Background: The Escherichia coli β-barrel assembly machinery complex (BamABCDE) facilitates outer membrane protein assembly.
Results: The unstructured N terminus of BamC binds and blocks the proposed substrate-binding pocket on BamD.
Conclusion: The unstructured N terminus of BamC is essential for BamCD interaction.
Significance: The first Bam lipoprotein complex structure reveals how BamC and BamD interact.
Keywords: Bacteria, Lipoprotein Structure, Membrane Biogenesis, Membrane Proteins, Protein Assembly, Protein Complexes, Protein Conformation, Protein Structure, Protein-Protein Interactions, X-ray Crystallography
Abstract
The β-barrel assembly machinery (BAM) complex of Escherichia coli is a multiprotein machine that catalyzes the essential process of assembling outer membrane proteins. The BAM complex consists of five proteins: one membrane protein, BamA, and four lipoproteins, BamB, BamC, BamD, and BamE. Here, we report the first crystal structure of a Bam lipoprotein complex: the essential lipoprotein BamD in complex with the N-terminal half of BamC (BamCUN (Asp28–Ala217), a 73-residue-long unstructured region followed by the N-terminal domain). The BamCD complex is stabilized predominantly by various hydrogen bonds and salt bridges formed between BamD and the N-terminal unstructured region of BamC. Sequence and molecular surface analyses revealed that many of the conserved residues in both proteins are found at the BamC-BamD interface. A series of truncation mutagenesis and analytical gel filtration chromatography experiments confirmed that the unstructured region of BamC is essential for stabilizing the BamCD complex structure. The unstructured N terminus of BamC interacts with the proposed substrate-binding pocket of BamD, suggesting that this region of BamC may play a regulatory role in outer membrane protein biogenesis.
Introduction
In Escherichia coli, β-barrel outer membrane proteins (OMPs)2 are synthesized in the cytosol and follow a pathway that directs them through the inner membrane and periplasm to the outer membrane, where they are finally folded and inserted (1, 2). This last step is catalyzed by the β-barrel assembly machinery (BAM) complex, which consists of one OMP, known as BamA, along with four lipoproteins, BamB, BamC, BamD, and BamE (see Fig. 1a) (3). Homologous systems can be found in all Gram-negative species, as well as in the mitochondria and chloroplasts of eukaryotes (4, 5). Although only BamA and BamD are essential for cell viability, understanding the structure and function of all components will provide greater insight into how outer membrane biogenesis occurs and could reveal the BAM complex as a new drug target (4–6).
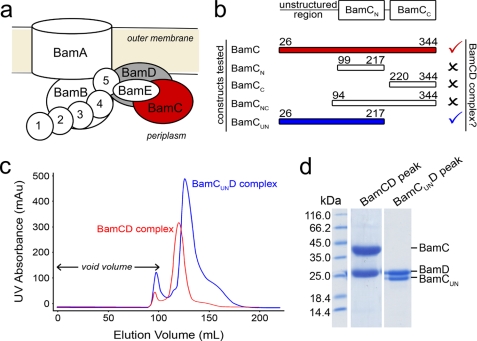
FIGURE 1.
Formation of the BamCD complex. a, schematic diagram of the E. coli BAM complex. Two of the lipoprotein components (BamC and BamD) that are the focus of this study are colored in red and gray, respectively. b, summary of BamC truncations made and their ability to interact with BamD to form the BamCD complex. Check marks signify formation of the complex, whereas X marks signify inability to form the complex. The N- and C-terminal boundaries of each BamC construct are given. c, overlaid size exclusion chromatograms of co-purified BamCD and BamCUND complexes. mAU, milli-absorbance units. d, SDS-PAGE confirmed co-elution of BamC (or BamCUN) with BamD from the major chromatogram peaks shown in c.
BamA consists of a β-barrel domain embedded in the outer membrane, as well as an N-terminal periplasmic domain composed of five polypeptide transport-associated (POTRA) motifs (7–9). BamB–E are lipoproteins anchored to the periplasmic face of the outer membrane via an N-terminally attached lipid (3). Several structures of the POTRA motifs of BamA and of the lipoproteins have been solved recently (10–26). With structures of individual BAM proteins available to supplement functional studies, one can start to piece together how the BAM complex forms.
