Abstract
The pro-survival kinase Akt requires phosphorylation at two conserved residues, the activation loop site (Thr-308) and the hydrophobic motif site (Ser-473), for maximal activation. Previous reports indicate that mTORC2 is necessary for phosphorylation of the hydrophobic motif and that this site is not phosphorylated in cells lacking components of the mTORC2 complex, such as Sin1. Here we show that Akt can be phosphorylated at the hydrophobic motif site (Ser-473) in the absence of mTORC2. First, increasing the levels of PIP3 in Sin1−/− MEFs by (i) expression of a constitutively active PI3K or (ii) relief of a negative feedback loop on PI3K by prolonged inhibition of mTORC1 or S6K is sufficient to rescue hydrophobic motif phosphorylation of Akt. The resulting accumulation of PIP3 at the plasma membrane results in Ser-473 phosphorylation. Second, constructs of Akt in which the PH domain is constitutively disengaged from the kinase domain are phosphorylated at the hydrophobic motif site in Sin1−/− MEFs; both myristoylated-Akt and Akt lacking the PH domain are phosphorylated at Ser-473. Thus, disruption of the interface between the PH and kinase domains of Akt bypasses the requirement for mTORC2. In summary, these data support a model in which Akt can be phosphorylated at Ser-473 and activated in the absence of mTORC2 by mechanisms that depend on removal of the PH domain from the kinase domain.
Introduction
The Akt/protein kinase B Ser/Thr protein kinases play a central role in signaling downstream of phosphatidylinositol 3-kinase (PI3K)2 (1). The three isoforms (Akt1, Akt2, and Akt3) share a similar domain structure: an N-terminal pleckstrin homology (PH) domain, followed by an α-helical linker, and a C-terminal kinase domain that is controlled by phosphorylation (2). Once activated by phosphorylation, Akt phosphorylates defined substrates throughout the cell, ultimately inducing pro-proliferation and anti-apoptotic signaling pathways (1). The activation state of Akt is tightly controlled, and its dysregulation is implicated in the development of a variety of diseases, most notably cancer (3).
One key regulator of Akt phosphorylation is the mammalian target of rapamycin (mTOR), an evolutionarily conserved Ser/Thr protein kinase that forms two distinct protein complexes in cells. These complexes are differentially regulated and perform distinct cellular functions (4). The mTOR complex 1 (mTORC1) is composed of mTOR, Raptor, and mLST8 and is sensitive to inhibition by rapamycin. mTORC1 is a crucial regulator of cell growth in response to nutrients, stress, and growth factors (5, 6). A second complex, mTOR complex 2 (mTORC2), consists of mTOR, Rictor, mLST8, and Sin1 and is generally considered to be rapamycin-insensitive (7, 8). It is this complex, mTORC2, that regulates Akt and, additionally, protein kinase C (9, 10).
Upon biosynthesis, the nascent Akt polypeptide is phosphorylated at the ribosome by mTORC2 (10–12) at a conserved C-terminal site originally identified in protein kinase C and named the turn motif site (13). Phosphorylation of this residue (Thr-450 in Akt1) (14) is important for the stability of AGC kinases (15, 16). Thus, Akt is heavily ubiquitinated and degraded in cells lacking mTORC2 because the turn motif site is not phosphorylated (10, 11). This processing phosphorylation of Akt is constitutive and its dephosphorylation in cells has not been reported.
Once processed by phosphorylation at the turn motif site, Akt localizes to the cytosol, where it is maintained in an inactive conformation through the interaction between its PH and kinase domains (17). In the presence of proliferative signals, PI3K is activated and generates the lipid second messenger phosphatidylinositol 3,4,5-trisphosphate (PIP3). Akt is subsequently recruited from the cytosol to the plasma membrane through binding of its PH domain to PIP3, resulting in a conformational change that separates the PH and kinase domains and unmasks two key regulatory residues whose phosphorylations are required for maximal activation of the kinase. The first site, the activation loop (Thr-308 on Akt1), is phosphorylated by PDK-1 (18). The second site is termed the hydrophobic motif and corresponds to Ser-473 on Akt1. Several candidates have been proposed as potential hydrophobic motif kinases, including Akt itself by autophosphorylation (19), DNA-dependent protein kinase (DNA-PK) (20), integrin-linked kinase (ILK) (21), and, most recently, mTORC2 (22–24). In support of the latter, genetic ablation of the mTORC2-specific subunits, Rictor or Sin1, results in the loss of Akt phosphorylation at both the turn motif and hydrophobic motif sites, resulting in impaired signaling toward certain substrates (22–24). However, it has not been clearly established whether the primary role of mTORC2 in regulation of the hydrophobic motif of Akt is through direct phosphorylation of this site or by facilitating phosphorylation on Ser-473 by other mechanisms.
In this report, we show that Akt can be phosphorylated at the hydrophobic motif site in the absence of mTORC2. In Sin1−/− cells, activation of PI3K by overexpression of p110α or prolonged inhibition of mTORC1 and/or S6 kinase (S6K) rescues phosphorylation of Akt at Ser-473 and restores its ability to signal to downstream effectors such as FOXO1/3a. Furthermore, expression of a myristoylated Akt or Akt lacking the PH domain is sufficient to rescue hydrophobic motif phosphorylation in Sin1−/− MEFs. Our results reveal that the hydrophobic motif site of Akt can be phosphorylated independently of mTORC2 by disrupting the interface between the PH and kinase domains.
