Abstract
The thylakoid membrane system is a complex membrane system that organizes and reorganizes itself to provide plants optimal chemical energy from sunlight under different and varying environmental conditions. Grana membranes are part of this system and contain the light-driven water-splitting enzyme Photosystem II (PSII) and light-harvesting antenna complexes. Here, we present a direct visualization of PSII complexes within grana membranes from spinach. By means of jumping mode atomic force microscopy in liquid, minimal forces were applied between the scanning tip and membrane or protein, allowing complexes to be imaged with high detail. We observed four different packing arrangements of PSII complexes, which occur primarily as dimers: co-linear crystalline rows, nanometric domains of straight or skewed rows, and disordered domains. Upon storing surface-adhered membranes at low temperature prior to imaging, large-scale reorganizations of supercomplexes between PSII and light-harvesting complex II could be induced. The highest resolution images show the existence of membrane domains without obvious topography extending beyond supercomplexes. These observations illustrate the possibility for diffusion of proteins and smaller molecules within these densely packed membranes.
Keywords: Atomic Force Microscopy, Membrane Biophysics, Membrane Structure, Photosynthesis, Photosystem II, Jumping Mode AFM, Supramolecular Organization
Introduction
Photosynthetic membranes are specialized structures that contain the various protein complexes that transform sunlight into an energy-rich compound. The majority of complexes within these membranes are light-harvesting complexes (LHCs)2 that absorb sunlight and transfer energy and reaction centers, to where this energy is directed to initiate electron transfer reactions (1).
In green plants, the photosynthetic membranes reside inside the chloroplasts. Under physiological conditions, part of the membranes is tightly stacked along their flat cytoplasmic surfaces, resulting in grana membranes, interconnected by stroma lamellae. Grana contain primarily Photosystem II (PSII) and its associated major light-harvesting antenna (LHCII) as well as minor LHCs (2). Analyses of EM images of isolated complexes from grana have indicated PSII and major and minor LHCs to be present in PSII-LHCII supercomplexes (3, 4). Using the three-dimensional crystallographic structures of the PSII core complex from cyanobacteria and the trimeric LHCII complex from plants (5, 6), a detailed model has been constructed based on the structure of these PSII-LHCII supercomplexes (4).
Grana membranes can be easily prepared by solubilization of stacked chloroplasts with the relatively strong detergent Triton X-100 (7). Structural investigations have revealed that these membranes are paired membranes with the extrinsic lumenal parts facing outwards (8). However, to retain biological activity, the membranes are usually diluted in detergent-free solutions, resulting in a strong aggregation of the membranes and preventing structural analyses. For this reason, we have developed the preparation of grana membranes by a much milder detergent (α-dodecyl maltoside), which prevents the need for detergent removal. These preparations were extensively characterized by biochemical and structural methods, including EM (see, for example, Refs. 9–12).
Strong evidence exists that the PSII supercomplex is composed of a PSII core dimer with two to four LHCII trimers attached at specific binding positions. Depending on the light conditions during plant growth, about four to eight additional LHCII complexes are not directly bound to a PSII-LHCII supercomplex and reside in the so-called LHCII-only membrane domains (see Ref. 2 and references therein). EM studies have furthermore indicated that the majority of supercomplexes are organized in a random fashion and that a small amount is organized in regular arrays. Under certain conditions such as low temperature, low-light intensity during growth, or low pH, a tendency for ordering in crystalline arrays has been observed (10, 13, 14). A detailed analysis has furthermore indicated a large heterogeneity in the organization and content between opposing grana membranes (14–16).
Atomic force microscopy (AFM) imaging protocols derived for two-dimensional crystals of membrane proteins (17, 18) have been successfully applied to photosynthetic membranes (19–23). Images with unprecedented precision allowed the visualization of individual proteins and protein complexes also in other types of biological membranes, i.e. mitochondria (24), eye lens (25), and retina (26). An AFM study of grana membranes has confirmed (following biochemical evidence (see Ref. 27)) that these membranes also are densely packed with protein (28). However, in this study, AFM was conducted on dried membranes in air, which prompts concerns as to dehydration and damage and prohibits high-resolution imaging.
