Abstract
Dendritic cells (DCs) are the most potent antigen-presenting cells of the immune system. Depending on their maturation status, they prime T cells to induce adaptive immunity or tolerance. DCs express CD155, an immunoglobulin-like receptor binding CD226 present on T and natural killer (NK) cells. CD226 represents an important co-stimulator during T cell priming but also serves as an activating receptor on cytotoxic T and NK cells. Here, we report that cells of the T and NK cell lineage of CD155−/− mice express markedly elevated protein levels of CD226 compared with wild type (WT). On heterozygous CD155+/− T cells, CD226 up-regulation is half-maximal, implying an inverse gene-dosis effect. Moreover, CD226 up-regulation is independent of antigen-driven activation because it occurs already in thymocytes and naïve peripheral T cells. In vivo, neutralizing anti-CD155 antibody elicits up-regulation of CD226 on T cells demonstrating, that the observed modulation can be triggered by interrupting CD155-CD226 contacts. Adoptive transfers of WT or CD155−/− T cells into CD155−/− or WT recipients, respectively, revealed that CD226 modulation is accomplished in trans. Analysis of bone marrow chimeras showed that regulators in trans are of hematopoietic origin. We demonstrate that DCs are capable of manipulating CD226 levels on T cells in vivo but not in vitro, suggesting that the process of T cells actively scanning antigen-presenting DCs inside secondary lymphoid organs is required for CD226 modulation. Hence, a CD226 level divergent from WT may be exploited as a sensor to detect abnormal DC/T-cell cross-talk as illustrated for T cells in mice lacking CCR7.
Keywords: Cell Surface Receptor, Cell-Cell Interaction, Dendritic Cells, Immunology, Receptor Regulation, T Cell
Introduction
CD155 (poliovirus receptor) and its ligand CD226 (DNAX accessory molecule 1) belong to a subfamily of Ig-like receptors. CD155 is expressed on many tissue cells such as fibroblasts, epithelia, and endothelia and on a variety of hematopoietic cell types, e.g. T cells and dendritic cells (DCs)4 (1, 2). Accordingly, CD155 is implicated in diverse cellular functions. It mediates cell-cell or cell-matrix contacts and was found to support proliferation, motility, and migration of tissue cells lacking such contacts (3–5). In addition, CD155 participates in the establishment of humoral immune responses of the gastrointestinal tract and is required for proper terminal maturation of CD8+ thymocytes (1, 6, 7). CD226 was assigned a co-stimulatory capacity for CD4+ T cells as well as CD8+ T cells and may be involved in directing naïve CD4+ T cells into the T helper 1 differentiation pathway (8–13). Moreover, on NK cells and cytotoxic T cells CD226 contributes to killing of target cells (14–16). By interacting with CD155 on endothelia, CD226 may also contribute to transendothelial migration of monocytes (17). Most recently, CD226 and CD155 were assigned a role in graft-versus-host disease (18, 19).
It is well documented that CD155 is overexpressed by a variety of tumors (20–27). It appears that an aberrant expression of CD155 on cancer cells correlates with metastatic potency and poor prognosis probably because CD155 increases the invasive capacity of tumor cells (28–32). However, CD155 expression also renders tumor cells more susceptible to killing by NK and cytotoxic T cells that express the activating receptor CD226 (20–23,26,27,33–35). Indeed, tumor growth is accelerated in mice lacking CD226 (36). Therefore, it is widely accepted that the conducive effects of CD155 on tumor spread and the concurrent drawback by stimulating NK cell activity build up a delicate balance determining growth and metastasis of several types of cancer. This view was refined by recent findings showing that tumor cells overexpressing CD155 can cause a substantial down-modulation of CD226 on NK cells (23, 33) thereby probably dampening their killing efficiency. Similarly, it was observed that chronic HIV infection correlates with a down-regulation of CD226 on antigen-specific cytotoxic CD8+ T cells that may help render these cells dysfunctional (37).
Here, we demonstrate that the modulation of CD226 expression on T and NK cells is part of a natural rheostat driven in trans by CD155 present on contacting cells. Thus, naïve T cells residing in a CD155-deficient environment up-regulate CD226 regardless of whether they express CD155 themselves or not. T cells isolated from CD155-deficient mice possessing high CD226 levels down-modulate the CD226 amount present on their cell surface back to wild-type (WT) levels upon transfer into WT recipients. We provide evidence that the cell type(s) expressing CD155 and capable of regulating CD226 in trans on T cells are of hematopoietic origin. We finally demonstrate that DCs modulate CD226 surface expression on T cells upon interaction within a peripheral lymph node (PLN).
EXPERIMENTAL PROCEDURES
Mice
WT BALB/c and C57BL/6 mice were either purchased from Charles River Laboratories or bred in the Hannover Medical School animal facility. BALB/c-Pvrtm1Gbn mice were described before and are referred to as CD155−/− mice throughout the paper (6). BALB/c-Ccr7tmRfor mice were bred in the Hannover Medical School animal facility and are referred to as CCR7−/− mice (38). All experiments including animals were conducted according to the regulations of the local government and institutional guidelines.
Flow Cytometry
Single-cell suspensions of lymphoid organs were prepared as described and stained in 96-well plates in a 50–100-μl format (6). The following antibodies were used: DX5-PE, αβTCR-FITC (Invitrogen), CD11c (clone HL3, BD Biosciences), and MHCII I-Ad (BD Biosciences). Home-made antibodies (clone name) CD3 (17A2), CD4 (RMCD4-2), CD8β (RMCD8-2), CD62L (MEL-14), CD155 (3F1), and CD226 (3B3, 5G8) were either used directly labeled as indicated or detected with fluorochrome-labeled standard secondary antibodies. In some experiments, anti-CD226 mAb Tx42 kindly provided by A. Shibuya (University of Tsukuba, Japan) was used. For intracellular staining of CD226, cells isolated from secondary lymphoid organs (SLOs) were first enriched by negative isolation kits (Invitrogen) to obtain either CD4+ T cells or CD8+ T cells. The purity of the preparations was >90%. Part of the cell preparations was then subjected to fixation and permeabilization using the BD Cytofix/Cytoperm kit (BD Biosciences). Flow cytometry was performed on a FACSCalibur or a LSRII flow cytometer (BD Biosciences); the data were evaluated using WinList 5.0. Cell populations were identified by the gating strategies as indicated in the figures. In particular, DCs isolated from PLNs were identified based on their CD11c/MHCII expression as LN resident DCs (CD11c+/MHCIIint), and a small population of semi-mature DCs immigrated from the periphery via afferent lymph (CD11c+MHCIIhi) (1, 39). DAPI or propidium iodide was included in all samples prepared for flow cytometry to allow for exclusion of dead cells (except for permeabilized cells).
Antibody Treatment of Mice
WT BALB/c mice were injected intravenously with 400 μg of anti-CD155 mAb 3F1 or an isotype control mAb every 5 days. The animals were sacrificed 25 days following the first injection.
FTY720 Application
CD155−/− BALB/c mice received FTY720 (40 μg/mouse; Calbiochem) by gavage 6–8 h following DC transfer. Gavage was repeated 2 days later. Drug effectiveness was confirmed by analyzing lymphopenia induction in the blood of treated animals. A group of mice not receiving DCs was used to analyze the effect of FTY720 on CD226 levels of T cells isolated from PLNs.
Real-time PCR
Sample preparation and real-time PCR was done as described (15) on CD4+CD8−CD62L+ cells and CD4−CD8+CD62L+ BALB/c cells obtained by FACS sorting of lymph node cells. Thymocytes were sorted as CD4+CD8− single positive (SP) cells and CD4−CD8+SP cells.