EXPERIMENTAL PROCEDURES
Plasmids
HA-tagged constructs of wild-type Akt (WT-Akt) and myristoylated Akt (Myr-Akt) were generous gifts from Dr. Alex Toker. HA-tagged constructs of WT-p70S6K and p70-S6K (T389E) were kind gifts from Drs. John Blenis and Tianyan Gao. A QuikChange Site-directed mutagenesis kit (Qiagen) was used to make single amino acid changes in the following Akt constructs according to the manufacturer's protocol. K179M was introduced into Myr-Akt to create a kinase-dead construct (Myr-KD-Akt), and T450D was introduced into WT-Akt to create a phosphomimetic at the turn motif of Akt (Akt-T450D). Amino acids 1–107 of WT-Akt were deleted to remove the PH domain (Akt-ΔPH). Myristoylated PI3K p110α was a kind gift from Dr. Peter Vogt.
Materials and Antibodies
LY294002, PF-4708671, wortmannin, and Akt inhibitor VIII were purchased from Calbiochem and dissolved in dimethyl sulfoxide (DMSO). Torin1 was a kind gift from Drs. Nathanael Gray and David Sabatini. Antibodies to PHLPP1 and PHLPP2 were purchased from Bethyl Laboratories. The following antibodies were purchased from Cell Signaling: phospho-antibodies for Thr-308 (p-T308), Thr-450 (p-T450), and Ser-473 (p-S473) of Akt, phospho-GSK-3α/β (Ser21/9), phospho-S6 ribosomal protein (Ser235/236), phospho-FOXO1/3a (Thr24/32), phospho-PI3K p85 (Tyr-458), phospho-P70S6K (Thr-389), total PI3K p85, total, p70S6K total S6, total IRS-2, PI3K p110α, and total Akt antibodies. An anti-HA monoclonal antibody was purchased from Covance. A monoclonal antibody to actin was purchased from Sigma. Protein A/G-agarose beads were obtained from Santa Cruz Biotechnology. All other materials and chemicals were reagent grade.
Cell Transfection and Immunoblotting
293T, Sin1+/+ MEF, and Sin1−/− MEF cell lines were maintained in DMEM (Cellgro) containing 10% FBS (Hyclone) and 1% penicillin/streptomycin at 37 °C in 5% CO2. Transient transfection of Sin1−/− MEFs was carried out using Lipofectamine PLUS transfection reagent (Invitrogen) following the manufacturer's protocol. For immunoblotting, cultured cells were lysed in Buffer A (50 mm Na2HPO4, 1 mm sodium pyrophosphate, 20 mm NaF, 2 mm EDTA, 2 mm EGTA, 1% Triton X-100, 1 mm DTT, 200 μm benzamidine, 40 μg ml−1 leupeptin, and 1 mm PMSF, pH 7.4), sonicated for 5 s, and protein yield was determined using Coomassie BCA Protein Assay (Pierce). Lysates containing equal amounts of protein were analyzed by SDS-PAGE, and individual blots were probed using the indicated antibodies. Densitometric analysis was performed with AlphaView analysis software (version 1.3.0.6) by Alpha Innotech Corporation.
Immunoprecipitation
Sin1−/− MEFs were transfected with the indicated DNA constructs for ∼30 h prior to harvest in Buffer B (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 5 mm EDTA, 1% Triton X-100, 10 mm sodium pyrophosphate, 1 mm phenylmethylsulfonyl fluoride, 1 mm sodium vanadate). Ten percent of the total detergent-solubilized cell lysate was quenched in SDS sample buffer for further analysis, and the remaining lysate was cleared by centrifugation at 13,000 × g for 5 min. The resulting supernatants were incubated with anti-HA antibody overnight at 4 °C and then with protein A/G-agarose beads for an additional 2 h. The immunocomplexes were washed three times with Buffer B, separated by SDS-PAGE, and analyzed by immunoblotting.
RESULTS
Akt Can Be Phosphorylated at Ser-473 in the Absence of mTOR Kinase Activity
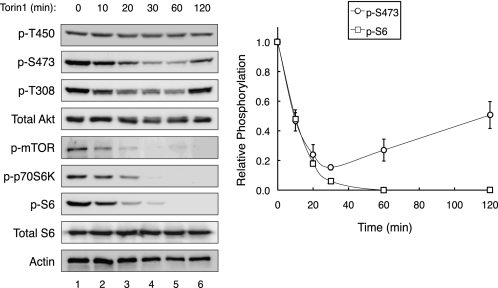
To determine whether phosphorylation of Akt on Ser-473 absolutely depends on mTOR kinase activity, we asked whether phosphorylation at this site was abolished by Torin1, a selective ATP-competitive inhibitor of mTOR that blocks the activity of both mTOR complexes (25). 293T cells were treated with 50 nm Torin1 and the relative phosphorylation of Akt at each of its three regulatory sites (Thr-450, Thr-308, Ser-473) was monitored using phosphospecific antibodies (Fig. 1). Inhibition of mTOR was assessed by examining the phosphorylation of S6 ribosomal protein, a downstream substrate of mTORC1. Fig. 1 shows that Torin1 treatment effectively abolished the phosphorylation of S6, with a half-time of 8.9 ± 0.1 min; this inhibition was maintained throughout the course of the experiment, consistent with quantitative and sustained inhibition of mTOR. Phosphorylation of Akt at the turn motif (Thr-450) was insensitive to acute mTOR inhibition (no significant decrease in phosphorylation following 120 min of Torin1 treatment, compare lanes 1 and 6). In contrast, Torin1 treatment caused the dephosphorylation of the hydrophobic motif (Ser-473) and activation loop (Thr-308) with kinetics that mirrored the inactivation of mTORC1 (Fig. 1, graph). However, unlike S6, this inhibition was transient, and the steady-state levels of Ser-473 and Thr-308 phosphorylation returned to half-maximal and basal levels, respectively, by the 120-min time point, despite the absence of mTOR kinase activity (Fig. 1). These data confirm a role for mTOR kinase activity in sustaining the steady-state phosphorylation of the hydrophobic motif of Akt, but also establish that this site can be phosphorylated in the absence of mTOR kinase activity.
