Abstract
Aims: Increased oxidative stress and mitochondrial dysfunction in obese adipocytes contribute to adipokine dysregulation, inflammation, and insulin resistance. Results: Through an advanced proteomic analysis, we found that peroxiredoxin 3 (Prx3), a thioredoxin-dependent mitochondrial peroxidase, is highly expressed in 3T3-L1 adipocytes compared to preadipocytes. Interestingly, in obese db/db mice and human subjects, adipose Prx3 levels were significantly decreased, indicating its association with obesity. We therefore employed Prx3 knockout (KO) mice and transfected 3T3-L1 cells to examine the role of endogenous Prx3 in adipocyte metabolism. Prx3 KO mice had increased fat mass compared to wild-type due to adipocyte hypertrophy. Increased adipogenic transcription factors and lipogenic gene expression during differentiation of adipose tissue-derived stem cells from Prx3-deficient mice confirmed that these adipocytes are likely to accumulate fat. Mitochondrial protein carbonylation in Prx3 KO adipose tissue and mitochondrial superoxide level in Prx3 knockdown 3T3-L1 cells were increased showing aberrant regulation of oxidative stress. Proteomic analysis and gene expression analysis of Prx3 KO mice adipocytes also showed defect in mitochondria biogenesis along with enzymes involved in glucose/lipid metabolism and oxidative phosphorylation. In addition, expression level of adiponectin was downregulated and plasminogen activator inhibitor-1 was upregulated in Prx3 KO adipocytes. Impaired glucose tolerance and insulin resistance further implied metabolic dysregulation in Prx3 KO mice. Innovation and Conclusion: These data suggest that endogenous Prx3 may play an essential role in maintaining normal characteristics of adipocytes and that defect in Prx3 alters mitochondrial redox state and function, and adipokine expression in adipocytes leading to metabolic alteration. Antioxid. Redox Signal. 16, 229–243.
Introduction
Obesity is one of the most prevalent chronic diseases worldwide (37). Obesity is defined as excessive fat accumulation in the adipose tissue that evokes a cluster of diseases such as insulin resistance, type 2 diabetes mellitus, and cardiovascular diseases referred to as ‘metabolic syndrome’ (39). Recent studies have implicated systemic oxidative stress as a major contributing factor for obesity-related metabolic complications (20) and that oxidative stress in adipose tissue results in inflammation, adipokine dysregulation, and insulin resistance (10, 16). Since adipose tissue plays a crucial role in regulating whole body insulin sensitivity (3), a balanced regulation of pro-oxidative and antioxidative mechanism in adipocytes is an important matter.
Adipocytes are the primary cells that compose adipose tissue, and these cells function in maintaining energy homeostasis by buffering lipid metabolites and secreting adipokines such as leptin, adiponectin, and plasminogen activator inhibitor-1 (PAI-1). Dysregulation of adipokine production affects the function of peripheral tissues, including liver and muscle, which increases the risk of metabolic complications (38). Therefore, a thorough understanding of adipocyte differentiation, expansion, and its endocrine function is necessary for developing therapies against obesity.
Innovation.
Accumulating evidence indicates that adipocyte oxidative stress plays an important role in development of obesity-related complications. However, adipocyte antioxidant mechanism and its control are poorly understood. The present study has identified Prx3, a thioredoxin-dependent mitochondrial antioxidant, as a new target for regulating adipocyte function, including adipokine expression, and glucose/lipid metabolism through diverse measures, including proteomics and employing Prx3 KO mice, adipocyte-derived stem cells isolated from Prx3 KO mice, and 3T3-L1 adipogenic model. Importantly, decreased Prx3 expression in peri-renal adipose tissue of overweight subjects provides a possible clinical implication on the role of adipocyte Prx3 in metabolic syndrome.
Proteomics have been widely applied recently for high-throughput analysis of proteins. Up to date, few investigations have attempted to characterize the proteins that change during adipogenesis (1, 2, 7, 21, 40) or those secreted from adipocytes (24, 42). However, the methodologies had limitations in detecting differences without gel separation and identifying proteins with low resolution ion trap MS. In this study, we employed an advanced proteomic methodology to detect the changes in protein expression profiles during differentiation of 3T3-L1 cells, a common in vitro model for studying adipogenesis (11).
Our novel finding is that peroxiredoxin 3 (Prx3) exists abundantly in mature 3T3-L1 adipocytes compared to preadipocytes and is reduced in fat tissues of obese mice and human subjects. Prxs are a family of cellular thiol peroxidases scavenging peroxides. Among six distinct members, Prx3 is unique in that it is exclusively localized in the mitochondria (8). Studies have reported that Prx3 knockout (KO) mice are hypersensitive to lipopolysaccharide-induced lung inflammation (30) and that overexpression of Prx3 has protective effects in neurons (13) and myocardial infarction (31). The role of endogenous Prx3 in adipocytes, however, has not been demonstrated.
Mitochondria are major source of cellular oxidative stress, and ‘mitochondrial fitness' plays a crucial role in the maintenance of adipocyte function (9). Since Prx3 is a mitochondrial antioxidant, our study focused on the role of endogenous Prx3 linking mitochondrial oxidative stress and regulation of adipocyte function. We found that Prx3-deficient mice displayed increased fat mass. Adipose tissue-derived stem cells from Prx3 KO mice and Prx3 knockdown 3T3-L1 adipocytes expressed higher levels of adipogenic genes. Furthermore, increased mitochondrial oxidative stress as well as downregulation in mitochondrial metabolic proteins was observed in Prx3-deficient adipocytes. These adipocytes also showed adipokine dysregulation, impaired glucose tolerance and insulin sensitivity, all of which suggest that Prx3 plays an essential role in maintaining adipose tissue integrity and sustaining whole body metabolism.
Results
Comparative proteomic analysis of 3T3-L1 preadipocytes and mature adipocytes reveals major changes in mitochondrial proteins related to metabolism
Protein profiles of 3T3-L1 preadipocytes and mature adipocytes were compared by proteomic analysis (Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/ars). Among 57 protein spots that showed significant changes (>1.5-fold) between preadipocytes and mature adipocytes (Supplementary Tables S1A and S1B), 16 proteins were significantly (p<0.05) upregulated and 6 proteins downregulated in mature adipocytes (Table 1). The identified proteins were classified and grouped based on their known functions using gene ontology (www.godatabase.org, accessed June 2010, Supplementary Fig. S2). More than 50% of the identified proteins function in cellular metabolism, while proteasome, chaperone, oxido-reduction, and transporter/channel related proteins each comprise about 10% of total identified proteins. Cellular location showed that mitochondrial proteins account for 38% of identified proteins, nearly equivalent to cytosolic proteins.
Table 1.
