Background: Bone homeostasis is maintained by balancing the activities of bone-resorbing osteoclasts and bone-forming osteoblasts.
Results: GSK-3β is inactivated by receptor activator of NF-κB ligand stimulation via serine phosphorylation during osteoclastogenesis.
Conclusion: GSK-3β is crucial for receptor activator of NF-κB ligand-mediated signaling as a negative regulator of osteoclast differentiation.
Significance: GSK-3β acts as a novel negative regulator of osteoclast biology.
Keywords: Bone, Differentiation, Glycogen Synthase Kinase-3, NFAT Transcription Factor, Signal Transduction, Osteoclasts
Abstract
Glycogen synthase kinase-3β (GSK-3β) is a serine/threonine kinase originally identified as a regulator of glycogen deposition. Although the role of GSK-3β in osteoblasts is well characterized as a negative regulator of β-catenin, its effect on osteoclast formation remains largely unidentified. Here, we show that the GSK-3β inactivation upon receptor activator of NF-κB ligand (RANKL) stimulation is crucial for osteoclast differentiation. Regulation of GSK-3β activity in bone marrow macrophages by retroviral expression of the constitutively active GSK-3β (GSK3β-S9A) mutant inhibits RANKL-induced osteoclastogenesis, whereas expression of the catalytically inactive GSK-3β (GSK3β-K85R) or small interfering RNA (siRNA)-mediated GSK-3β silencing enhances osteoclast formation. Pharmacological inhibition of GSK-3β further confirmed the negative role of GSK-3β in osteoclast formation. We also show that overexpression of the GSK3β-S9A mutant in bone marrow macrophages inhibits RANKL-mediated NFATc1 induction and Ca2+ oscillations. Remarkably, transgenic mice expressing the GSK3β-S9A mutant show an osteopetrotic phenotype due to impaired osteoclast differentiation. Further, osteoclast precursor cells from the transgenic mice show defects in expression and nuclear localization of NFATc1. These findings demonstrate a novel role for GSK-3β in the regulation of bone remodeling through modulation of NFATc1 in RANKL signaling.
Introduction
Bone homeostasis is maintained by balancing the activities of bone-resorbing osteoclasts and bone-forming osteoblasts, and imbalances in bone remodeling cause a variety of bone diseases (1, 2). Two cytokines are essential for osteoclast differentiation: receptor activator of NF-κB ligand (RANKL),5 which belongs to the tumor necrosis factor (TNF) family, and macrophage colony-stimulating factor (M-CSF) (3, 4). RANKL mediates its signal transduction through binding to RANK, which is expressed by osteoclast precursors. RANK recruits the adapter molecule, TNF receptor-associated factor 6 (TRAF6), which activates NF-κB and mitogen-activated protein kinases (MAPKs), including c-Jun N-terminal kinase and p38 (5, 6). RANKL-induced activation of NF-κB and c-Fos is required for initial expression of the key transcription factor, NFATc1 (7, 8).
NFATc1 is an NFAT family member, which is activated by the Ca2+/calmodulin-regulated phosphatase calcineurin (9). In osteoclast precursors, calcium signaling activates the existing NFATc1, and an AP-1 complex containing c-Fos may cooperate with NFATc1 to trigger NFATc1 autoamplification (8). Autoamplification enables robust induction of NFATc1 on activation of RANKL signaling (10). Upon activation, NFATc1 proteins are dephosphorylated by calcineurin, and then they translocate from the cytoplasm into the nucleus, where they direct transcription of osteoclast-specific genes, such as TRAP, cathepsin K, Oscar, and Atp6v0d2 (11, 12). Nuclear export of NFAT members is facilitated by phosphorylation, and several kinases have been suggested to regulate NFAT function, including GSK-3 (13), CK1 (14), p38 (15), and JNK1 (16).
Glycogen synthase kinase-3 (GSK-3) is a serine/threonine kinase originally identified for its role in the regulation of glycogen deposition. GSK-3 has two isoforms, GSK-3α and GSK-3β (17), both of which are implicated in many different biological processes including metabolism, transcription, translation, cell growth, and apoptosis (18). With respect to transcription, GSK-3 regulates a wide variety of transcription factors, including cyclin D1, c-Jun, NFATc, and β-catenin (13, 19, 20). In resting cells, GSK-3 is constitutively active, and its activity is inhibited by various kinases via phosphorylation of a serine residue, Ser-21 in GSK-3α and Ser-9 in GSK-3β in response to different stimuli (21). Serine phosphorylation on GSK-3 blocks the access of substrate to the GSK-3 catalytic domain, thus inhibiting substrate phosphorylation (22).
Of the two isoforms of GSK-3, GSK-3β is a more likely candidate for being an NFATc1 kinase, influencing NFATc1 subcellular localization through phosphorylation (13). However, the significance of the ability of GSK-3β to regulate NFATc1 during osteoclastogenesis has not yet been demonstrated. In addition, because GSK-3β-deficient mice die in utero (23), the in vivo relevance of GSK-3β in osteoclast precursors has not been well characterized. Therefore we investigated the role of GSK-3β in RANKL-mediated osteoclast differentiation and also clarified the relevance of GSK-3β and NFATc1. In addition, to understand the physiological role of GSK-3β in vivo, we generated transgenic mice that express a constitutively active GSK-3β (GSK3β-S9A) mutant under the control of the mouse tartrate-resistant acid phosphatase (TRAP) gene promoter, which is specific to the osteoclast lineage. Our results demonstrate that GSK-3β suppresses osteoclastogenesis through regulation of NFATc1 expression. We also show that GSK-3β is inactivated by RANKL stimulation via serine phosphorylation. Remarkably, transgenic mice expressing the GSK3β-S9A mutant show a defect in NFATc1 and osteoclast-specific gene expression and therefore show a severe osteopetrotic phenotype. Based on these results, we suggest that GSK-3β is crucial for RANKL-mediated signaling as a negative regulator of osteoclast differentiation.