Previous research has shown that the POTRA motifs of BamA interact with the BamB–E lipoproteins, and it is believed that the POTRA motifs are also the docking site for the unfolded OMP substrates (3, 10). Although BamB and BamD independently interact with the POTRA motifs directly, BamC and BamE require BamD to co-purify with BamA (6, 10). Direct interaction between BamC and BamD has also been shown, and the mutagenesis data are consistent with the C terminus of BamD (residues 227–245) being necessary for the association (6). However, the region of BamC involved in the interaction with BamD was not determined.
To gain insight into the nature of the BamC-BamD interaction, a series of BamC truncation mutations were made and screened for interaction with BamD. This resulting in vitro interaction data, along with the crystal structure, reveal that the long, unstructured N terminus of BamC is required to stabilize the BamCD complex by forming a lasso-like structure that binds to BamD.
EXPERIMENTAL PROCEDURES
Cloning
The DNAs coding for the full-length BamC and BamD constructs and the truncation mutants (BamCNC, BamCN, BamCC, and BamCUN) were amplified from E. coli K12 using the primers listed in supplemental Table 1. All forward and reverse primers contained the restriction sites NdeI and XhoI, respectively (except for BamCNC, which had NdeI and HindIII). The PCR products of full-length BamC, full-length BamD, and BamCNC were ligated into vector pET28a (Novagen), and the resulting constructs had cleavable N-terminal hexahistidine affinity tags. The PCR products of BamCN, BamCC, and BamCUN were ligated into pET24a (Novagen), resulting in C-terminal hex-histidine tags. Subsequent DNA sequencing (Macrogen) confirmed that the BamC and BamD inserts matched the sequences reported in the Swiss-Prot Database (P0A903 and P0AC02, respectively).
Protein Overexpression
Each expression plasmid coding for BamC, BamD, BamCNC, BamCN, BamCC, or BamCUN was transformed into E. coli BL21(λDE3) cells and used to inoculate (1:100 back-dilution) 2 liters of LB medium containing kanamycin (50 μg/ml). Cultures were grown at 37 °C until A600 nm = 0.6. The culture was then induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h.
Purification of BamCD Complex
Cells overexpressing BamC and BamD were separately harvested by centrifugation and subsequently combined prior to lysis. The combined cell pellet was lysed using an Avestin EmulsiFlex-3C cell homogenizer in buffer A (20 mm Tris-HCl (pH 8.0) and 100 mm NaCl). The resulting lysate was clarified by centrifugation at 45,000 × g for 30 min at 4 °C, and the overexpressed proteins were initially purified by Ni2+ affinity chromatography. BamC and BamD were co-eluted from the nickel-nitrilotriacetic acid-agarose column (Qiagen) with a step gradient method (100–500 mm imidazole in buffer A in 100 mm increments). The fractions containing both proteins were pooled and concentrated to ∼10 mg/ml using an Amicon ultracentrifugal filter device (Millipore). The concentrated BamCD sample was then further purified by size exclusion chromatography (Sephacryl S-100 HiPrep 26/60 column) in buffer A on an ÄKTAprime system (GE Healthcare).
Protein-Protein Interaction Studies
To test whether or not different truncation mutant forms of BamC can form a BamCD heterodimer, each truncation mutant was co-lysed with BamD and purified by Ni2+ affinity and gel filtration chromatography as described above. The oligomeric state of the purified sample was further confirmed by gel filtration chromatography (Superdex 200 column, GE Healthcare) in-line with a multiangle light-scattering system (Wyatt Technologies Inc.). A sample of 100 μl of purified sample (5 mg/ml) was injected and resolved at a flow rate of 0.5 ml/min in buffer A. The molecular masses of proteins in each sample were determined by a multiangle light-scattering DAWN EOS instrument with a 684 nm laser (Wyatt Technologies Inc.) coupled to a refractive index instrument (Optilab rEX, Wyatt Technologies Inc.). The molar mass of the protein was calculated from the observed light-scattering intensity and differential refractive index using ASTRA v5.1 software (Wyatt Technologies Inc.) based on the Zimm fit method using a refractive index increment of dn/dc = 0.185 ml/g.
