Background: The goal is to understand the molecular mechanism of metastasis and the roles of IL-6/OSM, STAT3, and fascin.
Results: STAT3 binds to the fascin promoter and is essential for its expression and cell migration in response to IL-6/OSM.
Conclusion: STAT3 plays a central role in cell migration through direct control of fascin expression.
Significance: Drug targets are identified to block tumor metastasis.
Keywords: Cytokine, Gene Regulation, Metastasis, STAT3, Transcription Regulation, Tumor, IL-6, Fascin
Abstract
The cytokines oncostatin M (OSM) and IL-6 promote breast cancer cell migration and metastasis. Both cytokines activate STAT3, a member of the STAT (signal transducers and activators of transcription) family of transcription factors. Through transcriptional regulation of its target genes, STAT3 controls a wide range of cellular processes, including cellular proliferation, oncogenesis, and cancer metastasis. Fascin is an actin-bundling protein involved in cell migration. Elevated levels of fascin expression are found in many metastatic cancers, and inhibition of fascin function by small chemical compounds leads to a block of tumor metastasis. In this work, we demonstrate that fascin is a direct STAT3 target gene in response to OSM and IL-6 in both mouse and human breast cancer cells. We show that NFκB also binds to the fascin promoter in response to cytokine treatment and this binding is STAT3-dependent. Both STAT3 and NFκB are required for the cytokine-induced expression of fascin in cancer cells. Furthermore, we demonstrate that STAT3, in directly controlling fascin expression, is both necessary and sufficient for breast cancer cell migration.
Introduction
Cancer metastasis is a multistep process that is influenced by many cellular signaling pathways (1). It has been shown that the cytokines interleukin-6 (IL-6) and oncostatin M (OSM)2 play important roles in breast cancer metastasis (2–6). They not only promote cell migration but also induce an epithelial-mesenchymal transition phenotype in breast cancer cells (3–7). IL-6 has long been associated with cancer metastasis (2, 8). Increased levels of IL-6 have been shown to promote cancer metastasis, and many tumors including breast and prostate cancers produce high levels of IL-6 (2, 8). Overexpression of IL-6 in organs such as the brain or lungs can also recruit circulating tumor cells to form metastatic tumors (8).
OSM and IL-6 are both members of the gp130 family (9, 10) and utilize the JAK-STAT signaling pathway to effect downstream cellular events (11, 12). Upon cytokines binding to their cell surface receptors, the tyrosine kinase JAK becomes activated and phosphorylates the STAT molecules associated with the receptor tail. The phosphorylated STAT molecules form dimers and then translocate to the nucleus where they bind to the promoters of their target genes to regulate transcription. A wide range of cellular processes are controlled by the JAK-STAT pathway through transcriptional regulation of STAT target genes in response to various cytokines and growth factors (11, 12). One member of the STAT family, STAT3, is specifically activated by the gp130 family of cytokines, including IL-6 and OSM (9, 10). STAT3 is directly involved in regulating many cellular processes including proliferation, postnatal survival, oncogenesis, and breast cancer metastasis (12–17). For its involvement in cancer, STAT3 has been shown to promote tumor growth as well as tumor metastasis (14–16). Constitutively activated STAT3 enhances metastasis of mammary tumors, and a STAT3 knockdown blocks breast cancer cell migration and metastasis (14, 15). There is evidence suggesting that IL-6 and OSM promote breast cancer metastasis through activation of STAT3 (3, 6). However, the mechanism by which cytokine-induced STAT3 activation promotes breast cancer metastasis is not well defined.
Recently, an actin-bundling protein, fascin, was identified as a key molecule in tumor metastasis (18). Fascin is a highly conserved actin-bundling protein that localizes to microspikes and filopodia and functions in cell adhesion and motility (19). Three isoforms of fascin exist in mammalian cells: fascin-1, fascin-2, and fascin-3 (20). Fascin-1 (from here and throughout referred to as fascin) is expressed during embryonic development and in a few normal cell types such as neuronal and dendritic cells (20). In contrast, the level of fascin expression is elevated in large numbers of metastatic cancers, including gastric, breast, colon, and ovarian (19, 20). Recently, it was discovered that fascin plays a key role in the process of tumor metastasis (18). Knocking down of fascin expression blocks breast cancer metastasis in mice, and inhibition of fascin-actin bundling activity by small chemical compounds results in blocking breast cancer metastasis (18). However, it is not known how fascin expression is elevated in metastatic tumor cells. It has been reported that IL-6 increases the expression of fascin in glioblastoma cells to promote cell invasion (21). This observation suggests a correlation between STAT3 activation and an increase in fascin expression.
In this report, we demonstrate that STAT3 directly regulates transcription of fascin to promote breast cancer cell migration. STAT3 binds specifically to the fascin promoter, and fascin expression is induced in response to IL-6 or OSM in both human and mouse breast cancer cell lines. This cytokine-induced fascin expression is dependent on STAT3. In addition, the transcription factor NFκB is also recruited to the fascin promoter in a STAT3-dependent manner. Furthermore, we show that STAT3 is essential for cell migration of metastatic breast cancer cells.
