Background: The mechanism underlying sperm flagellar development is unknown.
Results: Mammalian MARCH10 is a microtubule-associated E3 ubiquitin ligase expressed in the tail of developing spermatids.
Conclusion: MARCH10 is suggested to have a role in the organization and maintenance of the sperm flagella.
Significance: This is additional evidence for the involvement of the ubiquitin-proteasome system in mammalian spermatogenesis.
Keywords: Cytoskeleton, E3 Ubiquitin Ligase, Microtubules, Spermatogenesis, Ubiquitination, Three-dimensional SIM, Membrane-associated RING-CH, Microtubule Association, Sperm Flagella
Abstract
Spermiogenesis is a complex and dynamic process of the metamorphosis of spermatids into spermatozoa. There is a great deal that is still unknown regarding the regulatory mechanisms for the formation of the sperm flagellum. In this study, we determined that the membrane-associated RING-CH 10 (March10) gene is predominantly expressed in rat testis. We isolated two March10 isoforms encoding MARCH10a and MARCH10b, which are generated by alternative splicing. MARCH10a is a long RING finger protein, and MARCH10b is a short RING finger-less protein. Immunohistochemical staining revealed that the MARCH10 proteins are specifically expressed in elongating and elongated spermatids, and the expression is absent in epididymal spermatozoa. MARCH10 immunoreactivity was observed in the cytoplasmic lobes as well as the principal piece and annulus of the flagella. When overexpressed in COS7 cells, MARCH10a was localized along the microtubules, whereas MARCH10b was distributed throughout the cytoplasm. An in vitro microtubule cosedimentation assay showed that MARCH10a is directly associated with microtubules. An in vitro ubiquitination assay demonstrated that the RING finger domain of MARCH10a exhibits an E3 ubiquitin ligase activity along with the E2 ubiquitin-conjugating enzyme UBE2B. Moreover, MARCH10a undergoes proteasomal degradation by autoubiquitination in transfected COS7 cells, but this activity was abolished upon microtubule disassembly. These results suggest that MARCH10 is involved in spermiogenesis by regulating the formation and maintenance of the flagella in developing spermatids.
Introduction
Mammalian spermatozoa are composed of two major parts, the head and the flagellum. Proper assembly of the flagellum is essential for sperm motility and fertilization. Morphologically, the flagellum is subdivided into the midpiece, the principal piece, and the end piece (1, 2). Common to the three parts is the central axoneme, which has a 9 + 2 arrangement of microtubules (9 doublets and 2 central singlets). The force for flagellar motility is driven by the active sliding of the doublet microtubules powered by dynein ATPase motors. In the midpiece, the axoneme is surrounded by nine longitudinal cytoskeletal structures, the outer dense fibers (ODF),2 on the outside of the doublet microtubules. The ODF are tightly wrapped, with helically arranged mitochondria (the mitochondrial sheath) that serve as the sites of the ATP production required for flagellar movement. In the principal piece, the ODF are extended, but two of them are replaced by the longitudinal columns of the fibrous sheath (FS) that are bridged by numerous circumferential ribs. The ODF and FS have been proposed to provide the necessary stiffness and elastic recoil for the flagellum, thereby modulating its beating pattern. The end piece is a very short terminal segment, in which the axoneme is surrounded only by the plasma membrane. Although a number of the components of the flagellum have been identified and characterized (3–5), the molecular mechanisms for the formation of the flagella remain to be established.
Ubiquitination is a post-translational modification in which the 76-amino acid polypeptide ubiquitin (Ub) is covalently attached to lysine residues in target proteins. Ubiquitination is catalyzed by the sequential actions of E1 Ub-activating, E2 Ub-conjugating, and E3 Ub ligase enzymes. The E3 enzymes are essential for specific substrate recognition. Ubiquitination is not only required for proteasomal degradation but is also involved in proteasome-independent functions such as protein localization and signaling (6). Recent investigations have suggested the involvement of the Ub-proteasome system in flagellar assembly and function as follows. 1) Sperm mitochondria are highly ubiquitinated and thereby undergo degradation after fertilization, which enables maternal mitochondrial inheritance (7–9). 2) Spermatozoa in mice lacking the E2 enzyme Ube2b or the E3 Ub ligase Herc4 display morphological abnormalities and impaired flagellum motility (10–13). 3) PSMC3 (also known as TBP-1), a subunit of the 26 S proteasome, is present in the ODF of elongating spermatids and spermatozoa (14–16). Therefore, the identification and characterization of the flagellar Ub-proteasome system should provide novel insight into molecular mechanisms regulating the assembly and function of the mammalian sperm flagella.
The membrane-associated RING-CH (MARCH) family is a RING finger protein family of E3 Ub ligases, consisting of 11 members in mammals (17–19). Nine MARCH members (i.e. MARCH1–6, -8, -9, and -11) contain hydrophobic transmembrane spans and are localized to the plasma membrane and intracellular organelle membrane (20). Transmembrane MARCH proteins mediate the ubiquitination and subsequent down-regulation of cell-surface immune regulatory molecules, such as major histocompatibility complex class II and CD86 (17, 18, 21). Other proposed functions include endoplasmic reticulum-associated degradation (22), endosomal protein trafficking (23, 24), mitochondrial dynamics (25, 26), and spermatogenesis (19). MARCH7 (also known as Axotrophin) and MARCH10 are predicted to have no transmembrane spanning region. Indeed, MARCH7 has been shown to localize to the cytosol and nucleus in transfected cultured cells (27). Studies of March7-null mice have suggested that MARCH7 has important roles in T-cell proliferation and immune tolerance (28, 29). However, the cellular localization and function of MARCH10 remain to be determined.
In this study, we identified and characterized rat MARCH10 as a novel E3 Ub ligase of developing spermatids. We show that MARCH10 is localized to the principal piece of elongating spermatids and is associated with microtubules, which is important for its E3 activity. These results suggest that MARCH10 may play a role in the formation of the sperm flagellum.
EXPERIMENTAL PROCEDURES
Animals
Male Wistar rats and Japanese White rabbits were purchased from Tokyo Laboratory Animals (Tokyo, Japan). The animal protocols and procedures were approved by the Institutional Animal Care and Use Committee of the Tokyo Institute of Technology (Yokohama, Japan).