The proposed functional consequences of segregation and aggregation of photosynthetic complexes include long-distance electron transfer, regulation of energy distribution by preventing energy spillover, supplying ways to help protect vulnerable PSII from light overexposure, and allowing repair of photodamaged PSII units (2). The packing of protein complexes observed in all photosynthetic membranes raises questions as to how diffusive processes can take place on physiological time scales under such crowded conditions (29).
To address these questions, detailed knowledge on the supramolecular organization of the protein complexes and the possibility for dynamic rearrangements is indispensable. AFM images may provide information on the protein organization within a single membrane in a close to native state and with high detail without image manipulation. Here, we present high-resolution AFM images of grana membranes from spinach. Adhered membranes were immersed in a liquid ionic buffer, which prevents dehydration and greatly enhances resolution. To minimize damage further, we employed a most delicate AFM mode, the so-called “jumping mode” (JM-AFM). The advantage of this method is that the lateral movement of the scanning tip with respect to the sample surface is performed when the two are not in contact. In this manner, lateral shear forces on the specimen are minimized (30, 31). This is particularly important for a fragile system such as a pair of membranes, in which the lower membrane is fixed to the surface, the upper membrane is probed, and both membranes are attached to each other by weak electrostatic forces.
EXPERIMENTAL PROCEDURES
Membrane Preparation
Grana membranes from spinach thylakoids were prepared fresh by solubilization with the mild detergent n-dodecyl α-d-maltoside according to van Roon et al. (9).
AFM
The samples obtained were then deposited on freshly cleaved mica in adsorbing buffer (10 mm Hepes (pH 8), 25 mm MgCl2, and 150 mm KCl) for at least 1 h in the dark at room temperature or at 4 °C. Subsequently, the samples were washed with imaging buffer (10 mm Hepes (pH 8) and 50 mm KCl) and repeatedly imaged in a liquid cell with a commercial AFM microscope (Nanotec) by jumping or tapping mode. The nominal spring constant of the cantilevers (Olympus) employed varied from 0.1 to 0.03 newtons/m, and their resonance frequency in liquid was 11–30 kHz. All measurements and the following image analysis were performed using the WsXM program (32). The elevation of grana stacks is measured with respect to the mica surface, and the height of core complexes is assessed with respect to the plane of the membrane. The width of and distances between masses is obtained only when masses protrude from an area of equal height or as indicated in the figures and text.
RESULTS
Membranes Incubated at Room Temperature
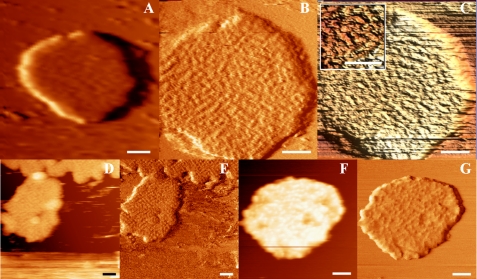
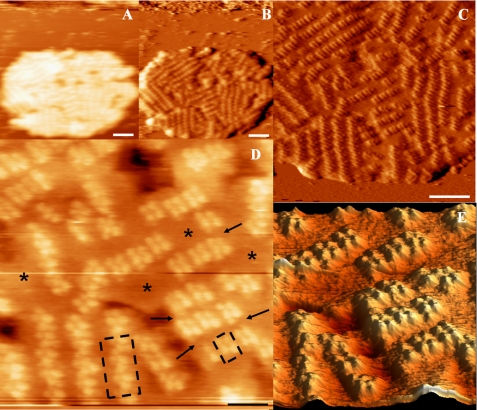
Freshly collected gel filtration fractions containing solubilized grana membranes were adhered to mica, immersed within a liquid ionic buffer, and directly imaged. Our adhesion and rinsing protocols resulted in the firm attachment of flat membranes. Membrane sizes varied between 200 and 600 nm in diameter and 18 and 60 nm in height. Fig. 1 A depicts a typical membrane imaged at low resolution by JM-AFM. Grana appear to be packed with protein, indicated as the most bright topographical masses. A more clear view of these masses is shown in Fig. 1B, displaying the same membrane at higher magnification. Fig. 1C depicts a three-dimensional image of Fig. 1B, clearly visualizing the unorganized but dense configuration of proteins within grana. The inset presents a digital zoom. The length of the individual masses appears to be between 30 and 60 nm, and their width is ∼15 nm, consistent with the size of the extrinsic parts of PSII dimers (2, 4). Fig. 1(D–G) shows AFM height (D and F) and accompanying error (E and G) images of similarly prepared membranes but incubated between 1 and 3 h on mica at room temperature. An error image shows the non-resolved protrusions of the height image and is particularly sensitive to regular structures. Clearly, the dense packing of protein is a common characteristic of these membranes. Although longer incubation results in a slightly better resolution, it does not allow further analyses.