Differentially Expressed Proteins in Mature 3T3-L1 Adipocytes
| No. | P value | Ratio (Adipo/3T3) | Score | %Cov. | Mr | pI | Accession No. | Symbol | Protein Name | Cellular Component |
|---|---|---|---|---|---|---|---|---|---|---|
| Proteins that function in metabolism | ||||||||||
| 8 | 0.00283 | −2.3 | 1.42E+03 | 26 | 39503 | 5.0 | Q9JHJ0 | TMOD3 | Tropomodulin-3 | Cyto, CK |
| 10 | 0.0445 | −2.0 | 649 | 43 | 28785.1 | 4.7 | P17918 | PCNA | Proliferating cell nuclear antigen;Cyclin | Nu |
| 13 | 0.0476 | −1.5 | 7.87E+03 | 40 | 20678.6 | 5.3 | Q9CPU0 | LGUL | Lactoylglutathione lyase | |
| 19 | 0.0097 | 2.6 | 2.40E+17 | 40 | 129685.7 | 6.3 | Q05920 | PYC | Pyruvate carboxylase, mitochondrial precursor | Mito |
| 20 | 0.0223 | 2.5 | 1.01E+11 | 31 | 129685.7 | 6.3 | Q05920 | PYC | Pyruvate carboxylase, mitochondrial precursor | Mito |
| 28 | 0.0267 | 2.7 | 6.46E+03 | 24 | 47874.5 | 8.0 | Q9DBL1 | ACDSB | Acyl-CoA dehydrogenase, short/branched chain specific, mitochondrial precursor | Mito |
| 29 | 0.000722 | 2.1 | 6.52E+05 | 36 | 43232.1 | 8.5 | P35486 | ODPA | Pyruvate dehydrogenase E1 component alpha subunit, somatic form, mitochondrial precursor | Mito |
| 30 | 0.0157 | 2.1 | 4.37E+03 | 23 | 102536.1 | 5.8 | O08528 | HXK2 | Hexokinase type II | Mito |
| 35 | 0.0496 | 1.6 | 4.89E+05 | 41 | 36346.3 | 6.2 | P14152 | MDHC | Malate dehydrogenase, cytoplasmic | Cyto |
| 38 | 0.0114 | 3.2 | 3.07E+04 | 41 | 32667.4 | 5.4 | Q9D819 | IPYR | Inorganic pyrophosphatase | Cyto |
| 42 | 0.0332 | 1.5 | 1.22E+05 | 59 | 29820.3 | 5.0 | P67778 | PHB | Prohibitin | Memb |
| 45 | 7.98E-06 | 2.3 | 9.31E+03 | 42 | 26000 | 6.1 | P47968 | RPIA | Ribose-5-phosphate isomerase | |
| Proteins that relate with proteasome | ||||||||||
| 3 | 0.0138 | −1.8 | 4.21E+10 | 38 | 80752.3 | 5.4 | Q9QUR6 | PPCE | Prolyl endopeptidase | Cyto |
| 24 | 0.00189 | 2.4 | 6.93E+03 | 16 | 61977 | 4.9 | Q8R317 | UBQL1 | Ubiquilin 1 | Cyto, Nu, Proteasome |
| Proteins that act as transporter/channel | ||||||||||
| 40 | 0.0124 | 2.0 | 1.05E+07 | 48 | 32892.3 | 6.1 | Q99J99 | THTM | 3-mercaptopyruvate sulfurtransferase | Cyto, Mito |
| 50 | 0.00779 | 2.1 | 5.57E+05 | 65 | 18618.4 | 5.5 | Q9DCX2 | ATP5H | ATP synthase D chain, mitochondrial | Mito |
| Proteins that act in oxido-reduction | ||||||||||
| 41 | 0.00981 | 1.6 | 2.50E+08 | 65 | 35440.3 | 8.4 | Q99L13 | 3HIDH | 3-hydroxyisobutyrate dehydrogenase, mitochondrial precursor | Mito |
| 47 | 0.000177 | 4.0 | 120 | 24 | 28109 | 7.2 | P20108 | PRDX3 | Thioredoxin-dependent peroxide reductase, mitochondrial precursor | Mito |
| 49 | 0.000176 | 2.1 | 3.35E+04 | 56 | 28127.3 | 7.2 | P20108 | PRDX3 | Thioredoxin-dependent peroxide reductase, mitochondrial precursor | Mito |
| 53 | 0.044 | 1.9 | 2.78E+03 | 43 | 15811.6 | 6.0 | P08228 | SODC | Superoxide dismutase [Cu-Zn] | Cyto |
| Unknown | ||||||||||
| 9 | 0.024 | −1.7 | 5.72E+09 | 59 | 35752.7 | 4.8 | P48036 | ANXA5 | Annexin A5 | |
| 11 | 0.0003 | −2.0 | 4.77E+04 | 30 | 37280.2 | 5.6 | P10605 | CATB | Cathepsin B precursor | Lys |
Mitochondrial biogenesis is a fundamental aspect of adipocyte differentiation (41). In our data, mitochondrial proteins were all significantly increased in mature adipocytes. Mitochondrial biogenesis was confirmed by increased mRNA expression of mitochondrial transcription factor A (mtTFA) and PPARγ coactivator-1 alpha (PGC1α) and MitoTracker staining (Supplementary Fig. S3).
Prx3 is highly expressed in mature adipocytes and decreased in obese mice and human subjects
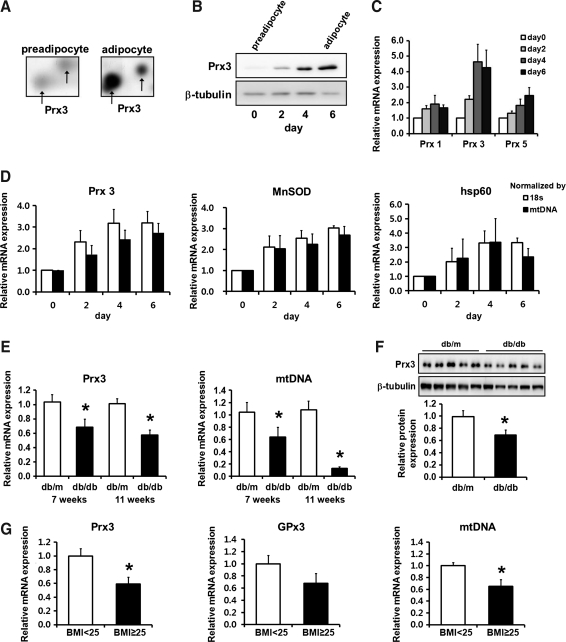
Proteomic analysis has revealed that mitochondrial specific antioxidant Prx3 is dramatically upregulated during adipogenesis (Fig. 1A). The mRNA and protein expression examined at day 0, 2, 4, and 6 of differentiation showed a gradual increase in Prx3 expression. Compared to Prx3, cytoplasmic Prx1 and cytoplasmic/mitochondrial Prx5 levels were increased to a lesser extent, indicating importance of Prx3 in mature adipocytes (Figs. 1B and 1C).
FIG. 1.
Prx3 levels are increased during adipogenesis and decreased in white adipose tissues of obese mice and human subjects. Prx3 levels in preadipocytes and mature adipocytes were evaluated by (A) proteomic analysis; (B) Western blot analysis. (C) real-time PCR analysis of Prx1, 3, and 5 during adipogenesis. (D) MnSOD and hsp60 mRNA levels are shown as a comparison. (E) mRNA levels of Prx3 and mtDNA and (F) protein levels of Prx3 in db/m and db/db mice white adipose tissues. (G) mRNA levels of Prx3, GPx3, and mtDNA were measured by real-time PCR in 28 human peri-renal white adipose tissues. Quantifications were normalized to 18S in each reaction, and to mtDNA or tubulin where indicated. Values are means±SE. *p<0.05 vs. db/m or BMI<25. BMI, body mass index.
Interestingly, mRNA and protein expression of Prx3 in white adipose tissues (WAT) of db/db mice was greatly decreased compared to db/m mice (Figs. 1E and 1F), whereas in liver and kidney Prx3 protein levels were significantly increased (Supplementary Fig. S4). Concurrently, results from human peri-renal WAT showed significantly decreased Prx3 mRNA expression in subjects with body mass index (BMI) higher than 25 (Fig. 1G). In line with our result, association between genetic variations of Prx3 and obesity risk has been recently reported (14). Also, glutathione peroxidase 3 (GPx3), which tended to decrease in our data, is substantially diminished in obese human subjects (BMI>30) which is linked with increased local and systemic oxidative stress (26). These results suggest a novel finding that decreased expression of Prx3 is closely associated with obesity.
Prx3 KO mice display larger fat pads with hypertrophied adipocytes
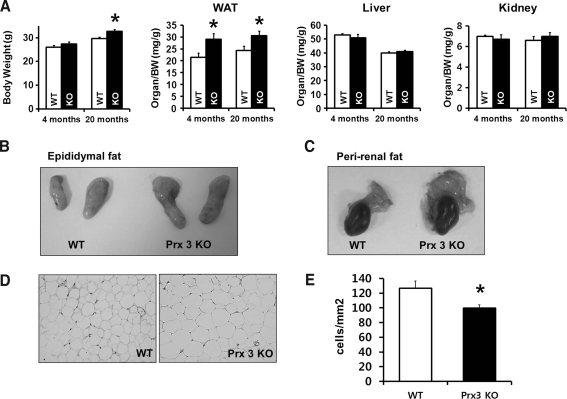
Since reactive oxygen species (ROS) produced by mitochondria are implicated in development of metabolic diseases (9, 33), Prx3 may play a protective role in regulating adipocytes. The body weights of wild-type (WT) and Prx3 KO mice were indistinguishable at the 4-month period. However, by 20 months of age, the body weights of Prx3 KO mice were higher than that of WT. A comparison of organ weights revealed a significant increase in epididymal WAT (Fig. 2A). Morphology and quantitative analysis further indicated that adipocytes were larger in Prx3 KO mice (Figs. 2D and 2E).