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmids, and Reagents
4T1 and MDA-MB-231 cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS. The STAT3 and NFκB antibodies for chromatin immunoprecipitation (ChIP) assays were from Santa Cruz Biotechnology. Antibodies for Western blot analyses were anti-phosphotyrosine STAT3 (Cell Signaling Technology), anti-STAT3 (BD Transduction Laboratories), and anti-tubulin (Sigma). The plasmid RcCMV (pCMV) was from Invitrogen. The plasmid STAT3c was constructed by introducing point mutations that changed residues Ala-661 and Asn-663 to cysteine in the wild-type STAT3 gene as described previously (16). Ligands were used at the following concentrations: 25 ng/ml OSM (R&D Systems) and 20 ng/ml IL-6 (Millipore) combined with 200 ng/ml soluble IL-6R (R&D Systems).
ChIP Assays
ChIP assays were performed as described previously (22, 23). 4T1 or MDA-MB-231 cells were treated with either OSM or IL-6 for 30 min followed by cross-linking with formaldehyde and sonication. The resulting cell lysates (Input) were subjected to immunoprecipitations with 2.5 μg of the STAT3 antibody. The precipitated STAT3-DNA complexes (IP) were subjected to proteinase treatment. DNA was purified from both Input and IP. The purified DNA was used for PCRs with α-32P-labeled dCTP and ChIP primers. PCR products were separated on a 6% polyacrylamide gel and subjected to autoradiography. Radioactive signals from the gels were quantitated with a PhosphorImager (Molecular Dynamics Typhoon Scanner). ChIP primers for the mouse fascin promoter were 5′-gtagggccagcattccag and 5′-tccagaccagagggacattc and for the human fascin promoter were 5′-accttgtgggcagcctgt and 5′-attccctgcagacaccacct.
Quantitative Real-time Reverse Transcription (RT)-PCR
RT-PCR analysis was performed as described previously (24). RNA was extracted using the TRIzol method (Invitrogen), reverse transcribed using qScript cDNA SuperMix (Quanta Biosciences), and subjected to quantitative real-time PCR using PerfeCTa SYBR Green FastMix, ROX (Quanta Biosciences). RT-PCR primers for mouse fascin were 5′-acgcatccgctagtagcttg and 5′-gacgagaaagcggcagtc, and human fascin primers were 5′-aaaagtgtgccttccgtacc and 5′-cccattcttcttggaggtca. RT-PCR primers for mouse GAPDH were 5′-acgaccccttcattgacc and 5′-agacaccagtagactccacg and human GAPDH, 5′-atcaagaaggtggtgaagca and 5′-gtcgctgttgaagtcagagga.
RNAi
RNAi was performed with STAT3 siRNA oligonucleotides from Qiagen for mouse (catalogue numbers SI01435294 and SI01435287), for human (SI02662898 and SI02662338), and a negative control siRNA (catalogue number 1022563) (22, 24, 25). NFκB siRNAs were from Qiagen (catalogue numbers SI02662618 and SI00300958). Cells were transfected using HiPerFect transfection reagent (Qiagen).
Cell Migration Assays
Wound healing assays were performed as described previously (26). Cells were cultured to confluence in RPMI 1640 medium supplemented with 10% FBS in 24-multiwell plates. Wounds were made with a sterile pipette tip, and cells were washed with PBS. Cells were cultured for the indicated time points either in medium without FBS, medium supplemented with 10% FBS, or medium without FBS supplemented with either OSM or IL-6 and IL-6R. Chamber assays were performed as described previously (26).
RESULTS
Fascin Is a Direct STAT3 Target Gene in Response to OSM in 4T1 Breast Cancer Cells
OSM and IL-6 have been shown to promote breast cancer metastasis through activation of STAT3 (3–6, 14, 15). However, the downstream targets of this signaling pathway are not well defined. Recently, the actin-bundling protein fascin was reported to be the key player for cancer metastasis (18). To see whether fascin could be a STAT3 target gene, we compared the human and mouse DNA sequences of the fascin promoter region. A conserved region of ∼160 bp was identified in both the human and mouse fascin promoter (Fig. 1A). Within this conserved region, a sequence motif similar to the canonical STAT3 site and an NFκB site were identified (Fig. 1A).
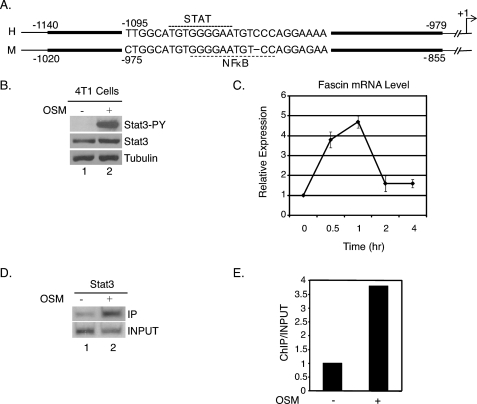
FIGURE 1.