FIGURE 1.
Akt can be phosphorylated at Ser-473 in the absence of mTOR kinase activity. 293T cells were treated with Torin1 (50 nm), and lysates were collected over 120 min. Immunoblotting with phosphospecific antibodies was used to determine the relative phosphorylation state of Akt on Thr-450, Thr-308, and Ser-473. The relative phosphorylation of S6 and Akt (Ser-473) normalized to total protein levels is shown in the graph; data represent the mean ± S.E. of three independent experiments.
Activation of PI3K in Cells Lacking mTORC2 Restores Akt (Ser-473) Phosphorylation
One possible explanation for the recovery of Akt (Ser-473) phosphorylation observed in response to Torin1 is that inhibition of mTORC1 relieves well-documented negative feedback inhibition of PI3K, thus promoting recruitment of Akt to the plasma membrane and poising it for phosphorylation. Phosphorylation of insulin receptor substrates (IRS) by S6K decreases their function and stability, acting as a negative feedback loop to dampen PI3K signaling in response to mTORC1 activation (26–28). Previous studies in cell lines and clinical trials demonstrate that prolonged inhibition of mTORC1 by rapamycin and its analogs leads to activation of PI3K, accumulation of PIP3 at the plasma membrane, and ultimately increased phosphorylation and activation of Akt (29–31). Thus, we hypothesized that relief of this feedback loop would promote recruitment of Akt to the plasma membrane, poising the enzyme in an active conformation that enables phosphorylation of the hydrophobic motif site in the absence of mTORC2.
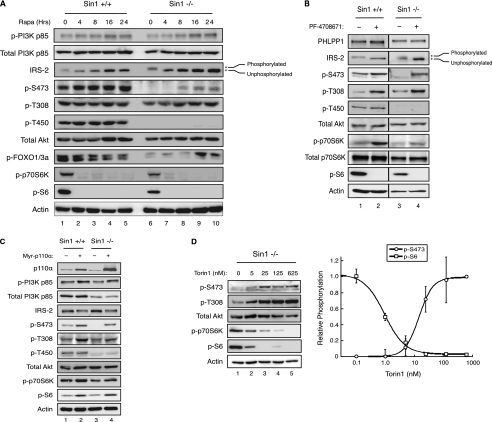
To test whether activation of PI3K via relief of negative feedback unmasks mTOR-independent Ser-473 phosphorylation, we took advantage of a cell system in which mTORC2 is absent. Previous studies have shown that Sin1 is an essential component of mTORC2, and genetic ablation results in the loss of phosphorylation at the turn motif and hydrophobic motif sites of Akt as well as loss of activity toward downstream substrates such as FOXO1/3a (22). WT and Sin1−/− MEFs were treated with 100 nm rapamycin for up to 24 h to inhibit mTORC1 and relieve negative feedback on PI3K (Fig. 2a). In both cell lines, rapamycin treatment effectively inhibited mTORC1 and S6K within 4 h, as determined by loss of S6 phosphorylation. Simultaneously, we observed an increase in the mobility of IRS-2, reflecting the loss of phosphorylation by S6K (32). As expected, over time the total levels of IRS-2 protein increased due to a decrease in proteasomal degradation that is normally promoted by S6K-mediated phosphorylation (33, 34). As a direct readout of PI3K activity, we monitored phosphorylation of p85 on Tyr-458, a site that has previously been reported to track with the activation of this enzyme (35, 36): rapamycin treatment resulted in an ∼2.5-fold increase in phosphorylation on Tyr-458 within 16 h (Fig. 2a, p-PI3K p85). These data indicate that prolonged treatment with rapamycin relieves feedback inhibition and activates PI3K in WT and Sin1−/− MEFs (Fig. 2a).
FIGURE 2.
Activation of PI3K is sufficient to restore Akt (Ser-473) phosphorylation in cells lacking mTORC2. Sin1−/− MEFs were (a) treated with rapamycin (100 nm) for the indicated times, (b) treated with DMSO or an active site inhibitor of p70S6K, PF-4708671 (5 μm) for 24 h, (c) transfected with Myr-p110α for ∼30 h, or (d) treated with the indicated doses of Torin1 for 24 h prior to harvest. Immunoblotting was used to determine total and phosphorylated protein levels, and the relative phosphorylation of S6 and Akt (Ser-473), normalized to actin, was determined by densitometry and fit to a dose response curve (kaleidagraph). Data represent the mean ± S.E. of three independent experiments.