FIGURE 1.
JM-AFM images of grana membranes adhered to mica and stored at room temperature prior to rinsing and imaging. A, typical image of a grana membrane directly measured after deposition on mica. B, same membrane as in A at higher magnification. C, three-dimensional enhanced representation of B. The inset shows a digital zoom of C. D and E, JM-AFM height and error images of a similarly prepared membrane but incubated for 1 h prior to imaging. F and G, same as D and E but after 3 h of incubation. Scale bars = 100 nm.
Membranes Incubated at 4 °C
We proceed by incubating membranes at 4 °C after adhesion to mica. Storage at lower temperature is known to induce a higher degree of order in grana membranes (33), although this has never before been tested on membranes adhered to a mica surface. Usually, we incubated the membranes overnight.
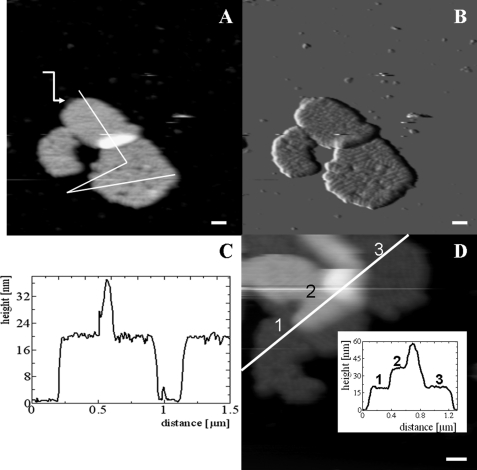
Typical JM-AFM images of the membranes prepared this way are presented in Fig. 2. Fig. 2A displays the height, and Fig. 2B shows the error image. The height image shows that a small part of the upper left membrane is located on another; this is likely a result of incubation and rinsing. Especially in the error image, already at this low resolution, domains of protein protrusions regularly arranged in rows can be clearly distinguished. The height profile of the membranes (Fig. 2C) reveals the height of a single granum as ∼20 nm, whereas the height of the two partially overlapping membranes is 38 nm. Fig. 2D shows a different piece of membrane. Here, three layers are visible on top of each other. The total height of this stack is 59 ± 1 nm. Fig. 2A also shows a piece of membrane with reduced height, pointing to a piece of membrane with only a single layer. This feature was observed previoiusly in analyses of these types of membranes by EM (10) and may indicate a fragment of stroma lamellae. We conclude that the attachment of the membranes on a mica surface does not prevent a structural rearrangement and a higher degree of order in at least the membrane facing the medium and the AFM tip.
FIGURE 2.
AFM images of grana membranes adhered to mica and stored at 4 °C prior to rinsing and imaging. A, height image. The white line indicates the position of the height profile presented in C. The arrow points to an empty piece of membrane. B, error image of A. C, height profile as indicated in A. The zero level is taken at the mica surface. D, height image and profile (inset) of three pieces of membrane on top of each other. Scale bars = 100 nm.