Fascin is a direct STAT3 target gene in 4T1 cells. A, schematic representation of the fascin promoter. The dark lines represent the conserved region between the human (top) and mouse (bottom) sequences of the fascin promoter. The STAT and NFκB sites are highlighted by dotted lines. The arrow represents the transcription start site. Accession numbers for the compared sequences are EU486847 (human) and AJ318756 (mouse). B, 4T1 cells cultured in RPMI 1640 medium supplemented with 10% FBS and treated with OSM for 15 min. Whole cell extracts were subjected to Western blot analyses for phosphotyrosine STAT3 (Stat3-PY), total STAT3, or tubulin. C, cells were treated with OSM for 0.5, 1, 2, or 4 h. RNA was subjected to real-time quantitative RT-PCR analyses for fascin and GAPDH. Results were standardized to GAPDH in each sample with the untreated sample set at 1. Results represent three independent experiments performed in triplicate. Error bars, S.D. D, 4T1 cells cultured in RPMI 1640 medium supplemented with 10% FBS and treated with OSM for 30 min. ChIP assays were performed with a STAT3 antibody and primers flanking a potential STAT3 site in the mouse fascin promoter. E, results from D quantitated with a PhosphorImager and expressed as ChIP/Input with the untreated sample set at 1.
To determine whether OSM-induced STAT3 activation promotes breast cancer metastasis through regulation of the fascin gene, STAT3 phosphorylation and fascin mRNA expression were first analyzed in 4T1 mouse breast cancer cells treated with OSM. 4T1 cells treated with OSM showed a rapid increase in STAT3 tyrosine phosphorylation at 15 min of treatment (Fig. 1B, lane 2) compared with untreated cells, where there is a very low level of STAT3 phosphorylation (Fig. 1B, lane 1). Real-time quantitative RT-PCR analyses were performed on mRNA samples from untreated cells and cells treated with OSM for 30 min, 1 h, 2 h, and 4 h. Fascin mRNA expression increased rapidly and peaked at 5-fold after 1 h of treatment and then decreased (Fig. 1C). This kinetic expression pattern of transient and rapid induction is typical of STAT target genes (24, 25, 27–29). These results suggest a correlation between STAT3 activation and fascin expression in mouse breast cancer cells.
To demonstrate that fascin is a direct STAT3 target gene of STAT3, ChIP assays (23) were performed with a STAT3 antibody and PCR primers flanking the potential STAT3 site to see whether STAT3 binds to the fascin promoter in response to OSM treatment. In 4T1 cells treated with OSM, there was a significant increase in STAT3 binding to the fascin promoter compared with untreated cells (Fig. 1D). Quantitation of the bands using a PhosphorImager showed an approximately 4-fold increase in STAT3 binding (Fig. 1E). These results demonstrate that STAT3 binds directly to the fascin promoter to regulate fascin transcription in response to OSM treatment.
STAT3 Is Essential for Fascin Expression and Cell Migration in Response to OSM Treatment
To determine whether STAT3 is required for fascin expression in response to OSM treatment, the RNAi technique was utilized to knock down STAT3. Two separate siRNAs targeting two different regions of STAT3 were utilized for RNAi experiments (22, 24, 25). 4T1 cells were transfected with either STAT3 siRNA separately (Fig. 2A, 1 or 2) or both together (Fig. 2A, 1+2). Western blot analyses demonstrated that either STAT3 siRNA used separately (Fig. 2A, lanes 3 and 4) or both together (Fig. 2A, lane 2) efficiently knocked down STAT3 protein levels compared with a control siRNA (Fig. 2A, lane 1).
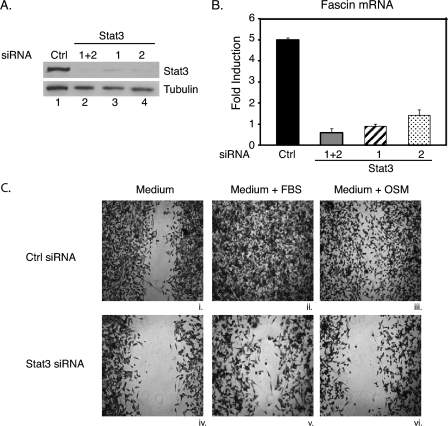
FIGURE 2.
STAT3 is required for OSM-induced expression of fascin and cell migration in 4T1 cells. 4T1 cells were cultured in RPMI 1640 medium supplemented with 10% FBS. Cells were transfected with a control siRNA (Ctrl) or two different STAT3 siRNA oligonucleotides either separately (1 or 2) or together (1+2) and cultured for 3 days. A, portions of the transfected cells were used for whole cell extracts and subjected to Western blot analyses with antibodies against total STAT3 or tubulin. B, portions of the cells were treated with OSM for 1 h or left untreated, and RNA from these cells was subjected to real-time quantitative RT-PCR analyses for fascin and GAPDH. Results are presented as -fold induction with untreated samples set at 1 for each transfection condition. Three independent experiments were performed in triplicate. Error bars, S.D. C, cells were cultured in RPMI 1640 medium supplemented with 10% FBS and transfected with either a control or STAT3 siRNAs (1+2) on day 1 and again on day 2 to maintain efficient knockdown of STAT3. On day 3, wound healing assays were performed in RPMI 1640 medium without FBS (i and iv), RPMI 1640 medium supplemented with 10% FBS (ii and v), or RPMI 1640 medium with OSM (iii and vi). Images of cells were taken after 19 h of culturing in the indicated conditions.