EXPERIMENTAL PROCEDURES
Cells and Reagents
Primary cells used in this study, including bone marrow macrophages (BMMs) and osteoblasts, were generated from murine bone marrow precursors of 4–6-week-old C57BL/6 mice (The Jackson Laboratory) and cultured in α-minimum essential medium (α-MEM; HyClone, Logan, UT) supplemented with 10% fetal bovine serum (FBS) and antibiotics. RAW264.7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; HyClone) with 10% FBS and antibiotics. To generate stable cell lines, RAW264.7 cells were infected with recombinant virus expressing GSK3β-S9A and then selected with puromycin (2 μg/ml) after 2 weeks of growth. Kenpaullone was purchased from Biomol International (Plymouth Meeting, PA). SB216763 and SB415286 were from Sigma.
Protein Analysis
For Western blot analysis, equal amounts of cell lysates harvested at the indicated conditions were subjected to analysis using specific antibodies against NFATc1, phospho-NFATc1 (Santa Cruz Biotechnology), GSK-3α/β (Invitrogen), GSK-3β, phospho-GSK-3β (Cell Signaling), OSCAR (R&D), hemagglutinin (HA; Roche Applied Science), and β-actin (Sigma). Anti-Atp6v0d2 (24) antibody was kindly provided by Y. Choi (University of Pennsylvania).
Retroviral Infection
HA epitope-tagged cDNAs of wild-type GSK-3β and two mutants, catalytically inactive GSK-3β (GSK3β-K85R) and constitutively active GSK-3β (GSK3β-S9A) mutant plasmids, were kindly provided by J. Chung (Seoul National University, Korea). V5 epitope-tagged cDNAs of GSK-3α and catalytically inactive GSK-3α (GSK3α-K148A) plasmids were kindly provided by J. R. Woodgett (Samuel Lunenfeld Research Institute). cDNAs of wild-type and mutant GSK-3β or GSK-3α were cloned into the retroviral vector, pMX-puro, as described previously (25). The plasmids were transfected into PLAT-E cells using LipofectamineTM 2000 (Invitrogen), and the supernatant was collected 24–36 h after transfection. The pMX-puro vector and PLAT-E cells were kindly provided by T. Kitamura (University of Tokyo, Japan). The supernatant including retroviruses was used to infect BMMs or RAW 264.7 cells as described previously (25). BMMs were cultured with virus supernatant for 1 day and then changed to the medium with M-CSF (100 ng/ml) and puromycin (2 μg/ml) for 2 days. Puromycin-resistant BMMs were used for osteoclast differentiation and Ca2+ measurement experiments.
In Vitro Osteoclast Differentiation
Osteoclasts were prepared from bone marrow cells using a standard method (26). In brief, bone marrow cells were cultured with 30 ng/ml M-CSF (R&D Systems) for 3 days to obtain osteoclast precursor cells of the monocyte/macrophage lineage. The precursors were cultured with 30 ng/ml M-CSF and 100 ng/ml RANKL for the indicated time periods. The various GSK-3 inhibitors including kenpaullone, SB216763, and SB415286 were added at the time of RANKL addition. TRAP+ cells with more than three nuclei or cells larger than 100 μm in diameter that contained more than 20 nuclei were counted as TRAP+ multinucleated cells (MNCs). Co-culture experiments were performed as described previously (27). In brief, primary calvarial osteoblast precursors were obtained from newborn C57BL/6 mice by digesting isolated calvariae with 0.8 units/ml dispase (Roche Applied Science, Penzberg, Germany) and 0.1% collagenase (Sigma) and incubating the dissociated cells in α-MEM (HyClone). The BMMs (2 × 105) infected with retrovirus expressing either GSK-3β or its mutants were seeded on top of the calvarial osteoblast precursors (2 × 104) on each well of 48-well plates and cultured for 7 days in the presence of 10 nm 1,25(OH)2D3 (Sigma) and 1 μm prostaglandin E2 (Sigma). Osteoclasts were identified by TRAP staining and counted.
Transfection and Reporter Assay
Stable RAW264.7 cells were seeded at a density of 105 cells/well in a 12-well plate 1 day prior to transfection using LipofectamineTM 2000 (Invitrogen) according to the manufacturer's protocol. Luciferase reporter constructs driven by the NFAT-responsive elements were described previously (28). Luciferase activity was measured by the luciferase assay system (Promega) and normalized relative to β-galactosidase activity. The data were obtained from three independent transfections and presented as the -fold induction in luciferase activity (mean ± S.D.) relative to the control.
GSK-3β Activity Measurements
For measurements of GSK-3β activity, BMMs were stimulated by RANKL at the indicated times and immunoprecipitated by anti-GSK-3β antibody. The immunoprecipitated beads were washed with kinase buffer (20 mm Tris, pH 7.5, 5 mm MgCl2, 1 mm dithiothreitol) and then resuspended in 30 μl of kinase buffer containing 250 μm ATP, 1.4 μCi of [γ-32P]ATP, and 50 μm phospho-glycogen synthase peptide (Upstate Biotech Millipore, Charlottesville, VA). The samples were incubated at 30 °C for 30 min and spotted on P81 filter paper circles (Whatman International Ltd., Maidstone, UK). The filter paper was dried, washed three times in 0.5% phosphoric acid for 1 h, and finally washed with 95% ethanol for 10 min. The air-dried filter was counted by scintillation counter, and activity was normalized to the amount of GSK-3β, as determined by immunoblotting.