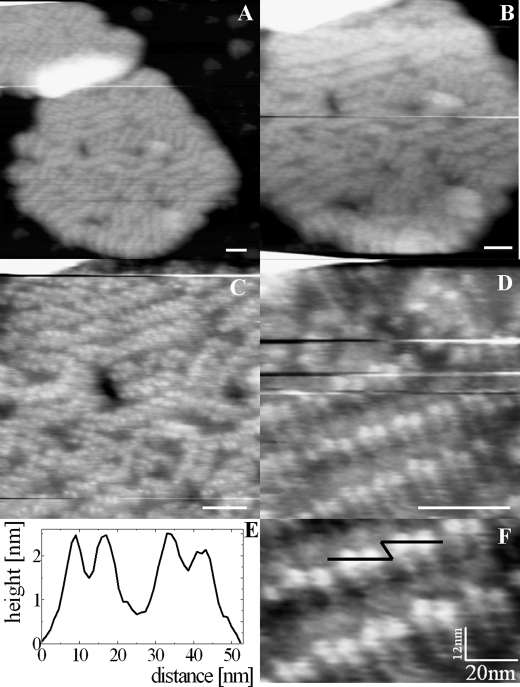
A view at higher magnification of one of the membranes from Fig. 2 is shown in Fig. 3. The protruding proteins are now more clearly visible as brighter parts in the image. Clearly, these membranes are packed with protein, organized as rows residing in nanometric subdomains (Fig. 3, A–D). In several cases, the integrity of membrane surface is broken in a form of holes, visible as black spots in Fig. 3 (B and C). The presence of such damage is likely due to the detergent treatment during the isolation procedure and was also observed in EM images of preparations of the same kind (10). In Fig. 3 (D and F (digital zoom of C)), close-up views of the protruding proteins are shown. At this magnification, we observe the dimeric arrangement of masses arranged in rows. A cross-section is depicted in Fig. 3F (black line) and shown in Fig. 3E. The two main closest protrusions form a unit spanning 20 nm and have individual maxima separated by 8 nm and a height of 2 nm. The distance between units is ∼10 nm. We measured similar values for our highest resolution images (see below).
FIGURE 3.
AFM images at higher magnification of the double-membrane image from Fig. 2. A–D, consecutive recorded images at progressively higher magnifications. E, height profile as indicated by the black line in F. F, digital zoom of C. Scale bars = 50 nm.
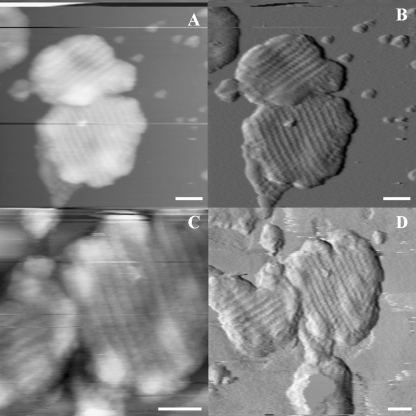
Occasionally, we could switch between JM-AFM and tapping mode AFM (TM-AFM). In Fig. 4, two double membranes were imaged initially by jumping mode and then reimaged by tapping mode. Fig. 4A (height image) and Fig. 4B (first derivative image) were recorded by JM-AFM, and Fig. 4C (height image) and Fig. 4D (error) by TM-AFM. Although most membranes show disordered rows of proteins as in Fig. 2, here, proteins are very regularly packed in co-linear rows spanning the entire membrane. The size of protruding masses is 20 nm, with a gap of 10 nm between the rows. Five consecutive rows span 180 nm; thus, each row spans 36 nm. Thirty min had passed between recording the two images, and in this time, the upper membrane has been repositioned to the left, indicative of the loose adhesion to the mica surface. The TM-AFM image shows similar detail, but the membrane appears to be distorted. We stress that generally the same membrane could be imaged consecutively many times without any changes. Also in previous TM-AFM investigations on bacterial photosynthetic membranes, the membranes could be consecutively imaged, exhibiting similar results (20).
FIGURE 4.
JM- and TM-AFM images of grana membranes from spinach. A and B, JM-AFM height and error images of a grana membrane. C and D, TM-AFM images of the same membrane obtained after switching from jumping to tapping mode 40 min after the images in A and B were obtained. Scale bars = 100 nm.