Quantitative real-time RT-PCR analyses were performed for fascin mRNA expression in the same cells from above. Cells transfected with the control siRNA showed a 5-fold increase in fascin mRNA expression when treated with OSM for 1 h (Fig. 2B). In contrast, fascin mRNA expression was not significantly induced by OSM in cells transfected with either STAT3 siRNA separately or both together (Fig. 2B). These results show that STAT3 is required for OSM-induced expression of fascin.
To determine further whether STAT3 is required for 4T1 cell migration, wound healing assays were performed on 4T1 cells transfected with the control siRNA or both STAT3 siRNAs together. Because the RNAi procedure uses transiently transfected siRNA oligonucleotides, the STAT3 knockdown is transient. Therefore, to be sure that STAT3 levels remained low for the wound healing assay, cells were subjected to two rounds of siRNA transfections on day 1 and day 2. On day 3, wound healing assays were performed, and the cells were further cultured in medium alone, medium with FBS, or medium with OSM. In the medium-alone condition, 4T1 cells could not migrate (Fig. 2C, i and iv). 4T1 cells transfected with a control siRNA migrated efficiently in medium supplemented with FBS (Fig. 2Cii) or OSM (Fig. 2Ciii). In contrast, migration of cells transfected with STAT3 siRNAs was significantly reduced in medium supplemented with FBS (Fig. 2Cv) or OSM (Fig. 2Cvi). These results demonstrate that STAT3 is required for breast cancer cell migration in response to FBS or OSM treatment.
STAT3 Is Essential for IL-6-induced Fascin Expression to Promote Cell Migration in MDA-MB-231 Human Breast Tumor Cells
The above results demonstrate that fascin is a direct STAT3 target gene in mouse breast cancer cells. To demonstrate further the specific regulation of fascin expression by STAT3 in response to cytokines, we analyzed the expression of fascin in a human breast cancer cell line, MDA-MB-231 and treated these cells with another cytokine that activates STAT3, IL-6, which has been shown to promote breast cancer metastasis (3, 4). Similar to the results obtained for OSM-treated mouse breast tumor cells, IL-6 induced STAT3 tyrosine phosphorylation in MDA-MB-231 cells (Fig. 3A, lane 2), and this correlated with a rapid transient induction of fascin mRNA expression after 30 min of cytokine treatment (Fig. 3B). These results show that in response to IL-6, STAT3 is activated, and fascin expression is induced in human breast cancer cells. ChIP analysis also demonstrated that STAT3 binds to the fascin promoter in MDA-MB-231 cells in response to IL-6 (data not shown).
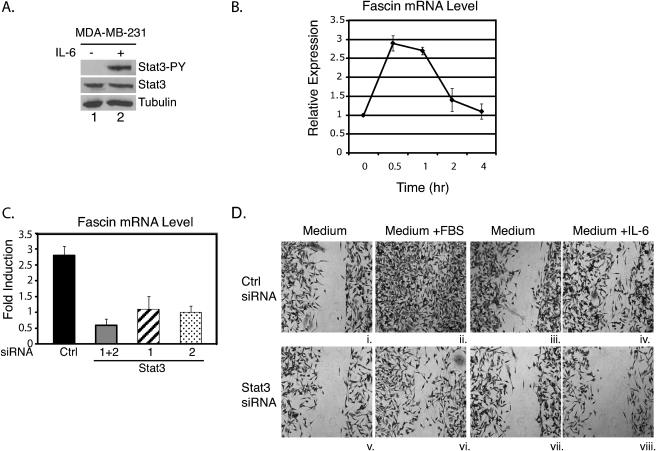
FIGURE 3.
STAT3 is required for IL-6-induced expression of fascin and cell migration in MDA-MB-231 cells. A, MDA-MB-231 cells were cultured in RPMI 1640 medium supplemented with 10% FBS and treated with IL-6 for 15 min. Whole cell extracts were subjected to Western blot analyses for phosphotyrosine STAT3 (Stat3-PY), total STAT3, or tubulin. B, cells were treated with IL-6 for 0.5, 1, 2, or 4 h. RNA was subjected to real-time quantitative RT-PCR analyses for fascin and GAPDH. Results were standardized to GAPDH with the untreated sample set at 1. Results represent three independent experiments performed in triplicate. Error bars, S.D. C, cells were transfected with a control siRNA (Ctrl) or two STAT3 siRNA oligonucleotides separately (1 or 2) or together (1+2) and cultured for 3 days. RNA from these cells was analyzed by real-time quantitative RT-PCR analyses for fascin and GAPDH. Results represent the -fold induction of IL-6-treated cells compared with untreated cells from three independent experiments. D, cells were transfected with either a control or both STAT3 siRNAs (1+2). 3 days after transfection, wound healing assays were performed in RPMI 1640 medium only (i, iii, v, and vii), RPMI 1640 medium supplemented with 10% FBS (ii and vi), or IL-6 (iv and viii). Cells were cultured in the indicated conditions for 24 h (i, ii, v, and vi) or for 30 h (iii, iv, vii, and viii).