Adoptive Transfer of Lymphocytes
BALB/c lymphocytes were prepared from lymph nodes (LNs) and purified using Lympholyte M (Cedarlane). WT cells were labeled at 37 °C for 10 min in 10 μm 5-carboxytetramethylrhodamine succinimidyl ester (TAMRA) whereas CD155−/− cells were labeled with 75 nm carboxyfluorescein diacetate succinimidyl ester (CFSE). Labeling with dyes was also done vice versa in some experiments to exclude dye-specific influences. WT and CD155−/− dye-labeled cells were mixed 1:1 and 5 × 106 injected intravenously into WT or CD155−/− recipients. Spleens were harvested 3 days later and the expression of CD226 determined. For some adoptive transfers (kinetics of CD226 regulation), T cells were purified using a T cell purification kit (Dynal mouse T cell negative isolation kit; Invitrogen) before labeling with TAMRA. For adoptive transfer into CD3ϵ−/− mice (C57BL/6) lacking T cells, 2 × 107 WT or CD155−/− lymphocytes were injected intravenously, and the spleen as well as PLNs were harvested 7 days later. T cells were analyzed by flow cytometry as indicated.
Bone Marrow Chimeras
Bone marrow was prepared from femur and tibia of BALB/c donors, and the cells were counted and washed in RPMI 1640 medium. For intravenous injection, 107cells were resuspended in PBS. Recipient mice were irradiated twice with 4 Grays each at an interval of 4 h. Mice were analyzed 6–8 weeks later.
Generation of DCs in Vitro
Bone marrow was harvested from femur and tibia of BALB/c mice, and 2 × 106 cells were cultivated in a 10-cm bacterial dish along with 10 ml of RPMI 1640 medium/10% FCS/50 μm 2-mercaptoethanol in the presence of GM-CSF. GM-CSF was obtained as culture supernatant of a GM-CSF-producing cell line (40). After 3 days 10 ml of medium was added, and after 6 days 10 ml of medium was replaced by fresh medium. At day 7 the immature DCs were collected from the plate and used for the experiments. In the case of DC transfer in vivo, DCs were stimulated for 24 h with TNFα (30 ng/ml) and prostaglandin E2 (1 μg/ml; Sigma) prior to injection.
Co-culture in Vitro of DCs and T Cells
In vitro differentiated DCs and T cells were mixed to obtain a total of 5 × 104 to 105 cells at the ratios indicated. Cells were seeded into round bottom 96-well plates in a volume of 200 μl of RPMI 1640 medium/10% FCS per well. Cells were incubated for 3 days, harvested, and subjected to expression analysis.
Intralymphatic Injection of DCs and Immunohistology
Intralymphatic injection was done as described elsewhere (41). In short, mice were anesthetized by injection of ketamine/medetomidine. The calves to be used for intralymphatic injection were shaved and depilated before a skin incision was made. A borosilicate glass capillary was filled with 2 × 105 DCs in 5 μl of PBS using a Microinjector PLI-100 (Harvard Apparatus). After locating an afferent lymphatic vessel of the popliteal LN (popLN), a micromanipulator (Harvard Apparatus) was used to guide the capillary and penetrate the lymphatic vessel. Following surgery, topical tissue adhesive (Nexabond) was used to close the incision. Four days later, the popLN and a nondraining LN were collected and cells isolated for flow cytometry. Alternatively, DCs were labeled with TAMRA prior to transfer and the popLN embedded after 4 days in TissueTek for cryosectioning. Staining of cryosections was done as described (42) using Cy5-labeled anti-CD3 (clone 2C11) and DAPI for detecting nuclei. Immunofluorescence imaging was performed using a BX61 microscope and analySIS P software (Olympus). The determination of immigration efficiency of WT or CD155−/− DCs into a given LN was done by counting the numbers of TAMRA+ DCs found to locate within the CD3+ areas of eight or nine randomly chosen cryosections representing the entire LN.
RESULTS
Expression of CD226 Is increased on T, NK, and NKT cells of CD155−/− Origin
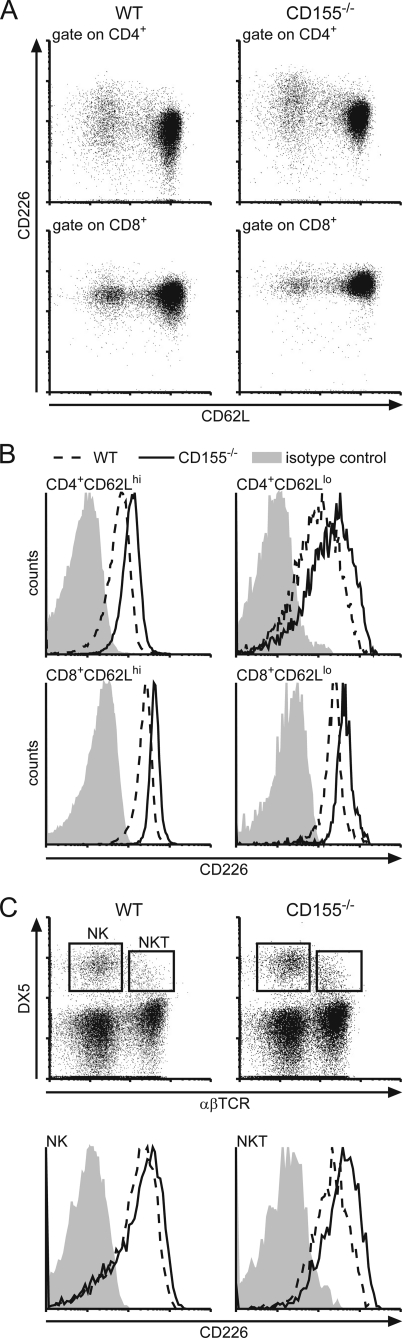
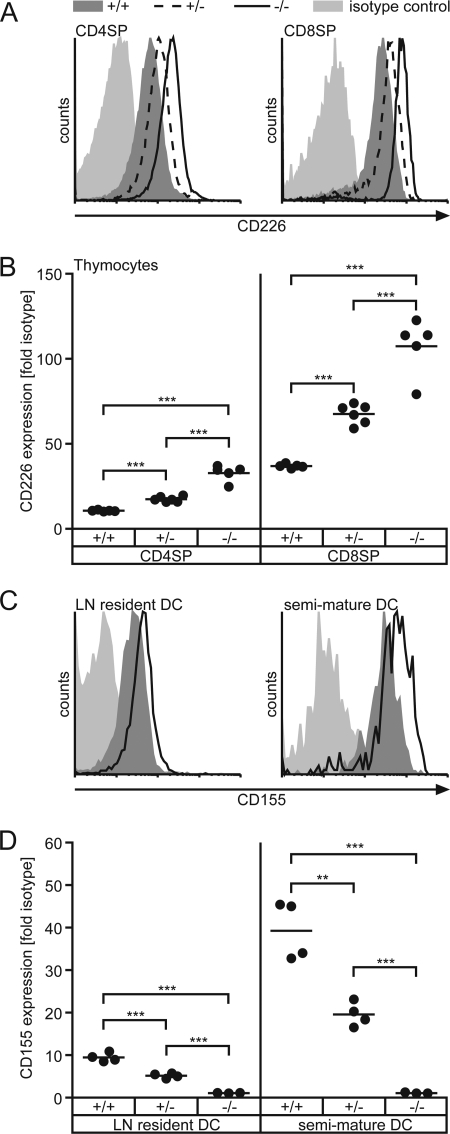
When analyzing leukocytes isolated from CD155-deficient mice for their surface expression of CD226 by flow cytometry, we observed substantially increased levels of CD226 on naïve (CD62Lhi) as well as antigen-experienced (CD62Llo) CD4+ T cells and CD8+ T cells isolated from spleen or peripheral lymph nodes (Fig. 1, A and B). Calculated as the ratio of mean fluorescence elicited by binding of anti-CD226 mAb to that of an isotype control antibody ± S.D. we found for T cells of PLNs (n = 5): WT CD4+CD62Lhi (6.00 ± 0.69), CD155−/− CD4+CD62Lhi (11.70 ± 1.88), WT CD4+CD62Llo (12.90 ± 2.07), CD155−/− CD4+CD62Llo (24.50 ± 3.77), WT CD8+CD62Lhi (7.30 ± 2.45), CD155−/− CD8+CD62Lhi (12.90 ± 5.20), WT CD8+CD62Llo (7.60 ± 2.80), CD155−/− CD8+CD62Llo (13.00 ± 5.66). The extent of up-regulation of CD226 due to CD155 absence was found to be virtually identical for naïve and antigen experienced T cells. Additionally, this effect was also observed on DX5+αβTCR− NK as well as DX5+αβTCR+ NKT cells (Fig. 1C). Whereas CD226 was up-modulated to a similar extent on CD155−/− NKT cells and on CD155−/− T cells (WT, 24.00 ± 4.88; CD155−/−, 38.80 ± 15.96; n = 5 mice) up-regulation on NK cells was marginal yet represented a consistent feature whenever CD155−/− NK cells were analyzed. It was shown before that upon T cell activation, CD226 is up-regulated (8, 15). However, on CD155−/− cells increased levels of CD226 could be observed already in naïve CD62Lhi T cells compared with WT. This indicates that a previous or ongoing encounter with antigen causing full stimulation via TCR is not required for CD226 modulation. In line with this, a significant up-regulation of CD226 was already evident in SP thymocytes (Fig. 2, A and B). Including heterozygous CD155+/− animals in the analyses, we observed that SP thymocytes of these mice displayed a degree of CD226 up-regulation approaching approximately half the extent as seen in SP thymocytes derived from CD155−/− animals. CD155 expression is gene-dosis-dependent on heterozygous cells possessing half the amount of CD155 as found on WT cells (Fig. 2, C and D for lymph node DCs) (6). These results suggest that the degree of CD226 up-modulation responds in a linear fashion to the amount of CD155 present on either the cells themselves or on cells contacting the thymocytes (see below). The described gene-dosis effect of CD226 up-regulation was not only found on thymocytes but also on peripheral T cells (data not shown).
