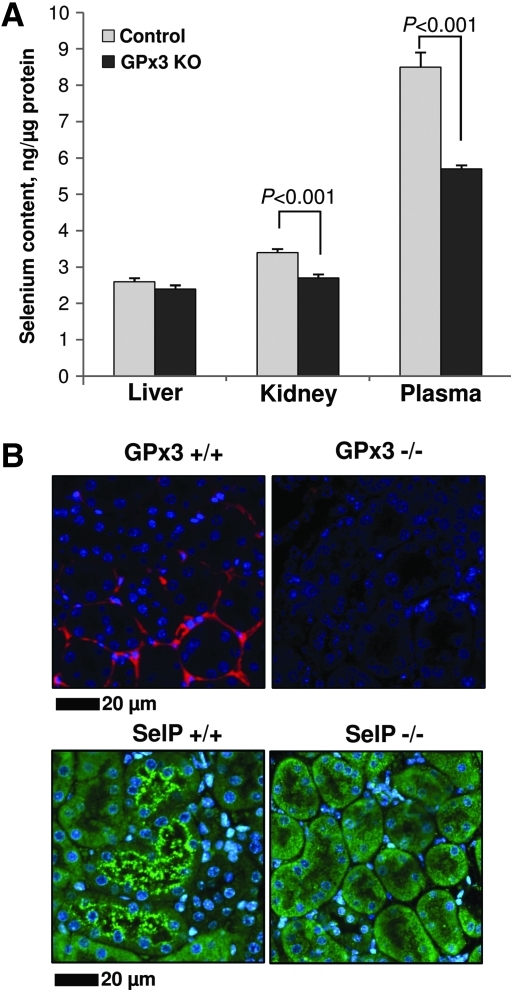
GPx3 is a plasma selenoprotein primarily expressed in the kidney. XFM showed that Se in the basement membrane of the kidney was significantly altered in GPx3 knockout mice (Table 2, Fig. 3B). ICP-MS analysis also showed a distinctive 20% decrease of Se in kidney and plasma (P<0.001), but not in the liver of GPx3-/- mice (Fig. 4A). We further confirmed the localization of GPx3 in the kidney by employing immunohistochemistry to stain paraffin sections with anti-GPx3 antibodies (Fig. 4B). The protein was mostly detected in basement membranes of proximal tubules and, therefore, co-localized with the distribution of Se. In addition, immunohistochemistry of kidney sections with anti-SelP antibodies revealed a luminal distribution of this protein in epithelial cells of proximal tubules (Fig. 4B).
FIG. 4.
ICP-MS and immunohistochemical analyses of GPx3 knockout mice. (A) ICP-MS analysis of Se in GPx3 knockout mice. Lysates from freshly frozen tissues were normalized for protein content, digested in 15% nitric acid, 15% hydrogen peroxide, and analyzed by ICP-MS. 50 ppb gallium was used as an internal control. Statistical difference was validated by the unpaired Student t-test (n=6). Significance is indicated above the bars. (B) Immunohistochemical analysis of SelP and GPx3 in mouse kidney. Upper panels: Paraffin sections were incubated with anti-GPx3 (1:2000 dilution) and detected with Alexa-647 conjugated secondary antibodies. GPx3 is localized to the basement membrane of proximal tubules in kidney. Lower panels: Paraffin sections were incubated with anti-SelP (1:1000 dilution). SelP was visualized with Alexa-488 antibodies. SelP is localized to the lumen of proximal tubules. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Discussion
The mechanisms of Se distribution in mammals have not been comprehensively investigated. Humans have 25 selenoprotein genes, and quantification of absolute levels of selenoproteins is difficult, considering their low expression levels. While ICP-MS can quantify Se in various organs, it does not address its spatial distribution within cells and tissues, which is the most useful information. To deal with this challenge, we used synchrotron XFM to image directly and quantify Se and other elements in mammalian tissues, focusing on liver and kidney, which are organs with high Se levels. This method has a lower sensitivity than ICP-MS and only allows imaging of concentrated and localized Se pools in tissues. However, it provides important morphological information necessary for understanding functional relations involving selenoproteins. Using ICP-MS and XFM data and assuming the weight of 0.5 g and a volume of 600 mm3 in the mouse kidney, we quantified the total amount of kidney Se. Both methods gave very similar estimations (0.3–0.5 μg of Se). XFM provided important high-resolution information about the distribution and identity of high Se pools in mouse tissues; however, this approach had a limited applicability in the mouse liver, where Se was more uniformly distributed.