Immunofluorescence Staining
BMMs were plated on poly-l-lysine-coated coverslips in 12-well plates and treated with RANKL and M-CSF for 3 days. Cells were then washed with phosphate-buffered saline (PBS), fixed with 3.8% paraformaldehyde in PBS, and permeabilized by incubation for 10 min with 0.1% Triton in PBS. The cells were blocked in 4% bovine serum albumin (BSA)-PBS for 1 h at room temperature. The cells were incubated with anti-NFATc1 antibody overnight, washed, and stained with Alexa Fluor 488-conjugated anti-mouse immunoglobulin (IgG) antibody (Alexa Fluor 488 goat anti-mouse IgG, Invitrogen) for 1 h. The coverslips were washed three times with PBS and then incubated with DAPI (4′,6-diamidino-2-phenylindole; Sigma) for 5 min. After washing three times, the coverslips were mounted on slides and visualized using an LSM 510 META (Carl Zeiss) confocal microscope.
RNA Interference
Custom SMARTpool plus small interfering RNA (siRNA) to target mouse GSK-3β (catalogue number M-041080) was designed and synthesized by Dharmacon (Lafayette, CO). siRNA (10 nmol) was transfected into BMMs using LipofectamineTM 2000 (Invitrogen) according to the manufacturer's protocol. After transfection, BMMs were cultured with M-CSF and RANKL for 4 days and then differentiated into osteoclasts.
Measurement of Intracellular Ca2+
Ca2+ measurements were performed as described previously (10). Infected BMMs were incubated with M-CSF and RANKL for 3 days and then incubated in the presence of 5 μm fluo-4 acetoxymethyl (fluo-4AM; Invitrogen), 5 μm fura red AM (Invitrogen), and 0.05% plutonic F127 for 30 min. Cells were washed twice and post-incubated in DMEM with 10 ng/ml M-CSF for 20 min. For measurement of [Ca2+]c (cytosolic Ca2+ concentration), single cells were viewed with a laser-scanning confocal system (FluoView 500, Olympus, Tokyo, Japan) attached to an upright microscope (BX51WI, Olympus). An argon laser (488 nm) was used for excitation, a green emission filter (505–525 nm) was used for fluo-4, and a red emission filter (<660 nm) was used for fura red to observe the fluorescent images. The ratio of the fluorescence intensity of fluo-4 to fura red was calculated. The maximum intensity of [Ca2+]c was obtained with the addition of 10 μm ionomycin at the end of each experiment. The ratio of increase from the basal level was expressed as the percentage of maximum ratio increase.
Generation of Transgenic Mice
The constitutively active GSK-3β (GSK3β-S9A) mutant cDNA was fused to the mouse TRAP gene promoter as described previously (29, 30). For generating transgenic mice, we used the standard pronuclear injection method with C57BL/6 mice (The Jackson Laboratory). Genomic DNA isolated from the tail was analyzed by polymerase chain reaction (PCR) using the specific primers (GT-F, 5′-TAGCCATCAACAGCCGTCAGT; GT-R, 5′-CTTCTGCCCCAGAGAATAAAG; GP-F, 5′-CAGGGTACAGTTTAGAATGGG; GP-R, 5′-GTACTAGGCAGACTGTGTAAAG) to detect the transgene. All the mouse experiments were performed with 4–6-week-old mice under the animal protocol approved by the Animal Care Committee of the Ewha Laboratory Animal Genomics Center.
Bone Histomorphometry and Microcomputed Tomography Analysis
Bones were fixed in 10% formaldehyde, decalcified in 0.5 m EDTA, pH 7.4, embedded in paraffin, and then cut into 4-μm sections. Hematoxylin and eosin (H&E) or TRAP staining was performed according to a standard protocol (24). The histomorphometric data were analyzed by Osteomeasure XP (OsteoMetrics Inc.). Quantitative microcomputed tomography was performed with Skyscan 1076 (Skyscan N.V.). The data from scanned slices were used for the three-dimensional analysis to calculate femoral morphometric parameters by CT-AN 1.10 (Skyscan N.V.). The nomenclature and units were according to the recommendation of the Nomenclature Committee of the American Society for Bone and Mineral Research (31).
RANKL-induced Bone Loss
Five-week-old female mice were administered with a local calvarial injection of RANKL at 2 mg/kg of body weight. After 5 days, osteoclast number per millimeter of trabecular bone surface and the percentage of bone surface covered by osteoclasts (eroded surface) were measured as described (32).
Statistics
Data are expressed as mean ± S.D. from at least three independent experiments. Statistical analyses were performed using the two-tailed Student's t test to analyze differences among groups. p < 0.05 was considered statistically significant.
RESULTS
GSK-3β Is Inactivated upon RANKL Treatment
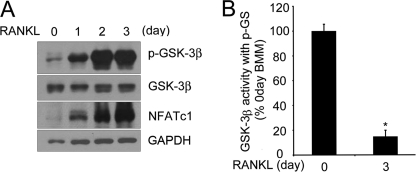
To examine the role of GSK-3β in RANKL-mediated osteoclast differentiation, we first assessed the time course of GSK-3β Ser-9 phosphorylation, which results in inhibition of GSK-3β activity in response to RANKL. Phosphorylation of GSK-3β was increased by RANKL stimulation in BMMs, indicating GSK-3β inactivation (Fig. 1A). Interestingly, the phosphorylation of GSK-3β was highly increased on day 3 after RANKL stimulation without any change in the level of GSK-3β protein during osteoclastogenesis. To confirm that RANKL stimulation in BMMs affects the kinase activity of GSK-3β, the kinase activity was measured using a phospho-glycogen synthase peptide as a primed substrate. Cell lysates from 3 days after RANKL stimulation retained GSK-3β activity less than 20% when compared with the day 0 activity (Fig. 1B). Taken together, GSK-3β was inactivated by RANKL stimulation through phosphorylation.