FIGURE 1.
Comparative expression analysis of CD226 on leukocytes of WT and CD155−/− origin. Cells were isolated from spleen and stained for CD226 expression using mAb 3B3. A, dot plots showing the levels of CD226 on WT and CD155−/− T cells as indicated. B, representative histograms of CD226 levels on naïve T cells (CD62Lhi) and antigen-experienced T cells (CD62Llo). Dashed line, WT; solid line, CD155−/−; shaded area, isotype control stain. C, upper, expression of DX5 and αβTCR on splenocytes of WT and CD155−/− origin. Lower, determination of CD226 expression on NK cells (DX5+αβTCR−) and NKT cells (DX5+αβTCR+) as indicated in B.
FIGURE 2.
Gene-dosis dependent up-regulation of CD226. SP thymocytes (A and B) or LN-derived DCs (C and D) of WT (CD155+/+), heterozygous (CD155+/−), and CD155-deficient mice (CD155−/−) were stained to detect expression of CD226 or CD155, respectively. A, representative CD226 stains of SP thymocytes. Light shaded area, isotype control; dark shaded area, WT; dashed line, CD155+/−; solid line, CD155−/−. B, quantitative determination of CD226 levels in SP thymocytes. C, representative CD155 stains of LN resident DCs (CD11c+MHCIIint) and recently immigrated semi-mature DCs (CD11c+MHCIIhi). Light shaded area, CD155−/−; dark shaded area, CD155+/−; solid line, WT. D, quantitative determination of CD155 levels in LN-derived DCs as indicated. Each dot represents cells from one animal of indicated origin. WT, +/+; CD155+/−, +/−; CD155−/−: −/−; **, p < 0.01; ***, p < 0.001.
CD226 Modulation Occurs at a Post-transcriptional Level
It was of interest to investigate whether the increased levels of CD226 observed on CD155−/− T cells are accompanied by elevated amounts of mRNA coding for CD226. An initial analysis using gene arrays indicated that mRNA levels specific for CD226 did not differ between peripheral WT and CD155−/− T cells. To investigate this more thoroughly, we applied real-time PCR on cDNA established from SP thymocytes and naïve peripheral T cells sorted by flow cytometry (15). Interestingly, we failed to observe statistically significant differences in the amount of CD226-specific mRNA when comparing WT and CD155-deficient cells (Fig. 3A). This demonstrates that the modulation of CD226 expression occurs at a post-transcriptional step. An altered level of cell surface-bound CD226 as observed in CD155−/− cells may be due to an imbalance between CD226 present on the surface and receptor that is possibly stored inside the cells. Therefore, we subjected T cells from WT and CD155−/− mice to flow cytometry and compared the mean fluorescence intensities specific for CD226 on either native cells or those that underwent fixation and permeabilization prior to mAb staining. The latter allows for detection of surface-bound as well as cell internal CD226, but the intensity of antibody staining suffers from chemical modification and/or denaturation of epitopes thus dampening signal intensities. Yet, an internal comparison of WT and CD155−/− T cells stained by either procedure indicates that cells lacking CD155 indeed possess more CD226 total protein compared with their WT counterparts (Fig. 3B).
FIGURE 3.
Determination of total versus cell surface-bound CD226 protein and CD226-specific mRNA. A, real-time PCR analysis of CD226-specific mRNA present in SP thymocytes and naïve peripheral T cells. Cells were sorted by flow cytometry and the CD226 mRNA quantified as described under “Experimental Procedures.” Each dot represents cells isolated from one animal. n.s., not significant. B, peripheral CD4+ T cells or CD8+ T cells purified by magnetic bead separation as described under “Experimental Procedures.” Cells were then either stained directly for detection of CD226 (surface staining) or fixed and permeabilized prior to CD226 staining (total CD226 levels; permeabilized cells). Shown are mean fluorescence intensities (MFI) because staining with isotype control mAb caused a uniform but elevated level of background when applied on fixed/permeabilized cells. Each dot represents cells isolated from one animal. The summarized result from two experiments is shown. The mean fluorescence elicited by staining of CD4+ T cells was set at 100 in each experiment. WT, +/+; CD155−/−, −/−; ***, p < 0.001.
CD226 Modulation Can Be Induced in Vivo by a CD155-neutralizing Antibody
The observed up-regulation of CD226 may have occurred during T cell development and subsequently maintained throughout T cell life. Therefore, we made use of the anti-CD155 mAb 3F1 known to interrupt the CD155-CD226 interaction (1, 15). This mAb was shown earlier to provoke loss of mature CD8 SP from WT thymi thus mimicking a CD155 knock-out phenotype (6). Upon injection of mAb 3F1 into WT animals, CD4+ T cells as well as CD8+ T cells up-regulated their CD226 level to an extent observed in the CD155−/− cells (Fig. 4A). This is particularly true also for antigen-experienced cells (not depicted separately). This result demonstrates that a disruption of the CD155-CD226 interaction is responsible for CD226 up-modulation on T cells. Based on these observations, it appears unlikely that absence of CD155 in cis and a concurrent lack of CD155-triggered signaling events caused CD226 up-regulation in CD155−/− T cells.
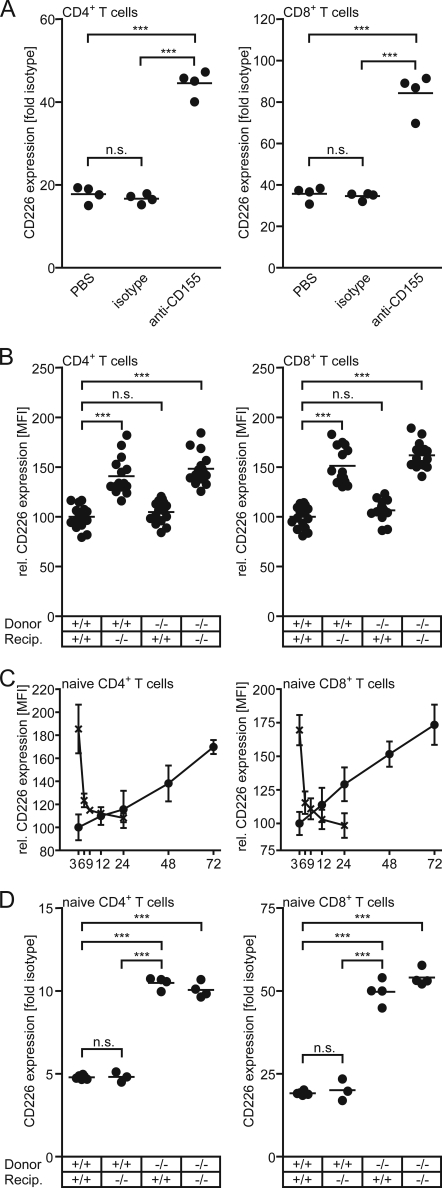
FIGURE 4.