It should be noted that we were able to use fixed tissue samples embedded in paraffin for high resolution XFM imaging since the vast majority of Se in mammals is bound to proteins (6). Paraffin has very low levels of trace element contamination and, thus, sections can be directly imaged without additional preparation (23). For some other trace elements, such deep tissue processing may lead to decreased detection and/or nonspecific signals (10).
Our study demonstrated that Se can be directly imaged and quantified in liver and kidney. This trace element is uniformly distributed in mouse liver, a finding that was verified by collecting its single point spectra. Furthermore, we proved that the liver Se pool is associated with selenoproteins (i.e., livers from liver-specific Sec tRNA[Ser]Sec knockout mice maintained on a Se-deficient diet showed no detectable Se). GPx1 is the major mammalian selenoprotein and it is expressed at a particularly high level in the liver. The uniform distribution of Se in hepatocytes is consistent with the concept that XFM largely detects GPx1.
Unlike liver, kidney had a highly localized Se-component surrounding the proximal tubules. Other kidney structures, such as glomeruli and distal tubules, only showed the uniformly distributed Se pattern. The specific enrichment of Se in proximal tubules was associated with the basement membranes (Fig. 1B). We also examined the phosphorus and sulfur maps to better understand tissue morphology. To investigate the origin of the specific Se signal in kidney, we analyzed mouse knockout models of two selenoproteins involved in Se metabolism. SelP, the main Se transport protein, is highly expressed in liver (32). In kidney, SelP localized to the lumen of proximal tubules (Fig. 4B) (21). GPx3 is another extracellular selenoprotein which is secreted primarily by proximal tubule cells. It has been proposed that SelP is the Se source for GPx3 expression (1); however, the Se content of basement membranes was not altered in the kidney of SelP knockout mice (Fig. 3A, Table 2). By contrast, the Se signal was eliminated in GPx3 knockout mice, indicating that this Se signal is derived from GPx3 (Fig. 3B). This protein was previously localized to tubular cells in mouse kidney (20), and we confirmed this GPx3 localization by immunohistochemistry (Fig. 4B). The exact source of Se for high GPx3 expression in SelP knockout mice is not known, but it is likely that low-molecular-weight Se compounds compensate for SelP deficiency by providing Se to various organs, including kidney. SelP knockout mice have increased amounts of Se excreted in the urine (7). Thus, considering the amount of urine produced each day and the total Se lost in SelP knockout mice, an alternative SelP-independent pathway must supply Se for GPx3 expression.
In another model organism examined in the current study, the naked mole rat, kidney had lower Se levels compared to that in mice, as measured by ICP-MS (Table 1). However, XFM of naked mole rat kidney revealed high Se levels on the basement membrane of proximal tubules and low levels of uniformly distributed Se within the proximal tubules (Table 1). These data suggest that the naked mole rat is characterized by low GPx1 expression, while its GPx3 expression is fully preserved, if not increased, compared to that in mice. This observation points to different mechanisms and Se sources involved in GPx1 and GPx3 expression. Our ICP-MS analysis showed that, in GPx3 knockout mice, Se was decreased by 20% in plasma and kidneys, but not in liver (Fig. 4A). In addition, naked mole rats had lower Se in the kidney than mice (Table 1). These data further support the relationships described above between Se levels and expression of GPx3, GPx1, and SelP.