FIGURE 1.
GSK-3β is inactivated during RANKL-induced osteoclastogenesis. A, phosphorylation of GSK-3β is increased upon RANKL treatment during osteoclastogenesis. Cell lysates were analyzed by Western blotting with anti-phospho-GSK-3β (p-GSK-3β), GSK-3β, or NFATc1 antibody. β-Actin was used as a loading control. B, the kinase activity of GSK-3β upon RANKL treatment was measured using a phospho-glycogen synthase peptide (P-GS). Cell lysates were immunoprecipitated with anti-GSK-3β antibody, and then the activity of GSK-3β was measured as described under “Experimental Procedures.” The activity of each sample was normalized to the amount of GSK-3β. Data represent means ± S.D. *, p < 0.01.
GSK-3β Inhibits Osteoclast Differentiation
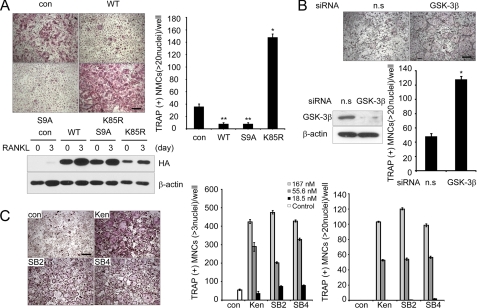
To better understand the role of GSK-3β in osteoclast differentiation, primary BMMs were infected with retroviruses expressing wild-type GSK-3β or its two mutants, constitutively active GSK-3β (GSK3β-S9A) or catalytically inactive GSK-3β (GSK3β-K85R). RANKL stimulation of control vector-infected BMMs increased the number of TRAP+ MNCs, whereas overexpression of wild-type GSK-3β or GSK3β-S9A mutant in BMMs suppressed the formation of TRAP+ MNCs (>20 or >3 nuclei) (Fig. 2A and supplemental Fig. S1). Conversely, BMMs infected with GSK3β-K85R showed a marked increase in the number of TRAP+ MNCs (Fig. 2A). Notably, the formation of large (>100 μm) osteoclasts containing 20 nuclei was greatly increased in BMMs expressing the GSK3β-K85R mutant, suggesting that the terminal differentiation of osteoclasts, characterized by multinucleation, was regulated by GSK-3β activity. We further analyzed the effect of GSK-3β using an siRNA-mediated knockdown experiment. RANKL-induced osteoclast formation was enhanced by knocking down GSK-3β, similar to the BMMs infected with the GSK3β-K85R mutant (Fig. 2B). Taken together, these data suggest that GSK-3β plays a critical role in the RANKL-mediated osteoclast differentiation process.
FIGURE 2.
GSK-3β activity regulates osteoclast formation. A, in vitro differentiation of osteoclasts from BMM cells infected with pMX-puro (con), HA-GSK3β (WT), HA-GSK3β-S9A (S9A), or HA-GSK3β-K85R (K85R), respectively. Because there was a large difference in numbers of large osteoclasts (>20 nuclei) among samples, we counted TRAP+ MNCs containing more than 20 nuclei. Of note, TRAP+ MNCs (more than three nuclei) were counted (supplemental Fig. S1). GSK-3β expression following infection was confirmed by Western blot analysis using the HA antibody and then reprobed with β-actin as a loading control. Data represent means ± S.D. *, p < 0.05, **, p < 0.01. Scale bar, 200 μm. B, effect of siRNA against GSK-3β on RANKL-induced osteoclastogenesis. Nonspecific RNA duplex was used as a negative control. Knockdown of GSK-3β was confirmed by Western blot analysis. Data represent means ± S.D. *, p < 0.01. Scale bar, 200 μm. n.s., not significant. C, effects of the GSK-3 inhibitors kenpaullone (Ken), SB216763 (SB2), and SB415286 (SB4) on osteoclastogenesis. Data represent means ± S.D. Scale bar, 200 μm.
To further confirm the role of GSK-3β in the differentiation of osteoclasts, we investigated the effects of pharmacological inhibition of GSK-3β on osteoclast formation. To augment the positive effect of GSK-3 inhibitors, kenpaullone, SB216763, or SB415286, BMMs were treated with a suboptimal concentration of RANKL (20 ng/ml). Consistently, the inhibitors remarkably enhanced the formation of TRAP+ MNCs (>3 or >20 nuclei) in a dose-dependent manner (Fig. 2C). When BMMs expressing GSK3β-S9A were co-cultured with primary osteoblasts in the presence of 1,25(OH)2D3 and prostaglandin E2, the number of TRAP+ MNCs was reduced when compared with empty vector control, whereas BMMs expressing GSK3β-K85R showed increased TRAP+ MNCs (supplemental Fig. S2). Collectively, these data point to the importance of GSK-3β in the differentiation process.
GSK-3β Blocks NFATc1 Transcription and Ca2+ Oscillation
Because it has been reported that GSK-3β causes phosphorylation and nuclear export of NFATc proteins as proven by overexpression of GSK-3β in COS cells (13), we examined the relevance of GSK-3β and NFATc1 in RANKL-stimulated osteoclast formation. For this, we performed a reporter assay with a luciferase reporter plasmid driven by tandem NFAT binding sites in the RAW264.7 cells stably expressing the GSK3β-S9A mutant. The GSK3β-S9A-expressing cells neither showed the NFAT transcriptional activity nor expressed a high level of NFATc1 protein after RANKL stimulation, whereas control cells effectively elevated transcriptional activity of NFATc1 and showed induction of NFATc1 at 3 day of RANKL treatment (Fig. 3A).