Under basal conditions in WT MEFs, Akt was fully phosphorylated at all three regulatory sites (Fig. 2a). As previously reported, Akt was phosphorylated only on the activation loop site but not the turn motif or hydrophobic motif sites in Sin1−/− MEFs under basal conditions (8, 9, 22). However, we observed that prolonged rapamycin treatment rescued phosphorylation of Akt on the hydrophobic motif site in Sin1−/− MEFs but had no effect on phosphorylation of the turn motif site. Similar results were observed in Rictor−/− MEFs, indicating that the rescue of Ser-473 phosphorylation is not specific to the loss of Sin1 (data not shown). The rescue of Akt hydrophobic motif site phosphorylation correlated with increased levels of IRS-2 and resultant activation of PI3K (Fig. 2a). Importantly, while prolonged treatment of Sin1−/− MEFs with rapamycin did not restore hydrophobic motif site phosphorylation to WT levels (∼30%, compare lanes 5 and 10), phosphorylation of FOXO1/3a was rescued in concert with phosphorylation of Akt (Ser-473), indicating that this pool of dually-phosphorylated Akt is capable of signaling to downstream substrates (Fig. 2a). To verify that the rescue of hydrophobic motif site phosphorylation in response to long-term rapamycin treatment was a direct result of p70S6K inhibition, we asked whether a rapamycin-insensitive construct of p70S6K, containing a phosphomimetic at the mTOR site (T389E), would prevent rescue. Sin1−/− MEFs transfected with vector, WT p70S6K, or p70S6K (T389E) were treated with rapamycin for 24 h. As expected, Ser-473 phosphorylation was restored by rapamycin treatment in cells expressing both vector and WT p70S6K. However, expression of p70S6K (T389E) negated the ability of rapamycin to cause an increase in IRS-2 levels and restore Akt (Ser-473) phosphorylation, indicating that inactivation of S6K is necessary to restore phosphorylation of the hydrophobic motif site (supplemental Fig. S1). These data indicate that inhibition of mTORC1 is sufficient to rescue hydrophobic motif site phosphorylation of Akt in the absence of mTORC2.
Next, we reasoned that if relief of the negative feedback loop on PI3K in response to rapamycin was responsible for promoting hydrophobic motif phosphorylation of Akt, then direct inhibition of S6K should elicit the same effect as rapamycin. To this end, we treated WT and Sin1−/− MEFs with PF-4708671, a potent, selective inhibitor of S6K, for 24 h (37). As was observed with rapamycin, inhibition of S6K resulted in the activation and accumulation of IRS-2, which correlated with rescue of Akt phosphorylation at the hydrophobic motif site (Fig. 2b). Thus, inhibition of S6K is able to restore Ser-473 phosphorylation of Akt in the absence of mTORC2.
We have previously identified a novel phosphatase, PHLPP, which selectively dephosphorylates the hydrophobic motif sites on Akt and protein kinase C (38, 39). It was recently demonstrated that the expression of PHLPP1 is reduced by rapamycin via inhibition of S6K- and 4EBP-1-mediated protein translation (40). Thus, we asked whether the increased phosphorylation of Ser-473 resulted from reduced expression of PHLPP1. Fig. 2b shows that PHLPP1 expression levels were unaffected by the S6K inhibitor PF-4708671, under conditions in which a robust increase in Ser-473 phosphorylation was observed. Furthermore, knockdown of PHLPP1 and PHLPP2 did not alter the biphasic kinetics of Ser-473 phosphorylation following Torin1 treatment in 293T cells, although it did result in higher basal phosphorylation of Ser-473 (data not shown). These results indicate that the increase in Ser-473 phosphorylation following S6K inhibition is not due to the suppression of PHLPP phosphatase activity.
To verify that activation of PI3K, as is observed in response to rapamycin, is sufficient to restore hydrophobic motif phosphorylation of Akt in the absence of mTORC2, a myristoylated, and thus constitutively-active, construct of the p110α subunit of PI3K (Myr-p110α) was expressed in WT and Sin1−/− MEFs. Overexpression of Myr-p110α in both cell lines increased phosphorylation of Akt on the activation loop site (T308) as well as S6 phosphorylation, indicative of heightened PI3K signaling. Importantly, expression of a constitutively-active PI3K in Sin1−/− MEFs was able to restore phosphorylation of Akt at the hydrophobic motif site (Ser-473), but not the turn motif site (Fig. 2c). These results indicate that activation of PI3K alone is sufficient to restore phosphorylation of the hydrophobic motif site of Akt in the absence of mTORC2.
Furthermore, to verify that phosphorylation of Akt on the hydrophobic motif site in cells lacking mTORC2 was independent of mTOR kinase activity, we treated Sin1−/− MEFs with increasing concentrations of Torin1 and monitored the relative phosphorylation state of Akt (Ser-473) and S6 by immunoblotting. In response to increasing concentrations of Torin1, we observed a gradual decrease in S6 phosphorylation, which preceded a dose-dependent increase in phosphorylation of Akt (Ser-473) (Fig. 2d, left). Quantification of Western blots revealed an inverse correlation between the phosphorylation state of Akt (Ser-473) and S6. Akt (Ser-473) phosphorylation was detected upon nearly complete inhibition of S6 phosphorylation, and maximal phosphorylation of Ser-473 was only reached once S6K activity was entirely blocked (Fig. 2c, right). Taken together, these data indicate that activation of PI3K through multiple mechanisms, independent of mTOR kinase activity, is sufficient to restore hydrophobic motif site phosphorylation of Akt.