CD226 expression on T cells is governed in trans by cells of hematopoietic origin. A, mice were treated with either PBS, an isotype control mAb, or mAb 3F1 neutralizing CD155 for 25 days as specified under “Experimental Procedures” before the expression of CD226 was determined on T cells harvested from PLNs. Data of a representative experiment are shown. B, a mixture of 5 × 106 WT and CD155−/− lymphocytes, labeled with either CFSE or TAMRA, was injected intravenously into WT or CD155−/− recipients. Three days later, the spleens were harvested, and the expression of CD226 was determined on CD4+ T cells and CD8+ T cells. To allow summarizing the results from several experiments, the data were normalized: the CD226 expression on CD4+ T cells, and CD8+ T cells, respectively, was set at 100 for WT animals receiving WT cells. C, dynamics of CD226 regulation is shown. CD226 levels on WT or CD155−/− T cells were determined following their adoptive transfer into CD155−/− or WT recipients, respectively, for the indicated periods of time (hours). Data were collected in two experiments in which a total of five or six PLNs/time point were analyzed. Shown are means of mean fluorescence intensity (MFI) ± S.D. (error bars). Gates were set to identify transferred naïve CD4+ T cells (left) and naïve CD8+ T cells (right). Data were normalized by setting the average mean fluorescence intensity of CD226 observed in WT CD4+ T cells and WT CD8+ T cells, respectively, at 100. D, syngeneic bone marrow chimeric mice were generated as described under “Experimental Procedures.” Four groups of mice were established based on WT or CD155−/− donor bone marrow and WT or CD155−/− recipients. PLNs were harvested 6–8 weeks later and the CD226 expression determined on the cell types indicated. Each dot represents cells obtained from one mouse. Note that differences in the -fold isotype control stain in T cell subsets as seen in A versus D are for the most part due to the use of different batches of secondary antibody conjugates. WT, +/+; CD155−/−, −/−; *, p < 0.05; ***, p < 0.001; n.s., not significant.
CD226 Modulation Is Induced by a CD155-CD226 Interaction in Trans
Because the leukocytes under investigation express both CD155 and CD226, respectively, mAb-triggered up-regulation is not suitable to discriminate whether CD226 modulation is due to interrupted interaction of CD155 with CD226 in cis or in trans. Therefore, we adoptively transferred fluorescently labeled lymphocytes isolated from WT into CD155−/− recipients or vice versa, CD155-deficient lymphocytes into WT animals. As controls, transfers from WT donors to WT recipients as well as from CD155−/− donors to CD155−/− recipients were done. When analyzing the T cells 3 days later, we found that the transferred cells adopted the CD226 level characteristic for the recipient, i.e. WT cells up-regulated CD226 in CD155−/− recipients whereas CD155−/− cells in WT recipients down-modulated their CD226 expression to exactly that level typically found for nontransferred recipient cells (Fig. 4B). Thus, CD226 expression adjusts to the level specific of the recipient's genotype irrespective of the cells' own CD155 expression. This indicates that cells present in the recipient govern in trans the level of CD226 on T cells. Adoptive transfers were also used to determine the kinetics of CD226 regulation in vivo. To this end, fluorescently labeled WT or CD155−/− T cells were injected into CD155−/−, or WT mice, respectively. Mice were sacrificed at the time points (hours) indicated in Fig. 4C, and the transferred T cells were analyzed following their reisolation. Up-modulation of CD226 on WT cells proceeded at a slow but constant rate in both CD4+ and CD8+ T cells, reaching levels of CD226 characteristically found on CD155−/− T cells after ∼3 days. In contrast, CD226 was rapidly down-regulated on CD155−/− cells approaching WT levels already 3–6 h following transfer into WT mice.
CD226 Modulation in Trans Is Induced by a Cell of Hematopoietic Origin
To dissect in more detail which cell type(s) are capable of regulating CD226 on T cells in trans, we established bone marrow chimera. WT or CD155−/− animals were reconstituted with WT or CD155−/− bone marrow following lethal irradiation. Six to 8 weeks later, the T cells grown out of the transplant were analyzed for their CD226 expression. Contrary to the results obtained following adoptive cell transfers, the T cells displayed a CD226 expression strength typical for the donor whereas the recipients genetic background (WT or CD155−/−) was irrelevant (Fig. 4D). The very same result was obtained when CD226 levels on SP thymocytes were investigated (data not shown). This suggests that cells differentiating from the donor bone marrow manipulate the CD226 levels on T cells and SP thymocytes. Because T cells from reconstituted animals completely adapted the CD226 phenotype of the donor, these results also imply that nonhematopoietic cells do not participate noticeably in regulation of T-expressed CD226 regardless of whether they express CD155.
A Non-T Cell Type Is Capable of Regulating CD226 on T Cells in Trans
The results presented thus far narrow the potential cell type(s) regulating CD226 on T cells in trans to those arising from bone marrow and residing in SLOs and thymus. Inside these organs, T cells are in constant movement, coming into close contact with each other. To investigate whether interactions between T cells trigger CD226 regulation, T cells of WT and CD155−/− origin were mixed in various ratios and co-cultured for up to 3 days before their CD226 levels were determined. We failed to observe significant regulation of CD226 on WT or CD155−/− T cells as observed in vivo (data not shown). We also adoptively transferred WT or CD155−/− T cells into CD3ϵ knock-out mice and determined the CD226 expression on the T cells reisolated 7 days later. CD3ϵ knock-out mice lack endogenous T cells, ensuring that all T cells recovered following transfer are of donor origin. If T cells control each others' CD226 expression in trans, it would be expected that WT and CD155−/− T cells keep their divergent original levels during their in vivo passage in the CD3ϵ knock-out hosts. However, analyzing the CD226 levels following reisolation we did not find differences any more between transferred T cells of WT and CD155−/− origin (Fig. 5A). This indicates that the recipient's milieu actively manipulated CD226 expression on T cells to achieve a uniform level. Even though the unique environment stimulating homeostatic proliferation (see below) precludes a comparison with the levels found on regular WT cells, this result indicates that a non-T cell type expressing CD155 and present in the donor is capable of adjusting CD226 levels on the transferred T cells.
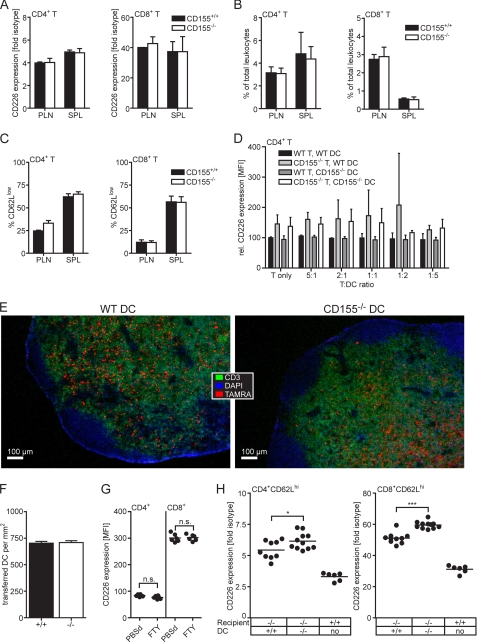
FIGURE 5.