FIG. 2.
Prx3 KO mice have larger amounts of adipose tissue compared to WT mice. (A) Body weight and organ weight of 4- and 20-month-old WT and Prx3 KO mice are shown. (B and C) Representative pictures of WT and Prx3 KO mice epididymal fat pads and peri-renal fat. (D) Histological sections of WAT stained with hematoxylin and eosin. (E) Quantitative analysis of cell numbers per mm2 in the sections of WAT. For each mouse, four random pictures were used. Values are means±SE of 5 (4 months old) or 9 (20 months old) mice per group. *p<0.05 vs. WT. WAT, white adipose tissue.
Adipogenic genes are increased in Prx3-deficient adipocytes and decreased in Prx3 overexpressed adipocytes
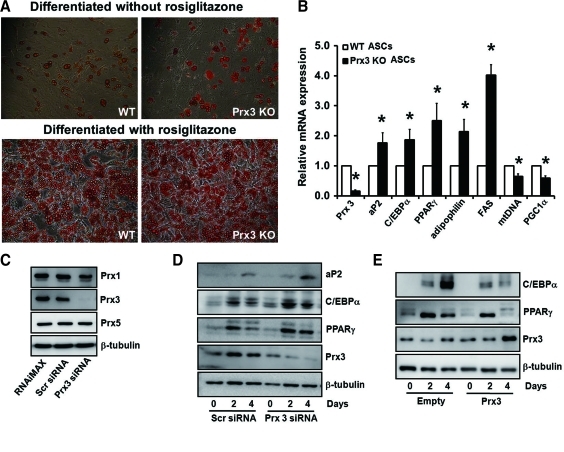
ROS production increases during differentiation of 3T3-L1 cells (10), likely due to increase in metabolic activity of the cell required for lipid accumulation and excessive ROS stimulate adipogenesis (15, 25). It can therefore be translated that increase in Prx3 is an adaptive response for fine adjustment of ROS levels during adipogenesis. To evaluate the role of Prx3 on adipogenesis, we compared the adipocyte differentiation capacity of adipose tissue-derived stem cells (ASCs) isolated from WT and Prx3 KO mice. Rosiglitazone-induced differentiation of ASCs showed dramatic accumulation of lipids, but we used conventional differentiation medium with 3-isobutyl-1-methylxanthine (IBMX) instead, since rosiglitazone is a PPARγ activator that forces preadipocytes to differentiate. Although little difference was detected by Oil Red O staining, possibly due to overall low differentiation rate, expression of aP2, C/EBPα, and PPARγ, the transcriptional factors regulating adipocyte differentiation, were significantly upregulated in differentiated ASCs from Prx3 KO mice compared to WT (Figs. 3A and 3B). Consistently, increased mRNA expression of adipophilin and fatty acid synthase (FAS) showed that Prx3 deficiency increases adipogenicity. Conversely, mitochondrial DNA (mtDNA) and PGC1α mRNA expressions were downregulated in Prx3 KO ASCs, implying that Prx3 is needed in the normal adipogenic process that includes a massive mitochondrial biogenesis. Data from siRNA-transfected 3T3-L1 cells confirmed that knockdown of endogenous Prx3 at day 0, 2, and 4 after induction of differentiation increased aP2, C/EBPα, and PPARγ expressions compared to scrambled siRNA-transfected adipocytes (Fig. 3D), whereas Prx3 overexpression induced opposite effect (Fig. 3E). These results indicate that deficiency in Prx3 increases the potential of preadipocytes to differentiate and accumulate lipids which may lead to increased fat mass in Prx3 KO mice.
FIG. 3.
Prx3 deficiency results in increased adipogenic gene expression during adipogenesis. (A) Adipose tissue-derived stem cells (ASCs) were isolated from subcutaneous fat pads of WT and Prx3 KO mice and differentiated with or without rosiglitazone. Differentiated cells were stained with Oil Red O and representative pictures were taken. (B) Genes related to adipogenesis and mitochondrial biogenesis were examined in ASCs from WT and Prx3 KO mice by real-time PCR. Quantifications were normalized to 18S in each reaction. Values are means±SE of six experiments. *p<0.05 vs. WT ASCs. (C) Transfection efficiency was validated by reduced Prx3 expression at day 4 after differentiation. There was no change in Prx1 or Prx5 levels by Prx3 siRNA transfection. Western blot analysis of 3T3-L1 cells transfected with (D) Prx3 siRNA or (E) Prx3 vector. 3T3-L1 cells were differentiated 48 h after transfection. Cells were harvested at days 0, 2, and 4 after differentiation. Scrambled (Scr) siRNA or empty vector was used as transfection control. Tubulin was used as an internal loading control. (To see this illustration in color, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Comparative proteomic analysis of Prx3 KO adipocytes shows dramatic decrease in mitochondrial proteins
To further validate the role of Prx3 in adipocytes, we again performed proteomic analysis with adipocytes isolated from epididymal WAT of WT and Prx3 KO mice. Among proteins that differed between the two groups (Fig. 4), 10 proteins significantly increased and 16 proteins decreased (Table 2). Interestingly, the majority of proteins which decreased in Prx3 KO mice were mitochondrial proteins mainly involved in oxidative phosphorylation and fatty acid metabolism, including ATP synthase subunit d (ATP5H) which is important for producing energy; NADH-ubiquinone oxidoreductase (NDUS1) working in respiratory electron transport; dihydrolipoyllysine-residue acetyltransferase component (ODP2) as a component of pyruvate dehydrogenase complex; dienoyl-CoA isomerase (ECH1) involved in fatty acid metabolism; and 3-hydroxyisobutyrate dehydrogenase (3HIDH) as an enzyme to generate acetoacetate in fatty acid synthesis.
FIG. 4.
Protein expression profiles of adipocytes obtained from WT and Prx3 KO mice. (A) Representative 2-DE gel images of WT and Prx3 KO mice adipocytes. Adipocytes isolated from the epididymal fat pads of WT and Prx3 KO mice were separated on 2D-PAGE (pH 4–7), detected with silver staining, and semi-quantitatively analyzed with image analysis. Differentially expressed proteins were excised and analyzed by in-gel digestion and UPLC-ESI-q-TOF tandem MS. Identified protein list is shown in Table 2. (B) Enlarged image of each spot identified by tandem MS.
Table 2.