To demonstrate that STAT3 is required for IL-6-induced fascin expression and cell migration in MDA-MB-231 cells, two siRNAs targeting human STAT3 were utilized. STAT3 levels were significantly decreased when transfected with either STAT3 siRNA or both together, similar to Fig. 2A (data not shown). Real-time RT-PCR analyses were performed on cells transfected with the STAT3 siRNAs or a control siRNA. Cells transfected with a control siRNA showed a 3-fold increase in fascin mRNA expression in response to IL-6 treatment (Fig. 3C). In cells transfected with either STAT3 siRNA (1 or 2) or both together (1+2), fascin mRNA levels were not significantly induced when treated with IL-6 (Fig. 3C). These results demonstrate that STAT3 is required for fascin expression in response to IL-6 in human breast cancer cells.
Cell migration was also analyzed for MDA-MB-231 cells transfected with control or STAT3 siRNAs. The knockdown effect lasted longer in these cells, therefore only one round of siRNA transfection was necessary. Because control cells in medium supplemented with FBS migrated faster than those in medium supplemented with IL-6, two different time points were utilized. One set of cells was cultured for 24 h (Fig. 3D, i, ii, v, and vi). Another set was cultured for 30 h (Fig. 3D, iii, iv, vii, and viii). Cells did not migrate in medium alone conditions (Fig. 3D, i, iii, v, and vii). Cells transfected with STAT3 siRNAs migrated less efficiently in medium supplemented with FBS compared with control cells (Fig. 3D, ii and vi). In medium supplemented with IL-6, migration of STAT3 RNAi knockdown cells was significantly reduced compared with control cells (Fig. 3D, iv and viii).
Altogether, these results demonstrate in human breast cancer cells, STAT3 directly regulates fascin expression in response to IL-6 treatment. Furthermore, STAT3 is essential for fascin expression and breast cancer cell migration.
STAT3 Is Required for NFκB Recruitment to the Fascin Promoter in Response to OSM
To understand further the role of STAT3 in transcription of the fascin gene, we analyzed the fascin promoter for other transcription factors that could be recruited by STAT3. We identified a NFκB binding site adjacent to the STAT3 binding site in the fascin promoter (Fig. 1A). To determine whether the NFκB p50 protein binds to the fascin promoter along with STAT3 in response to OSM, ChIP assays were performed in 4T1 cells. There was a significant increase in NFκB bound to the fascin promoter when cells were treated with OSM (Fig. 4, A and B). NFκB also bound to the fascin promoter in response to IL-6 treatment in human MDA-MB-231 cells (data not shown).
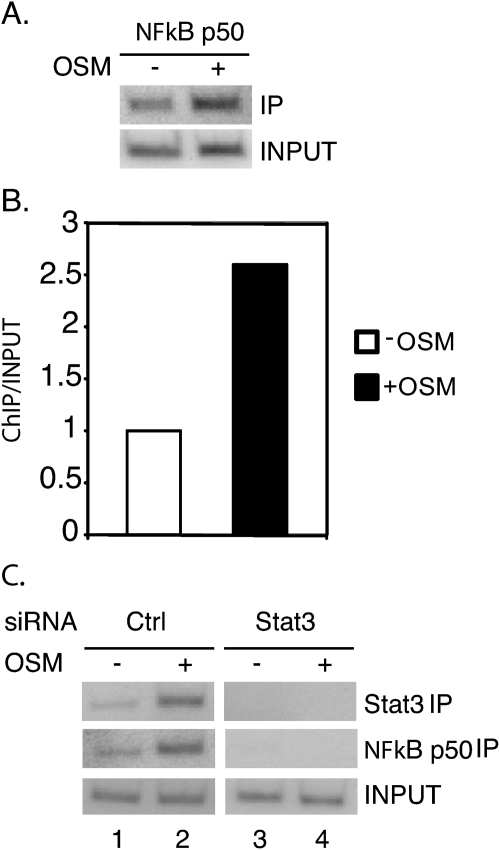
FIGURE 4.
NFκB binds to the fascin promoter in response to OSM in a STAT3-dependent manner. A, 4T1 cells were cultured in RPMI 1640 medium supplemented with 10% FBS and treated with OSM for 30 min. ChIP assays were performed with an NFκB antibody and primers flanking the potential NFκB binding site. B, results from A were quantitated with a PhosphorImager and expressed as ChIP/Input with the untreated sample set at 1. C, 4T1 cells were transfected with either a control siRNA or both STAT3 siRNAs (1+2) together and cultured for 3 days. Cells were treated with OSM for 30 min and ChIP assays were performed with STAT3 or NFκB antibodies and primers flanking the potential STAT3/NFκB binding site.
To determine whether STAT3 is required for NFκB recruitment to the fascin promoter in response to OSM, STAT3 was knocked down as described in Fig. 2. ChIP assays were performed with either STAT3 or NFκB antibodies in both control and STAT3 knockdown cells. In control cells, there was a significant increase in STAT3 and NFκB bound to the fascin promoter in response to OSM treatment (Fig. 4C, lane 2). In contrast, in the STAT3 siRNA knockdown cells, there was no STAT3 or NFκB binding to the fascin promoter in response to OSM treatment (Fig. 4C, lane 4). NFκB protein levels were not affected in the STAT3 siRNA knockdown cells (data not shown). These results demonstrate that in response to OSM treatment, NFκB binds to the fascin promoter in a STAT3-dependent manner.