DCs can modulate CD226 levels on T cells in trans. A–C, CD3ϵ-deficient mice were adoptively transferred with 2 × 107 lymphocytes of WT or CD155−/− origin. Seven days later, lymphocytes were prepared from PLNs and spleen (SPL). A, expression levels of CD226 on CD4+ T cells and CD8+ T cells. B, frequency of recovered CD4+ T cells and CD8+ T cells in PLN and spleen. C, frequency of CD62lo cells among the recovered T cells. Shown is one representative experiment of two where the results obtained from two WT and four CD155−/− recipients are summarized. D, in vitro differentiated DCs and T cells mixed in the indicated ratios and seeded into 96-well plates in duplicate. Cells were stained 3 days later to determine their CD226 expression. Shown are the combined results of two experiments where the mean fluorescence intensity (MFI) of the T cells incubated in the absence of DCs was set at 100. E, fluorescence microscopic images (4× objective) of LNs 4 days following transfer of TAMRA-labeled DCs of WT or CD155−/− origin. Cryosections were stained to detect the T cell area (CD3) and counterstained with DAPI allowing identification of nuclei. F, immigration efficiency of transferred WT and CD155−/− DCs determined as described under “Experimental Procedures” and expressed as numbers of TAMRA+ DCs detected/mm2 T cell area. Shown are the combined results (± S.D.; error bars) from two transfers of WT DCs into CD155−/− animals and three transfers of CD155−/− DCs into WT recipients. G, CD4+ T cells and CD8+ T cells of PLN of mice treated for 4 days with FTY720 or PBS were analyzed for their CD226 expression. Summary results from two experiments are shown with each dot representing cells from one PLN. H, determination of CD226 expression done 4 days following transfer of the indicated matured DCs into CD155−/− recipients. All mice were treated with FTY720 regardless of whether they received DCs or not. Gates were set to identify naïve CD4+CD62Lhi T cells and CD8+CD62Lhi T cells. Shown are the combined data from two experiments where each dot represents cells isolated from one LN. n.s., not significant; *, p < 0.05; ***, p < 0.001.
As shown in Fig. 5B, the sizes of the T cell pools of both CD4+ and CD8+ cells were identical regardless of whether WT or CD155−/− cells were transferred. In chronically lymphopenic hosts, most transferred T cells undergo a rapid proliferation within 1 week, and a huge proportion of these cells is CD62Llo (43). We observed a considerable fraction of such cells proving that spontaneous proliferation occurred in the CD3ϵ knock-out mice (Fig. 5C). This suggests that survival of the T cells and/or their spontaneous proliferation in the lymphopenic mice is not influenced by CD155 deficiency. All in all, in vitro as well as in vivo evidence indicated that despite expressing CD155, T cells lack the capacity to regulate CD226 levels on other T cells contacting them.
DCs Can Influence CD226 Levels on T Cells in Vivo but Not in Vitro
To test the potential of DCs to regulate CD226 expression upon contacting T cells we generated DCs in vitro from bone marrow and co-cultivated them along with CD4+ T cells at varying DC:T cell ratios. Three days later, the expression of CD226 on the CD4+ T cells was determined. We failed to observe an influence of the DCs on CD226 expression because WT and CD155−/− T cells kept their original CD226 level regardless of whether they were co-incubated in the absence or presence of DCs of whatever origin (Fig. 5D). Experiments similar to those for CD4+ T cells could not be performed using CD8+ T cells that do not survive long enough under the nonstimulatory in vitro conditions applied. The possible influence of DCs on CD226 levels of T cells was then explored in vivo. To this end, in vitro differentiated WT or CD155−/− DCs were injected into the afferent lymph vessel connecting to the popLN of CD155−/− recipients. By this route, the DCs gain immediate access to the subcapsular sinuses of the targeted popLN from where they subsequently migrate into the T cell zone and make contact with the endogenous T cells present therein. To prevent the loss of cells that underwent contact with injected DCs, T cells were trapped inside the LN by blocking their egress following FTY720 treatment (44). By blocking exit of lymphocytes from LN, FTY720 causes lymphopenia thereby also interrupting the constant influx of T cells from the blood into LN. Therefore, this strategy allows monitoring of the CD226 expression of those T cells already present at the time the DCs arrived in the popLN and excludes a significant contribution of newly immigrated (CD226hi) CD155−/− T cells that were not yet exposed to the putative influence of the transferred DCs. Because FTY720 also inhibits DC migration (40), drug treatment was started only 6–8 h after DC transfer when the vast majority of DCs reached their final destination, the T cell zone. The efficient immigration of the transferred DCs into the popLN was controlled by immunohistology, also demonstrating that DCs of WT or CD155−/− origin settle the T cell zone similarly well (Fig. 5, E and F). T cells from the popLNs as well as WT control PLNs were isolated 4 days following DC transfer and analyzed for their CD226 expression. A significant reduction in CD226 levels was observed for both CD4+ and CD8+ T cells upon transfer of WT DCs compared with that of CD155−/− DCs (Fig. 5H). A complete accommodation to WT levels was not found presumably due to an insufficient extent of DC/T contacts and/or the competition of the T cells interacting with endogenous DCs that are of CD155−/− origin. These DCs would counteract the influence of adoptively transferred WT DCs by instructing the contacting T cells to keep a high level of CD226. As an additional control, WT mice were treated with FTY720 to test whether the drug manipulates CD226 expression. Levels of CD226 are unchanged in T cells from PLNs of FTY720-treated mice, suggesting that the drug does not influence CD226 expression (Fig. 5G). In summary, our observations demonstrate that DCs are capable of influencing in trans the levels of CD226 on T cells but can do so only in an in vivo environment.
CD226 Expression Is Altered on T cells in CCR7−/− Mice
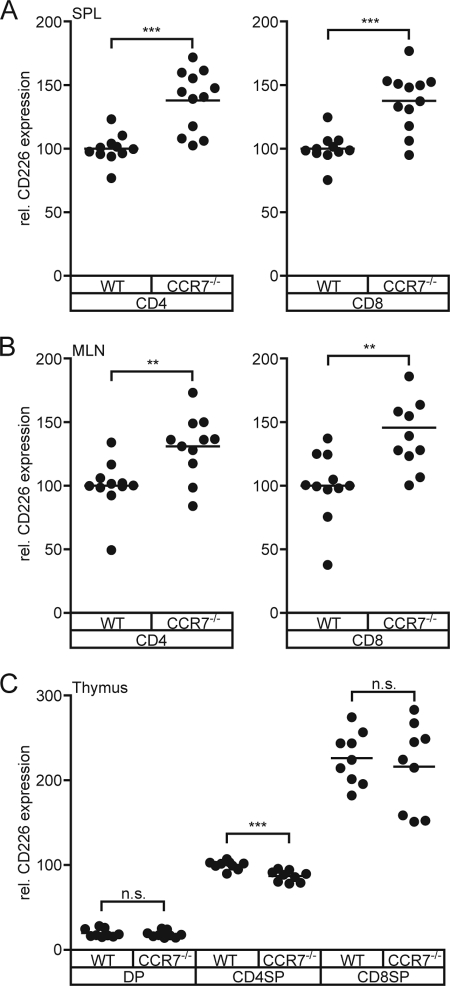
It appears that ongoing contacts between CD155-expressing DCs and T cells are of importance to keep the expression of CD226 at exactly that level that is observed in WT. Once the frequency or quality of these contacts is out of balance, the “system” reacts with changing the amount of CD226 detectable on T cells. Taken the other way around, a non-WT expression level of CD226 on T cells may represent a hint for an irregular cross-talk between DCs and T cells. To test this, we analyzed the expression of CD226 on peripheral T cells of CCR7-deficient (CCR7−/−) mice. These mice display a severely disturbed recirculation of lymphocytes into LNs and almost abrogated migration of DCs into SLOs via afferent lymphatics. Additionally, the internal architecture of their SLOs is disturbed (38, 39). In spleen, CCR7−/− T cells reside to a large extent in the red pulpa instead in the white pulpa where lymphocytes accumulate normally. Indeed, it is evident that T cells isolated from CCR7−/− spleens possess markedly elevated levels of CD226 (Fig. 6A). Similarly, up-regulated levels of CD226 on T cells were also observed in the mesenteric LN of CCR7−/− mice (Fig. 6B).
FIGURE 6.
CD226 is aberrantly expressed on CCR7−/− T cells. Cells were isolated from CCR7−/− mice and stained for CD226 expression. A and B, CD226 levels on CD4+ T cells and CD8+ T cells from spleen (A) and mesenteric LNs (B). The level found in WT was set at 100. C, CD226 levels on CD4+ SP and CD8+ SP thymocytes. The level found on WT CD4+ thymocytes was set at 100. Summary results are from four experiments. Each dot represents cells from one mouse. n.s., not significant; **, p < 0.01; ***, p < 0.001.