Crystallization
BamCD crystals were grown by the sitting drop vapor diffusion method. A final v/v concentration of 0.03% n-dodecyl β-maltoside was added to the protein sample prior to setting up crystallization plates. The crystallization drops were prepared by mixing 1 μl of protein (30 mg/ml) suspended in buffer A with 1 μl of reservoir solution and then equilibrating the drop against 1 ml of reservoir solution. The BamCD construct yielded crystals in the space group I121 with unit cell dimensions of 73.8, 133.4, and 145.0 Å. The optimal crystallization reservoir condition was 0.2 m K2HPO4 and 20% PEG-3350. Crystallization was performed at room temperature (∼22 °C). The cryosolution contained 0.2 m K2HPO4, 20% PEG-3350, and 30% glycerol. Crystals were washed in the cryosolution before being flash-cooled in liquid nitrogen.
Data Collection
Diffraction data were collected on the BamCD crystals at beamline X25 at the National Synchrotron Light Source, using an ADSC Q315 CCD x-ray detector. The crystal-to-detector distance was 375 mm. A total of 360 images were collected with 1° oscillations, and each image was exposed for 1 s. The diffraction data were processed with the programs iMosflm (27), POINTLESS (28), and SCALA (28). See supplemental Table 2 for data collection statistics.
Structure Determination and Refinement
Although the full-length BamCD complex was used for crystallization, BamC was cleaved to a smaller fragment corresponding to BamCUN (Asp28–Ala217) in the crystallization drop during incubation. BamC has been previously shown to be susceptible to degradation in the linker region connecting the N- and C-terminal domains (29). Phases were obtained by molecular replacement using the program PHASER 2.1 (30). Previously solved E. coli BamCN (Protein Data Bank code 2YH6) and BamD (code 2YHC) structures were used as search models (11). The N-terminal unstructured region of BamC was manually fit into the difference electron density map using the program COOT (31). The structure was refined using restrained refinement in REFMAC5 (32), and further manual adjustments to the atomic coordinates, especially in regions of BamD, were performed with the program COOT. The final model was obtained by running restrained refinement in REFMAC5 with TLS restraints obtained from the TLS motion determination server (33). The refinement statistics are shown in supplemental Table 2.
Structural Analysis
Secondary structural analysis was performed with the program DSSP (34). The program COOT was used to overlap coordinates for structural comparison. The stereochemistry of the structure was analyzed with the program PROCHECK (35). The on-line servers PISA (36) and PROTORP (37) were used to identify protein-protein interaction interfaces.
RESULTS AND DISCUSSION
The results from size exclusion chromatography and multiangle light-scattering analysis are consistent with both BamC and BamD existing in a monomeric state in solution when purified separately (supplemental Fig. 1). However, when cells overexpressing each protein were combined prior to lysis, a large population of BamC and BamD was observed to co-elute from a size exclusion column as a BamCD heterodimeric complex (Fig. 1). The molar mass of the complex was verified by multiangle light-scattering analysis (60.1 ± 1.8 kDa) (supplemental Fig. 2), which is consistent with the sum of the calculated molecular masses (64.4 kDa) of the BamC (36.4 kDa) and BamD (28.0 kDa) constructs used in this study.