Differentially Expressed Proteins in Adipocytes of Prx3 KO Mouse
|
Upregulated Proteins in Prx3 KO Mouse | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Spot No | Score | % Cov. | Queries Matched | Theor.Mr | Theor.pI | Accession No. | Symbol | Protein Name | Cellular Component |
| Proteins that function in metabolism | |||||||||
| 19 | 171 | 19 | 11 | 48963 | 9.11 | Q9D2G2 | ODO2 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | Mito |
| 22 | 171 | 28 | 12 | 48324 | 6.19 | P29758 | OAT | Ornithine aminotransferase, mitochondrial | Mito |
| Proteins that act in oxido-reduction | |||||||||
| 20 | 168 | 22 | 7 | 43069 | 5.95 | Q62465 | VAT1 | Synaptic vesicle membrane protein VAT-1 homolog | |
| 23, 25 | 320 | 31 | 28 | 52592 | 6.52 | Q91WD5 | NDUS2 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial | Mito |
| Proteins that function in structural molecules | |||||||||
| 17 | 413 | 10 | 28 | 226232 | 5.54 | Q8VDD5 | MYH9 | Myosin-9 | |
| Proteins that act as chaperone/mediators of protein folding | |||||||||
| 16 | 739 | 41 | 38 | 80159 | 6.25 | Q9CQN1 | TRAP1 | Heat shock protein 75 kDa, mitochondrial | Mito |
| 18 | 800 | 52 | 59 | 56643 | 5.88 | P27773 | PDIA3 | Protein disulfide-isomerase A3 | ER |
| Proteins that act as transporter/channel | |||||||||
| 21 | 328 | 59 | 39 | 50505 | 5.93 | Q61598 | GDIB | Rab GDP dissociation inhibitor beta | |
| 26 | 655 | 53 | 35 | 15232 | 4.96 | P56395 | CYB5 | Cytochrome b5 | |
| Downregulated Proteins in Prx3 KO Mouse | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Proteins that function in metabolism | |||||||||
| 1 | 443 | 17 | 27 | 129602 | 6.25 | Q05920 | PYC | Pyruvate carboxylase, mitochondrial | Mito |
| 7 | 1206 | 27 | 51 | 67899 | 8.81 | Q8BMF4 | ODP2 | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | Mito |
| 9 | 744 | 63 | 26 | 29802 | 5.57 | P67778 | PHB | Prohibitin | Memb, Mito |
| 10 | 475 | 19 | 15 | 36095 | 7.6 | O35459 | ECH1 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial | Mito |
| Proteins that act in oxido-reduction | |||||||||
| 2, 3, 4 | 380 | 31 | 28 | 79698 | 5.51 | Q91VD9 | NDUS1 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | Mito |
| 11 | 425 | 42 | 25 | 35417 | 8.37 | Q99L13 | 3HIDH | 3-hydroxyisobutyrate dehydrogenase, mitochondrial | Mito |
| 12 | 257 | 49 | 16 | 27268 | 7.0 | Q9D6J6 | NDUV2 | NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial | Mito |
| 13, 14 | 82 | 17 | 3 | 28109 | 7.15 | P20108 | PRDX3 | Thioredoxin-dependent peroxide reductase, mitochondrial | Mito |
| Proteins that act as transporter/channel | |||||||||
| 5, 6 | 545 | 36 | 26 | 75838 | 5.34 | P14824 | ANXA6 | Annexin A6 | Cyto |
| 8 | 180 | 14 | 5 | 40578 | 7.63 | Q99LC3 | NDUAA | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial | Mito |
| 15 | 540 | 85 | 26 | 18738 | 5.52 | Q9DCX2 | ATP5H | ATP synthase subunit d, mitochondrial | Mito |
| Proteins that act as signal transducer | |||||||||
| 24 | 419 | 16 | 28285 | 4.8 | P61982 | 1433G | 14-3-3 protein gamma | Cyto | |
| 24 | 310 | 18 | 28069 | 4.77 | Q9CQV8 | 1433B | 14-3-3 protein beta/alpha | Cyto | |
Mitochondrial biogenesis, lipid metabolism, and antioxidant systems are impaired in Prx3 KO mice adipocytes
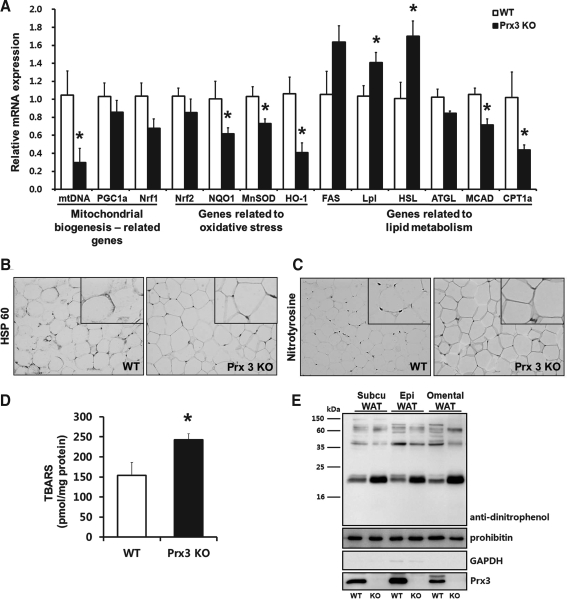
Consistent with Prx3 KO ASCs, mtDNA was significantly reduced in Prx3 KO adipocytes compared to WT (Fig. 5A). PGC1α and nuclear respiratory factor 1 (Nrf1), genes responsible for mitochondrial biogenesis, tended to be downregulated in Prx3 KO adipocytes. Immunostaining of WAT showed that mitochondrial marker heat shock protein 60 (hsp60) was decreased in Prx3 KO mice (Fig. 5B). As previously reported in ob/ob mice (32), C/EBPα and PPARγ were significantly downregulated in Prx3 KO adipocytes as well as db/db mice (Supplementary Fig. S5), which could be explained that in these terminally differentiated fat cells, some degree of dedifferentiation has occurred.
FIG. 5.
Prx3 KO mice adipocytes show impaired mitochondrial biogenesis, lipid metabolism, and antioxidant systems. (A) Genes related to mitochondrial biogenesis, oxidative stress, and lipid metabolism were examined in adipocytes isolated from WT and Prx3 KO mice by real-time PCR. Quantifications were normalized to 18S in each reaction. (B and C) Immunohistochemical analysis of white adipose tissue stained with heat shock protein 60 and nitrotyrosine. Enlarged inset pictures are shown for better viewing. (D) Plasma lipid peroxidation levels were measured as TBARS concentration in WT and Prx3 KO mice. (E) Protein carbonylation in mitochondria isolated from WT and Prx3 KO subcutaneous (Subcu), epididymal (Epi), and omental WATs. Values are means±SE of 5 mice per group. *p<0.05 vs. WT. TBARS, thiobarbituric acid reactive substances.
Gene expressions of antioxidant enzymes showed that manganese superoxide dismutase (MnSOD) and heme oxygenase-1 (HO-1) were significantly downregulated in Prx3 KO adipocytes. NAD(P)H: quinone oxidoreductase 1 (NQO1), a downstream target of nuclear factor E2-related factor 2 (Nrf2), was also significantly downregulated. Plasma thiobarbituric acid reactive substances (TBARS) levels and nitrotyrosine staining in WAT confirmed increased oxidative stress in Prx3 KO mice (Fig. 5C and 5D).
Lipid metabolism related gene expression showed that FAS, lipoprotein lipase (Lpl), and hormone-sensitive lipase (HSL) levels were upregulated, suggesting that overall lipid metabolism is increased in Prx3 KO adipocytes. Conversely, mRNA levels of medium-chain acyl-CoA dehydrogenase (MCAD) and carnitine palmitoyl transferase 1a (CPT1a), mitochondrial enzymes involved in fatty acid oxidation, were downregulated, indicating that Prx3 KO adipocytes lack their ability to burn excessive fat. Taken together with the proteomics data, metabolic enzymes are impaired in Prx3 KO mice adipocytes and as a result fatty acids are likely to accumulate in WAT.
Prx3 deficiency increases mitochondrial oxidative stress and decreases mitochondrial viability in 3T3-L1 cells
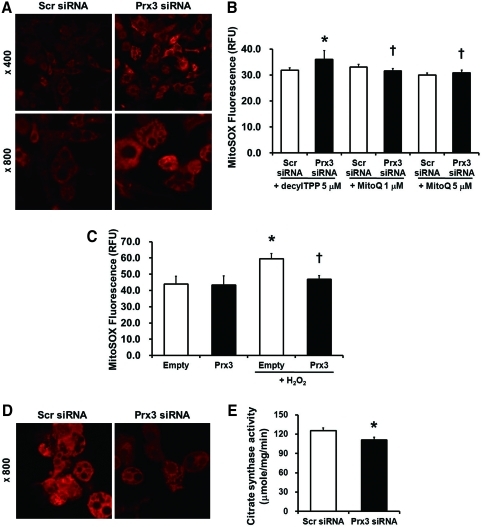
To evaluate whether the source of ROS are mitochondria specific, Prx3 knockdown 3T3-L1 adipocytes were loaded with MitoSOX. Confocal image showed that mitochondrial superoxide level was significantly increased in Prx3-deficient 3T3-L1 cells (Fig. 6A). Similar results were observed with DCF-DA implying increased cellular H2O2 in Prx3-deficient cells (data not shown). Moreover, increased mitochondrial ROS in Prx3-deficient 3T3-L1 adipocytes were reversed by treatment of mitochondria-targeted antioxidant MitoQ (Fig. 6B). As shown in Figure 6C, Prx3-overexpressed cells were protected from H2O2-induced mitochondrial ROS production. Moreover, dysfunctional ROS metabolism resulted in increased protein carbonylation of mitochondrial proteins isolated from subcutaneous, epididymal, and omental Prx3 KO WAT (Fig. 5E). To determine the effect of Prx3 on mitochondrial function, we measured the accumulation of MitoTracker CMXRos, which is dependent on mitochondrial membrane potential, and citrate synthase activity, a key component of the TCA cycle. Prx3 knockdown of 3T3-L1 adipocytes showed decreased mitochondrial potential and citrate synthase activity (Figs. 6D and 6E).