Further analyses included the thymus of CCR7−/− mice because we found that CD155−/− SP thymocytes displayed a CD226 up-regulation similar to that observed for peripheral CD155−/− T cells. In addition, DCs in thymus express CD155 (6) and participate in negative selection proving that thymocytes come into close contact with DCs (45, 46). The thymi of CCR7−/− animals are distinguished by an abnormal architecture regarding the size and arrangement of cortical and medullary areas. Moreover, CCR7−/− T cells are prone to provoke autoimmunity that is at least in part due to a skewed negative selection of thymocytes (47, 48). Contrary to the expectation and to the results shown for peripheral CD8+ T cells, we found that CCR7−/− CD8+ SP thymocytes express WT levels of CD226 (Fig. 6C). Interestingly, CCR7−/− CD4+ SP thymocytes possess slightly but significantly lower CD226 levels. This would suggest that despite the observed abnormalities, CCR7−/− SP thymocytes meet a “normal CD155 environment” during their stay in thymus, excluding aberrant DC/SP thymocyte contacts as a cause for defects in negative selection. In support of this, we found that DCs show a WT-like distribution pattern in CCR7−/− thymi (data not shown), confirming a very recent report (49).
The results described here unravel the existence of a regulatory mechanism that contributes to determine the levels of cell surface-exposed receptor. Regulation takes place with different kinetics: Up-regulation of CD226 proceeds slowly whereas its down-modulation occurs rather rapidly. Intriguingly, the control is exerted in trans and requires an in vivo context, demonstrating that the presence of CD155 on contacting cells is necessary but not sufficient for CD226 regulation. Notably, not any cell expressing CD155 is licensed to participate in CD226 regulation on T cells. We found that DCs but not T cells or stromal cells are capable of instructing contacting T cells to modulate their level of CD226.
DISCUSSION
We demonstrated that the CD226/CD155 system represents a dynamic unit where cells expressing CD155 have the capacity of regulating in trans the level of CD226 on T cells. We identified DCs to be at least partially involved in this regulation. The underlying mechanism that executes CD226 regulation on the surface of naïve T cells remains elusive. The finding that CD226 modulation requires an in vivo environment prevented classical experiments exploring the effect of diverse drugs on cellular pathways in in vitro assays. However, the rather rapid down-regulation of CD226 would be in favor of endocytosis and is reminiscent of descriptions in the literature documenting, for example, that MHC class I chain-related molecules overexpressed by cancer cells down-regulate surface-expressed NKG2D on T and NK cells with similar kinetics (4–24 h) (50, 51). Endocytosis was shown to be involved in this process (51, 52), but other mechanisms such as trogocytosis or receptor shedding may also contribute to receptor down-regulation. In contrast, up-regulation of CD226 proceeds slowly but at a constant rate, requiring approximately 3 days to reach a plateau characterized by the level of CD226 observed in CD155−/− T cells. It is assumed that this represents a simple receptor accumulation over time due to the absence of CD155 on cells contacting the T cells. We showed that mRNA levels coding for CD226 are unaffected in T cells of CD155−/− mice but that the total protein content as well as the amount of CD226 detectable on the cell surface are increased. This CD226 tuning in response to interaction with CD155 reflects a completely independent mechanism from that observed for T cells differentiating into follicular helper T cells where CD226 regulation occurs at the level of transcription (7). In addition, we never observed a cell type naturally devoid of CD226 becoming CD226+ when CD155 is absent (e.g. B cells; data not shown), suggesting that CD155 can only operate on a CD226 level already existing on other cells. These findings are in line with the idea that CD226 modulation represents a post-transcriptional event. Interestingly, loss of CD226 on cytotoxic T cells of HIV-patients is also most likely accomplished at a post-transcriptional level (37).
Because mAb neutralizing CD155 exerted the same effect as the CD155 knock-out, we conclude that an ongoing CD155-CD226 interaction is required to keep the CD226 expression at the level typically found in WT. Moreover, anti-CD226 mAb blocking binding to CD155 exerted no influence on CD155 levels present on DCs, T cells, or NK cells when administered in vivo (data not shown). It is therefore likely that the dynamic CD226-CD155 unit represents a one-way system where CD155 influences CD226 expression on contacting cells, whereas CD226 has no impact on CD155 levels. We could elicit spontaneous CD226 regulation in vivo not only by treatment of mice with mAb blocking the CD155-CD226 interaction but also by adoptive transfer of T cells or DCs. However, we failed to observe regulation in vitro by co-culturing DCs and T cells, indicating that a still unknown in vivo parameter is also required to accomplish modulation. Such a factor may be a biologically active compound like a cytokine. Another parameter may consist of the active migration of T cells inside the LN, enabling their recurring contact formation to DCs, processes that require a continuous rearrangement of the cytoskeleton. Interestingly, CD226 was shown to form a complex with protein 4.1G that allows T cells to connect surface-bound CD226 to the actin skeleton upon activation (53). Similarly to our findings, down-regulation of CD226 on HIV-specific cytotoxic T cells or on NK cells in ovarian cancer patients requires a physical contact with CD155-expressing cells (33, 37). However, in these particular settings, regulation can be achieved in vitro, indicating that different minimal requirements exist for adjustment of CD226 levels on afflicted cells.
The physiological role or consequences of increased CD226 levels on CD155-deficient T cells remain unknown. So far, there is no evidence that changed levels of CD226 expressed by naïve T cells can influence their proliferative capacity or differentiation pathways (7 and this report). However, it should be noted that most of the observations using CD155−/− cells refer to the role of DCs in priming T cells and mainly addressed CD4+ T cells. Gilfillan et al. reported that transgenic CD8+ T cells (OT-I) lacking CD226 are stimulated like WT cells when DCs were used to activate them (9). Intriguingly, when a nonprofessional antigen-presenting cell was used for this purpose, CD226-deficient OT-I cells were much less efficiently stimulated compared with WT. Therefore, CD226 is probably dispensable or at least functionally redundant in settings when DCs prime T cells. Considering this, it is possible that ongoing contacts between DCs and T cells permanently adjust CD226 levels on T cells to fine tune the sensitivity toward antigens presented by antigen-presenting cells other than DCs. Consequently, an immunological effect of elevated CD226 levels on T cells may become apparent only in pathways bypassing DCs as initiators of T cell responses.
The results obtained from the analyses of bone marrow chimeric animals revealed that only cells of hematopoietic origin are capable of manipulating CD226 expression in trans on peripheral T cells or SP thymocytes. This was an unexpected finding because T cells inside the T area of SLOs make recurrent contacts with stromal cells (54). The latter are important because their dendritic network represents a guiding path for T cells quickly moving inside SLOs (55). Stromal cells express CD155 (data not shown), thus providing ample opportunity for CD155-CD226 complex formation. Likewise, during negative selection, SP thymocytes contact medullary epithelial cells (56). These nonhematopoietic cells also express CD155 (6). Therefore, naïve T cells as well as SP thymocytes discriminate between CD155 expressed by various other cell types. CD226 modulation as a response to cell-cell contact may occur only when the CD155-CD226 complex is part of the synapse otherwise the presence of CD155 on the attaching cell is ignored by the T cell. Apart from this speculation, our findings document that CD226 recognizes selectively the presence of CD155 on other cells. Thus, the widespread expression pattern of CD155 is narrowed and split up into functionally relevant subpatterns depending on its diverse biological activities.