To determine which region of BamC is important for forming the BamCD complex, a series of BamC truncations were created and examined for their ability to interact with BamD. BamC can be divided into three domains: the unstructured region at the far N terminus of the protein (BamCU), the N-terminal domain (BamCN), and the C-terminal domain (BamCC) (13, 21–23). The truncated forms of BamC created for this study are as follows: 1) BamCN (residues 99–217), 2) BamCC (residues 220–344), 3) BamCNC (both the N- and C-terminal domains, residues 94–344), and 4) BamCUN (the N-terminal unstructured region followed by the N-terminal domain, residues 26–217). Size exclusion chromatography analysis showed that any truncated form of BamC missing the unstructured N terminus (i.e. BamCN, BamCC, and BamCNC) was unable to form the BamCD complex (Fig. 1b and supplemental Fig. 1). On the other hand, BamCUN co-purified with BamD throughout the entire purification process despite missing the C-terminal domain (Fig. 1, c and d). Multiangle light-scattering analysis confirmed that BamCUN and BamD formed a complex with a molecular mass of 55.6 ± 1.7 kDa, which is comparable with the sum (49.9 kDa) of the molecular masses of BamCUN (21.9 kDa) and BamD (28.0 kDa) (supplemental Fig. 2). Together, these results show that the unstructured N terminus of BamC is required for the formation of the BamCD complex.
Although the structures of the individual BamC and BamD monomers have been solved previously, that of the BamCD complex has not. In fact, no Bam lipoprotein complex structures have yet been solved. BamC forms two globular domains (N- and C-terminal domains) both with the “helix-grip” fold (38) that are connected by a flexible linker (11, 23). On the other hand, the BamD structure consists of 10 α-helices that form five tetratricopeptide repeats (TPRs) (Fig. 2a) (11, 21). In this study, we crystallized BamD in a heterodimeric complex with BamCUN, and its structure was solved and refined to 2.9 Å resolution (supplemental Table 2). The most outstanding structural feature of the BamCD complex is the 73-residue-long unstructured N terminus of BamC that has not been observed in previously reported structures (11, 23, 39); it folds into an elongated U-shaped loop structure that interacts extensively with BamD by fitting into a trail of crevices that run along the longitudinal axis of BamD (Fig. 2b and supplemental Fig. 3). The globular N-terminal domain of BamC that follows this unstructured region lies adjacent to the N-terminal half of the BamD molecule. Although a previous study has predicted that BamC interacts with the C-terminal end of BamD (Met227–Thr245) (6), our structure shows that the N-terminal region of BamD is also an important site of interaction, as BamC binds along the entire length of the BamD molecule.
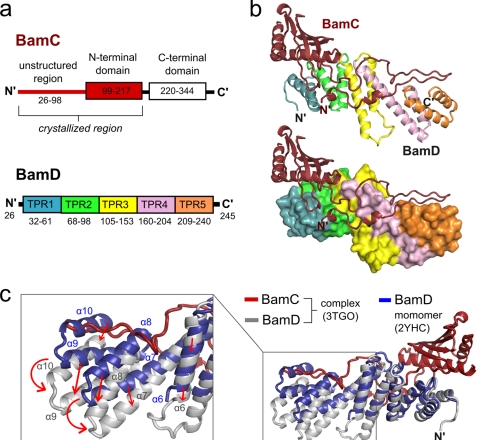
FIGURE 2.
Structure of the BamCD complex and conformational changes in BamD upon binding. a, domain organization of BamC and BamD. Each TPR motif of BamD is shown in a different color. b, structure of the BamCD complex. Ribbon (upper) and surface (lower) diagrams of BamD are shown with the TPR motifs colored as in a. c, BamCD complex (BamC in red and BamD in gray) superimposed onto monomeric BamD (blue). The boxed inset shows a close-up view of the BamD region that undergoes the greatest conformation change upon binding to BamC, with arrows indicating direction of movement.
To determine whether the BamC and BamD molecules undergo conformational changes upon binding to and forming the stable BamCD complex, the structures of both proteins were individually compared with the previously solved structures of the monomeric form of each protein. Although the N-terminal domain structure of BamC superimposes very closely with the previously reported crystal structure (root mean square deviation of 0.54 Å) and the NMR structure (root mean square deviation of 1.24 Å) (supplemental Fig. 4), BamD in the BamCD complex shows a significantly different conformation compared with the monomeric structure (root mean square deviation of 2.3 Å). In the BamCD complex, the positions of α-helices in TPR motifs 3 (α6), 4 (α7 and α8), and 5 (α9 and α10) of BamD are shifted to better accommodate the binding of BamC (Fig. 2c). The four C-terminal α-helices of TPR motifs 4 and 5, in particular, show the greatest change in conformation.