FIG. 6.
Prx3 deficiency increases mitochondrial oxidative stress and decreases mitochondrial viability in 3T3-L1 cells. (A) Mature 3T3-L1 adipocytes transfected with siRNA were loaded with 5 μM MitoSOX, a mitochondrial superoxide specific dye and examined by confocal microscopy. (B) Transfected 3T3-L1 adipocytes were treated with or without MitoQ (1 and 5 μM) for 1 h before loading with MitoSOX. DecylTPP was used as control compound. MitoSOX fluorescence levels were quantified by fluorometer. (C) Prx3 overexpressed cells were treated with 1 mM H2O2 for 15 min and loaded with MitoSOX. (D) siRNA transfected 3T3-L1 cells were loaded with 100 nM MitoTracker Red CMXRos and examined by confocal microscopy. (E) Citrate synthase activity was measured in transfected 3T3-L1 adipocytes. Values are means±SE of 3–4 experiments. *p<0.05 vs. Scr siRNA or empty vector, †p<0.05 vs. Prx3 siRNA or empty vector treated with H2O2. (To see this illustration in color, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Adipokine levels are impaired in Prx3 KO mice
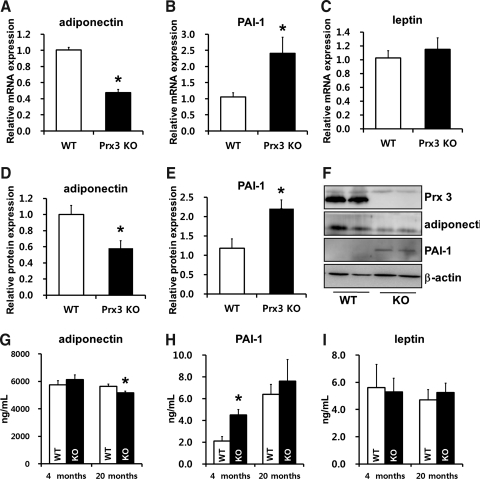
Data achieved from proteomics and gene expression analysis imply that Prx3 KO adipocytes may be functionally impaired. Since adipokine dysregulation is a hallmark of adipocyte impairment, we measured adipokine expression in Prx3 KO adipocytes. Adiponectin mRNA and protein expression was significantly downregulated and PAI-1 expression was upregulated in Prx3 KO adipocytes compared to WT (Figs. 7A–7F). Plasma levels of adiponectin was significantly decreased in 20-month-old Prx3 KO mice, and PAI-1 was increased in both 4- and 20-month-old Prx3 KO mice but only had significance at 4-month-old mice. Adipocyte and plasma leptin levels were not significantly different between two groups (Figs. 7G–7I).
FIG. 7.
Deficiency in Prx3 results in dysregulation of adipokines. (A–C) Real-time PCR analysis of adipocytes isolated from epididymal WAT. (D–F) Western blot analysis of adipocytes isolated from epididymal WAT and its quantification. (G–I) Plasma levels of adiponectin, PAI-1, and leptin were measured by ELISA in 4- and 20-month-old WT and Prx3 KO mice. Values are means±SE of 5 (4 months old) or 9 (20 months old) mice per group. *p<0.05 vs. WT. PAI-1, plasminogen activator inhibitor-1.
Metabolic profiles show that Prx3 KO mice are glucose intolerant and insulin resistant
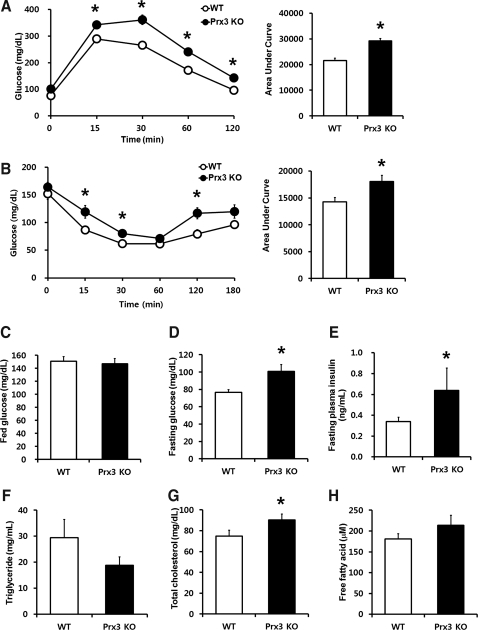
Increased fat mass, impaired mitochondrial enzymes, and adipokine dysregulation suggest that deficiency in Prx3 may result in altered whole body metabolism. To assess the impact of Prx3 on glucose homeostasis, we conducted oral glucose tolerance test (OGTT). Administration of glucose led to a more rapid increase of blood glucose in Prx3 KO mice which remained significantly higher throughout the study period (Fig. 8A). Interestingly, loss in Prx3 was effective enough to raise the fasting glucose level, whereas there was no change in fed glucose level (Figs. 8C and 8D). Likewise, Prx3 KO mice exhibited significantly decreased sensitivity to insulin as measured by insulin tolerance test (ITT) with elevated fasting plasma insulin levels (Figs. 8B and 8E). In addition, plasma cholesterol was increased in Prx3 KO mice (Fig. 8G). Of note, decreased pattern of triglycerides (TG) may be explained by increased level of Lpl mRNA levels in Prx3 KO adipocytes (Fig. 5A). These results indicate that a mild metabolic dysregulation has occurred in Prx3 KO mice.
FIG. 8.
Prx3 KO mice are glucose intolerant and insulin insensitive with higher levels of fasting glucose and total cholesterol. (A) Oral glucose tolerance test of WT and Prx3 KO mice were performed by injecting 2 g glucose per kg body weight after overnight fasting. (B) Insulin tolerance test of WT and Prx3 KO mice were performed by injecting 0.75 IU insulin per kg body weight after 6 h fasting. (C and D) Glucose levels of WT and Prx3 KO mice in a fed and fasted state. (E) Fasting plasma insulin levels of WT and Prx3 KO mice. (F–H) Plasma triglyceride, total cholesterol, and free fatty acid levels of WT and Prx3 KO mice. Values are means±SE of 5 (4 months old) or 9 (20 months old) mice per group. *p<0.05 vs. WT.
Discussion
Our study demonstrated that Prx3 is highly expressed in mature adipocytes and decreased in obesity. Employing Prx3 KO mice and Prx3 knockdown and overexpressed 3T3-L1 cells, we further demonstrated that Prx3 may play a key role in regulation of mitochondrial ROS and function, adipokine expression in adipocytes and that Prx3 deficiency leads to accumulation of WAT and metabolic alteration.
Comparative proteomic analysis allowed detection of proteins that changed during adipogenesis, some of which had not been previously reported (Supplementary Fig. S6). MS/MS analysis combined with 2D-PAGE has the ability to pick up protein changes that might go undetected by other approaches. For example, many of the mitochondrial proteins which we have identified escape detection in gel-based comparative proteomics due to inherent methodological limitations.
To our knowledge, this is the first study that focused on the role of endogenous Prx3 in adipocytes. In line with the previous reports (28–30), Prx3 KO mice exhibit no overt phenotype. However, histology showed that adipocytes were significantly larger in Prx3 KO mice. Hypertrophied adipocytes due to enlarged lipid droplets are frequently observed in mice models of obesity (17) and these cells exhibit impaired mitochondrial function and insulin signaling along with higher ROS levels (10, 18, 33).
The key question is how Prx3 deficiency induces obesity. We believe that the most plausible mechanism could be the effect of Prx3 on mitochondrial fitness. Proteomic analysis of Prx3 KO adipocytes confirmed major changes in mitochondrial proteins associated with glucose and lipid metabolism. Decreased mtDNA levels observed in Prx3 KO mice adipocytes and ASCs further support the defect in mitochondrial biogenesis. Along with this, significantly decreased mitochondrial membrane complex I proteins, including NDUS1 which oxidizes NADH to regenerate NAD+; NADH dehydrogenase flavoprotein 2 (NDUV2); NADH dehydrogenase 1 alpha subcomplex subunit 10 (NDUAA); NDUS2, NDSU3; and dihydrolipoyllysine-residue acetyl and succinyltransferase (ODP2, ODO2) functioning in pyruvate dehydrogenase complex to convert glycolysis to TCA cycle, were observed. These results suggest that Prx3 affects mitochondrial complex I and conversion of glycolysis to TCA cycle. Also, downregulation of ATP5H suggests that mitochondrial respiratory chain may also be disturbed by Prx3 deficiency in adipocytes, supporting that lack of Prx3 may result in impaired metabolism by accumulating NADH, blocking TCA cycle and then raising fatty acid synthesis.