Rescue of Ser-473 Phosphorylation in Sin1−/− MEFs Is Dependent upon PI3K Activity and the Conformation of Akt
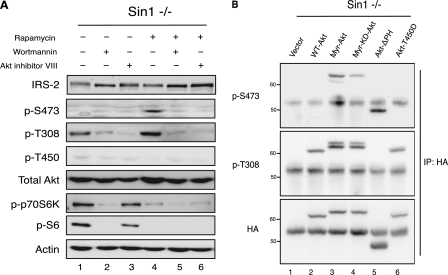
The finding that PI3K activation is sufficient to rescue hydrophobic motif phosphorylation of Akt in cells lacking mTORC2 led us to ask whether binding to PIP3 at the plasma membrane mediated this effect. Specifically, we tested whether rapamycin-induced phosphorylation of the hydrophobic motif site in Sin1−/− MEFs was prevented by 1) an inhibitor of PI3K (wortmannin) or 2) an allosteric inhibitor of Akt (Akti VIII) that locks Akt in an inactive conformation and prevents the PH domain from disengaging and binding to membranes (17). As reported above, treatment with rapamycin rescued Ser-473 phosphorylation of Akt (Fig. 3a, lane 4). However, when combined with wortmannin, rapamycin was no longer able to induce phosphorylation of Akt at the hydrophobic motif site (Fig. 3a, lane 5), revealing that PI3K activity is essential for the rescue. Similarly, treatment with Akti VIII blocked phosphorylation of Ser-473 in response to rapamycin (Fig. 3a, lane 6), indicating that translocation of Akt to the plasma membrane and/or a conformational change disrupting the PH and kinase domain interface is also required (Fig. 3a, lane 6). Similar to our results using Torin1 (Fig. 2c), we observed that the induction of Ser-473 phosphorylation in Sin1−/− MEFs in response to rapamycin tracked with inactivation of S6K (Fig. 3a, compare lanes 1 and 4). Furthermore, treatment with increasing concentrations of either LY294002 or Akti VIII led to a dose-dependent decrease in rapamycin-induced hydrophobic motif site phosphorylation, correlating with the extent of PI3K inactivation or Akt inhibition, respectively (supplemental Fig. S1). Taken together, these data indicate that PI3K activity and the ability of Akt to attain an active conformation are essential for phosphorylation of the hydrophobic motif site in the absence of mTORC2.
FIGURE 3.
Disruption of the PH and kinase domain interface of Akt is sufficient for Ser-473 phosphorylation in the absence of mTORC2. a, Sin1−/− MEFs were treated with rapamycin (100 nm), wortmannin (500 nm), or Akt inhibitor VIII (5 μm), alone or in combination, for 24 h. Immunoblotting was used to monitor the relative level of total and phosphorylated proteins. B, Sin1−/− MEFs were transfected with the indicated Akt constructs for ∼30 h prior to harvest. Exogenous Akt was immunoprecipitated and the phosphorylation state of Akt was determined using phosphospecific antibodies.
Disruption of the PH and Kinase Domain Interface of Akt Is Sufficient for Ser-473 Phosphorylation in the Absence of mTORC2
We next sought to determine whether we could bypass the necessity for mTORC2 in hydrophobic motif site phosphorylation of Akt by expression of Akt constructs having altered localization, activity, or structure. Sin1−/− MEFs were transfected with the following: vector control, WT Akt, myristoylated Akt (Myr-Akt), myristoylated kinase-dead Akt (Myr-KD-Akt), Akt lacking the PH domain (Akt-ΔPH), or Akt with a phosphomimetic at the turn motif site (Akt-T450D). The transfected Akt was then immunoprecipitated to examine the phosphorylation of Ser-473 and Thr-308. All constructs were phosphorylated to a similar extent on Thr-308 (Fig. 3b), with the exception of Akt-ΔPH, which exhibited markedly less phosphorylation at this site. Phosphorylation of Thr-450 was not detected on any of these constructs (data not shown). WT-Akt, similar to our results for endogenous Akt, was not phosphorylated on Ser-473 (lane 2). Similarly, replacement of the turn motif Thr with a phosphomimetic (T450D) failed to restore Ser-473 phosphorylation. Verifying previously published data; Myr-Akt was robustly phosphorylated on Ser-473 in the absence of mTORC2 (10). This restoration of phosphorylation largely depended on the intrinsic catalytic activity of Akt, as significantly less phosphorylation (∼30%) accumulated at Ser-473 on a myristoylated construct of Akt rendered catalytically inactive by a point mutation (K179M) in its ATP-binding pocket (lane 4). Note that two species are labeled with the p-T308 antibody, but only the slower mobility species is labeled with the p-S473 antibody, reflecting a minor pool of Akt that is dually phosphorylated (upper band, lanes 3 and 4). Importantly, Akt was robustly phosphorylated at Ser-473 when the PH domain was removed from the catalytic domain of Akt by genetic truncation (lane 5). Therefore, disruption of the PH domain from the kinase domain by either targeting Akt to the membrane and thus forcing the open conformation or by genetic truncation of the PH domain is sufficient to relieve autoinhibition and rescue hydrophobic motif site phosphorylation in the absence of mTORC2. However, phosphorylation of the hydrophobic motif site is severely compromised in an Akt construct with impaired catalytic activity.