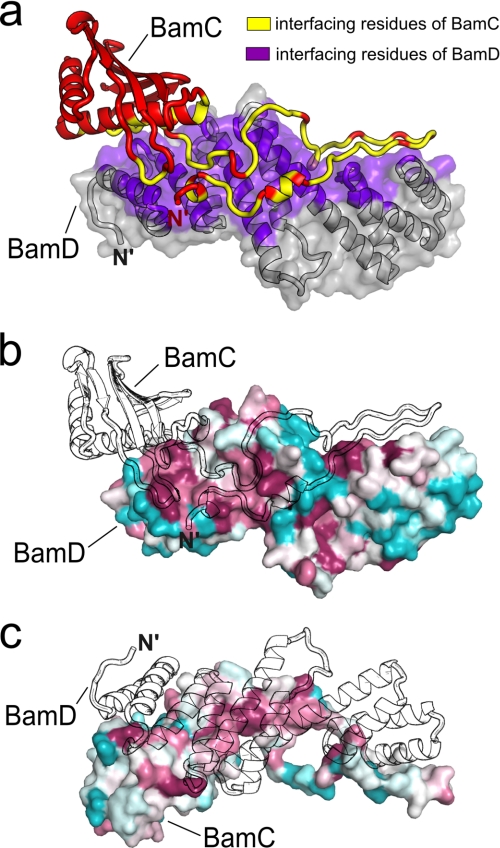
The interaction between BamC and BamD is created predominantly by the direct contact between the N-terminal unstructured region of BamC and all five TPR motifs of BamD (Fig. 2b). This interaction between the two proteins is mediated by numerous hydrogen bonds, salt bridges, and van der Waals forces and has an average interface area of 2249.4 Å2 (Fig. 3a and supplemental Table 3). Many conserved residues are found concentrated at the interaction interfaces on both proteins (Fig. 3, b and c), suggesting that the interaction between these two proteins has important biological and functional implications. For BamC, a multiple sequence alignment (supplemental Fig. 5) shows that its unstructured N terminus is the most well conserved region of the protein, which is not surprising considering its essential role in stabilizing the BamCD complex structure. For BamD, about half of the conserved residues are found at the BamCD interaction interface, but the other half are found clustered on the opposite side of the protein and are solvent-exposed in this heterodimeric complex (Fig. 3c and supplemental Figs. 6 and 7).
FIGURE 3.
BamC-BamD interface. a, ribbon diagram of BamC (red) with ribbon and semitransparent surface diagrams of BamD (gray). The interfacing residues are colored yellow (BamC) and purple (BamD). b, conserved residues mapped onto BamD with absolutely conserved residues in maroon and highly variable residues in cyan. An outline of BamC is drawn to show its position relative to BamD. c, conserved residues mapped onto BamC and colored as in b. An outline of BamD is also shown.
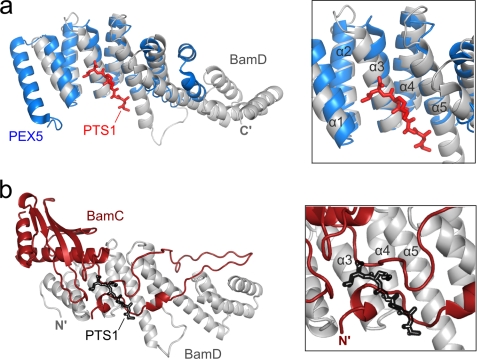
Although this structural information has revealed how BamC and BamD interact with each other, the insights gained in this study raise many questions regarding the function and structure of the BAM complex. For instance, a pocket present in the N-terminal region of BamD (formed by TPR motifs 1 and 2) has been predicted to recognize and bind to the C-terminal targeting sequence of unfolded OMP substrates (11, 21). In fact, this binding pocket of BamD has been previously shown to resemble closely that of PEX5 (Protein Data Bank code 3CVP) (21, 40), a peroxisomal targeting signal receptor (root mean square deviation of 1.7 Å) (Fig. 4a). In the BamCD complex structure, however, this proposed binding pocket is occupied by part of the unstructured region of BamC that has no sequence similarity to the C-terminal targeting sequences of OMP substrates or to PTS1, which is a C-terminal peroxisomal targeting sequence recognized by PEX5 (Fig. 4b). If the function of BamD is indeed recognition of OMP substrates via their C-terminal targeting sequences, then perhaps one of the roles of BamC (more specifically, the unstructured N terminus of BamC) is that of a regulatory one, where it may block or expose the targeting sequence-binding site of BamD depending on need.