Plasmids
The rat March10a cDNA was amplified from rat testis by reverse transcription (RT)-PCR with the primers 5′-atcgatgttgcatgaagcaagggacaggca-3′ and 5′-atcctagatgacccggcctgggtaaacgtt-3′ and then inserted into the EcoRV site of pBluescript II SK− (Stratagene, La Jolla, CA) yielding pBS-March10a. To generate FLAG-MARCH10a and enhanced green fluorescent protein (EGFP)-MARCH10a, the EcoRV fragment of pBS-March10a was inserted into the EcoRV site of p3× FLAGCMV-10 (Sigma) and into the blunt-ended EcoRI site of pEGFP-C2 (Clontech), respectively. FLAG-RINGmut was generated by introducing C641S and C644S point mutations into the FLAG-MARCH10a plasmid by site-directed mutagenesis with the primers 5′-gggagacttgtctcggatctctcagatagc-3′ and 5′-gctatctgagagatccgagacaagtctccc-3′. To generate FLAG-MARCH10b, a cDNA encoding MARCH10b was amplified from rat testis by RT-PCR with the primers 5′-atcgatgttgcatgaagcaagggacaggc-3′ and 5′-atcttaccacatgaggtaaattttactgg-3′ and then inserted into the EcoRV site of p3× FLAGCMV-10. To generate deletion mutants of EGFP fusion constructs containing the residues 1–127, 1–370, 1–536, and 1–703 of MARCH10a, the following pairs of oligonucleotides were annealed and then inserted into the EGFP-MARCH10a plasmid digested with PstI, SmaI, SpeI, and SalI, respectively: 5′-gtgtgatatcggcagatcag-3′ and 5′-gatcctgatctgccgatatcacactgca-3′ for residues 1–127; 5′-gggtgatatcggcagatcag-3′ and 5′-gatcctgatctgccgatatcaccc-3′ for residues 1–370; 5′-ctagttgatatcggcagatcag-3′ and 5′-gatcctgatctgccgatatcaa-3′ for residues 1–536; and 5′-tcgactgatatcggcagatcag-3′ and 5′-gatcctgatctgccgatatcag-3′ for residues 1–703. To generate EGFP fusion constructs containing residues 482–790 of MARCH10a, the EGFP-MARCH10a plasmid was digested with HindIII and then self-ligated. Prokaryote expression plasmids for glutathione S-transferase (GST) fusion proteins containing residues 1–900 (GST-MAR10N) and 900–1701 (GST-MAR10M) were constructed by cloning the corresponding cDNA fragments into the BamHI and EcoRI sites of pGex4T2 (GE Healthcare). GST-RINGmut was generated by site-directed mutagenesis as described above. His6-tagged rat UBE2B was constructed by inserting the corresponding cDNA fragment into the BamHI and XhoI sites of pRSETA (Invitrogen). The cDNA sequences of all of the constructs were verified by sequencing.
Northern Blot Analysis
Northen blot analysis was carried out as described previously (30). The EcoRV-digested fragment of pBS-March10a was used as a probe, as shown in Fig. 1B. The cDNA fragments encompassing either exons 1–3, 4–5, 6–10, or exon 4′ were amplified by PCR and then used as probes, as shown in Fig. 1C.
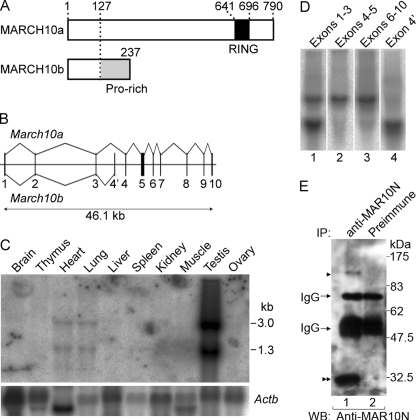
FIGURE 1.
Structural features and tissue expression of rat MARCH10a and MARCH10b. A, schematic representations of MARCH10a and MARCH10b protein structures. The RING finger domain and the proline-rich region are indicated in black and gray, respectively. B, schematic representation of alternative splicing of the rat March10 gene. Exons are indicated by the vertical lines and boxes and are numbered. The introns and flanking regions are indicated by horizontal lines. March10b is generated through alternative usage of exon 4′. C, Northern blot of 20 μg of total RNA prepared from the indicated rat tissues and that was hybridized with specific probes for March10 (top) and Actb (bottom). D, Northern blots of 20 μg of total RNA prepared from rat testis were hybridized with specific probes for exons 1–3 (lane 1), exons 4–5 (lane 2), exons 6–10 (lane 3), and exon 4′ (lane 4). E, lysates of rat testis (400 μg of protein) were immunoprecipitated (IP) with anti-MAR10N (lane 1) or preimmune serum (lane 2). The immunoprecipitates were analyzed by Western blotting (WB) with anti-MAR10N antiserum. Arrowhead and double arrowhead indicate the bands corresponding to MARCH10a and MARCH10b, respectively.
Preparation of Recombinant Proteins and Antibodies
Preparation of the recombinant proteins and the immunization protocol for the production of rabbit antisera were performed as described previously (19). Polyclonal anti-MAR10N (808) and anti-MAR10M (810) antisera were obtained by immunizing rabbits with GST-MAR10N and GST-MAR10M, respectively.
In Vitro Ubiquitination Assay
In vitro ubiquitination assays were performed with GST-RING or GST-RINGmut (1 μg) as described previously (19), with the exception that the reaction mixtures were incubated for 24 h.
Cell Culture and Fluorescence Microscopy
Maintenance of COS7 cells, transfection with plasmids, and immunofluorescence staining were performed as described previously (23).
Immunoprecipitation
Immunoprecipitation of endogenous MARCH10 proteins from the testis was performed as described previously (19). In Fig. 9B, COS7 cells were transfected with plasmids encoding hemagglutinin (HA)-tagged Ub (0.5 μg) alone or together with FLAG-MARCH10a (1 μg). After incubation with or without 20 μm nocodazole for 12 h at 37 °C, the cells were lysed in PBS containing 1% Triton X-100 and protease inhibitors (10 mm leupeptin, 1 mm pepstatin, 5 mg/ml aprotinin, and 1 mm PMSF). The lysates were incubated with 15 μl of anti-FLAG antibody beads (Sigma) overnight at 4 °C. After extensive washing with PBS containing 1% Triton X-100, the bound materials were eluted with 25 μl of 0.1 m glycine-HCl, pH 2.5, and 1% Nonidet P-40.
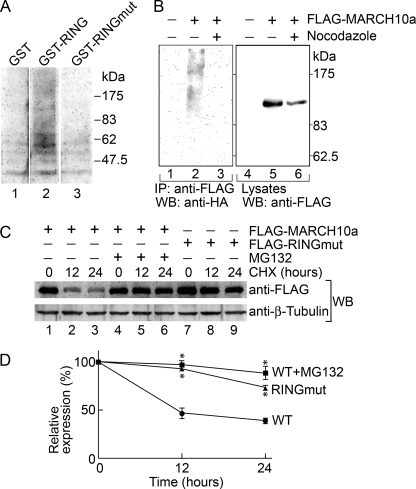
FIGURE 9.