Materials and Methods
X-ray fluorescence microscopy
C57BL/6 mice, fed with standard rodent chow (0.4 ppm Se), were sacrificed and their tissues dissected according to approved protocols. Liver-specific Sec tRNA[Ser]Sec knockout mice were fed a Se-deficient diet (0 ppm) for 3 months. Tissues from SelP knockout mice (13) and the corresponding wild-type tissues were kindly provided by Drs. Raymond Burk and Kristina Hill (Vanderbilt University). GPx3 knockout mice and Sec tRNA[Ser]Sec knockout mice (8) were maintained at the Brigham and Women's Hospital. All mice in the study were 8–12-weeks old. Six-year old naked mole rats (Heterocephalus glaber) were maintained at the University of Illinois at Chicago. Freshly dissected tissues were either frozen in liquid nitrogen and stored at −80°C or fixed in 4% formaldehyde in PBS for elemental imaging and immunohistochemical studies. After fixation for 12 h in formaldehyde solution, samples were processed in a Leica tissue processor (Leica Microsystems, Vienna, Austria) and embedded in paraffin. Five-micron sections from formalin-fixed, paraffin-embedded tissues were transferred to a clean water bath, mounted on 2×2 mm silicon nitride windows (Silson, Blisworth, UK), and stored at room temperature until analysis.
Trace elements were mapped at beamline 2-ID-E at the Advanced Photon Source (APS), Argonne National Laboratory (Argonne, IL) using a hard X-ray microprobe (6–30 keV). Prior to XFM analysis, light microscopy pictures were taken for all samples using a Leica DMXRE microscope (Leica Microsystems, Bannockburn, IL). In-house software was used to convert positions of the microscope's motorized stage to coordinates of the XFM microprobe, allowing us to locate and image areas of interest. Phase contrast images were also taken for each XFM image area. Elemental imaging was performed using 13 keV X-rays generated by an APS Sector 2 undulator. Fresnel zone plate optics (X-radia, Concord, CA) were used to focus X-rays to a measured spot size of 0.4×0.4 μm. For high resolution images, specimens were raster-scanned with 1 μm or 0.4 μm steps. Full X-ray fluorescence spectra were recorded for each pixel using energy dispersive Vortex detector (SII Nanotechnologies, Northridge, CA). Excitation at this energy with the collection of full fluorescence spectra generated data sets with quantitative information for elements from S through Se in atomic number. Exposure times varied from 1.4 sec to 2 sec per pixel, depending on the intensity of the incident beam. X-ray fluorescence spectra for each sample were normalized and fitted to Gaussians at characteristic emission energies for each element using MAPS software (30). The images were quantified by fitting the sample spectra against the signal derived from the thin film standards NBS-1832 and NBS-1833 (National Bureau of Standards, MD), which were collected after each set of experiments.
Morphological analysis of tissue samples
In order to evaluate tissue morphology, samples were stained with hematoxylin and eosin (H&E) following XFM analyses. Paraffin tissue sections mounted on the silicon nitride windows were deparaffinized in xylene (Sigma, St. Louis, MO) (3 changes, 5 min each) and rehydrated in histology grade ethanol (Fisher Scientific, Hampton, NH) (100%, 95%, 70%, 3 min each). Each window was washed in water and stained with Zymed hematoxylin (Zymed Laboratories, Carlsbad, CA) for 10 min and developed in PBS (15 min). Counterstaining was performed by dipping samples in alcohol eosin (Eosin Y) for 30 sec (Sigma). Samples were dehydrated in ethanol (10 min) and cleared in xylene (5 min). For storage and imaging, samples were immersed in a glycerol-vinyl-alcohol (GVA) mount (Zymed Laboratories). Imaging was performed with an Olympus (Shinjuku, Japan) AX70 upright microscope using a high-resolution digital camera.