DISCUSSION
Akt is crucial to maintaining the proper balance between cell survival and apoptosis, and its hyperactivation is strongly correlated with the onset and progression of human tumors (3). Previous studies have clearly established that PDK-1 is responsible for phosphorylation of Akt at the activation loop (Thr-308) site. Similarly, mTORC2 has been shown to serve a critical function in controlling the phosphorylation of Akt at the turn motif (Thr-450) and hydrophobic motif (Ser-473) sites (7, 8, 10, 22). Phosphorylation at the turn motif site has recently been established to occur at the ribosome by direct phosphorylation of the nascent polypeptide by mTORC2 (11, 12). In this report we reveal that Akt can be phosphorylated and activated in the absence of mTORC2. Specifically, the requirement for mTORC2 for hydrophobic motif site, but not turn motif site, phosphorylation can be bypassed by disengaging the PH domain from the kinase domain, suggesting a novel role for mTORC2 in controlling the hydrophobic motif site is to promote this disengagement, allowing phosphorylation by alternative mechanisms.
Treatment of Sin1−/− MEFs with inhibitors of mTOR, mTORC1, or S6K relieves negative feedback on IRS and drives the activation of PI3K. Similar to overexpression of a constitutively active PI3K, enhancing PIP3 signaling engages Akt at the plasma membrane, altering its conformation, exposing the hydrophobic motif site, and permitting phosphorylation through mTORC2-independent mechanisms. Interestingly, phosphorylation of Akt (Ser-473) in response to rapamycin was not observed until 8 h after treatment in Sin1−/− MEFs (Fig. 2a). Given that IRS-2 levels continued to increase up to 24 h, we speculate that a certain threshold of PIP3 must be obtained to rescue Ser-473 phosphorylation in the absence of mTORC2. Indicative of the importance of a conformational change in Akt induced by binding to PIP3, inhibition of PI3K or treatment with an allosteric inhibitor of Akt, which locks Akt in an inactive conformation, prevented phosphorylation of the hydrophobic motif site in response to rapamycin (Fig. 3a). Thus, binding to PIP3 and attaining an active conformation are essential for hydrophobic motif site phosphorylation of endogenous Akt in cells lacking mTORC2. Furthermore, under basal conditions, we observe Ser-473 phosphorylation of both a myristoylated Akt and Akt lacking the PH domain in Sin1−/− MEFs (Fig. 3b). Both of these Akt constructs have a common characteristic; the PH domain is no longer interacting with the kinase domain. These data demonstrate that disruption of the inhibitory interaction between PH domain and the kinase domain of Akt is sufficient to rescue the defect in hydrophobic motif site phosphorylation observed in the absence of mTORC2 (Fig. 4).
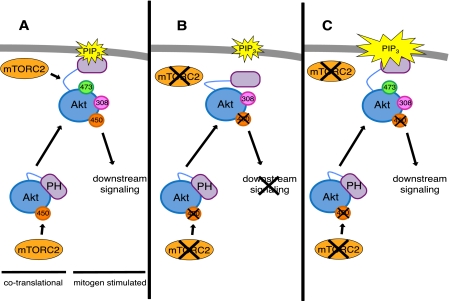
FIGURE 4.
Disruption of the interaction between the PH and kinase domains of Akt overcomes the necessity for mTORC2 for hydrophobic motif site phosphorylation. Model depicting two distinct roles for mTORC2 in controlling Akt phosphorylation: First, direct, co-translational, phosphorylation of the turn motif, and second, facilitation of disengagement of the PH domain from the kinase domain to expose Ser-473 for phosphorylation by either mTORC2 itself or by other mechanisms. a, in wild-type cells, mTORC2 promotes an active confirmation of Akt, allowing Ser-473 phosphorylation and downstream signaling. b, in Sin1−/− MEFs, Akt is only phosphorylated at Thr-308, low levels of PIP3 are not sufficient to disrupt the PH and kinase domain interaction, and Ser-473 is not phosphorylated. c, in Sin1−/− MEFs, increased and/or sustained levels of PIP3 at the plasma membrane bind Akt to elicit a conformational change allowing for phosphorylation at Ser-473 in the absence of mTORC2.
The finding that the hydrophobic motif site can be phosphorylated in an mTORC2-independent manner raises the question of which kinases modify this site. A recent report identifies two membrane-associated kinases, IκB kinase epsilon and TANK-binding kinase 1, that are able to phosphorylate membrane-bound Akt at Ser-473 in the absence of mTORC2 (41). However, additional mechanisms are also likely to control Akt because constructs of Akt that do not readily localize to the membrane (e.g. Akt-ΔPH) are robustly phosphorylated at Ser-473 in Sin1−/− MEFs. An additional mechanism to account for the mTORC2-independent phosphorylation is autophosphorylation, which accounts for phosphorylation of protein kinase C at its hydrophobic motif site (13, 42). Indeed, Akt efficiently autophosphorylates at this site in vitro (19). In this report, the ability of a myristoylated Akt to bypass the mTORC2 requirement for Ser-473 phosphorylation was in large part dependent on the intrinsic catalytic activity of Akt, a result previously noted by Jacinto and co-workers (10). Thus, numerous kinases are likely to phosphorylate the hydrophobic motif site of Akt when the enzyme is poised in the proper conformation, an event that is facilitated by mTORC2.