Adipocyte mitochondrial dysfunction in Prx3 KO mice is interconnected with increased mitochondrial ROS. Among mitochondrial antioxidant enzymes, Prx3 has been estimated to decompose approximately 90% of hydrogen peroxide owing to its higher abundance than GPx1 (8), suggesting that Prx3 has a potential to play a major role in mitochondrial redox signaling. Increased mitochondrial protein carbonylation in Prx3 KO mice WAT and increased MitoSOX intensity in Prx3 knockdown adipocytes substantiate this hypothesis. Reversal by MitoQ further supports that Prx3 regulates mitochondrial ROS metabolism. Future studies regarding in vivo effect of antioxidants such as MitoQ on Prx3 KO mice are needed.
Imhoff et al. have recently reported that thioredoxin 2 (Trx2), an electron supplier for the catalytic cycle of Prx3, becomes oxidized in correlation with oxidative mitochondrial environment during adipogenesis (15). In our setting, oxidized (sulfonic acid) form of Prx3 which presumably moves to acidic site as shown in MDA-MB231 cell and in WT mouse adipocytes (Supplementary Figs. S7D and S7E) was not detected in 3T3-L1 adipocytes (Supplementary Figs. S7A and S7B). Instead, other post-translational modifications of Prx3, including modifications at Lys107 to allysine and at Asp84 to methylester, were identified. It is possible that the redox state of Trx2 is more sensitive than Prx3 and therefore important in regulation of Prx3 activity.
As described above, adipogenesis is accompanied by increased ROS levels. However, excessive ROS stimulation increases adipogenicity of adipocytes (25) and use of antioxidants inhibits adipogenesis (19, 36). Likewise, knockdown of Prx3 resulted in accelerated adipogenesis with increased lipid metabolism which may result in adipocyte hypertrophy and reversely in Prx3 overexpressed cells, adipogenic markers were downregulated. Therefore, Prx3 is needed in ‘normal’ differentiation of adipocytes.
Increased oxidative stress is a major factor that alters adipokine levels in obese or diabetic condition (10, 16). Consistently, loss of Prx3 resulted in decreased adiponectin and increased PAI-1 expression in adipocytes. It was previously reported that impaired mitochondrial function sequentially activates JNK and ATF3, mechanisms involved in ER stress, to decrease adiponectin synthesis (23). Therefore, decreased mitochondrial vitality and diminished mtDNA in Prx3 KO adipocytes may explain reduced adiponectin expression.
Impact of Prx3 on mitochondrial ROS and oxidative phosphorylation might eventually contribute to altered lipid metabolism in the KO mice. As the main site of cellular energy, mitochondria generate ATP, NADPH, and substrates that are essential to synthesize fatty acids from glucose and other substrates. In adipose tissue, de novo lipogenesis accounts for up to 40% of whole-body lipogenesis, suggesting the importance of mitochondrial metabolic pathway in regulation of lipid contents (5). Increased FAS and decreased MCAD and CPT1a suggest that adipocytes are likely to accumulate fat in Prx3 KO mice.
Based on our data on adipocytes, it is not difficult to link our observations with whole body metabolism. Chen et al. (6) reported that Prx3 overexpressed mice were resistant to diet-induced elevations in blood glucose, displayed reduced production of mitochondrial ROS, and improved glucose homeostasis. Likewise, our results showed that dissipation in Prx3 influences glucose and insulin homeostasis. We cannot rule out the possibility of insulin resistance in liver and skeletal muscle, which are major sites for glucose metabolism. Intriguingly, changes in mRNA levels were somewhat different in the liver with only MnSOD being significantly increased (Supplementary Fig. S8), suggesting that the function of Prx3 might be context dependent. Although partly demonstrated in Prx3 knockdown 3T3-L1 cells, genetic manipulations specific to adipocytes in mice will help assess whether defects shown here are intrinsic to fat cells or secondary to functional disturbances in other tissues. Also, Prx3 overexpression in adipocytes in vitro and in vivo would be interesting to confirm the protective role of Prx3. In our study, Prx3 overexpression has been performed to observe its effect on adipogenesis and ROS regulation to strengthen the involvement of Prx3 on adipocyte regulation.
Since Prx3 deficiency has been shown to increase the susceptibility to inflammation or oxidative stress (4, 30), we recognize that under conditions of metabolic stress such as high-fat diet, Prx3 KO mice would exhibit a more clear difference demonstrating the role of Prx3. Body composition studies and assessment of food intake or energy expenditure are also needed to understand the protective role of Prx3 in whole body metabolism. Exploring the role of other Prx isoforms such as Prx1 and 2 (cytoplasmic/nucleus) in adipocytes and obesity would be interesting since cytoplasmic ROS induced by NADPH oxidase still remain to be the major contributor of adipocyte oxidative stress (10). It has been reported that Prx1 protects against atherosclerosis and Prx2 overexpression inhibits lipid accumulation during adipogenesis of mesenchymal stem cells (19), which is similar to our Prx3 overexpression data (Fig. 3E). Prx5, which resides in the mitochondria as well as cytoplasm, may have similar effect on mitochondrial ROS in adipocytes.
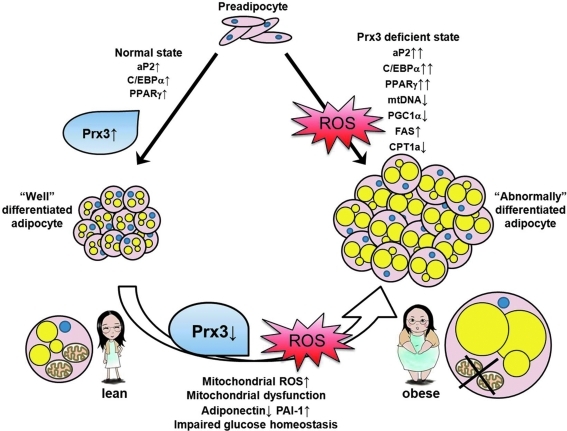
In summary, our data suggest an important biological role for endogenous Prx3 in regulation of adipocytes. Our efforts to unravel the complex mechanism of adipogenesis through proteomics have led to identification of Prx3. Analysis of Prx3 KO mice and Prx3 transfected 3T3-L1 cells demonstrated the critical role of Prx3 in maintaining the function of mature adipocytes. Although the cause and effect relationships are yet unknown, this study has provided the first evidence that defective Prx3 expression in adipose tissue is associated with reduced antioxidant/mitochondrial function, adipokine dysregulation, and impaired glucose/insulin homeostasis (Fig. 9). We propose that Prx3 may play an important role in the regulation of obesity and metabolic syndrome.
FIG. 9.
Suggested model for Prx3 in regulation of adipocytes. Increased Prx3 is associated with normal adipocyte differentiation. In Prx3-deficient state, adipogenic genes are upregulated, resulting in abnormally differentiated adipocytes, possibly due to increased ROS. In mature adipocytes, Prx3 levels are decreased in obese individuals, which results in accumulation of mitochondrial ROS. This in turn causes defect in mitochondrial function, lipid accumulation, and adipokine dysregulation. As a result of adipocyte dysregulation, glucose and insulin homeostasis is impaired. Therefore, Prx3 is necessary in regulation of redox state and metabolism in adipocytes. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Materials and Methods
Materials and methods for Prx3 KO mice generation, 3T3-L1 adipocyte culture, proteomic analysis, gene expression analysis, Western blot analysis, histology, immunohistochemistry, and statistical analysis are described in the Supplementary Methods, available online.
Prx3 KO mice
Male homozygous Prx3 KO mice and their littermates (WT) were fed a standard chow and water ad libitum. Plasma cholesterol and TG were assayed at Korea Clinical Laboratories (Siheung, Gyeonggi-do, Korea). Plasma free fatty acids, adiponectin, PAI-1, leptin, and insulin levels were assayed using commercial kits (from BioAssay Systems, Hayward, CA; R&D systems, Minneapolis, MN; Innovative Research, Novi, MI; and Millipore, Billerica, CA, respectively) according to the manufacturer's instructions. Plasma levels of TBARS were measured as a marker of lipid peroxidation as previously described (12). OGTT and ITT were performed using a standard protocol (34). All animal studies were conducted in accordance with protocols approved by the institutional animal care and use committee of Ewha Womans University.