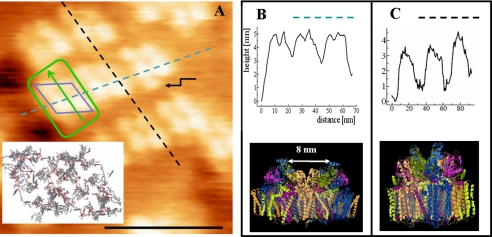
Our AFM method occasionally allowed membranes to be imaged to a resolution at which individual protein protrusions could be visualized. A high-resolution image of a partially ordered grana membrane is shown in Fig. 5. Fig. 5 (A and B) shows height and error images of this membrane adhered to a mica surface. Domains of regular rows of protein mass are clearly visible. A closer inspection of this membrane is shown in Fig. 5C, and at this magnification, the grouping of domains and the spacing between rows are clearly visible. Only at the highest magnification achievable (Fig. 5D) did we observe the masses to consist of four protrusions close together. The lower panel of Fig. 5D shows the two prevalent dimeric interactions indicated by dashed boxes: a skewed dimer (left) and straight dimer (right). The dimensions of the dimer are ∼25 × 16 nm, slightly larger than the extrinsic protrusions of PSII supercomplexes in the plant model presented by Nield et al. (34). The highest protrusions extend 4–6 nm from the membrane, consistent with the same structural model. Only at the highest magnification (Fig. 5, C and D) could the space between the protruding PSII units of ordered arrays be observed (see arrows). Also other, larger areas devoid of protruding masses are observed between membrane domains containing ordered arrays of PSII dimers (indicated by asterisks). Fig. 5D allows us to count the number of supercomplexes: 70 ± 2. The total area of this panel is 260 × 260 nm, which amounts to 67,600 nm2. Assuming we are viewing the most common C2S2M supercomplex in spinach with an area of 540 nm2 (2, 4, 10), a total of 125 supercomplexes could have been packed within this area. Thus, the packing density of C2S2M supercomplexes in this entire area is ∼56%. As shown in all images shown here, the protein density varies considerably within one membrane. The densest area is occupied by co-linear rows of supercomplexes. In Fig. 5 (A and B), we estimate that half of the membrane shown is occupied by such rows; here, the packing fraction of C2S2M supercomplexes is ∼95%, confirming that the crystalline arrays consist of C2S2M supercomplexes.
FIGURE 5.
High-resolution JM-AFM images of spinach grana membranes adhered to mica and stored at 4 °C prior to rinsing and imaging. A and B, height and error images of a grana membrane. C and D, higher magnification images of the same membrane. Asterisks and arrows depict large and small areas without topography, respectively. E, computer-generated zoom of D (three-dimensional enhanced image). Scale bars = 100 nm (A–C) and 50 nm (D and E).
In Fig. 6, we show analyses of the lower right part of the membrane AFM image of Fig. 5D: a domain of rows consisting of four protruding masses as a basic unit. The cross-section within a row is indicated by a dashed blue line in Fig. 5B. The width of a unit is 20 nm, and within such a unit, the two protrusions have a height of 5 nm and are separated by 8 nm. The size of each protrusion is 5 nm. These values closely match those expected for the extrinsic PsbO, PsbU, and PsbV proteins from the oxygen-evolving complex (OEC) of the PSII core complex from Thermosynechococcus elongatus (6). The cross-section between rows is indicated by the dashed black line in Fig. 6C. The length of a unit is 30 nm, extending from topological mass to the next mass. As we will discuss further below, this indicates that we are observing proximate PSII supercomplexes, most likely of the C2S2M variety, with their long axes along the dashed black line. At this particular domain, we also observe an extra mass, associated with a supercomplex (indicated by the arrow) of unknown origin but reported previously (35).
FIGURE 6.
Comparison of topography of grana membranes and PSII crystal structure. A–C, analyses of height profiles of part of the image shown in Fig. 5D (A here). The height profile of the dashed blue line is shown in B, and that of the dashed black line is shown in C. The blue diamond represents the unit of topographic masses in this area, consistent with the diamond-like arrangement of cofactors of PSII core dimers, which is shown in the inset of A. The green outline depicts the repeating length scale within this area following the cross-sections in B and C and the corresponding side views of the PSII core dimer. The Protein Data Bank code for the x-ray crystallographic structures is 2AXT. Scale bar = 50 nm.