Inductively-coupled plasma mass spectrometry
Se in animal tissues was quantified using ICP-MS. All analyses were performed at the University of Nebraska-Lincoln spectroscopy core facility equipped with Agilent Technologies ICP-MS 7500cx series (Agilent Technologies, Santa Clara, CA) and SC autosampler (Elemental Scientific, Omaha, NE). Tissues were homogenized and sonicated in PBS containing Complete Protease Inhibitor Cocktail (Roche, Basel, Switzerland). Total protein content was measured in lysates using the Bradford Protein Assay (Bio-Rad, Hercules, CA) for sample normalization. Samples were digested in 20% nitric acid, 15% hydrogen peroxide for 2 h at 70°C. Fifty ppb gallium was added as an internal standard. Prior to ICP-MS analysis, samples were diluted 10 times and stored at −80°C. In addition, before analysis of each series of samples, the ICP-MS instrument was calibrated using custom elemental standards covering the range of concentrations in the samples. Each sample was analyzed 3 times (0.3 ms each time) and in triplicate. The analysis used a collision cell filled with 3.5 mL/min of H2 and 1.5 mL/min of He with an Ar carrier flow of 0.9 L/min and Ar make-up flow of 0.15 L/min, and RF power of 1500 W.
Immunohistochemistry of tissue samples
Five-micron sections from formalin-fixed, paraffin-embedded mouse kidney were deparaffinized in xylene followed by rehydration in a series of ethanol washes followed by water. Antigen retrieval was performed for 1 min in a decloaking chamber in Tris-EDTA buffer (10 mM Tris Base, 1 mM EDTA, 0.05% Tween 20, pH 9.0) at 124°C. Samples were blocked with TBST (20 mM Tris·HCl, pH 8.0, 150 mM sodium chloride, 0.025% Triton X100) containing 1% BSA and 10% goat serum. Sections were incubated with anti-SelP (1:1000 dilution) and anti-GPx3 (1:2000 dilution) antibodies (Abcam, Cambridge, MA). Control sections were incubated with non-immune IgG. Sections were next washed in TBST and incubated with secondary antibodies Alexa-488 and Alexa-647 (Invitrogen, Carlsbad, CA). 4′,6-Diamidino-2-phenylindole (DAPI) was added to the secondary antibody solution to stain DNA. Slides were washed in TBS and mounted with glycerol-based FluorMount (Thermo, Pittsburgh, PA). Slides were imaged using a Zeiss Observer Z1 inverted confocal microscope (Carl Zeiss, Jena, Germany) using a 40x objective. Images were processed and prepared using Zeiss ZEN software.
Supplementary Material
Abbreviations Used
- GPx
glutathione peroxidase
- ICP-MS
inductively-coupled plasma mass spectrometry
- Se
selenium
- Sec
selenocysteine
- SECIS
selenocysteine insertion sequence
- TGR
thioredoxin-glutathione reductase
- XFM
X-ray fluorescence microscopy
Acknowledgments
We thank Drs. Raymond Burk and Kristina Hill (Vanderbilt University) for providing tissues of SelP knockout mice. This work was supported by National Institutes of Health grants to VNG (GM061603 and CA080946) and JL (HL061795, HL070819, and HL048743), and the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, to DLH. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
Author Disclosure Statement
The authors have no competing financial interests to declare.
References
- 1.Avissar N. Ornt DB. Yagil Y. Horowitz S. Watkins RH. Kerl EA. Takahashi K. Palmer IS. Cohen HJ. Human kidney proximal tubules are the main source of plasma glutathione peroxidase. Am J Physiol. 1994;266:C367–75. doi: 10.1152/ajpcell.1994.266.2.C367. [DOI] [PubMed] [Google Scholar]
- 2.Bellinger FP. Raman AV. Reeves MA. Berry MJ. Regulation and function of selenoproteins in human disease. Biochem J. 2009;422:11–22. doi: 10.1042/BJ20090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birringer M. Pilawa S. Flohe L. Trends in selenium biochemistry. Nat Prod Rep. 2002;19:693–718. doi: 10.1039/b205802m. [DOI] [PubMed] [Google Scholar]