Whether mTORC2 activity directly controls the phosphorylation of Ser-473 of Akt remains unresolved. Here we show that the requirement for mTORC2 can be bypassed by any of several methods to disengage the PH domain from the kinase domain. However, we also show that acute inhibition of mTOR results in the dephosphorylation of Akt with kinetics that mirror the inactivation of mTOR. Furthermore, regulation of Ser-473 is specifically mediated by mTORC2 because Torin1 acutely reduces Ser-473 phosphorylation on the constitutively phosphorylated Akt-ΔPH construct in Sin1+/+ cells but not in Sin1−/− cells (supplemental Fig. S2). Thus, mTORC2 kinase activity maintains phosphate on Ser-473. The loss of phosphate on the hydrophobic motif following mTORC2 inhibition could reflect 1) suppression of the kinase component (e.g. if catalyzed directly by mTORC2 or any other mTORC2-controlled kinase) or 2) enhancement of the phosphatase component (e.g. if a hydrophobic motif phosphatase is suppressed by mTORC2) in maintaining steady-state levels of phosphate on the HM.
Note that the biphasic effect of mTOR inhibitors on Akt phosphorylation has recently been reported by Rosen and co-workers (43). Curiously, using different catalytic site inhibitors of mTOR, they observed dephosphorylation of both the activation loop and HM, but phosphate was only restored on the activation loop site; this was sufficient to rescue phosphorylation of FOXO1/3a, an Akt substrate previously shown to be dependent upon phosphorylation of the hydrophobic motif site (22). One possibility is that a small pool of Akt is being phosphorylated at the hydrophobic motif site; consistent with this, we observed that rescue of Ser-473 phosphorylation in several cell lines was minimal (below 10% of WT Ser-473 phosphorylation), yet sufficient to restore signaling to FOXO1/3a. Thus, we find that phosphorylation of both the activation loop and hydrophobic motif sites is effectively restored following prolonged mTOR inhibition, regardless of mTORC2 status.
This study reveals a novel function for mTORC2 in the regulation of Akt phosphorylation and activation. Taken together with previous findings, our data are consistent with a model in which mTORC2 has two distinct roles: 1) co-translational phosphorylation of Akt (Thr-450), which depends on mTORC2 (11, 12) and cannot be bypassed, and 2) facilitation of hydrophobic motif site phosphorylation of Akt, which can be effectively bypassed by disengaging the PH domain.
Supplementary Material
Acknowledgments
We thank Dr. Peter Vogt for the gift of the pCAGGS-Myr-p110α construct and Drs. Estella Jacinto and Bing Su for the gift of Sin1+/+ MEFs and Sin1−/− MEFs. Regarding Grant BC093021, the U.S. Army Medical Research Acquisition Activity, 820 Chandler Street, Fort Detrick, MD 21702-5014 is the awarding and administering acquisition office. The content of this article does not necessarily reflect the position or the policy of the U.S. Government.
This work was supported, in whole or in part, by National Institutes of Health Grants GM43154 and NIH GM067946 (to A. C. N.) and DoD BCRP award BC093021 (to N. A. W.). This work was also supported by the UCSD Graduate Training Program in Cellular and Molecular Pharmacology through an institutional training grant from the National Institute of General Medical Sciences T32 GM007752 (to N. A. W. and M. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- PI3K
- phosphatidylinositol 3-kinase
- PH
- pleckstrin homology
- mTOR
- mammalian target of rapamycin
- DNA-PK
- DNA-dependent protein kinase.
REFERENCES
- 1. Cantley L. C. (2002) Science 296, 1655–1657 [DOI] [PubMed] [Google Scholar]
- 2. Alessi D. R., Cohen P. (1998) Curr. Opin. Genet. Dev. 8, 55–62 [DOI] [PubMed] [Google Scholar]
- 3. Testa J. R., Tsichlis P. N. (2005) Oncogene 24, 7391–7393 [DOI] [PubMed] [Google Scholar]
- 4. Sarbassov D. D., Ali S. M., Sabatini D. M. (2005) Curr. Opin. Cell Biol. 17, 596–603 [DOI] [PubMed] [Google Scholar]
- 5. Sengupta S., Peterson T. R., Sabatini D. M. (2010) Mol. Cell 40, 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guertin D. A., Sabatini D. M. (2005) Trends Mol. Med. 11, 353–361 [DOI] [PubMed] [Google Scholar]
- 7. Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 8. Yang Q., Inoki K., Ikenoue T., Guan K. L. (2006) Genes Dev. 20, 2820–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ikenoue T., Inoki K., Yang Q., Zhou X., Guan K. L. (2008) EMBO J. 27, 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Facchinetti V., Ouyang W., Wei H., Soto N., Lazorchak A., Gould C., Lowry C., Newton A. C., Mao Y., Miao R. Q., Sessa W. C., Qin J., Zhang P., Su B., Jacinto E. (2008) EMBO J. 27, 1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oh W. J., Wu C. C., Kim S. J., Facchinetti V., Julien L. A., Finlan M., Roux P. P., Su B., Jacinto E. (2010) EMBO J. 29, 3939–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zinzalla V., Stracka D., Oppliger W., Hall M. N. (2011) Cell 144, 757–768 [DOI] [PubMed] [Google Scholar]