FIGURE 4.
Proposed C-terminal targeting sequence-binding pocket of BamD. a, superimposed ribbon diagrams of BamD (gray) and the C-terminal domain of PEX5 (blue), a peroxisomal targeting sequence receptor. The PTS1 peptide, the peroxisomal targeting sequence that is recognized and bound by PEX5, is shown in red. b, in the BamCD complex, part of the unstructured region of BamC (red) blocks the proposed C-terminal targeting sequence-binding pocket of BamD (gray). PTS1 (black outline) is shown for reference.
Many aspects of the BAM complex structure and function remain to be elucidated. How does BamD interact with the POTRA motifs of BamA? It is possible that the conserved region of BamD that is not binding to BamC may serve as a binding surface for POTRA5 of BamA. Where is the C-terminal domain of BamC positioned relative to the rest of the BamC and BamD molecules? Does the C-terminal domain of BamC also interact with BamD, or does it have a separate binding partner? Further biochemical and structural investigation of different interactions formed between the BAM proteins will be required to answer these questions. Continued progress in the structural and functional analysis of the BAM complex and its individual components will not only enhance our understanding of how β-barrel proteins assemble and insert into the outer membrane but may also ultimately contribute to the development of novel antibiotics.
Supplementary Material
Acknowledgments
We thank Dr. R. M. Sweet and the RapiData staff at beamline X25 of the National Synchrotron Light Source, Brookhaven National Laboratory.
This work was supported in part by the Canadian Institutes of Health Research (to M. P.), the National Science and Engineering Research Council of Canada (to M. P.), the Michael Smith Foundation for Health Research (to M. P.), and the Canadian Foundation of Innovation (to M. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–7 and Tables 1–3.
The atomic coordinates and structure factors (code 3TGO) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- OMP
- outer membrane protein
- BAM
- β-barrel assembly machinery
- POTRA
- polypeptide transport-associated
- TPR
- tetratricopeptide repeat.
REFERENCES
- 1. Silhavy T. J., Kahne D., Walker S. (2010) Cold Spring Harbor Perspect. Biol. 2, a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bos M. P., Robert V., Tommassen J. (2007) Annu. Rev. Microbiol. 61, 191–214 [DOI] [PubMed] [Google Scholar]
- 3. Hagan C. L., Silhavy T. J., Kahne D. (2011) Annu. Rev. Biochem. 80, 189–210 [DOI] [PubMed] [Google Scholar]
- 4. Voulhoux R., Bos M. P., Geurtsen J., Mols M., Tommassen J. (2003) Science 299, 262–265 [DOI] [PubMed] [Google Scholar]
- 5. Paschen S. A., Neupert W., Rapaport D. (2005) Trends Biochem. Sci. 30, 575–582 [DOI] [PubMed] [Google Scholar]
- 6. Malinverni J. C., Werner J., Kim S., Sklar J. G., Kahne D., Misra R., Silhavy T. J. (2006) Mol. Microbiol. 61, 151–164 [DOI] [PubMed] [Google Scholar]
- 7. Voulhoux R., Tommassen J. (2004) Res. Microbiol. 155, 129–135 [DOI] [PubMed] [Google Scholar]
- 8. Misra R. (2007) ACS Chem. Biol. 2, 649–651 [DOI] [PubMed] [Google Scholar]
- 9. Gentle I. E., Burri L., Lithgow T. (2005) Mol. Microbiol. 58, 1216–1225 [DOI] [PubMed] [Google Scholar]
- 10. Kim S., Malinverni J. C., Sliz P., Silhavy T. J., Harrison S. C., Kahne D. (2007) Science 317, 961–964 [DOI] [PubMed] [Google Scholar]