NFκB Is Required for Fascin Expression in Response to Cytokine Treatment
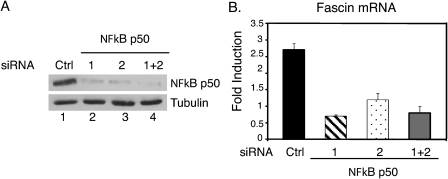
To determine whether NFκB is required for fascin expression in response to cytokine treatment, NFκB p50 was knocked down using the RNAi technique, and fascin expression was examined. Two separate siRNAs were used to knock down NFκB p50 in MDA-MB-231 cells. Both siRNAs together (1 + 2) or separately (1 or 2) efficiently knocked down NFκB p50 (Fig. 5A, lanes 2, 3, and 4) compared with a control siRNA (Fig. 5A, lane 1). Cells were treated with IL-6, and fascin mRNA expression was analyzed. Fascin was induced approximately 3-fold in cells transfected with a control siRNA whereas fascin was not significantly induced in cells transfected with either NFκB p50 siRNA (1 or 2) or both together (1+2) (Fig. 5B). These results demonstrate that NFκB is required for fascin mRNA expression in response to cytokine treatment.
FIGURE 5.
NFκB is required for cytokine-induced fascin expression. A, MDA-MB-231 cells were transfected with either a control siRNA or two NFκB p50 siRNAs either together (1+2) or separately (1 or 2) and cultured for 4 days. Whole cell extracts were performed, and Western blot analyses were performed with antibodies against NFκB p50 or tubulin. B, portions of the cells in A were treated with IL-6 and IL-6R for 30 min, and RNA was subjected to quantitative real-time RT-PCR for fascin and GAPDH. Results were standardized to GAPDH and expressed as -fold induction with untreated samples set as 1.
Constitutively Activated STAT3 Increases Fascin Expression and Cell Migration
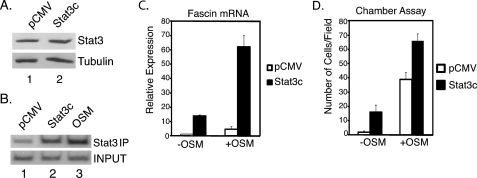
To analyze further the role of STAT3 in fascin transcriptional regulation, we utilized a constitutively activated STAT3 (STAT3c) construct. STAT3c is constitutively dimerized and translocated to the nucleus (16). In 4T1 cells transfected with STAT3c, there was a slight increase in total STAT3 protein levels compared with cells transfected with an empty vector (pCMV) (Fig. 6A).
FIGURE 6.
Constitutively activated STAT3 induces fascin mRNA expression and cell migration in 4T1 cells. A, 4T1 cells were cultured in RPMI 1640 medium supplemented with 10% FBS and transfected with pCMV or a STAT3c construct. Cells were cultured for 24 h, and whole cell extracts were subjected to Western blot analyses for total STAT3 or tubulin. B, 4T1 cells were transfected with pCMV or STAT3c and cultured for 24 h. Transfected cells were used for ChIP analyses along with untransfected cells treated with OSM for 30 min. ChIP analyses were performed with STAT3 antibodies and primers flanking the potential STAT3 site in the fascin promoter. C, portions of the cells from A were treated with OSM for 1 h or left untreated, and quantitative real-time RT-PCR analyses were performed for fascin. Results were standardized to GAPDH and with untreated pCMV samples set at 1. D, 4T1 cells were transfected with pCMV or STAT3c and cultured for 24 h. Chamber cell migration assays were performed in medium alone or medium containing OSM for 6 h. The number of migrated cells was examined microscopically and counted in three random fields. Results represent the average number of cells/field. Error bars, S.D.
To demonstrate that the STAT3c binds to the fascin promoter without cytokine stimulation, ChIP assays were performed with cells transfected with pCMV or STAT3c. Cells transfected with the STAT3c construct showed a significant increase in STAT3 bound to the fascin promoter without cytokine treatment (Fig. 6B, lane 2) compared with cells transfected with pCMV (Fig. 6B, lane 1). ChIP analysis was also performed on untransfected cells treated with OSM for 30 min as a positive control (Fig. 6B, lane 3). These results demonstrate that STAT3c efficiently binds to the fascin promoter in the absence of cytokine treatment.
Fascin mRNA expression was also analyzed in cells transfected with pCMV or STAT3c. Without OSM, very little fascin expression was detected in cells transfected with pCMV (Fig. 6C). In contrast, cells transfected with STAT3c showed an approximately 15-fold increase in fascin expression without OSM treatment (Fig. 6C). When cells were treated with OSM for 1 h, cells transfected with pCMV showed an approximately 5-fold increase in fascin expression whereas STAT3c-transfected cells showed a much higher induction of fascin at approximately 60-fold (Fig. 6C). These results demonstrate that STAT3c significantly induces fascin expression in the absence of cytokine treatment, and OSM treatment further increases this expression.
To determine whether STAT3c induced cell migration, chamber cell migration assays were performed. Cells transfected with pCMV and left untreated did not migrate efficiently. In contrast, untreated cells transfected with STAT3c showed a 15-fold increase in the number of cells migrating (Fig. 6D). Cells transfected with pCMV and treated with OSM showed a 40-fold increase in cell migration whereas cells transfected with STAT3c and treated with OSM showed a greater increase at almost 70-fold (Fig. 6D). Wound healing assays also demonstrated that STAT3c induces cell migration in the absence of OSM (data not shown).