Characterization of E3 activity of MARCH10a. A, GST (lane 1), GST-RING (lane 2), and GST-RINGmut (lane 3) were incubated in reaction mixtures containing Ub, E1, and His6-UBE2B for 24 h at 30 °C. Ubiquitinated materials were analyzed by Western blotting with anti-Ub antibody. B, COS7 cells were transfected with HA-tagged Ub alone (lanes 1 and 4) or together with FLAG-MARCH10a (lanes 2, 3, 5, and 6). The cells were incubated in the absence (lanes 1, 2, 4, and 5) or presence (lanes 3 and 6) of nocodazole for 12 h. To the cells were added 25 μm MG132, and they were then further incubated for 3 h. Whole cell lysates (300 μg of proteins) were then subjected to immunoprecipitation (IP) with anti-FLAG antibody beads. The immunoprecipitates (lanes 1–3) and cell lysates (5 μg of proteins; lanes 4–6) were analyzed by Western blotting (WB) with anti-HA and anti-FLAG antibodies, respectively. C, COS7 cells transfected with FLAG-MARCH10a (lanes 1–6) or FLAG-RINGmut (lanes 7–9) were treated with 1 mg/ml CHX in the presence (lanes 4–6) or absence (lanes 1–3 and 7–9) of 5 μm MG132. At the indicated time points, the cells were lysed, and the lysates (5 μg of proteins) were analyzed by Western blotting (WB) with anti-FLAG and anti-β-tubulin antibodies. D, graph shows the relative expression levels of FLAG-MARCH10a (WT) and FLAG-RINGmut (RINGmut) at 0–24 h post-CHX treatments. Data are expressed as the mean ± S.E. of three independent experiments. *, p < 0.002 compared with the levels of FLAG-MARCH10a (filled circle) at the same time points.
Cycloheximide (CHX) Chase Assay
COS7 cells transfected with either FLAG-MARCH10a or FLAG-RINGmut were treated for different times with 1 mg/ml CHX in the presence or absence of 5 μm MG132. Whole cell lysates (5 μg of proteins) were subjected to Western blotting with anti-FLAG antibody and with anti-β-tubulin. The band intensity was quantified using ImageJ software (rsbweb.nih.gov), and a graph was created using GraphPad Prism (GraphPad Software, San Diego). Student's t test was performed to evaluate statistical differences.
Immunohistochemistry
Preparation and staining of cryosections (31) and smear samples (32) of rat testis and epididymis were performed according to the previously reported methods with the exception that rats were perfused with PBS containing 2% paraformaldehyde. Dilutions of anti-MARCH10 antisera and preimmune serum were 1:500. The testicular sections were observed under Axiovert 200 M inverted fluorescence microscope (Carl Zeiss, Thornwood, NY) or Leica TCS SPE confocal microscope (Leica Microsystems, Wetzlar, Germany). For super resolution imaging, the immunostained smear samples were imaged with a DeltaVision Optical Microscope eXperimental (OMX; Applied Precision, Issaquah, WA). Three-dimensional reconstructions were performed from 23 z-stack images at intervals of 0.125 μm using softWoRx Explorer 2.0 software (Applied Precision).
Microtubule Cosedimentation Assay
MARCH10a was synthesized by in vitro transcription and translation using the TnT Quick-Coupled reticulocyte lysate system (Promega, Madison, WI) according to the manufacturer's instructions. After the reaction, the lysate (20 μl) was added to 180 μl of PEM buffer (80 mm PIPES-KOH, pH 6.9, 1 mm EGTA, and 2 mm MgCl2) and then cleared of insoluble materials by centrifugation for 20 min at 50,000 rpm on a TLA100.3 rotor (Beckman Coulter, Fullerton, CA). Ninety microliters of the resulting supernatant or PEM buffer containing 10 μg of bovine serum albumin (BSA) were supplemented with purified tubulin (Cytoskeleton, Denver, CO), GTP, Taxol (Sigma), and glycerol to final concentrations of 0.3 mg/ml, 1 mm, 30 μm, and 5%, respectively. The samples (100 μl) were layered on 900 μl of 1 m sucrose in PEM buffer containing 0.1 mm GTP and 1 μm Taxol in a 1.5-ml microcentrifuge tube (Beckman Coulter), and were then incubated for 30 min at 37 °C. The polymerized microtubules were separated by centrifugation at 50,000 rpm on a TLA100.3 rotor for 30 min at 25 °C. The resulting supernatants (100 μl) and pellets (dissolved in 100 μl of Laemmli buffer) were subjected to Coomassie Brilliant Blue staining or Western blot analysis. The control experiments were performed in the absence of GTP and Taxol.
RESULTS
Expression of Two March10 Isoforms in the Rat Testis
The nucleotide sequences of mammalian March10 were found in the GenBankTM and Ensembl databases. Based on the sequence information, we designed a set of primers and obtained the entire open reading frame (ORF) of rat March10 by RT-PCR amplification from testis cDNA. The ORF encodes a protein of 790 amino acids with no characteristic structure other than the C-terminal RING finger domain (accession number AB552845; Fig. 1A). The overall structural features of MARCH10 resemble those of MARCH7 (27), but sequence homology is only present in the RING finger domains (64% identity). Data base searches indicated that the March10 gene is limited to mammals, whereas the March7 gene is present in vertebrates ranging from fish to humans. These facts suggest that MARCH10 may have a distinct function from that of MARCH7. By utilizing the Ensemble exon definitions, there are at least two March10 isoforms in the rat (designated here as March10a and March10b). March10a is identical to the isolated March10 sequence that is comprised of 10 exons (Ensemble transcript ID ENSRNOT0000009316; Fig. 1B). March10b is a shorter transcript that shares the first three exons but has a different exon between exons 3 and 4 (exon 4′; Ensemble transcript ID ENSRNOT00000055136; Fig. 1B). This variant transcript encodes a 237-amino acid protein, in which the first 126 amino acid residues are identical to MARCH10a, but the following 111 residues are a nonrelated proline-rich (accession number, AB552846; Fig. 1A).
To determine the tissue expression pattern of rat March10 mRNA, Northern blot analysis of rat tissue total RNA was performed using the March10 ORF as a probe. Two mRNA species of 1.3 and 3.0 kb were detected predominantly in the testis and to a much lesser extent in the heart and lung (Fig. 1C). We expected that the upper and lower bands correspond to March10a and March10b, respectively. To verify the identity of the two bands, Northern blot analysis of rat testis was performed with four different March10 cDNA probes for exons 1–3, 4–5, 6–10, or 4′. As shown in Fig. 1D, the two bands were detected again with the common exon probe (exons 1–3; lane 1). The 3.0-kb band was detected with the March10a-specific probes (exons 4–5 and 6–10; lanes 2 and 3), whereas the 1.3-kb band was detected with the March10b-specific probe (exon 4′; lane 4). These results indicate that at least two March10 isoforms are generated by alternative splicing and are predominantly expressed in the rat testis.