FIGURE 3.
GSK-3β inactivation is required for the induction of NFATc1 and Ca2+ oscillation by RANKL. A, left panel, inhibition of NFATc1 transcriptional activity by GSK-3β. The stable RAW264.7 cells expressing HA-GSK3β-S9A (S9A) or vector control (con) were transfected with an NFATc1 reporter construct, and then the transcriptional activity of NFATc1 was measured. Data represent means ± S.D. *, p < 0.01, **, p < 0.001. Right panel, expression of NFATc1 in RAW264.7 cells stably expressing either empty vector (con) or HA-GSK3β-S9A (S9A). The stable RAW264.7 cells were stimulated with RANKL and analyzed by Western blotting with NFATc1 antibody. B, [Ca2+]i changes were traced in BMMs infected with empty vector (con), GSK-3β K85R (K85R), or GSK3β-S9A (S9A) treated with RANKL and M-CSF for 3 days. Changes were estimated as the ratio of fluorescence intensity of fluo-4 to fura red. Each color indicates a different cell in the same field. Note that Ca2+ oscillations are impaired in BMMs infected with S9A but that BMMs infected with K85R were more sensitive to the Ca2+ oscillation than control cells. Max, maximum.
During osteoclastogenesis, RANKL stimulation induces sustained Ca2+ oscillation, which is necessary for NFATc1 activation and expression. To examine whether GSK-3β could regulate the activation of Ca2+ oscillation induced by RANKL, BMM cells were infected with retroviruses expressing the GSK3β-S9A or GSK3β-K85R mutants. Ca2+ oscillation was severely impaired in the BMMs infected with the GSK3β-S9A mutant when compared with empty vector controls, whereas BMMs infected with the GSK3β-K85R mutant were more sensitive to Ca2+ oscillation than control cells (Fig. 3B). Taken together, these data suggest that regulation of GSK-3β activity may modulate Ca2+ oscillation on RANKL stimulation.
Transgenic Mice Expressing GSK3β-S9A Show Osteopetrotic Phenotype due to Impaired Osteoclast Formation and NFATc1 Down-regulation
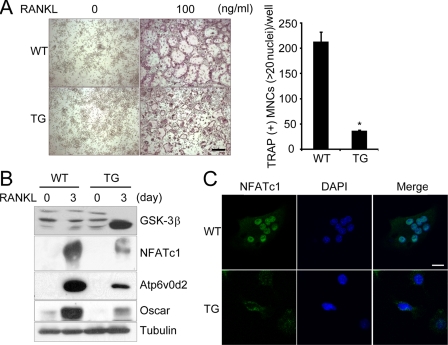
To investigate the in vivo physiological role of GSK-3β, we generated transgenic mice expressing GSK3β-S9A under control of the TRAP gene promoter, an enzyme that is highly expressed in osteoclasts (33) (supplemental Fig. S3, A and B). RT-PCR analysis indicated specific expression of the GSK3β-S9A mutant in the osteoclast lineage from Tg mice (supplemental Fig. S3C). Consistent with the previous findings (Fig. 2), osteoclast formation of BMMs from Tg mice expressing the GSK3β-S9A mutant was markedly lower than that of wild-type littermates (Fig. 4A). Expression of the GSK3β-S9A mutant was effectively induced by RANKL stimulation in Tg mice, which further confirmed successful control by the TRAP promoter in the Tg mice (Fig. 4B). Further, Western blot analysis revealed that NFATc1 protein was decreased 3 days after RANKL stimulation of the BMMs from the Tg mice, and the protein levels of NFAT-dependent genes, Atp6v0d2 and Oscar, were also impaired (Fig. 4B).
FIGURE 4.
Impaired osteoclastogenesis and NFATc1 induction in BMMs of Tg mice expressing GSK3β-S9A mutant. A, BMMs isolated from GSK3β-S9A Tg (TG) mice or wild-type littermates (WT) were cultured for 4 days in the presence of M-CSF and RANKL as indicated. A representative TRAP staining is shown in the left panel, and TRAP+ MNCs were counted in the right panel. Data represent mean ± S.D. *, p < 0.01. Scale bar, 100 μm. B, overexpression of GSK3β-S9A in mice down-regulates induction of NFATc1 itself and the NFAT-dependent genes, Atp6v0d2 and Oscar. BMMs isolated from TG mice and WT littermates were cultured for 3 days with or without RANKL. Cell lysates were analyzed by Western blotting using the indicated antibodies specific for GSK-3α/β, NFATc1, Atp6v0d2, and Oscar. C, immunofluorescent images of NFATc1 protein in BMMs isolated from TG mice and WT littermates. BMM cells were stimulated with RANKL and M-CSF for 3 days and analyzed for the subcellular localization of NFATc1 protein as described under “Experimental Procedures.” Nuclei were stained with DAPI. Scale bar, 10 μm.
We questioned whether the impairment of osteoclast formation and the defect in NFAT expression in the Tg mice may arise from NFATc1 phosphorylation by GSK-3β, thereby causing nuclear export of NFATc1. To answer this question, BMMs from Tg mice and wild-type littermates were incubated with M-CSF and RANKL for 3 days and then analyzed by immunofluorescence staining. Consistent with the Western blot data, NFATc1 immunoreactivity was barely detectable in the pre-osteoclasts from Tg mice, and further, most NFATc1 remained in the cytosol (Fig. 4C). In contrast, NFATc1 immunoreactivity in osteoclasts derived from wild-type littermates was readily detectable in the nuclei, but not the cytosol of multinucleated cells (Fig. 4C). Together, these results indicate that overexpression of GSK3β-S9A in Tg mice down-regulates NFATc1 expression and regulates nuclear localization of NFATc1, thereby suppressing osteoclast formation.