DISCUSSION
JM-AFM
The common protocol to obtain a high-resolution AFM image is the application of a liquid ionic buffer solution that counters repulsive van der Waals forces when the imaging tip of the AFM cantilever approaches the surface. The highest resolution images can be obtained with contact mode AFM, in which the force between the tip of the cantilever and the probe is kept constant, or TM-AFM, in which the tip oscillates with a certain frequency and only minimally interacts with the surface. Contact mode AFM provides the highest resolution images when applied to naturally flat membranes (19, 23), and TM-AFM when applied to naturally curved membranes (20). For both methods, high resolution is achieved only when a delicate interaction between the scanning tip and the topographic masses is achieved. Generally, this is possible only in the absence of large (more than a few nanometers) height differences. JM-AFM is a more sensitive scanning probe method, but, as such, it cannot deal with large height differences due to the fact that the withdraw-approach procedure of the piezo element must be repeated at every measuring point. This prolongs the scanning time and makes the cantilever liable to noise caused by the liquid solution (36). For comparison, we also applied TM-AFM in liquid. This resulted in a lowered resolution though: resolving the PSII core structure was not feasible, despite the fact that the organization of regular rows of complexes within the membrane remained clearly visible. Moreover, the membrane was affected by scanning, as part of it appeared to be moved and stretched. However, no other damage was noticeable. In contrast, TM-AFM has been applied successfully before to bacterial photosynthetic membranes (20). We speculate that TM-AFM is too harsh because the double grana membranes are more flexible structures compared with the singular bacterial membranes. Another difference with bacterial membranes is that the domains of photosynthetic complexes are completely packed without any areas containing only lipids or loosely connected LHCs. A lesser degree of packing density of protein complexes in subdomains may be responsible not only for the observed mass-less areas (Fig. 5D) but also for the lower sturdiness of the grana.
Visualizing Grana and PSII Complexes
We need to take into consideration that our purification method produced inside-out paired membrane discs cut out from the grana stacks by mild detergent treatment and prevented from aggregation by retaining small amounts of detergent in the final sample solution. Therefore, any multilayered structures observed on mica are purely coincidental and by no means reflect the real state of organization in native appressed grana. Nevertheless, the height of single and multilayered membranes can be informative concerning the protrusions of proteins. The average height of membranes throughout our study is 20 nm. According to the crystal structure of cyanobacterial PSII (20), the PSII core dimer has dimensions of 20.5 × 11 × 10.5 nm (length ×width × height). Bearing in mind that the transmembranal part of the complex measures 4.5 nm, the stromal protrusion is 1 nm, and the lumenal protrusion of OEC is up to 5 nm, one can assume that a double membrane containing intact PSII particles is at least 20 nm high. The observed height is in good agreement with estimates from thin sectioning (reviewed in Ref. 2) and recent electron microscope tomography of thylakoids (35, 37). The measured height is considerably longer than observed previously for AFM on air-dried grana membranes (28), which can be explained by a shrinking of the height due to drying. The height profile of the overlay of two or three double-membrane stacks shows a height of ∼40 and 60 nm, respectively. These values suggest that the lumenal protrusions of PSII complexes in paired grana are facing each other. Note that the extrinsic proteins of spinach PSII differ from those from cyanobacteria (38). The values found here do indicate a similar height above the membrane for both types.
Our most detailed images (Fig. 6A) reveal symmetrically arranged structures with a pronounced long (30 nm) and short (20 nm) side (Fig. 6, B and C). The repeating units consist of four separate masses protruding 5 nm above the membrane, arranged in a diamond shape (Fig. 6A). This arrangement matches closely that of the cyanobacterial PSII crystal structure (Fig. 6A, inset) and the extension of the OEC proteins of ∼5 nm from the lumenal side of PSII. On the basis of the topography of the OEC proteins from the crystal structure, we can also deduce the orientation of the PSII complexes. This topography is very different for the two directions in the crystal structure: in one direction, the OECs of the two PSII monomers are separated by a small gap of only 1 nm, and in the other direction by 8 nm. The latter separation fills precisely the gap between the masses observed in our images (Fig. 6, B and C). Thus, the long axis of the PSII core dimer lies along the 20-nm repeating unit. The repeating length scale on the other side is found to be 30 nm, and the topography appears regular within the rows with symmetrical units. We therefore expect an extra 5-nm mass on each side of the PSII core dimer. Such a mass is expected for PSII supercomplexes consisting of a PSII core dimer flanked by one LHCII and two minor antennae on each side (2, 4, 39). These structures are likely undetected here because their protrusions are maximally 1 nm from the membrane (5). In Fig. 6A, this tentative structure is outlined in green, with the arrow indicating the orientation of a supercomplex.