Protein Expression of MARCH10 in Testis
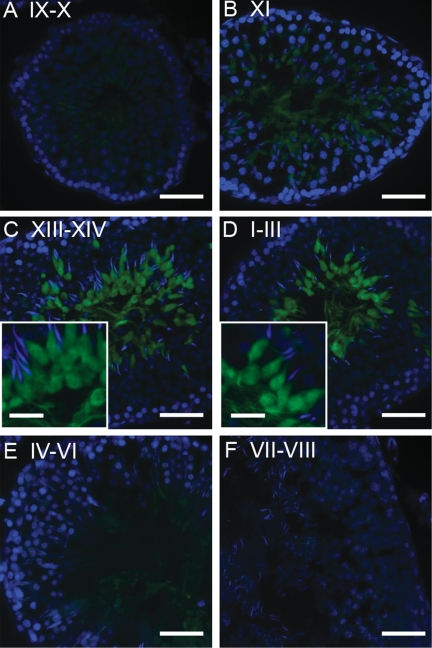
To analyze MARCH10 protein expression in the testis, we prepared rabbit polyclonal antisera as follows: one against the N-terminal region common to both MARCH10a and MARCH10b (anti-MAR10N) and the other against the middle portion of MARCH10a (anti-MAR10M). We were unable to produce antiserum specific to MARCH10b. To confirm their specificity, COS7 cells were transfected with either FLAG-tagged MARCH10a or MARCH10b and were then doubly stained with each of the anti-MARCH10 antisera and anti-FLAG antibodies. Anti-MAR10N antiserum detected both constructs, whereas anti-MAR10M antiserum recognized only FLAG-MARCH10a (supplemental Fig. S1). FLAG-MARCH10a staining showed a filamentous pattern within the cytoplasm in contrast to a diffuse cytoplasmic distribution of FLAG-MARCH10b, as described in detail below. To confirm the expression of MARCH10a and MARCH10b, the soluble fraction of rat testicular lysates was subjected to immunoprecipitation with anti-MAR10N antiserum or the preimmune serum. When the immunoprecipitates were analyzed by Western blotting with the anti-MAR10N antiserum, two immunoreactive bands were detected at 90 and 30 kDa, which were close to the estimated molecular masses of MARCH10a and MARCH10b, respectively (Fig. 1E, lane 1, arrowheads). Next, we determined the cell types expressing MARCH10 proteins by immunohistochemistry on rat testicular sections. As shown in Fig. 2, the anti-MAR10N antiserum reacted with the inner layers and luminal contents of a subset of seminiferous tubules where the spermatids were present. The signals were detected in the seminiferous tubules at stages I–III and XI–XIV. Negative control experiments with preimmune serum detected no such signal (supplemental Fig. S2).
FIGURE 2.
Immunofluorescence staining of MARCH10 in testis. Sections (5 μm) from 16-week-old rat testis were stained with anti-MAR10N antiserum (green). Nuclei were visualized using Hoechst 33342 (blue). Stages IX–X (A), XI (B), XIII–XIV (C), I–III (D), IV–VI (E), and VII–VIII (F) of the seminiferous tubules are shown. Bars, 50 and 20 μm in insets.
Localization of MARCH10 in Elongating Spermatids
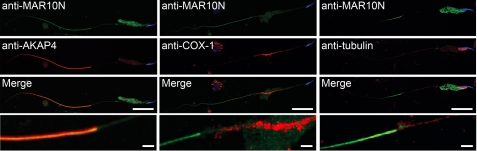
To further analyze the developmental expression profile in detail, we stained smear preparations of rat testis with anti-MAR10N antiserum. As shown in Fig. 3, A–D, the staining became detectable in the cytoplasmic lobe of spermatids at step 11 and was also steadily observed in the posterior portion of the tail. The staining was prominent at steps 13–15 and then gradually decreased. When the smear samples were stained with anti-MAR10M antiserum, signals were also detected in the cytoplasm and tail of elongating spermatids in steps 11–16, but the cytoplasmic staining was weaker than that with anti-MAR10N antiserum (Fig. 3, E and F). To analyze MARCH10 expression in mature spermatozoa, smear preparations of rat epididymis were stained with anti-MAR10N antiserum. No specific signal was observed in epididymal spermatozoa (Fig. 3G). Because the mammalian sperm tail consists of two different segments, the midpiece and the principal piece, the tail staining was most likely to be detected in the principal piece. To evaluate this, the smear samples were doubly stained with anti-MAR10N antiserum and antibodies against either cytochrome c oxidase subunit 1 (COX-1; a midpiece marker), protein kinase A-anchoring protein 4 (AKAP4; a principal piece marker (33)), or α- and β-tubulin. As shown in Fig. 4, the signal for MARCH10 overlapped with that of AKAP4 (left panels), but it was clearly separated from COX-1 (middle panels), confirming MARCH10 localization in the principal piece. The MARCH10 staining was observed along the flagellar microtubules but not in the manchette, a spermatid-specific microtubule structure (Fig. 4, right panels). Taken together, these results suggest that the MARCH10 proteins are specifically expressed in the cytoplasmic lobes and the principal piece of elongating spermatids.
FIGURE 3.
Immunofluorescence staining of MARCH10 in elongating spermatids and epididymal spermatozoa. Smear preparations of rat testis (A–F) and epididymis (G) were stained with anti-MAR10N (A–D and G, green) or anti-MAR10M antiserum (E and F, green). Right panels show the corresponding differential interference contrast images. Bars, 20 and 2 μm in insets.
FIGURE 4.
Double immunofluorescence staining of MARCH10 in elongating spermatids. Rat elongating spermatids in smear preparations were stained with anti-MAR10N antiserum (green) followed by mouse monoclonal antibodies to AKAP4 (left panels, red), COX-1 (middle panels, red), or α- and β-tubulin (right panels, red). Nuclei were visualized using Hoechst 33342 (blue). Images were obtained by confocal microscopy. Bottom panels show high magnification images of the boundary region of the midpiece and the principal piece. Bars, 20 μm in the 3 top panels and 2 μm in bottom panel.
Super-resolution Imaging of MARCH10 Localization with Three-dimensional Structured Illumination Microscopy (SIM)
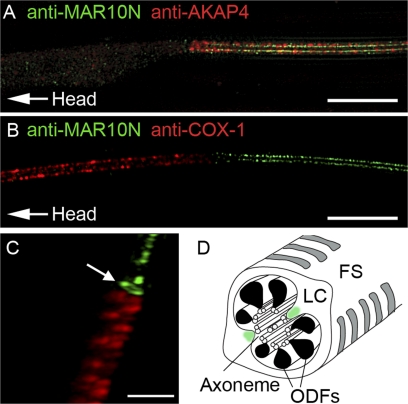
To analyze the MARCH10 localization at a higher resolution, we employed the three-dimensional SIM system, which provides twice the resolution of conventional confocal microscopy (i.e. ∼100 nm on the x and y axes and 220 nm on the z axis) (34, 35). A series of three-dimensional SIM, z-stack images of the fluorescent-labeled elongating spermatids, was captured and used to generate three-dimensional projections of the staining of MARCH10, COX-1, and AKAP4. In the confocal images of the elongating spermatids, the signal for the MARCH10 proteins has the appearance of a single longitudinal configuration in the principal piece (Fig. 4). Using three-dimensional SIM, we were able to resolve the MARCH10 signal, and it was detected as two dotted strands running parallel to each other (Fig. 5A). Although the AKAP4 signal appeared to have a spatial pattern similar to that of MARCH10, it was found to be located outside the area of MARCH10 staining (Fig. 5A and supplemental movie 1). We confirmed that the MARCH10 staining is clearly separated from that of COX-1 (Fig. 5, B and C). Moreover, a ring-shaped MARCH10 staining pattern was observed at the boundary of the midpiece and the principal piece, consistent with the staining in the annulus (Fig. 5C and supplemental movie 2).