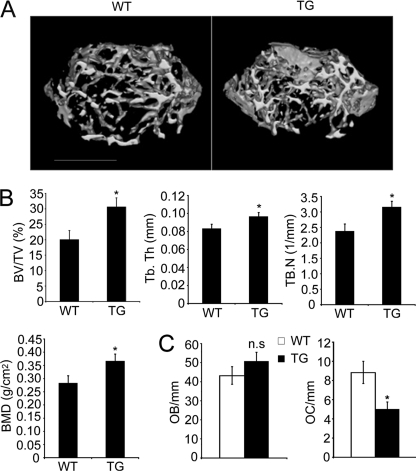
To evaluate the role of GSK-3β in vivo, we analyzed the bone phenotype of Tg mice by microcomputed tomography and histological analysis. Microcomputed tomography analysis of distal femur metaphyses revealed a significant increase in bone volume due to increased trabecular thickness and number in the Tg mice (Fig. 5, A and B). Histomorphometric analysis indicated an increase in bone volume associated with a decrease in osteoclast number, but no significant difference in the number of osteoblasts (Fig. 5C).
FIGURE 5.
Osteopetrotic phenotype of Tg mice expressing GSK3β-S9A mutant. A, three-dimensional microstructural analysis of the femurs of wild-type (WT) and Tg mice (TG) by microcomputed tomography. B, histograms represent the three-dimensional trabecular structural parameters in femurs: bone volume fraction (BV/TV), trabecular thickness (Tb.Th), trabecular number (Tb.N), and bone mineral densities (BMD). C, quantification of osteoclasts and osteoblasts form histological analysis of long bone of WT and Tg mice. OC/mm, osteoclast number per bone surface; OB/mm, osteoblast number per bone surface. Data represent means ± S.D. n = 6. *, p < 0.05. n.s., not significant.
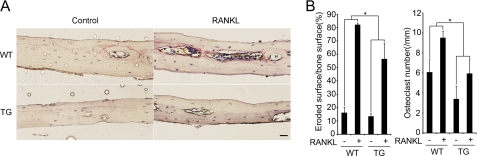
To investigate the role of GSK-3β in a RANKL-induced model of bone destruction, we treated Tg mice expressing the GSK3β-S9A mutant that were injected with RANKL or PBS (Control). The extent of bone erosion was notably reduced in the Tg mice, and the formation of TRAP+ MNCs was greatly suppressed (Fig. 6, A and B), suggesting that overexpression of GSK3β-S9A in Tg mice can inhibit RANKL-induced bone destruction. Thus, these results demonstrate that overexpression of GSK3β-S9A in Tg mice results in an osteopetrotic phenotype due to a defect in osteoclast formation.
FIGURE 6.
Protective phenotype of Tg mice expressing GSK3β-S9A mutant. The role of GSK-3β in RANKL-induced bone destruction in wild-type (WT) mice and Tg (TG) mice was studied. A, histology of the calvarial bone injected with PBS (Control) or RANKL in WT and Tg mice (TRAP and hematoxylin staining). Scale bar, 0.02 mm. B, eroded surface (left) and the number of osteoclasts (right) were analyzed. Data represent means ± S.D. n = 5. *, p < 0.05.
DISCUSSION
GSK-3β is a multitasking kinase that plays central roles in a diverse range of signaling pathways, including Wnts, hedgehog, growth factors, cytokines, and G protein-coupled ligands (34, 35). Specifically, in osteoblasts, it has been shown that GSK-3β acts as a negative regulator of the Wnt/β-catenin signaling pathways required for osteoblast differentiation (1, 36, 37). However, the role of GSK-3β in RANKL-induced osteoclast formation is unknown. Here, we demonstrated for the first time a physiological role for GSK-3β in osteoclast biology. Our findings through in vitro and in vivo analysis suggest a novel role for GSK-3β as a negative regulator of NFATc1 in RANKL-induced differentiation of monocytes/macrophages into osteoclasts.
Unlike other kinases, GSK-3β is constitutively active and is inactivated in response to cellular signal (34). We found that GSK-3β protein was expressed in osteoclast precursors and that the level of GSK-3β remained constant during the course of osteoclastogenesis. Remarkably, 3 days after RANKL treatment, GSK-3β was highly phosphorylated at Ser-9, causing its inactivation. We confirmed that the kinase activity was decreased at day 3 of RANKL treatment. It is likely that RANKL-induced GSK-3β inactivation is involved in osteoclast formation. Our findings in primary BMMs support this possibility in that overexpression of catalytically inactive GSK-3β mutant, pharmacological inhibition of GSK-3β, or RNA interference of GSK-3β accelerated efficient formation of osteoclasts. Conversely, overexpression of constitutively active GSK-3β mutant markedly abolished formation of osteoclasts. However, overexpression of GSK-3α in BMMs had no effect on RANKL-mediated osteoclastogenesis (supplemental Fig. S4). These findings suggest that kinase activity of GSK-3β is critical for suppression of osteoclast formation.