Supramolecular Organization of Grana
We imaged grana membranes isolated and treated using two different protocols. In all cases, the membranes appear densely packed with protein masses. The sizes of these masses correspond to dimeric PSII units. Adhering membranes at room temperature resulted in a random distribution of clustered supercomplexes (Fig. 1). A higher degree of organization was found when the membranes were incubated at 4 °C while fixed onto a mica surface (Fig. 5). Such a membrane reorganization is indicative of a disordered-ordered phase transition. Only at low temperature can the weak specific interactions between supercomplexes become a dominant organizational driving force. Besides, the possibility for protein reorganization to take place indicates large-scale diffusion to be possible despite dense packing. Moreover, this reorganization takes place despite adhesion to the mica surface. Most likely, this is due to the upper membrane being more loosely attached to the lower membrane, which in turn adheres to the surface. Recently, it has been shown that the primary photosynthetic reactions are maintained on surface-adhered membranes (40); here, we found that proteins can reorganize on a large-scale within a membrane despite adhesion to a surface.
In general, we have shown that PSII supercomplexes can cluster into membrane domains of ordered arrays. We have shown previously that protein clustering within membrane domains originates in the depletion-induced attraction within packed membranes (41). In that work, we demonstrated that size differences between core and peripheral LHCs, together with the intrinsic curvature of the protein complexes, can drive membrane domain formation of one type of protein complex. In the grana membranes, the size differences between the two main components, PSII supercomplexes and peripheral LHCII complexes, are much larger and may solely cause these domains to be formed.
In our images, four types of packing lattices are observed. In most cases, supercomplexes are arranged in rows, either co-linearly arranged through the membrane (Fig. 4) or within nanodomains (Figs. 3 and 5). In both cases, little mass-free space can be detected. Our highest resolution images of such a latter domain (Fig. 5) show another packing configuration of supercomplexes: skewed rows (Fig. 5C). Strikingly, between these rows and between the nanodomains of specific order, mass-less space is observed. This space is likely occupied by LHCII, undetected here because these complexes hardly protrude from the membrane. On the other hand, the z-resolution in AFM is generally <1 nm, especially in a tight packing configuration. Topography-less area may thus indicate a lipid-only area, densely packed LHCII complexes, or loosely interacting LHCII complexes; yet, on the other hand, the temperature-induced reorganizations observed here indicate the possibility for large-scale protein movements, arguing against dense packing. Generally, our data show that such highly specialized structures as grana membranes retain their high flexibility to adapt to external conditions even in vitro.
Conclusions
We have applied a delicate AFM imaging technique to stacked grana membranes from spinach immersed in an ionic buffer solution. Using this method, we were able to obtain the first high-resolution images of PSII and visualized their organization within these membranes. Surface-adhered membranes are capable of remodeling from random to row-like organizations when temporarily stored at 4 °C. We can discriminate four different packing lattices: co-linear rows of close laying PSII that span the entire membrane, nanometric domains of linear or skewed rows, and disordered domains. Although the membranes appear to be densely packed with protein at lower resolution, the highest resolution images reveal large domains without protrusions to be present. These areas span beyond the dimensions of the supercomplexes, also when unresolved topography is taken into account. The high-resolution images allow the packing density of PSII supercomplexes to be estimated, varying between 50% for random and 75% for close packing. Areas with loosely interacting LHCII, lipid only, and less tight packing likely allow the large-scale protein remodeling of grana membranes during low-temperature incubation. Most importantly, these features may also facilitate other diffusive processes necessary for sustainment and adaptation within chloroplasts.
Acknowledgment
R. N. F. gratefully acknowledges Josep Pàmies for helpful discussions.
This work was supported in part by the Netherlands Organization for Scientific Research (NWO).
- LHC
- light-harvesting complex
- PSII
- Photosystem II
- AFM
- atomic force microscopy
- JM
- jumping mode
- TM
- tapping mode
- OEC
- oxygen-evolving complex.