FIGURE 5.
Super resolution imaging of MARCH10 staining in elongating spermatids. A–C, three-dimensional SIM images of the flagella of the rat elongating spermatids labeled with anti-MAR10N antiserum (green) and either anti-AKAP4 (red in A) or anti-COX-1 (red in B and C) antibody. Arrow indicates the ring-like staining corresponding to the annulus. Bars, 5 μm in A and B and 2 μm in C. D, schematic illustration of the principal piece of the flagellum. MARCH10 (green) is likely to be expressed in the inner parts of the longitudinal columns (LC) of the fibrous sheath (FS).
Microtubule Association of MARCH10a
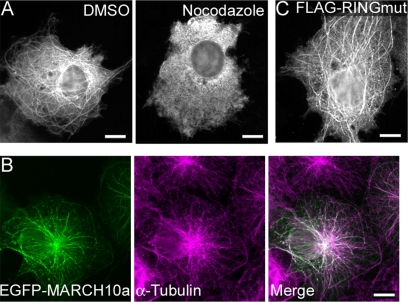
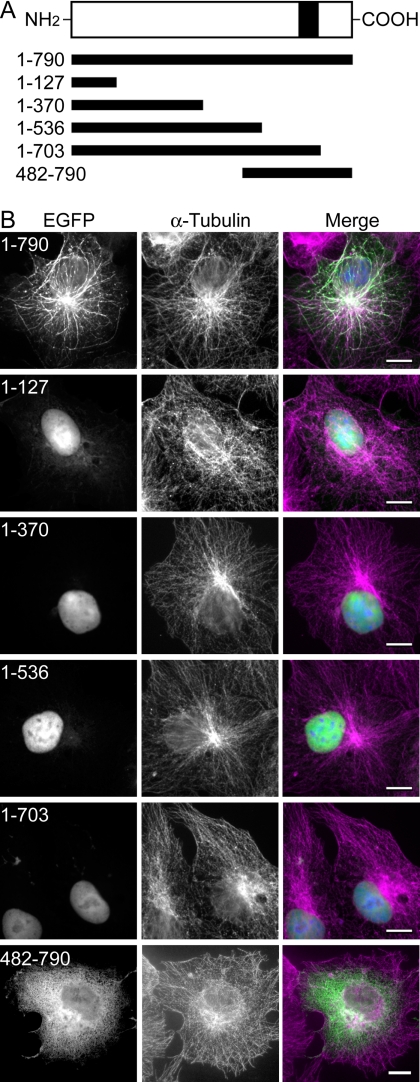
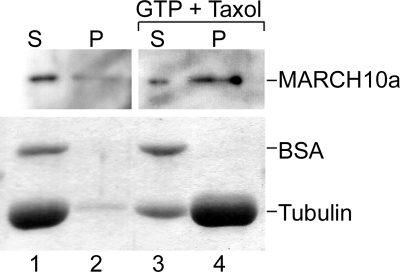
As described above, exogenously expressed FLAG-MARCH10a was localized to cytoplasmic filamentous structures in COS7 cells, whereas FLAG-MARCH10b was distributed throughout the cytoplasm (supplemental Fig. S1). We found that the filamentous distribution of FLAG-MARCH10a was abolished by treatment with the microtubule-depolymerizing agent nocodazole (Fig. 6A). Moreover, when overexpressed in COS7 cells, an EGFP fusion protein of MARCH10a (EGFP-MARCH10a) was colocalized with the microtubules (Fig. 6B). Thus, the MARCH10a-associated structures are likely to be microtubules. The E3 activity of several of the MARCH proteins has been shown to contribute to their subcellular localization (17, 24, 27). To test if the E3 activity of MARCH10a has such an effect, a mutant FLAG-MARCH10a containing serine substitutions at the conserved catalytic cysteine residues (C641S and C644S; FLAG-RINGmut) was transfected into COS7 cells, and the localization was determined by immunofluorescence microscopy. As shown in Fig. 6C, FLAG-RINGmut exhibited a filamentous pattern similar to that of FLAG-MARCH10a, suggesting that the E3 activity of MARCH10a is not essential for its subcellular localization. Next, to investigate the region responsible for the microtubule localization, we performed deletion analysis of MARCH10a. The localization of a series of MARCH10a fragments fused to EGFP was determined in COS7 cells by immunofluorescence microscopy (Fig. 7A). As shown in Fig. 7B, none of the deletion constructs tested exhibited microtubule localization; the N-terminal deletion mutants were preferably localized in the nucleus (top 4 panels) and the C-terminal deletion resulted in a cytosolic distribution pattern (bottom). These results suggest that the entire region of MARCH10a is required for its microtubule localization. To confirm the microtubule association of MARCH10a, we performed an in vitro microtubule cosedimentation assay. Purified tubulin and in vitro translated MARCH10a or BSA (used as a nonbinding control protein) were incubated at 37 °C in the presence or absence of GTP and Taxol, a microtubule-stabilizing reagent. The polymerized microtubules were then allowed to sediment through a sucrose cushion, after which the resulting supernatants and pellets were analyzed by Coomassie Brilliant Blue staining and Western blotting with anti-MAR10N antiserum. As shown in Fig. 8, MARCH10a was detected in the pellets in the presence of polymerized microtubules, whereas BSA was not found to be cosedimented. Taken together, these results suggest that MARCH10a directly associates with microtubules.
FIGURE 6.
Microtubule localization of MARCH10a in COS7 cells. A, COS7 cells transiently transfected with FLAG-MARCH10a were treated with DMSO (left) or 20 μm nocodazole (right) for 30 min and then processed for immunofluorescence microscopy with anti-FLAG antibodies. B, COS7 cells transiently transfected with EGFP-MARCH10a were processed for immunofluorescence microscopy with anti-α-tubulin antibodies. Signals for EGFP and α-tubulin are shown in green and magenta, respectively. C, COS7 cells transiently transfected with FLAG-RINGmut were processed for immunofluorescence microscopy with anti-FLAG antibodies. Bars, 10 μm.
FIGURE 7.
Deletion analysis of EGFP-MARCH10a localization. A, schematic representation of EGFP-MARCH10a and its deletion mutants used to analyze the subcellular localization. All constructs contained an N-terminal EGFP tag (not depicted). B, COS7 cells were transiently transfected with the indicated EGFP fusion MARCH10a constructs. The cells were labeled with Hoechst 33342 to visualize nuclei and then processed for immunofluorescence microscopy with anti-α-tubulin antibodies. Right panels show the merged signals for EGFP (green), α-tubulin (magenta), and nuclei (blue). Bars, 10 μm.