It is notable that increase in NFATc1 protein level follows GSK-3β phosphorylation. Because NFATc1 represents a master switch that regulates terminal differentiation of osteoclasts downstream of RANKL, induction of NFATc1 by RANKL is crucial for optimal differentiation of osteoclasts (7). In this context, it is likely that GSK-3β inactivation is a prerequisite for the NFATc1 amplification process. A previous study showed that GSK-3β causes phosphorylation and nuclear export of NFAT proteins in vitro (13), but the physiological importance of NFATc1 phosphorylation by GSK-3β has not yet been clarified. We found that overexpression of constitutively active GSK-3β showed a defect in NFATc1 expression and its transcriptional activity. Consistent with the finding from in vitro analysis, the formation of TRAP+ MNCs induced by RANKL was markedly inhibited in the Tg mice expressing a constitutively active GSK-3β mutant. Protein levels of NFATc1 as well as NFATc1-dependent genes such as Atp6v0d2 and Oscar induced by RANKL in the Tg mice were also decreased at 3 days after RANKL stimulation. More importantly, RANKL-dependent nuclear localization of NFATc1 was severely impaired in the Tg mice. Based on our data, we propose that GSK-3β may promote nuclear export of NFATc1 through phosphorylation, thereby resulting in down-regulation of NAFTc1 induction.
Interestingly, our results also showed that overexpression of constitutively active GSK-3β in osteoclast precursors decreased Ca2+ oscillation. In contrast, catalytically inactive GSK-3β promoted Ca2+ oscillation. It is well known that NFATc1 is activated by Ca2+ signaling induced by the activation of the immunoglobulin-like receptor signaling associated with immunoreceptor tyrosine-based activation motif-harboring adaptors, which is known to provide co-stimulatory signaling (38). The long lasting Ca2+ oscillation, which is evident during osteoclastogenesis, may ensure the robust induction of NFATc1 through an autoamplification mechanism (10). We currently have no evidence that GSK-3β directly regulates Ca2+ signaling cascades through its kinase activity. However, we cannot exclude the possibility that GSK-3β may directly regulate RANKL- or co-stimulatory receptor-induced Ca2+ signaling cascades by unidentified mechanism(s). Otherwise, it seems likely that the defect in robust induction of NFATc1 by overexpression of the constitutively active GSK-3β may affect Ca2+ oscillation. Specifically, our data showed that the protein level of OSCAR was markedly decreased in RANKL-stimulated pre-osteoclasts from Tg mice. Consistent with this finding, it was reported that the OSCAR-FcRγ (Fc receptor common γ subunit) complex regulates RANKL-mediated activation of Ca2+ signaling in pre-osteoclasts, which leads to the amplification of NFATc1 (38). Further studies will be required to elucidate mechanism(s) of how GSK-3β regulates Ca2+ oscillation.
Given the multiple processes regulated by GSK-3β, GSK-3β has been implicated in various diseases such as diabetes, cancer, bipolar disorder, and Alzheimer disease, and GSK-3β inhibitors are being actively developed as drugs for the treatment of the various disorders (19, 39). Moreover, because GSK-3β is a negative regulator of Wnt-β-catenin signaling that is known to stimulate bone formation, modulating its activity with small molecules is a promising strategy for increasing bone mass (36). However, it is important to note that GSK-3β is not a specialized repressor of β-catenin as it can participate in RANKL signaling as shown in our study. In the context of skeletal health, given the potential risk of long term inhibition of GSK-3β, a more comprehensive study with regard to targeting of drugs to the GSK-3β for the treatment of chronic disorders such as osteoporosis should be performed using animal models followed by human case study. Moreover, further mechanistic studies will be needed to understand the pathophysiological relevance of GSK-3β in bone remodeling and its disorders.
Supplementary Material
Acknowledgments
We thank T. Kitamura for providing PLAT-E cells and Y. Choi for the anti-Atp6v0d2 antibody.
This work was supported by National Research Foundation of Korea (NRF) grants funded by the Korea Government (Ministry of Education, Science, and Technology; Grants R0A-2008-000-20001-0; R31-2008-000-10010-0; 2011-0006244; and 2010-0020577).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- RANKL
- receptor activator of NF-κB ligand
- GSK-3β
- glycogen synthase kinase-3β
- NFAT
- nuclear factor of activated T-cells
- BMM
- bone marrow macrophage
- TRAP
- tartrate-resistant acid phosphatase
- MNC
- multinucleated cells
- Tg
- transgenic.