All together, these results demonstrate that STAT3c induces fascin expression and promotes breast cancer cell migration without cytokine treatment and strongly suggest STAT3 plays a central role in the induction of fascin expression.
DISCUSSION
It has been shown previously that IL-6 and OSM along with STAT3 activation promote cell migration and breast cancer metastasis (3–6, 14). The actin-bundling protein fascin functions in cell adhesion and motility, and high levels of fascin have been reported in many metastatic cancers (19, 20). Recently, fascin was identified as a key player in tumor metastasis, and knockdown of fascin expression or inhibition of fascin function blocks breast cancer metastasis (18). In this work, we demonstrate that STAT3 directly regulates expression of fascin in response to IL-6 or OSM treatment in breast cancer cells. Furthermore, we show that STAT3 is essential for NFκB recruitment to the fascin promoter and for fascin expression and breast cancer cell migration in response to cytokine treatment.
Fascin levels can be abnormally high in many cancers (18–20), and this work shows that IL-6 or OSM treatment can further increase these levels to promote cell migration (21). It is also possible that other factors, such as β-catenin and cAMP-response element-binding protein, contribute to the expression of fascin in cancer cells (30–32). It is likely that an enhancersome containing several transcription factors controls the precise regulation of fascin expression (33, 34). The transcription factor NFκB can recruit unphosphorylated STAT3 to promoters to activate transcription (35, 36). It has been shown previously that the fascin promoter contains several NFκB sites and NFκB has been shown to be involved in breast cancer metastasis (37, 38). However, the exact role of NFκB in fascin transcription has not been established. In this work we demonstrate that NFκB is recruited to the fascin promoter in a STAT3-dependent manner in response to cytokine treatment. Similar to STAT3, NFκB is required for cytokine-induced expression of fascin. These observations suggest that NFκB functions at the STAT3-dependent enhancersome to increase fascin expression and promote cancer metastasis. Together with the results from the STAT3c experiment (Fig. 6), it is most likely that STAT3 plays a central role in fascin expression. Studies of STAT3-regulated expression of fascin will provide new insights into the mechanism by which IL-6 and OSM promote breast cancer metastasis in vivo, where multiple factors including cytokines contribute to the critical step of metastasis of primary tumors. Further detailed analyses of the fascin promoter and the specific interactions between transcription factors such as STAT3 and NFκB could also identify potential drug targets to block metastasis.
In addition to fascin, STAT3 regulates other genes that function in breast cancer metastasis (15, 39). It has been shown previously that STAT3 regulates the Twist gene to promote breast cancer metastasis in mice (15). It is possible that STAT3 could regulate other genes that function in metastasis. Furthermore, several other pathways in addition to the JAK-STAT pathway function in cancer metastasis, including WNT, epidermal growth factor receptor, and Src signaling pathways (40, 41). STAT3 could function either independently or together with these pathways to promote breast cancer metastasis. Further study of the target genes regulated by these pathways will lead to greater understanding of the mechanisms that contribute to cancer metastasis.
Acknowledgments
We thank all laboratory members for helpful discussions and the Dept. of Microbiology and Immunology at Weill Cornell Medical College for the use of the Applied Biosystems PRISM 7900HT Sequence Detection Equipment.
This work was supported, in whole or in part, by National Institutes of Health Grant CA136837 (to X.-Y. H.).
- OSM
- oncostatin M
- STAT3c
- constitutively activated STAT3.
REFERENCES
- 1. Said N. A., Williams E. D. (2011) Cells Tissues Organs 193, 85–97 [DOI] [PubMed] [Google Scholar]
- 2. Knüpfer H., Preiss R. (2007) Breast Cancer Res. Treat. 102, 129–135 [DOI] [PubMed] [Google Scholar]
- 3. Sullivan N. J., Sasser A. K., Axel A. E., Vesuna F., Raman V., Ramirez N., Oberyszyn T. M., Hall B. M. (2009) Oncogene 28, 2940–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walter M., Liang S., Ghosh S., Hornsby P. J., Li R. (2009) Oncogene 28, 2745–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jorcyk C. L., Holzer R. G., Ryan R. E. (2006) Cytokine 33, 323–336 [DOI] [PubMed] [Google Scholar]
- 6. Queen M. M., Ryan R. E., Holzer R. G., Keller-Peck C. R., Jorcyk C. L. (2005) Cancer Res. 65, 8896–8904 [DOI] [PubMed] [Google Scholar]