FIGURE 8.
Microtubule association of MARCH10a in vitro. Porcine brain tubulin proteins were mixed with either in vitro translated MARCH10a or BSA in PEM buffer. The mixtures were incubated at 37 °C for 30 min in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of GTP and Taxol, after which polymerized microtubules were then sedimented through a 1 m sucrose cushion by ultracentrifugation. The resulting supernatants (100 μl) were recovered, and the pellets were dissolved with 100 μl of sample buffer. The supernatant (20 μl; S, lanes 1 and 3) and pellet samples (20 μl; P, lanes 2 and 4) were separated by SDS-PAGE followed by Western blotting with anti-MAR10N antiserum (top) and Coomassie Brilliant Blue staining to visualize BSA and tubulin (bottom).
Microtubule-dependent E3 Ub Ligase Activity of MARCH10a
To assess the E3 Ub ligase activity of the RING finger domain of MARCH10a, we performed an in vitro ubiquitination assay with E1, E2, and Ub in the presence or absence of a recombinant GST fusion protein of the RING finger (GST-RING). We used recombinant UBE2B as an E2 enzyme, because UBE2B is known to be expressed in spermatids (36, 37). Western blotting of the reaction samples revealed that the GST-RING catalyzed dense, polyubiquitinated products (Fig. 9A, lane 2). This polyubiquitin formation was abolished by the C641S and C644S mutations (GST-RINGmut; Fig. 9A, lane 3). Various MARCH members have been shown to mediate self-ubiquitination in transfected cultured cells (25, 27, 38). To obtain evidence on the in vivo E3 activity of MARCH10a, FLAG-MARCH10a was transfected into COS7 cells along with HA-tagged Ub. After cells were treated with the proteasome inhibitor MG132 for 3 h, the whole cell lysates were immunoprecipitated with anti-FLAG antibody followed by Western blotting with anti-HA antibody. As shown in Fig. 9B, a smear signal was detected in the anti-FLAG immunoprecipitates with the anti-HA antibody (lane 2), indicating that MARCH10a undergoes self-ubiquitination. Interestingly, when the cells were treated with nocodazole, this self-ubiquitination activity was strongly repressed (Fig. 9B, lane 3). These results suggest that the E3 activity of MARCH10a is dependent on intact microtubules. To determine whether the self-ubiquitination has the capacity to catalyze protein degradation, we compared the stability of FLAG-MARCH10a with FLAG-RINGmut using the protein synthesis inhibitor cycloheximide (CHX). COS7 cells transiently transfected with FLAG-MARCH10a or FLAG-RINGmut were treated with CHX, and their expression levels were analyzed by Western blotting with the anti-FLAG antibody. As shown in Fig. 9C, ∼50% of the FLAG-MARCH10a proteins were degraded within 12 h (lanes 1–3), whereas FLAG-RINGmut was much more stable (lanes 7–9). The degradation of FLAG-MARCH10a was blocked by MG132 (Fig. 9C, lanes 4–6). The protein quantification data is shown in Fig. 9D. These results indicate that self-ubiquitination of MARCH10a accelerates its proteasomal degradation.
DISCUSSION
In this study, we identified two isoforms of the March10 gene products that are predominantly expressed in the testis. March10a encodes a long RING finger protein having E3 Ub ligase activity, and March10b encodes a short proline-rich protein lacking the RING finger domain. The following observations suggest that MARCH10a is a microtubule-associated protein. First, when overexpressed in COS7 cells, MARCH10a was localized along the microtubules, and this pattern of distribution was dispersed by microtubule depolymerization. Second, MARCH10a formed a complex with microtubules in vitro. Deletion analysis failed to identify the region responsible for the microtubule binding but suggested that the tertiary structure of MARCH10a would likely be important. In this regard, it seems reasonable that MARCH10b exhibited diffuse cytoplasmic distribution in COS7 cells. We demonstrated that the E3 Ub ligase activity of MARCH10a is not essential for its subcellular localization, but the microtubule association does appear to be important to the E3 activity. Therefore, it is plausible that MARCH10a is a microtubule-associated E3 Ub ligase that is active on microtubule structures. The cytoplasmic localization and lack of the E3 activity of MARCH10b suggest that it has a function that is distinct from MARCH10a. Proline-rich sequences, which are found in the C-terminal region of MARCH10b, have been shown to mediate protein-protein interactions with, for example, the Src homology 3, WW, and EVH1 domains (39). MARCH10b may thus act as an adaptor or scaffold protein.
Immunohistochemical studies demonstrated that MARCH10 proteins are abundantly expressed in elongating and elongated spermatids and disappear in epididymal spermatozoa, suggesting that MARCH10 is required for spermatid maturation. Immunostaining of the smear samples of rat testis revealed that the signals for MARCH10 were detected in the cytoplasmic lobes and the principal piece of the flagella. In this experiment, two polyclonal antisera, which stained the same regions of the spermatids with different degrees of immunoreactivity, were used as follows: anti-MAR10N antiserum stained the cytoplasm and the principal piece quite evenly, whereas anti-MAR10M antiserum stained the principal piece more heavily. Therefore, MARCH10a and MARCH10b are preferentially localized in the tail and the cytoplasmic lobe, respectively. MARCH10 expression was initially observed in the cytoplasm at step 10, followed by the tail region. The delayed localization to the flagella suggests that the MARCH10 proteins are synthesized in the cytoplasm and then transported to the developing flagella via the intraflagellar transport system (40). Microscopic studies provided detailed information on the morphological development of the spermatid flagella, which takes place during the early stages of spermiogenesis (i.e. steps 1–3) and continues until step 19 (1). The longitudinal columns of the FS are deposited along the axoneme from the distal end to the proximal end of the principal piece at steps 2–10, and the ribs are assembled at steps 11–17 (41, 42). The ODF first appear in the most proximal portion of the axoneme at step 9 and continue to extend to the end of the principal piece until step 19 (42, 43). MARCH10 proteins are unlikely to be essential structural components of the flagellum, because their expression is considerably reduced in the late stages of spermiogenesis. Rather, they may help regulate the organization of the ODF and FS. In this context, it is interesting that MARCH10a exerted the E3 activity with the E2 enzyme UBE2B (also known as HR6B). It has been reported that mouse Ube2b mRNA is expressed during meiosis and spermiogenesis, and its disruption causes structural abnormalities in both the sperm head and flagella, in which the longitudinal columns are assembled in an incorrect manner (10–12). Our data thus suggest that MARCH10a might be an as-yet-unidentified E3 partner of the spermatid UBE2B in the course of FS formation.