The finding that CD226 modulation is dependent on the CD155 gene-dosis suggests that CD226 levels found on T cells reflect the extent of effective molecular CD155-CD226 interactions. The frequency of cell-cell contacts, their intensity (number of participating molecules in the synapse) as well as their duration might sum up the observed net result of CD226 expression. At any rate, the actual level of CD226 probably mirrors the history of active encounters of the cells with other cells expressing CD155. Because we can observe changes in the CD226 levels between 3 days (e.g. following mAb application in vivo), it seems likely that the CD226 status only reflects the very recent CD155-relevant cell-cell communication. Even though it is impossible to deduce from the actual level of CD226 expression the nature and frequency of cell-cell contacts, a level divergent from WT might well inform whether a T cell experienced a regular CD155 environment during the past days. Because in vivo cell-cell contacts are required for modulation and DCs are one (if not the only) cell type capable of modulating CD226 expression in a healthy mouse, a regular CD155 environment may translate into a normal contact scenario regarding DC-T cell interactions. Indeed, the CD226 levels found on T cells of CCR7−/− support such a hypothesis. Here, peripheral T cells show abnormally high CD226 expression. This can be explained by the irregular compartmentalization of the CCR7−/− SLOs where T cells are only loosely organized in T cell zones of abnormal size and location (38). T cell areas usually represent a DC-T cell interaction platform of utmost importance and because DCs also require CCR7 to immigrate from the periphery (39), it can be assumed that the frequency of DC-T contacts in CCR7−/− SLOs is well below average compared with WT. This provides a plausible explanation for the delayed primary immune response in CCR7−/− animals (38). In contrast, CCR7−/− thymocytes showed normal (CD8+ SP) or even lower CD226 levels (CD4+ SP). The latter findings suggest that CCR7−/− thymocytes do not suffer from a paucity of contacts with DCs despite the profound structural abnormalities of the CCR7−/− thymi.
The results presented here demonstrate that regulation of CD226 on T cells is not an exceptional case met in cancer or infection involving only effector cells. It rather represents a natural rheostat existing already in thymocytes and naïve peripheral T cells. It is assumed that the biological effect of a CD155-driven CD226 regulation is beneficial for the immune system and its host. From this perspective, cancer cells or viruses such as HIV exploit an existing system, helping them to escape immune surveillance. It will be interesting to learn whether other receptor-ligand interactions obey similar regulatory mechanisms as described for the CD155-CD226 system. In this case, such a receptor-driven ligand balancing may represent an important general tool in controlling the density of surface-exposed proteins complementing the array of mechanisms influencing for example post-Golgi transport and receptor endocytosis. Future work will also need to address the question of why such a modulatory pathway exists already in the steady state of an unchallenged immune system.
Acknowledgments
We thank Oliver Pabst (Institute of Immunology, Hannover Medical School) for critically reading the manuscript and Drs. Akira and Kazuko Shibuya (Department of Immunology, University of Tsukuba, Japan) for mAb Tx42.
This work was supported by Deutsche Forschungsgemeinschaft Grants BE1886/2-1 and BE1886/2-2 (to G. B.).
- DC
- dendritic cell
- CFSE
- carboxyfluorescein diacetate succinimidyl ester
- LN
- lymph node
- PLN
- peripheral LN
- popLN
- popliteal LN
- NK
- natural killer
- SLO
- secondary lymphoid organ
- SP
- single positive
- TAMRA
- carboxytetramethylrhodamine succinimidyl ester.
REFERENCES
- 1. Maier M. K., Seth S., Czeloth N., Qiu Q., Ravens I., Kremmer E., Ebel M., Müller W., Pabst O., Förster R., Bernhardt G. (2007) Eur. J. Immunol. 37, 2214–2225 [DOI] [PubMed] [Google Scholar]
- 2. Takai Y., Miyoshi J., Ikeda W., Ogita H. (2008) Nat. Rev. Mol. Cell Biol. 9, 603–615 [DOI] [PubMed] [Google Scholar]
- 3. Erickson B. M., Thompson N. L., Hixson D. C. (2006) Hepatology 43, 325–334 [DOI] [PubMed] [Google Scholar]
- 4. Ogita H., Ikeda W., Takai Y. (2008) J. Microsc. 231, 455–465 [DOI] [PubMed] [Google Scholar]
- 5. Takai Y., Ikeda W., Ogita H., Rikitake Y. (2008) Annu. Rev. Cell Dev. Biol. 24, 309–342 [DOI] [PubMed] [Google Scholar]
- 6. Qiu Q., Ravens I., Seth S., Rathinasamy A., Maier M. K., Davalos-Misslitz A., Forster R., Bernhardt G. (2010) J. Immunol. 184, 1681–1689 [DOI] [PubMed] [Google Scholar]
- 7. Seth S., Ravens I., Kremmer E., Maier M. K., Hadis U., Hardtke S., Förster R., Bernhardt G. (2009) Eur. J. Immunol. 39, 3160–3170 [DOI] [PubMed] [Google Scholar]
- 8. Dardalhon V., Schubart A. S., Reddy J., Meyers J. H., Monney L., Sabatos C. A., Ahuja R., Nguyen K., Freeman G. J., Greenfield E. A., Sobel R. A., Kuchroo V. K. (2005) J. Immunol. 175, 1558–1565 [DOI] [PubMed] [Google Scholar]
- 9. Gilfillan S., Chan C. J., Cella M., Haynes N. M., Rapaport A. S., Boles K. S., Andrews D. M., Smyth M. J., Colonna M. (2008) J. Exp. Med. 205, 2965–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shibuya K., Lanier L. L., Phillips J. H., Ochs H. D., Shimizu K., Nakayama E., Nakauchi H., Shibuya A. (1999) Immunity 11, 615–623 [DOI] [PubMed] [Google Scholar]
- 11. Shibuya K., Shirakawa J., Kameyama T., Honda S., Tahara-Hanaoka S., Miyamoto A., Onodera M., Sumida T., Nakauchi H., Miyoshi H., Shibuya A. (2003) J. Exp. Med. 198, 1829–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shirakawa J., Shibuya K., Shibuya A. (2005) Int. Immunol. 17, 217–223 [DOI] [PubMed] [Google Scholar]
- 13. Shirakawa J., Wang Y., Tahara-Hanaoka S., Honda S., Shibuya K., Shibuya A. (2006) Int. Immunol. 18, 951–957 [DOI] [PubMed] [Google Scholar]
- 14. Pende D., Castriconi R., Romagnani P., Spaggiari G. M., Marcenaro S., Dondero A., Lazzeri E., Lasagni L., Martini S., Rivera P., Capobianco A., Moretta L., Moretta A., Bottino C. (2006) Blood 107, 2030–2036 [DOI] [PubMed] [Google Scholar]
- 15. Seth S., Georgoudaki A. M., Chambers B. J., Qiu Q., Kremmer E., Maier M. K., Czeloth N., Ravens I., Foerster R., Bernhardt G. (2009) J. Leukocyte Biol. 86, 91–101 [DOI] [PubMed] [Google Scholar]
- 16. Shibuya A., Campbell D., Hannum C., Yssel H., Franz-Bacon K., McClanahan T., Kitamura T., Nicholl J., Sutherland G. R., Lanier L. L., Phillips J. H. (1996) Immunity 4, 573–581 [DOI] [PubMed] [Google Scholar]