Isolation of adipocytes and differentiation of ASCs
Adipocytes were isolated from epididymal WAT using the collagenase method (35) without BSA to avoid interference during 2D-PAGE. The suspension was passed through a sterile 250 μm nylon mesh (Nunc, Rochester, NY) and centrifuged at 500 g for 10 min. The floating adipocytes were used for mRNA and protein analysis. ASCs were isolated from the subcutaneous WAT, as previously described (22). Differentiation was induced with DMEM/F-12 containing 10% FBS, 1 μM dexamethasone, 2 μM insulin, and 0.25 mM IBMX. Three days later, medium was changed to DMEM/F-12 containing 5% FBS, 1 μM dexamethasone, and 2 μM insulin. Cells were harvested 4 days after induction of differentiation for further analysis.
Fluorescence detection of mitochondrial ROS and membrane potential
To detect mitochondria specific oxidative stress, mature 3T3-L1 adipocytes were loaded with 5 μM MitoSOX Red (Molecular Probes, Carlsbad, CA) for 10 min. Cells were either examined by confocal microscopy (LSM 510 META: Carl Zeiss, Thornwood, NY) or by fluorometer (Molecular Devices, Sunnyvale, CA) at 510 nm excitation and 580 nm emission. To evaluate the mitochondrial membrane potential, 3T3-L1 adipocytes were loaded with 100 nM MitoTracker Red CMXRos (Molecular Probes) for 30 min at 37°C and examined by confocal microscopy.
Protein carbonylation
Protein carbonylation was detected using DNPH-derivatization method as described previously (27). Briefly, 20 μg of protein lysate was denatured with 12% SDS, and derivatized with 10 mM DNPH solution for 15 min. Neutralized samples were immunoblotted using anti-DNP antibody (Sigma, St. Louis, MO).
Citrate synthase activity
Citrate synthase activity was measured in 3T3-L1 lysates, according to instructions provided with the citrate synthase kit (Sigma).
Supplementary Material
Abbreviations Used
- 3HIDH
3-hydroxyisobutyrate dehydrogenase
- ASCs
adipose tissue-derived stem cells
- ATP5H
ATP synthase subunit d
- BMI
body mass index
- CPT1a
carnitine palmitoyl transferase 1a
- ECH1
dienoyl-CoA isomerase
- ER
endoplasmic reticulum
- FAS
fatty acid synthase
- GPxs
glutathione peroxidases
- HO-1
heme oxygenase-1
- HSL
hormone-sensitive lipase
- IBMX
3-isobutyl-1-methylxanthine
- ITT
insulin tolerance test
- KO
knockout
- Lpl
lipoprotein lipase
- MCAD
medium-chain acyl-CoA dehydrogenase
- MnSOD
manganese superoxide dismutase
- mtDNA
mitochondrial DNA
- mtTFA
mitochondrial transcription factor A
- NDUS1
NADH-ubiquinone oxidoreductase
- NQO1
NAD(P)H: quinine oxidoreductase
- Nrf 1
nuclear respiratory factor
- Nrf 2
nuclear factor E2-related factor 2
- ODP2
dihydrolipoyllysine-residue acetyltransferase component
- OGTT
oral glucose tolerance test
- PAI-1
plasminogen activator inhibitor-1
- PGC1α
PPARγ coactivator-1 alpha
- Prx3
peroxiredoxin 3
- ROS
reactive oxygen species
- TBARS
thiobarbituric acid reactive substances
- TG
triglyceride
- Trx2
thioredoxin 2
- WAT
white adipose tissue
- WT
wild type
Acknowledgments
We wish to thank Ms. Jung Hwa Lee for her technical assistance. MitoQ was kindly provided by Professor Murphy at Cambridge University, UK. This work was supported by the Center for Cell Signaling Research and Drug Discovery Research (CCS & DDR) at Ewha Womans University from the National Core Research Center (NCRC) program (2011-0006244) and by WCU project (R31-2008-000-10010-0). Joo Young Huh was supported by the Brain Korea 21 Project and Jehyun Park by RP-Grant 2009 (2009-1949-1-1) from Ewha Womans University.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Adachi J. Kumar C. Zhang Y. Mann M. In-depth analysis of the adipocyte proteome by mass spectrometry and bioinformatics. Mol Cell Proteomics. 2007;6:1257–1273. doi: 10.1074/mcp.M600476-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Atiar Rahman M. Kumar SG. Lee SH. Hwang HS. Kim HA. Yun JW. Proteome analysis for 3T3-L1 adipocyte differentiation. J Microbiol Biotechnol. 2008;18:1895–1902. [PubMed] [Google Scholar]
- 3.Bays H. Mandarino L. DeFronzo RA. Role of the adipocyte, free fatty acids, and ectopic fat in pathogenesis of type 2 diabetes mellitus: Peroxisomal proliferator-activated receptor agonists provide a rational therapeutic approach. J Clin Endocrinol Metab. 2004;89:463–478. doi: 10.1210/jc.2003-030723. [DOI] [PubMed] [Google Scholar]
- 4.Chang TS. Cho CS. Park S. Yu S. Kang SW. Rhee SG. Peroxiredoxin III, a mitochondrion-specific peroxidase, regulates apoptotic signaling by mitochondria. J Biol Chem. 2004;279:41975–41984. doi: 10.1074/jbc.M407707200. [DOI] [PubMed] [Google Scholar]
- 5.Chascione C. Elwyn DH. Davila M. Gil KM. Askanazi J. Kinney JM. Effect of carbohydrate intake on de novo lipogenesis in human adipose tissue. Am J Physiol. 1987;253:E664–669. doi: 10.1152/ajpendo.1987.253.6.E664. [DOI] [PubMed] [Google Scholar]
- 6.Chen L. Na R. Gu M. Salmon AB. Liu Y. Liang H. Qi W. Van Remmen H. Richardson A. Ran Q. Reduction of mitochondrial H2O2 by overexpressing peroxiredoxin 3 improves glucose tolerance in mice. Aging Cell. 2008;7:866–878. doi: 10.1111/j.1474-9726.2008.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi KL. Wang Y. Tse CA. Lam KS. Cooper GJ. Xu A. Proteomic analysis of adipocyte differentiation: Evidence that alpha2 macroglobulin is involved in the adipose conversion of 3T3 L1 preadipocytes. Proteomics. 2004;4:1840–1848. doi: 10.1002/pmic.200300697. [DOI] [PubMed] [Google Scholar]
- 8.Cox AG. Winterbourn CC. Hampton MB. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem J. 2009;425:313–325. doi: 10.1042/BJ20091541. [DOI] [PubMed] [Google Scholar]
- 9.De Pauw A. Tejerina S. Raes M. Keijer J. Arnould T. Mitochondrial (dys)function in adipocyte (de)differentiation and systemic metabolic alterations. Am J Pathol. 2009;175:927–939. doi: 10.2353/ajpath.2009.081155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa S. Fujita T. Shimabukuro M. Iwaki M. Yamada Y. Nakajima Y. Nakayama O. Makishima M. Matsuda M. Shimomura I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J Clin Invest. 2004;114:1752–1761. doi: 10.1172/JCI21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green H. Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- 12.Ha H. Endou H. Lipid peroxidation in isolated rat nephron segments. Am J Physiol. 1992;263:F201–207. doi: 10.1152/ajprenal.1992.263.2.F201. [DOI] [PubMed] [Google Scholar]
- 13.Hattori F. Murayama N. Noshita T. Oikawa S. Mitochondrial peroxiredoxin-3 protects hippocampal neurons from excitotoxic injury in vivo. J Neurochem. 2003;86:860–868. doi: 10.1046/j.1471-4159.2003.01918.x. [DOI] [PubMed] [Google Scholar]
- 14.Hiroi M. Nagahara Y. Miyauchi R. Misaki Y. Goda T. Kasezawa N. Sasaki S. Yamakawa-Kobayashi K. The combination of genetic variations in the PRDX3 gene and dietary fat intake contribute to obesity risk. Obesity (Silver Spring) 2011;19:882–887. doi: 10.1038/oby.2010.275. [DOI] [PubMed] [Google Scholar]