- 4.Boitani C. Puglisi R. Selenium, a key element in spermatogenesis and male fertility. Adv Exp Med Biol. 2008;636:65–73. doi: 10.1007/978-0-387-09597-4_4. [DOI] [PubMed] [Google Scholar]
- 5.Buffenstein R. The naked mole-rat: A new long-living model for human aging research. J Gerontol A Biol Sci Med Sci. 2005;60:1369–1377. doi: 10.1093/gerona/60.11.1369. [DOI] [PubMed] [Google Scholar]
- 6.Burk RF. Hill KE. Selenoprotein P-expression, functions, and roles in mammals. Biochim Biophys Acta. 2009;1790:1441–1447. doi: 10.1016/j.bbagen.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burk RF. Hill KE. Motley AK. Austin LM. Norsworthy BK. Deletion of selenoprotein P upregulates urinary selenium excretion and depresses whole-body selenium content. Biochim Biophys Acta. 2006;1760:1789–1793. doi: 10.1016/j.bbagen.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson BA. Novoselov SV. Kumaraswamy E. Lee BJ. Anver MR. Gladyshev VN. Hatfield DL. Specific excision of the selenocysteine tRNA[Ser]Sec (Trsp) gene in mouse liver demonstrates an essential role of selenoproteins in liver function. J Biol Chem. 2004;279:8011–8017. doi: 10.1074/jbc.M310470200. [DOI] [PubMed] [Google Scholar]
- 9.Eckschlager T. Adam V. Hrabeta J. Figova K. Kizek R. Metallothioneins and cancer. Curr Protein Pept Sci. 2009;10:360–375. doi: 10.2174/138920309788922243. [DOI] [PubMed] [Google Scholar]
- 10.Farquharson MJ. Geraki K. Falkenberg G. Leek R. Harris A. The localization and micro-mapping of copper and other trace elements in breast tumors using a synchrotron micro-XRF system. Appl Radiat Isot. 2007;65:183–188. doi: 10.1016/j.apradiso.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Flohe L. Selenium in mammalian spermiogenesis. Biol Chem. 2007;388:987–95. doi: 10.1515/BC.2007.112. [DOI] [PubMed] [Google Scholar]
- 12.Hatfield DL. Gladyshev VN. The Outcome of Selenium and Vitamin E Cancer Prevention Trial (SELECT) reveals the need for better understanding of selenium biology. Mol Interv. 2009;9:18–21. doi: 10.1124/mi.9.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill KE. Zhou J. McMahan WJ. Motley AK. Atkins JF. Gesteland RF. Burk RF. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol Chem. 2003;278:13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 14.Jin RC. Mahoney CE. Zhang Y-Y. Handy DE. Loscalzo J. Abstract 5302: Glutathione peroxidase-3 deficiency promotes platelet-dependent thrombosis and thrombotic stroke in vivo. Circulation. 2009;120:S1080–1081. doi: 10.1161/CIRCULATIONAHA.110.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasaikina MV. Lobanov AV. Malinouski MY. Lee BC. Seravalli J. Fomenko DE. Turanov AA. Finney L. Vogt S. Park TJ. Miller RA. Hatfield DL. Gladyshev VN. Reduced utilization of selenium by naked mole rats due to a specific defect in GPx1 expression. J Biol Chem. 2011;286:17005–17014. doi: 10.1074/jbc.M110.216267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kehr S. Malinouski M. Finney L. Vogt S. Labunskyy VM. Kasaikina MV. Carlson BA. Zhou Y. Hatfield DL. Gladyshev VN. X-ray fluorescence microscopy reveals the role of selenium in spermatogenesis. J Mol Biol. 2009;389:808–818. doi: 10.1016/j.jmb.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kryukov GV. Castellano S. Novoselov SV. Lobanov AV. Zehtab O. Guigo R. Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 18.Liang S. Mele J. Wu Y. Buffenstein R. Hornsby PJ. Resistance to experimental tumorigenesis in cells of a long-lived mammal, the naked mole-rat (Heterocephalus glaber) Aging Cell. 2010;9:626–635. doi: 10.1111/j.1474-9726.2010.