REFERENCES
- 1. Harada S., Rodan G. A. (2003) Nature 423, 349–355 [DOI] [PubMed] [Google Scholar]
- 2. Teitelbaum S. L. (2000) Science 289, 1504–1508 [DOI] [PubMed] [Google Scholar]
- 3. Boyle W. J., Simonet W. S., Lacey D. L. (2003) Nature 423, 337–342 [DOI] [PubMed] [Google Scholar]
- 4. Suda T., Takahashi N., Udagawa N., Jimi E., Gillespie M. T., Martin T. J. (1999) Endocr. Rev. 20, 345–357 [DOI] [PubMed] [Google Scholar]
- 5. Wong B. R., Josien R., Lee S. Y., Vologodskaia M., Steinman R. M., Choi Y. (1998) J. Biol. Chem. 273, 28355–28359 [DOI] [PubMed] [Google Scholar]
- 6. Asagiri M., Takayanagi H. (2007) Bone 40, 251–264 [DOI] [PubMed] [Google Scholar]
- 7. Asagiri M., Sato K., Usami T., Ochi S., Nishina H., Yoshida H., Morita I., Wagner E. F., Mak T. W., Serfling E., Takayanagi H. (2005) J. Exp. Med. 202, 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wagner E. F., Eferl R. (2005) Immunol. Rev. 208, 126–140 [DOI] [PubMed] [Google Scholar]
- 9. Crabtree G. R. (1999) Cell 96, 611–614 [DOI] [PubMed] [Google Scholar]
- 10. Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., Taniguchi T. (2002) Dev. Cell 3, 889–901 [DOI] [PubMed] [Google Scholar]
- 11. Crabtree G. R., Olson E. N. (2002) Cell 109, (suppl.) S67–S79 [DOI] [PubMed] [Google Scholar]
- 12. Hogan P. G., Chen L., Nardone J., Rao A. (2003) Genes Dev. 17, 2205–2232 [DOI] [PubMed] [Google Scholar]
- 13. Beals C. R., Sheridan C. M., Turck C. W., Gardner P., Crabtree G. R. (1997) Science 275, 1930–1934 [DOI] [PubMed] [Google Scholar]
- 14. Okamura H., Garcia-Rodriguez C., Martinson H., Qin J., Virshup D. M., Rao A. (2004) Mol. Cell. Biol. 24, 4184–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang T. T., Xiong Q., Enslen H., Davis R. J., Chow C. W. (2002) Mol. Cell. Biol. 22, 3892–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chow C. W., Rincón M., Cavanagh J., Dickens M., Davis R. J. (1997) Science 278, 1638–1641 [DOI] [PubMed] [Google Scholar]
- 17. Woodgett J. R. (1990) EMBO J. 9, 2431–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frame S., Cohen P. (2001) Biochem. J. 359, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sugden P. H., Fuller S. J., Weiss S. C., Clerk A. (2008) Br. J. Pharmacol. 153, Suppl. 1, S137–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Logan C. Y., Nusse R. (2004) Annu. Rev. Cell Dev. Biol. 20, 781–810 [DOI] [PubMed] [Google Scholar]
- 21. Jope R. S., Johnson G. V. (2004) Trends Biochem. Sci. 29, 95–102 [DOI] [PubMed] [Google Scholar]
- 22. Frame S., Cohen P., Biondi R. M. (2001) Mol. Cell 7, 1321–1327 [DOI] [PubMed] [Google Scholar]
- 23. Hoeflich K. P., Luo J., Rubie E. A., Tsao M. S., Jin O., Woodgett J. R. (2000) Nature 406, 86–90 [DOI] [PubMed] [Google Scholar]
- 24. Lee S. H., Rho J., Jeong D., Sul J. Y., Kim T., Kim N., Kang J. S., Miyamoto T., Suda T., Lee S. K., Pignolo R. J., Koczon-Jaremko B., Lorenzo J., Choi Y. (2006) Nat. Med. 12, 1403–1409 [DOI] [PubMed] [Google Scholar]
- 25. Yoon K., Jung E. J., Lee S. R., Kim J., Choi Y., Lee S. Y. (2008) Cell Death Differ. 15, 730–738 [DOI] [PubMed] [Google Scholar]
- 26. Suda T., Jimi E., Nakamura I., Takahashi N. (1997) Methods Enzymol. 282, 223–235 [DOI] [PubMed] [Google Scholar]
- 27. Lee S. E., Woo K. M., Kim S. Y., Kim H. M., Kwack K., Lee Z. H., Kim H. H. (2002) Bone 30, 71–77 [DOI] [PubMed] [Google Scholar]
- 28. Kim J. H., Kim K., Youn B. U., Jin H. M., Kim N. (2010) Cell. Signal. 22, 1341–1349 [DOI] [PubMed] [Google Scholar]
- 29. Reddy S. V., Hundley J. E., Windle J. J., Alcantara O., Linn R., Leach R. J., Boldt D. H., Roodman G. D. (1995) J. Bone Miner. Res. 10, 601–606 [DOI] [PubMed] [Google Scholar]
- 30. Schwartzberg P. L., Xing L., Hoffmann O., Lowell C. A., Garrett L., Boyce B. F., Varmus H. E. (1997) Genes Dev. 11, 2835–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parfitt A. M., Drezner M. K., Glorieux F. H., Kanis J. A., Malluche H., Meunier P. J., Ott S. M., Recker R. R. (1987) J. Bone Miner. Res. 2, 595–610 [DOI] [PubMed] [Google Scholar]
- 32. Takayanagi H., Ogasawara K., Hida S., Chiba T., Murata S., Sato K., Takaoka A., Yokochi T., Oda H., Tanaka K., Nakamura K., Taniguchi T. (2000) Nature 408, 600–605 [DOI] [PubMed] [Google Scholar]
- 33. Hikata T., Takaishi H., Takito J., Hakozaki A., Furukawa M., Uchikawa S., Kimura T., Okada Y., Matsumoto M., Yoshimura A., Nishimura R., Reddy S. V., Asahara H., Toyama Y. (2009) Blood 113, 2202–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Doble B. W., Woodgett J. R. (2003) J. Cell Sci. 116, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu D., Pan W. (2010) Trends Biochem. Sci. 35, 161–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bodine P. V., Komm B. S. (2006) Rev. Endocr. Metab. Disord. 7, 33–39 [DOI] [PubMed] [Google Scholar]
- 37. Clément-Lacroix P., Ai M., Morvan F., Roman-Roman S., Vayssière B., Belleville C., Estrera K., Warman M. L., Baron R., Rawadi G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17406–17411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koga T., Inui M., Inoue K., Kim S., Suematsu A., Kobayashi E., Iwata T., Ohnishi H., Matozaki T., Kodama T., Taniguchi T., Takayanagi H., Takai T. (2004) Nature 428, 758–763 [DOI] [PubMed] [Google Scholar]
- 39. Cohen P., Goedert M. (2004) Nat. Rev. Drug Discov. 3, 479–487 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.