- 15.Imhoff BR. Hansen JM. Extracellular redox environments regulate adipocyte differentiation. Differentiation. 2010;80:31–39. doi: 10.1016/j.diff.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Iyer A. Fairlie DP. Prins JB. Hammock BD. Brown L. Inflammatory lipid mediators in adipocyte function and obesity. Nat Rev Endocrinol. 2010;6:71–82. doi: 10.1038/nrendo.2009.264. [DOI] [PubMed] [Google Scholar]
- 17.Johnson PR. Hirsch J. Cellularity of adipose depots in six strains of genetically obese mice. J Lipid Res. 1972;13:2–11. [PubMed] [Google Scholar]
- 18.Kahn BB. Flier JS. Obesity and insulin resistance. J Clin Invest. 2000;106:473–481. doi: 10.1172/JCI10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanda Y. Hinata T. Kang SW. Watanabe Y. Reactive oxygen species mediate adipocyte differentiation in mesenchymal stem cells. Life Sci. 2011;89:250–258. doi: 10.1016/j.lfs.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Keaney JF., Jr Larson MG. Vasan RS. Wilson PW. Lipinska I. Corey D. Massaro JM. Sutherland P. Vita JA. Benjamin EJ Framingham Study. Obesity and systemic oxidative stress: Clinical correlates of oxidative stress in the Framingham Study. Arterioscler Thromb Vasc Biol. 2003;23:434–439. doi: 10.1161/01.ATV.0000058402.34138.11. [DOI] [PubMed] [Google Scholar]
- 21.Kim KB. Kim BW. Choo HJ. Kwon YC. Ahn BY. Choi JS. Lee JS. Ko YG. Proteome analysis of adipocyte lipid rafts reveals that gC1qR plays essential roles in adipogenesis and insulin signal transduction. Proteomics. 2009;9:2373–2382. doi: 10.1002/pmic.200800811. [DOI] [PubMed] [Google Scholar]
- 22.Kim MH. Kim I. Kim SH. Jung MK. Han S. Lee JE. Nam JS. Lee SK. Bang SI. Cryopreserved human adipogenic-differentiated pre-adipocytes: A potential new source for adipose tissue regeneration. Cytotherapy. 2007;9:468–476. doi: 10.1080/14653240701358452. [DOI] [PubMed] [Google Scholar]
- 23.Koh EH. Park JY. Park HS. Jeon MJ. Ryu JW. Kim M. Kim SY. Kim MS. Kim SW. Park IS. Youn JH. Lee KU. Essential role of mitochondrial function in adiponectin synthesis in adipocytes. Diabetes. 2007;56:2973–2981. doi: 10.2337/db07-0510. [DOI] [PubMed] [Google Scholar]
- 24.Kratchmarova I. Kalume DE. Blagoev B. Scherer PE. Podtelejnikov AV. Molina H. Bickel PE. Andersen JS. Fernandez MM. Bunkenborg J. Roepstorff P. Kristiansen K. Lodish HF. Mann M. Pandey A. A proteomic approach for identification of secreted proteins during the differentiation of 3T3-L1 preadipocytes to adipocytes. Mol Cell Proteomics. 2002;1:213–222. doi: 10.1074/mcp.m200006-mcp200. [DOI] [PubMed] [Google Scholar]
- 25.Lee H. Lee YJ. Choi H. Ko EH. Kim JW. Reactive oxygen species facilitate adipocyte differentiation by accelerating mitotic clonal expansion. J Biol Chem. 2009;284:10601–10609. doi: 10.1074/jbc.M808742200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee YS. Kim AY. Choi JW. Kim M. Yasue S. Son HJ. Masuzaki H. Park KS. Kim JB. Dysregulation of adipose glutathione peroxidase 3 in obesity contributes to local and systemic oxidative stress. Mol Endocrinol. 2008;22:2176–2189. doi: 10.1210/me.2008-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine RL. Wehr N. Williams JA. Stadtman ER. Shacter E. Determination of carbonyl groups in oxidized proteins. Methods Mol Biol. 2000;99:15–24. doi: 10.1385/1-59259-054-3:15. [DOI] [PubMed] [Google Scholar]
- 28.Li L. Kaifu T. Obinata M. Takai T. Peroxiredoxin III-deficiency sensitizes macrophages to oxidative stress. J Biochem. 2009;145:425–427. doi: 10.1093/jb/mvp011. [DOI] [PubMed] [Google Scholar]
- 29.Li L. Shoji W. Oshima H. Obinata M. Fukumoto M. Kanno N. Crucial role of peroxiredoxin III in placental antioxidant defense of mice. FEBS Lett. 2008;582:2431–2434. doi: 10.1016/j.febslet.2008.05.050. [DOI] [PubMed] [Google Scholar]
- 30.Li L. Shoji W. Takano H. Nishimura N. Aoki Y. Takahashi R. Goto S. Kaifu T. Takai T. Obinata M. Increased susceptibility of MER5 (peroxiredoxin III) knockout mice to LPS-induced oxidative stress. Biochem Biophys Res Commun. 2007;355:715–721. doi: 10.1016/j.bbrc.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Matsushima S. Ide T. Yamato M. Matsusaka H. Hattori F. Ikeuchi M. Kubota T. Sunagawa K. Hasegawa Y. Kurihara T. Oikawa S. Kinugawa S. Tsutsui H. Overexpression of mitochondrial peroxiredoxin-3 prevents left ventricular remodeling and failure after myocardial infarction in mice. Circulation. 2006;113:1779–1786. doi: 10.1161/CIRCULATIONAHA.105.582239. [DOI] [PubMed] [Google Scholar]
- 32.Nadler ST. Stoehr JP. Schueler KL. Tanimoto G. Yandell BS. Attie AD. The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc Natl Acad Sci USA. 2000;97:11371–11376. doi: 10.1073/pnas.97.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicolson GL. Metabolic syndrome and mitochondrial function: Molecular replacement and antioxidant supplements to prevent membrane peroxidation and restore mitochondrial function. J Cell Biochem. 2007;100:1352–1369. doi: 10.1002/jcb.21247. [DOI] [PubMed] [Google Scholar]
- 34.Pfluger PT. Herranz D. Velasco-Miguel S. Serrano M. Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci USA. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodbell M. Metabolism of isolated fat cells. I. Effects of hormones on glucose metabolism and lipolysis. J Biol Chem. 1964;239:375–380. [PubMed] [Google Scholar]
- 36.Samuni Y. Cook JA. Choudhuri R. Degraff W. Sowers AL. Krishna MC. Mitchell JB. Inhibition of adipogenesis by Tempol in 3T3-L1 cells. Free Radic Biol Med. 2010;49:667–673. doi: 10.1016/j.freeradbiomed.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smyth S. Heron A. Diabetes and obesity: The twin epidemics. Nat Med. 2006;12:75–80. doi: 10.1038/nm0106-75. [DOI] [PubMed] [Google Scholar]
- 38.Trujillo ME. Scherer PE. Adipose tissue-derived factors: Impact on health and disease. Endocr Rev. 2006;27:762–778. doi: 10.1210/er.2006-0033. [DOI] [PubMed] [Google Scholar]
- 39.Van Gaal LF. Mertens IL. De Block CE. Mechanisms linking obesity with cardiovascular disease. Nature. 2006;444:875–880. doi: 10.1038/nature05487. [DOI] [PubMed] [Google Scholar]
- 40.Welsh GI. Griffiths MR. Webster KJ. Page MJ. Tavare JM. Proteome analysis of adipogenesis. Proteomics. 2004;4:1042–1051. doi: 10.1002/pmic.200300675. [DOI] [PubMed] [Google Scholar]
- 41.Wilson-Fritch L. Burkart A. Bell G. Mendelson K. Leszyk J. Nicoloro S. Czech M. Corvera S. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol. 2003;23:1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zvonic S. Lefevre M. Kilroy G. Floyd ZE. DeLany JP. Kheterpal I. Gravois A. Dow R. White A. Wu X. Gimble JM. Secretome of primary cultures of human adipose-derived stem cells: Modulation of serpins by adipogenesis. Mol Cell Proteomics. 2007;6:18–28. doi: 10.1074/mcp.M600217-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.