- 11. Albrecht R., Zeth K. (2011) J. Biol. Chem. 286, 27792–27803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clantin B., Delattre A. S., Rucktooa P., Saint N., Méli A. C., Locht C., Jacob-Dubuisson F., Villeret V. (2007) Science 317, 957–961 [DOI] [PubMed] [Google Scholar]
- 13. Gatzeva-Topalova P. Z., Warner L. R., Pardi A., Sousa M. C. (2010) Structure 18, 1492–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gatzeva-Topalova P. Z., Walton T. A., Sousa M. C. (2008) Structure 16, 1873–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heuck A., Schleiffer A., Clausen T. (2011) J. Mol. Biol. 406, 659–666 [DOI] [PubMed] [Google Scholar]
- 16. Kim K. H., Kang H. S., Okon M., Escobar-Cabrera E., McIntosh L. P., Paetzel M. (2011) Biochemistry 50, 1081–1090 [DOI] [PubMed] [Google Scholar]
- 17. Kim K. H., Paetzel M. (2011) J. Mol. Biol. 406, 667–678 [DOI] [PubMed] [Google Scholar]
- 18. Knowles T. J., Browning D. F., Jeeves M., Maderbocus R., Rajesh S., Sridhar P., Manoli E., Emery D., Sommer U., Spencer A., Leyton D. L., Squire D., Chaudhuri R. R., Viant M. R., Cunningham A. F., Henderson I. R., Overduin M. (2011) EMBO Rep. 12, 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Knowles T. J., Jeeves M., Bobat S., Dancea F., McClelland D., Palmer T., Overduin M., Henderson I. R. (2008) Mol. Microbiol. 68, 1216–1227 [DOI] [PubMed] [Google Scholar]
- 20. Noinaj N., Fairman J. W., Buchanan S. K. (2011) J. Mol. Biol. 407, 248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sandoval C. M., Baker S. L., Jansen K., Metzner S. I., Sousa M. C. (2011) J. Mol. Biol. 409, 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vanini M. M., Spisni A., Sforça M. L., Pertinhez T. A., Benedetti C. E. (2008) Proteins 71, 2051–2064 [DOI] [PubMed] [Google Scholar]
- 23. Warner L. R., Varga K., Lange O. F., Baker S. L., Baker D., Sousa M. C., Pardi A. (2011) J. Mol. Biol. 411, 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Renault M., Bos M. P., Tommassen J., Baldus M. (2011) J. Am. Chem. Soc. 133, 4175–4177 [DOI] [PubMed] [Google Scholar]
- 25. Zhang H., Gao Z. Q., Hou H. F., Xu J. H., Li L. F., Su X. D., Dong Y. H. (2011) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 734–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Knowles T. J., Sridhar P., Rajesh S., Manoli E., Overduin M., Henderson I. R. (2010) Biomol. NMR Assign. 4, 179–181 [DOI] [PubMed] [Google Scholar]
- 27. Powell H. R. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 1690–1695 [DOI] [PubMed] [Google Scholar]
- 28. Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 29. Albrecht R., Zeth K. (2010) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1586–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 33. Painter J., Merritt E. A. (2006) J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 34. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 35. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 36. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 37. Reynolds C., Damerell D., Jones S. (2009) Bioinformatics 25, 413–414 [DOI] [PubMed] [Google Scholar]
- 38. Iyer L. M., Koonin E. V., Aravind L. (2001) Proteins 43, 134–144 [DOI] [PubMed] [Google Scholar]
- 39. Knowles T. J., McClelland D. M., Rajesh S., Henderson I. R., Overduin M. (2009) Biomol. NMR Assign. 3, 203–206 [DOI] [PubMed] [Google Scholar]
- 40. Sampathkumar P., Roach C., Michels P. A., Hol W. G. (2008) J. Mol. Biol. 381, 867–880 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.