- 7. Sasser A. K., Sullivan N. J., Studebaker A. W., Hendey L. F., Axel A. E., Hall B. M. (2007) FASEB J. 21, 3763–3770 [DOI] [PubMed] [Google Scholar]
- 8. Ara T., Declerck Y. A. (2010) Eur. J. Cancer 46, 1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Akira S., Nishio Y., Inoue M., Wang X. J., Wei S., Matsusaka T., Yoshida K., Sudo T., Naruto M., Kishimoto T. (1994) Cell 77, 63–71 [DOI] [PubMed] [Google Scholar]
- 10. Heinrich P. C., Horn F., Graeve L., Dittrich E., Kerr I., Müller-Newen G., Grötzinger J., Wollmer A. (1998) Z. Ernahrungswiss. 37, 43–49 [PubMed] [Google Scholar]
- 11. Santos C. I., Costa-Pereira A. P. (2011) Biochim. Biophys. Acta 1816, 38–49 [DOI] [PubMed] [Google Scholar]
- 12. Levy D. E., Darnell J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651–662 [DOI] [PubMed] [Google Scholar]
- 13. Akira S. (2000) Oncogene 19, 2607–2611 [DOI] [PubMed] [Google Scholar]
- 14. Barbieri I., Pensa S., Pannellini T., Quaglino E., Maritano D., Demaria M., Voster A., Turkson J., Cavallo F., Watson C. J., Provero P., Musiani P., Poli V. (2010) Cancer Res. 70, 2558–2567 [DOI] [PubMed] [Google Scholar]
- 15. Ling X., Arlinghaus R. B. (2005) Cancer Res. 65, 2532–2536 [DOI] [PubMed] [Google Scholar]
- 16. Bromberg J. F., Wrzeszczynska M. H., Devgan G., Zhao Y., Pestell R. G., Albanese C., Darnell J. E., Jr. (1999) Cell 98, 295–303 [DOI] [PubMed] [Google Scholar]
- 17. Akira S. (1999) Stem Cells 17, 138–146 [DOI] [PubMed] [Google Scholar]
- 18. Chen L., Yang S., Jakoncic J., Zhang J. J., Huang X. Y. (2010) Nature 464, 1062–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Adams J. C. (2004) Curr. Opin. Cell Biol. 16, 590–596 [DOI] [PubMed] [Google Scholar]
- 20. Machesky L. M., Li A. (2010) Commun. Integr. Biol. 3, 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Q., Li G., Li R., Shen J., He Q., Deng L., Zhang C., Zhang J. (2010) J. Neurooncol. 100, 165–176 [DOI] [PubMed] [Google Scholar]
- 22. Snyder M., Huang X. Y., Zhang J. J. (2008) J. Biol. Chem. 283, 3791–3798 [DOI] [PubMed] [Google Scholar]
- 23. Aparicio O., Geisberg J. V., Sekinger E., Yang A., Moqtaderi Z., Struhl K. (2005) Curr. Protoc. Mol. Biol. 21, 3.1–3.33 [DOI] [PubMed] [Google Scholar]
- 24. Snyder M., Huang X. Y., Zhang J. J. (2010) J. Biol. Chem. 285, 23639–23646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Snyder M., Huang X. Y., Zhang J. J. (2011) FEBS Lett. 585, 148–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shan D., Chen L., Njardarson J. T., Gaul C., Ma X., Danishefsky S. J., Huang X. Y. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3772–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ramsauer K., Farlik M., Zupkovitz G., Seiser C., Kröger A., Hauser H., Decker T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2849–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Snyder M., He W., Zhang J. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14539–14544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun W., Snyder M., Levy D. E., Zhang J. J. (2006) FEBS Lett. 580, 5880–5884 [DOI] [PubMed] [Google Scholar]
- 30. Vignjevic D., Schoumacher M., Gavert N., Janssen K. P., Jih G., Laé M., Louvard D., Ben-Ze'ev A., Robine S. (2007) Cancer Res. 67, 6844–6853 [DOI] [PubMed] [Google Scholar]
- 31. Hashimoto Y., Loftis D. W., Adams J. C. (2009) PLoS One 4, e5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bros M., Ross X. L., Pautz A., Reske-Kunz A. B., Ross R. (2003) J. Immunol. 171, 1825–1834 [DOI] [PubMed] [Google Scholar]
- 33. Zhang X., Wrzeszczynska M. H., Horvath C. M., Darnell J. E., Jr. (1999) Mol. Cell. Biol. 19, 7138–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lerner L., Henriksen M. A., Zhang X., Darnell J. E., Jr. (2003) Genes Dev. 17, 2564–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang J., Liao X., Agarwal M. K., Barnes L., Auron P. E., Stark G. R. (2007) Genes Dev. 21, 1396–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoshida Y., Kumar A., Koyama Y., Peng H., Arman A., Boch J. A., Auron P. E. (2004) J. Biol. Chem. 279, 1768–1776 [DOI] [PubMed] [Google Scholar]
- 37. Grothey A., Hashizume R., Ji H., Tubb B. E., Patrick C. W., Jr., Yu D., Mooney E. E., McCrea P. D. (2000) Oncogene 19, 4864–4875 [DOI] [PubMed] [Google Scholar]
- 38. Liu M., Sakamaki T., Casimiro M. C., Willmarth N. E., Quong A. A., Ju X., Ojeifo J., Jiao X., Yeow W. S., Katiyar S., Shirley L. A., Joyce D., Lisanti M. P., Albanese C., Pestell R. G. (2010) Cancer Res. 70, 10464–10473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ranger J. J., Levy D. E., Shahalizadeh S., Hallett M., Muller W. J. (2009) Cancer Res. 69, 6823–6830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hu T., Li C. (2010) Mol. Cancer 9, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guarino M. (2010) J. Cell. Physiol. 223, 14–26 [DOI] [PubMed] [Google Scholar]