Given the microtubule association of MARCH10a, its E3 substrate protein(s) may be present in proximity to the axoneme and/or the inner sides of the ODF and FS. In support of this notion, our three-dimensional SIM imaging suggests that MARCH10 seems to be present in the inner part of the longitudinal columns of the FS (Fig. 5D). E3 Ub ligases have been shown to bind to microtubules and to regulate the function and maintenance of microtubules in somatic cells. For example, MID1, which is associated with Opitz syndrome, controls the phosphorylation of microtubule-associated proteins by targeting the catalytic subunit of protein phosphatase 2A (PPP2CA) for ubiquitination and subsequent degradation (44). PARK2 (also known as Parkin), which is mutated in Parkinson disease, participates in the quality control of microtubules by promoting tubulin ubiquitination (45, 46). MARCH10a may participate in this quality control activity by targeting damaged and defective flagellar proteins for Ub-mediated degradation, thereby ensuring proper flagellar assembly. Recent work by Huang et al. (47) has reported a role for ubiquitination in the resorption of the flagella of Chlamydomonas reinhardtii. Before cell division starts, ubiquitination occurs on flagellar proteins, including α-tubulin, a subunit of the outer dynein arm, and also on signaling molecules, which likely gives the cue for disassembly of the flagella without any need of proteasomal degradation. This ubiquitination has been hypothesized to facilitate the retrograde transport of disassembled flagellar proteins to the cell body. It is important to determine whether this activity is conserved in other species, including mammals. Another role of flagellar ubiquitination is known to prevent the paternal inheritance of mitochondrial DNA; sperm mitochondria are highly ubiquitinated, which takes place during the late stages of spermiogenesis, and thus undergo degradation after fertilization (7–9). The ODF and FS are also destroyed in fertilized eggs (48), but their ubiquitination has not been observed in spermatids or spermatozoa. Nevertheless, we cannot exclude the possibility that MARCH10a and/or other E3 Ub ligases might tag ODF/FS proteins, even if to a much lesser extent than mitochondria, for degradation after fertilization. Self-ubiquitination of MARCH10a promotes its turnover by proteasomal degradation. Thus, the elimination of MARCH10 expression before sperm maturation may be regulated by the Ub-proteasome system. If so, it is possible that the MARCH10a activity is suppressed during spermiogenesis in a manner dependent on microtubule binding.
Three-dimensional SIM imaging revealed the localization of MARCH10 in the annulus, an electron-dense structure surrounding the axoneme between the midpiece and the principal piece. The annulus is assembled from septins, which are polymerizing GTP-binding proteins, and serves as a physical barrier to prevent the diffusion of the components of the midpiece and principal piece (33, 49–53). This structure is essential for sperm flagellar morphology and motility (33, 49). Although less is known about the involvement of the Ub-proteasome system, MARCH10 may contribute to the assembly and integrity of the annulus.
These results provide supporting evidence for the importance of the Ub-proteasomal system in spermiogenesis. To clarify the exact mechanisms underlying the formation of the flagella, it will be necessary to determine the detailed subcellular localization and substrate protein(s) of MARCH10.
Supplementary Material
Acknowledgments
We thank Takaji Nemoto (Seki Technotoron, Tokyo, Japan) for access to the DeltaVision OMX system and for generous support for data analysis. We thank Yoko Yamamoto, Ayako Takada, Yuri Morokuma, and Yusuke Ito for technical assistance and Yuriko Ishii, Tomoko Okada, and Setsuko Sato for secretary assistance. Pacific Edit reviewed the manuscript prior to submission.
This work was supported by Grants-in-aid for Scientific Research 21026010, 22370029, and 22770123 and the 21st Century and Global Center of Excellence Program of the Ministry of Education, Culture, Sports, Science, and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Movies 1 and 2.
- ODF
- outer dense fiber
- FS
- fibrous sheath
- Ub
- ubiquitin
- EGFP
- enhanced green fluorescent protein
- HA-Ub
- HA-tagged Ub
- CHX
- cycloheximide
- SIM
- structured illumination microscopy.
REFERENCES
- 1. Oko R., Clermont Y. (1990) in Controls of Sperm Motility: Biological and Clinical Aspects (Gagnon C. ed) pp. 3–28, CRC Press, Ann Arbor, MI [Google Scholar]
- 2. Oko R. J., Clermont Y. (1991) Ann. N.Y. Acad. Sci. 637, 203–223 [DOI] [PubMed] [Google Scholar]
- 3. Eddy E. M. (2007) Soc. Reprod. Fertil. Suppl. 65, 45–62 [PubMed] [Google Scholar]
- 4. Cao W., Gerton G. L., Moss S. B. (2006) Mol. Cell. Proteomics 5, 801–810 [DOI] [PubMed] [Google Scholar]
- 5. Hermo L., Pelletier R. M., Cyr D. G., Smith C. E. (2010) Microsc. Res. Tech. 73, 320–363 [DOI] [PubMed] [Google Scholar]
- 6. Staub O., Rotin D. (2006) Physiol. Rev. 86, 669–707 [DOI] [PubMed] [Google Scholar]
- 7. Sutovsky P., Moreno R. D., Ramalho-Santos J., Dominko T., Simerly C., Schatten G. (1999) Nature 402, 371–372 [DOI] [PubMed] [Google Scholar]
- 8. Sutovsky P., Moreno R. D., Ramalho-Santos J., Dominko T., Simerly C., Schatten G. (2000) Biol. Reprod. 63, 582–590 [DOI] [PubMed] [Google Scholar]
- 9. Thompson W. E., Ramalho-Santos J., Sutovsky P. (2003) Biol. Reprod. 69, 254–260 [DOI] [PubMed] [Google Scholar]
- 10. Roest H. P., van Klaveren J., de Wit J., van Gurp C. G., Koken M. H., Vermey M., van Roijen J. H., Hoogerbrugge J. W., Vreeburg J. T., Baarends W. M., Bootsma D., Grootegoed J. A., Hoeijmakers J. H. (1996) Cell 86, 799–810 [DOI] [PubMed] [Google Scholar]
- 11. Escalier D. (2003) Biol. Reprod. 69, 373–378 [DOI] [PubMed] [Google Scholar]