- 17. Reymond N., Imbert A. M., Devilard E., Fabre S., Chabannon C., Xerri L., Farnarier C., Cantoni C., Bottino C., Moretta A., Dubreuil P., Lopez M. (2004) J. Exp. Med. 199, 1331–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nabekura T., Shibuya K., Takenaka E., Kai H., Shibata K., Yamashita Y., Harada K., Tahara-Hanaoka S., Honda S., Shibuya A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 18593–18598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seth S., Ravens I., Lee C. W., Glage S., Bleich A., Förster R., Bernhardt G., Koenecke C. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, E32–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carlsten M., Björkström N. K., Norell H., Bryceson Y., van Hall T., Baumann B. C., Hanson M., Schedvins K., Kiessling R., Ljunggren H. G., Malmberg K. J. (2007) Cancer Res. 67, 1317–1325 [DOI] [PubMed] [Google Scholar]
- 21. Castriconi R., Dondero A., Corrias M. V., Lanino E., Pende D., Moretta L., Bottino C., Moretta A. (2004) Cancer Res. 64, 9180–9184 [DOI] [PubMed] [Google Scholar]
- 22. Chan C. J., Andrews D. M., McLaughlin N. M., Yagita H., Gilfillan S., Colonna M., Smyth M. J. (2010) J. Immunol. 184, 902–911 [DOI] [PubMed] [Google Scholar]
- 23. El-Sherbiny Y. M., Meade J. L., Holmes T. D., McGonagle D., Mackie S. L., Morgan A. W., Cook G., Feyler S., Richards S. J., Davies F. E., Morgan G. J., Cook G. P. (2007) Cancer Res. 67, 8444–8449 [DOI] [PubMed] [Google Scholar]
- 24. Masson D., Jarry A., Baury B., Blanchardie P., Laboisse C., Lustenberger P., Denis M. G. (2001) Gut 49, 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Merrill M. K., Bernhardt G., Sampson J. H., Wikstrand C. J., Bigner D. D., Gromeier M. (2004) Neuro-oncology 6, 208–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pende D., Spaggiari G. M., Marcenaro S., Martini S., Rivera P., Capobianco A., Falco M., Lanino E., Pierri I., Zambello R., Bacigalupo A., Mingari M. C., Moretta A., Moretta L. (2005) Blood 105, 2066–2073 [DOI] [PubMed] [Google Scholar]
- 27. Toutirais O., Cabillic F., Le Friec G., Salot S., Loyer P., Le Gallo M., Desille M., de La Pintière C. T., Daniel P., Bouet F., Catros V. (2009) Eur. J. Immunol. 39, 1361–1368 [DOI] [PubMed] [Google Scholar]
- 28. Enloe B. M., Jay D. G. (2011) J. Neurooncol. 102, 225–235 [DOI] [PubMed] [Google Scholar]
- 29. Morimoto K., Satoh-Yamaguchi K., Hamaguchi A., Inoue Y., Takeuchi M., Okada M., Ikeda W., Takai Y., Imai T. (2008) Oncogene 27, 264–273 [DOI] [PubMed] [Google Scholar]
- 30. Nakai R., Maniwa Y., Tanaka Y., Nishio W., Yoshimura M., Okita Y., Ohbayashi C., Satoh N., Ogita H., Takai Y., Hayashi Y. (2010) Cancer Sci. 101, 1326–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sloan K. E., Eustace B. K., Stewart J. K., Zehetmeier C., Torella C., Simeone M., Roy J. E., Unger C., Louis D. N., Ilag L. L., Jay D. G. (2004) BMC Cancer 4, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Textor S., Dürst M., Jansen L., Accardi R., Tommasino M., Trunk M. J., Porgador A., Watzl C., Gissmann L., Cerwenka A. (2008) Int. J. Cancer 123, 2343–2353 [DOI] [PubMed] [Google Scholar]
- 33. Carlsten M., Norell H., Bryceson Y. T., Poschke I., Schedvins K., Ljunggren H. G., Kiessling R., Malmberg K. J. (2009) J. Immunol. 183, 4921–4930 [DOI] [PubMed] [Google Scholar]
- 34. Lakshmikanth T., Burke S., Ali T. H., Kimpfler S., Ursini F., Ruggeri L., Capanni M., Umansky V., Paschen A., Sucker A., Pende D., Groh V., Biassoni R., Höglund P., Kato M., Shibuya K., Schadendorf D., Anichini A., Ferrone S., Velardi A., Kärre K., Shibuya A., Carbone E., Colucci F. (2009) J. Clin. Invest. 119, 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tahara-Hanaoka S., Shibuya K., Kai H., Miyamoto A., Morikawa Y., Ohkochi N., Honda S., Shibuya A. (2006) Blood 107, 1491–1496 [DOI] [PubMed] [Google Scholar]
- 36. Iguchi-Manaka A., Kai H., Yamashita Y., Shibata K., Tahara-Hanaoka S., Honda S., Yasui T., Kikutani H., Shibuya K., Shibuya A. (2008) J. Exp. Med. 205, 2959–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cella M., Presti R., Vermi W., Lavender K., Turnbull E., Ochsenbauer-Jambor C., Kappes J. C., Ferrari G., Kessels L., Williams I., McMichael A. J., Haynes B. F., Borrow P., Colonna M. (2010) Eur. J. Immunol. 40, 949–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Förster R., Schubel A., Breitfeld D., Kremmer E., Renner-Müller I., Wolf E., Lipp M. (1999) Cell 99, 23–33 [DOI] [PubMed] [Google Scholar]
- 39. Ohl L., Mohaupt M., Czeloth N., Hintzen G., Kiafard Z., Zwirner J., Blankenstein T., Henning G., Förster R. (2004) Immunity 21, 279–288 [DOI] [PubMed] [Google Scholar]
- 40. Czeloth N., Bernhardt G., Hofmann F., Genth H., Förster R. (2005) J. Immunol. 175, 2960–2967 [DOI] [PubMed] [Google Scholar]
- 41. Braun A., Worbs T., Moschovakis G. L., Halle S., Hoffmann K., Bölter J., Münk A., Förster R. (2011) Nat. Immunol. 12, 879–887 [DOI] [PubMed] [Google Scholar]
- 42. Ravens I., Seth S., Förster R., Bernhardt G. (2003) Biochem. Biophys. Res. Commun. 312, 1364–1371 [DOI] [PubMed] [Google Scholar]
- 43. Min B., Yamane H., Hu-Li J., Paul W. E. (2005) J. Immunol. 174, 6039–6044 [DOI] [PubMed] [Google Scholar]
- 44. Matloubian M., Lo C. G., Cinamon G., Lesneski M. J., Xu Y., Brinkmann V., Allende M. L., Proia R. L., Cyster J. G. (2004) Nature 427, 355–360 [DOI] [PubMed] [Google Scholar]
- 45. Hogquist K. A., Baldwin T. A., Jameson S. C. (2005) Nat. Rev. Immunol. 5, 772–782 [DOI] [PubMed] [Google Scholar]
- 46. Wu L., Shortman K. (2005) Semin. Immunol. 17, 304–312 [DOI] [PubMed] [Google Scholar]
- 47. Davalos-Misslitz A. C., Rieckenberg J., Willenzon S., Worbs T., Kremmer E., Bernhardt G., Förster R. (2007) Eur. J. Immunol. 37, 613–622 [DOI] [PubMed] [Google Scholar]
- 48. Davalos-Misslitz A. C., Worbs T., Willenzon S., Bernhardt G., Förster R. (2007) Blood 110, 4351–4359 [DOI] [PubMed] [Google Scholar]
- 49. Lei Y., Ripen A. M., Ishimaru N., Ohigashi I., Nagasawa T., Jeker L. T., Bösl M. R., Holländer G. A., Hayashi Y., de Waal Malefyt R., Nitta T., Takahama Y. (2011) J. Exp. Med. 208, 383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Doubrovina E. S., Doubrovin M. M., Vider E., Sisson R. B., O'Reilly R. J., Dupont B., Vyas Y. M. (2003) J. Immunol. 171, 6891–6899 [DOI] [PubMed] [Google Scholar]
- 51. Groh V., Wu J., Yee C., Spies T. (2002) Nature 419, 734–738 [DOI] [PubMed] [Google Scholar]
- 52. Ogasawara K., Hamerman J. A., Hsin H., Chikuma S., Bour-Jordan H., Chen T., Pertel T., Carnaud C., Bluestone J. A., Lanier L. L. (2003) Immunity 18, 41–51 [DOI] [PubMed] [Google Scholar]
- 53. Ralston K. J., Hird S. L., Zhang X., Scott J. L., Jin B., Thorne R. F., Berndt M. C., Boyd A. W., Burns G. F. (2004) J. Biol. Chem. 279, 33816–33828 [DOI] [PubMed] [Google Scholar]
- 54. Bajénoff M., Egen J. G., Koo L. Y., Laugier J. P., Brau F., Glaichenhaus N., Germain R. N. (2006) Immunity 25, 989–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Worbs T., Förster R. (2009) Curr. Top. Microbiol. Immunol. 334, 71–105 [DOI] [PubMed] [Google Scholar]
- 56. Peterson P., Org T., Rebane A. (2008) Nat. Rev. Immunol. 8, 948–957 [DOI] [PMC free article] [PubMed] [Google Scholar]