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lippman SM. Klein EA. Goodman PJ. Lucia MS. Thompson IM. Ford LG. Parnes HL. Minasian LM. Gaziano JM. Hartline JA. Parsons JK. Bearden JD., 3rd Crawford ED. Goodman GE. Claudio J. Winquist E. Cook ED. Karp DD. Walther P. Lieber MM. Kristal AR. Darke AK. Arnold KB. Ganz PA. Santella RM. Albanes D. Taylor PR. Probstfield JL. Jagpal TJ. Crowley JJ. Meyskens FL., Jr. Baker LH. anad Coltman CA., Jr Effect of selenium and vitamin E on risk of prostate cancer and other cancers: The Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA. 2009;301:39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson GE. Whitin JC. Hill KE. Winfrey VP. Motley AK. Austin LM. Deal J. Cohen HJ. Burk RF. Extracellular glutathione peroxidase (Gpx3) binds specifically to basement membranes of mouse renal cortex tubule cells. Am J Physiol Renal Physiol. 2010;298:F1244–1253. doi: 10.1152/ajprenal.00662.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson GE. Winfrey VP. Hill KE. Burk RF. Megalin mediates selenoprotein P uptake by kidney proximal tubule epithelial cells. J Biol Chem. 2008;283:6854–6860. doi: 10.1074/jbc.M709945200. [DOI] [PubMed] [Google Scholar]
- 22.Palacios O. Encinar JR. Bertin G. Lobinski R. Analysis of the selenium species distribution in cow blood by size exclusion liquid chromatography-inductively coupled plasma collision cell mass spectrometry (SEC-ICP-MS) Anal Bioanal Chem. 2005;383:516–522. doi: 10.1007/s00216-005-0015-8. [DOI] [PubMed] [Google Scholar]
- 23.Ralle M. Lutsenko S. Quantitative imaging of metals in tissues. Biometals. 2009;22:197–205. doi: 10.1007/s10534-008-9200-5. [DOI] [PubMed] [Google Scholar]
- 24.Rayman MP. The importance of selenium to human health. Lancet. 2000;356:233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 25.Renko K. Werner M. Renner-Muller I. Cooper TG. Yeung CH. Hollenbach B. Scharpf M. Kohrle J. Schomburg L. Schweizer U. Hepatic selenoprotein P (SePP) expression restores selenium transport and prevents infertility and motor-incoordination in Sepp-knockout mice. Biochem J. 2008;409:741–749. doi: 10.1042/BJ20071172. [DOI] [PubMed] [Google Scholar]
- 26.Seluanov A. Hine C. Azpurua J. Feigenson M. Bozzella M. Mao Z. Catania KC. Gorbunova V. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc Natl Acad Sci USA. 2009;106:19352–19357. doi: 10.1073/pnas.0905252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surai PF. Karadas F. Pappas AC. Sparks NH. Effect of organic selenium in quail diet on its accumulation in tissues and transfer to the progeny. Br Poult Sci. 2006;47:65–72. doi: 10.1080/00071660500475244. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi K. Newburger PE. Cohen HJ. Glutathione peroxidase protein. Absence in selenium deficiency states and correlation with enzymatic activity. J Clin Invest. 1986;77:1402–1404. doi: 10.1172/JCI112449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voetsch B. Jin RC. Bierl C. Deus-Silva L. Camargo EC. Annichino-Bizacchi JM. Handy DE. Loscalzo J. Role of promoter polymorphisms in the plasma glutathione peroxidase (GPx-3) gene as a risk factor for cerebral venous thrombosis. Stroke. 2008;39:303–307. doi: 10.1161/STROKEAHA.107.490094. [DOI] [PubMed] [Google Scholar]
- 30.Vogt S. MAPS: A set of software tools for analysis and visualization of 3D X-ray fluorescence data sets. J Pysiscs IV France. 2003;104:635–638. [Google Scholar]
- 31.Walczak R. Westhof E. Carbon P. Krol A. A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA. 1996;2:367–379. [PMC free article] [PubMed] [Google Scholar]
- 32.Whitin JC. Bhamre S. Tham DM. Cohen HJ. Extracellular glutathione peroxidase is secreted basolaterally by human renal proximal tubule cells. Am J Physiol Renal Physiol. 2002;283:F20–28. doi: 10.1152/ajprenal.00014.2001. [DOI] [PubMed] [Google Scholar]
- 33.Yang GQ. Ge KY. Chen JS. Chen XS. Selenium-related endemic diseases and the daily selenium requirement of humans. World Rev Nutr Diet. 1988;55:98–152. doi: 10.1159/000415560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.