- 12. Escalier D., Bai X. Y., Silvius D., Xu P. X., Xu X. (2003) Mol. Reprod. Dev. 65, 298–308 [DOI] [PubMed] [Google Scholar]
- 13. Rodriguez C. I., Stewart C. L. (2007) Dev. Biol. 312, 501–508 [DOI] [PubMed] [Google Scholar]
- 14. Rivkin E., Cullinan E. B., Tres L. L., Kierszenbaum A. L. (1997) Mol. Reprod. Dev. 48, 77–89 [DOI] [PubMed] [Google Scholar]
- 15. Kierszenbaum A. L. (2000) Mol. Reprod. Dev. 57, 109–110 [DOI] [PubMed] [Google Scholar]
- 16. Mochida K., Tres L. L., Kierszenbaum A. L. (2000) Mol. Reprod. Dev. 57, 176–184 [DOI] [PubMed] [Google Scholar]
- 17. Goto E., Ishido S., Sato Y., Ohgimoto S., Ohgimoto K., Nagano-Fujii M., Hotta H. (2003) J. Biol. Chem. 278, 14657–14668 [DOI] [PubMed] [Google Scholar]
- 18. Bartee E., Mansouri M., Hovey Nerenberg B. T., Gouveia K., Früh K. (2004) J. Virol. 78, 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morokuma Y., Nakamura N., Kato A., Notoya M., Yamamoto Y., Sakai Y., Fukuda H., Yamashina S., Hirata Y., Hirose S. (2007) J. Biol. Chem. 282, 24806–24815 [DOI] [PubMed] [Google Scholar]
- 20. Nathan J. A., Lehner P. J. (2009) Exp. Cell Res. 315, 1593–1600 [DOI] [PubMed] [Google Scholar]
- 21. Ohmura-Hoshino M., Matsuki Y., Aoki M., Goto E., Mito M., Uematsu M., Kakiuchi T., Hotta H., Ishido S. (2006) J. Immunol. 177, 341–354 [DOI] [PubMed] [Google Scholar]
- 22. Zavacki A. M., Arrojo E., Drigo R., Freitas B. C., Chung M., Harney J. W., Egri P., Wittmann G., Fekete C., Gereben B., Bianco A. C. (2009) Mol. Cell. Biol. 29, 5339–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamura N., Fukuda H., Kato A., Hirose S. (2005) Mol. Biol. Cell 16, 1696–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fukuda H., Nakamura N., Hirose S. (2006) J. Biochem. 139, 137–145 [DOI] [PubMed] [Google Scholar]
- 25. Yonashiro R., Ishido S., Kyo S., Fukuda T., Goto E., Matsuki Y., Ohmura-Hoshino M., Sada K., Hotta H., Yamamura H., Inatome R., Yanagi S. (2006) EMBO J. 25, 3618–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakamura N., Kimura Y., Tokuda M., Honda S., Hirose S. (2006) EMBO Rep. 7, 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nathan J. A., Sengupta S., Wood S. A., Admon A., Markson G., Sanderson C., Lehner P. J. (2008) Traffic 9, 1130–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Metcalfe S. M., Muthukumarana P. A., Thompson H. L., Haendel M. A., Lyons G. E. (2005) FEBS Lett. 579, 609–614 [DOI] [PubMed] [Google Scholar]
- 29. Muthukumarana P. A., Lyons G. E., Miura Y., Thompson L. H., Watson T., Green C. J., Shurey S., Hess A. D., Rosengard B. R., Metcalfe S. M. (2006) Int. Immunopharmacol. 6, 1993–2001 [DOI] [PubMed] [Google Scholar]
- 30. Nakamura N., Hirose S. (2008) Mol. Biol. Cell 19, 1903–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aihara T., Nakamura N., Honda S., Hirose S. (2009) Biol. Reprod. 80, 762–770 [DOI] [PubMed] [Google Scholar]
- 32. Kato A., Nagata Y., Todokoro K. (2004) Dev. Biol. 269, 196–205 [DOI] [PubMed] [Google Scholar]
- 33. Miki K., Willis W. D., Brown P. R., Goulding E. H., Fulcher K. D., Eddy E. M. (2002) Dev. Biol. 248, 331–342 [DOI] [PubMed] [Google Scholar]
- 34. Gustafsson M. G. (2000) J. Microsc. 198, 82–87 [DOI] [PubMed] [Google Scholar]
- 35. Schermelleh L., Carlton P. M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M. C., Agard D. A., Gustafsson M. G., Leonhardt H., Sedat J. W. (2008) Science 320, 1332–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koken M. H., Hoogerbrugge J. W., Jasper-Dekker I., de Wit J., Willemsen R., Roest H. P., Grootegoed J. A., Hoeijmakers J. H. (1996) Dev. Biol. 173, 119–132 [DOI] [PubMed] [Google Scholar]
- 37. Baarends W. M., Wassenaar E., Hoogerbrugge J. W., van Cappellen G., Roest H. P., Vreeburg J., Ooms M., Hoeijmakers J. H., Grootegoed J. A. (2003) Mol. Cell. Biol. 23, 1151–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hassink G., Kikkert M., van Voorden S., Lee S. J., Spaapen R., van Laar T., Coleman C. S., Bartee E., Früh K., Chau V., Wiertz E. (2005) Biochem. J. 388, 647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zarrinpar A., Bhattacharyya R. P., Lim W. A. (2003) Sci. STKE 2003, RE8. [DOI] [PubMed] [Google Scholar]
- 40. Kierszenbaum A. L. (2002) Mol. Reprod. Dev. 63, 1–4 [DOI] [PubMed] [Google Scholar]
- 41. Irons M. J., Clermont Y. (1982) Am. J. Anat. 165, 121–130 [DOI] [PubMed] [Google Scholar]
- 42. Oko R., Clermont Y. (1989) Anat. Rec. 225, 46–55 [DOI] [PubMed] [Google Scholar]
- 43. Irons M. J., Clermont Y. (1982) Anat. Rec. 202, 463–471 [DOI] [PubMed] [Google Scholar]
- 44. Trockenbacher A., Suckow V., Foerster J., Winter J., Krauss S., Ropers H. H., Schneider R., Schweiger S. (2001) Nat. Genet. 29, 287–294 [DOI] [PubMed] [Google Scholar]
- 45. Ren Y., Zhao J., Feng J. (2003) J. Neurosci. 23, 3316–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang F., Jiang Q., Zhao J., Ren Y., Sutton M. D., Feng J. (2005) J. Biol. Chem. 280, 17154–17162 [DOI] [PubMed] [Google Scholar]
- 47. Huang K., Diener D. R., Rosenbaum J. L. (2009) J. Cell Biol. 186, 601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sutovsky P., Schatten G. (2000) Int. Rev. Cytol. 195, 1–65 [DOI] [PubMed] [Google Scholar]
- 49. Kissel H., Georgescu M. M., Larisch S., Manova K., Hunnicutt G. R., Steller H. (2005) Dev. Cell 8, 353–364 [DOI] [PubMed] [Google Scholar]
- 50. Kwitny S., Klaus A. V., Hunnicutt G. R. (2010) Biol. Reprod. 82, 669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Steels J. D., Estey M. P., Froese C. D., Reynaud D., Pace-Asciak C., Trimble W. S. (2007) Cell Motil. Cytoskeleton 64, 794–807 [DOI] [PubMed] [Google Scholar]
- 52. Lin Y. H., Lin Y. M., Wang Y. Y., Yu I. S., Lin Y. W., Wang Y. H., Wu C. M., Pan H. A., Chao S. C., Yen P. H., Lin S. W., Kuo P. L. (2009) Am. J. Pathol. 174, 1857–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chao H. C., Lin Y. H., Kuo Y. C., Shen C. J., Pan H. A., Kuo P. L. (2010) J. Assist. Reprod. Genet. 27, 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.