- 13. Keranen L. M., Dutil E. M., Newton A. C. (1995) Curr. Biol. 5, 1394–1403 [DOI] [PubMed] [Google Scholar]
- 14. Bellacosa A., Chan T. O., Ahmed N. N., Datta K., Malstrom S., Stokoe D., McCormick F., Feng J., Tsichlis P. (1998) Oncogene 17, 313–325 [DOI] [PubMed] [Google Scholar]
- 15. Edwards A. S., Faux M. C., Scott J. D., Newton A. C. (1999) J. Biol. Chem. 274, 6461–6468 [DOI] [PubMed] [Google Scholar]
- 16. Bornancin F., Parker P. J. (1996) Curr. Biol. 6, 1114–1123 [DOI] [PubMed] [Google Scholar]
- 17. Calleja V., Laguerre M., Parker P. J., Larijani B. (2009) PLoS Biol. 7, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Calleja V., Alcor D., Laguerre M., Park J., Vojnovic B., Hemmings B. A., Downward J., Parker P. J., Larijani B. (2007) PLoS Biol. 5, e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Toker A., Newton A. C. (2000) J. Biol. Chem. 275, 8271–8274 [DOI] [PubMed] [Google Scholar]
- 20. Feng J., Park J., Cron P., Hess D., Hemmings B. A. (2004) J. Biol. Chem. 279, 41189–41196 [DOI] [PubMed] [Google Scholar]
- 21. Delcommenne M., Tan C., Gray V., Rue L., Woodgett J., Dedhar S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11211–11216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jacinto E., Facchinetti V., Liu D., Soto N., Wei S., Jung S. Y., Huang Q., Qin J., Su B. (2006) Cell 127, 125–137 [DOI] [PubMed] [Google Scholar]
- 23. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 24. Guertin D. A., Stevens D. M., Thoreen C. C., Burds A. A., Kalaany N. Y., Moffat J., Brown M., Fitzgerald K. J., Sabatini D. M. (2006) Dev. Cell 11, 859–871 [DOI] [PubMed] [Google Scholar]
- 25. Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., Gray N. S. (2009) J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harrington L. S., Findlay G. M., Lamb R. F. (2005) Trends Biochem. Sci. 30, 35–42 [DOI] [PubMed] [Google Scholar]
- 27. Harrington L. S., Findlay G. M., Gray A., Tolkacheva T., Wigfield S., Rebholz H., Barnett J., Leslie N. R., Cheng S., Shepherd P. R., Gout I., Downes C. P., Lamb R. F. (2004) J. Cell Biol. 166, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shah O. J., Wang Z., Hunter T. (2004) Curr. Biol. 14, 1650–1656 [DOI] [PubMed] [Google Scholar]
- 29. Wan X., Harkavy B., Shen N., Grohar P., Helman L. J. (2007) Oncogene 26, 1932–1940 [DOI] [PubMed] [Google Scholar]
- 30. O'Reilly K. E., Rojo F., She Q. B., Solit D., Mills G. B., Smith D., Lane H., Hofmann F., Hicklin D. J., Ludwig D. L., Baselga J., Rosen N. (2006) Cancer Res. 66, 1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carracedo A., Ma L., Teruya-Feldstein J., Rojo F., Salmena L., Alimonti A., Egia A., Sasaki A. T., Thomas G., Kozma S. C., Papa A., Nardella C., Cantley L. C., Baselga J., Pandolfi P. P. (2008) J. Clin. Invest. 118, 3065–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hartley D., Cooper G. M. (2002) J. Cell Biochem. 85, 304–314 [DOI] [PubMed] [Google Scholar]
- 33. Zhande R., Mitchell J. J., Wu J., Sun X. J. (2002) Mol. Cell Biol. 22, 1016–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haruta T., Uno T., Kawahara J., Takano A., Egawa K., Sharma P. M., Olefsky J. M., Kobayashi M. (2000) Mol. Endocrinol. 14, 783–794 [DOI] [PubMed] [Google Scholar]
- 35. Kim J. H., Xu C., Keum Y. S., Reddy B., Conney A., Kong A. N. (2006) Carcinogenesis 27, 475–482 [DOI] [PubMed] [Google Scholar]
- 36. Lau C., Wang X., Song L., North M., Wiehler S., Proud D., Chow C. W. (2008) J. Immunol. 180, 870–880 [DOI] [PubMed] [Google Scholar]
- 37. Pearce L. R., Alton G. R., Richter D. T., Kath J. C., Lingardo L., Chapman J., Hwang C., Alessi D. R. (2010) Biochem. J. 431, 245–255 [DOI] [PubMed] [Google Scholar]
- 38. Gao T., Furnari F., Newton A. C. (2005) Mol Cell 18, 13–24 [DOI] [PubMed] [Google Scholar]
- 39. Gao T., Brognard J., Newton A. C. (2008) J. Biol. Chem. 283, 6300–6311 [DOI] [PubMed] [Google Scholar]
- 40. Liu J., Stevens P. D., Gao T. (2011) J. Biol. Chem. 286, 6510–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xie X., Zhang D., Zhao B., Lu M. K., You M., Condorelli G., Wang C. Y., Guan K. L. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 6474–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Behn-Krappa A., Newton A. C. (1999) Curr. Biol. 9, 728–737 [DOI] [PubMed] [Google Scholar]
- 43. Rodik-Outmezguine V. S., Chandarlapary S., Pagano N., Poulikakos P. I., Scaltriti M., Moskatel E., Baselga J., Guichard S., Rosen N. (2011) Cancer Discovery 1, 248–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.