REFERENCES
- 1. Nelson N., Yocum C. F. (2006) Annu. Rev. Plant Biol. 57, 521–565 [DOI] [PubMed] [Google Scholar]
- 2. Dekker J. P., Boekema E. J. (2005) Biochim. Biophys. Acta 1706, 12–39 [DOI] [PubMed] [Google Scholar]
- 3. Boekema E. J., van Roon H., Calkoen F., Bassi R., Dekker J. P. (1999) Biochemistry 38, 2233–2239 [DOI] [PubMed] [Google Scholar]
- 4. Caffarri S., Kouril R., Kereïche S., Boekema E. J., Croce R. (2009) EMBO J. 28, 3052–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., Chang W. (2004) Nature 428, 287–292 [DOI] [PubMed] [Google Scholar]
- 6. Zouni A., Witt H. T., Kern J., Fromme P., Krauss N., Saenger W., Orth P. (2001) Nature 409, 739–743 [DOI] [PubMed] [Google Scholar]
- 7. Berthold D. A., Babcock G. T., Yocum C. F. (1981) FEBS Lett. 134, 231–234 [Google Scholar]
- 8. Dunahay T. G., Staehelin L. A., Seibert M., Ogilvie P. D., Berg S. P. (1984) Biochim. Biophys. Acta 764, 179–193 [Google Scholar]
- 9. van Roon H., van Breemen J. F., de Weerd F. L., Dekker J. P., Boekema E. J. (2000) Photosynth. Res. 64, 155–166 [DOI] [PubMed] [Google Scholar]
- 10. Boekema E. J., van Breemen J. F., van Roon H., Dekker J. P. (2000) J. Mol. Biol. 301, 1123–1133 [DOI] [PubMed] [Google Scholar]
- 11. Yakushevska A. E., Keegstra W., Boekema E. J., Dekker J. P., Andersson J., Jansson S., Ruban A. V., Horton P. (2003) Biochemistry 42, 608–613 [DOI] [PubMed] [Google Scholar]
- 12. Ruban A. V., Wentworth M., Yakushevska A. E., Andersson J., Lee P. J., Keegstra W., Dekker J. P., Boekema E. J., Jansson S., Horton P. (2003) Nature 421, 648–652 [DOI] [PubMed] [Google Scholar]
- 13. Kirchhoff H., Haase W., Wegner S., Danielsson R., Ackermann R., Albertsson P. A. (2007) Biochemistry 46, 11169–11176 [DOI] [PubMed] [Google Scholar]
- 14. Staehelin L. A. (2003) Photosynth. Res. 76, 185–196 [DOI] [PubMed] [Google Scholar]
- 15. Yakushevska A. E., Jensen P. E., Keegstra W., van Roon H., Scheller H. V., Boekema E. J., Dekker J. P. (2001) Eur. J. Biochem. 268, 6020–6028 [DOI] [PubMed] [Google Scholar]
- 16. Kouril R., Dekker J. P., Boekema E. J. (June 23, 2011) Biochim. Biophys. Acta 10.1016/j.bbabio.2011.05.024 [DOI] [PubMed] [Google Scholar]
- 17. Möller C., Allen M., Elings V., Engel A., Müller D. J. (1999) Biophys. J. 77, 1150–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Müller D. J., Fotiadis D., Scheuring S., Müller S. A., Engel A. (1999) Biophys. J. 76, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scheuring S., Seguin J., Marco S., Lévy D., Robert B., Rigaud J. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100, 1690–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bahatyrova S., Frese R. N., Siebert C. A., Olsen J. D., Van Der Werf K. O., Van Grondelle R., Niederman R. A., Bullough P. A., Otto C., Hunter C. N. (2004) Nature 430, 1058–1062 [DOI] [PubMed] [Google Scholar]
- 21. Gonçalves R. P., Bernadac A., Sturgis J. N., Scheuring S. (2005) J. Struct. Biol. 152, 221–228 [DOI] [PubMed] [Google Scholar]
- 22. Scheuring S., Gonçalves R. P., Prima V., Sturgis J. N. (2006) J. Mol. Biol. 358, 83–96 [DOI] [PubMed] [Google Scholar]
- 23. Scheuring S., Sturgis J. N. (2005) Science 309, 484–487 [DOI] [PubMed] [Google Scholar]
- 24. Gonçalves R. P., Buzhynskyy N., Prima V., Sturgis J. N., Scheuring S. (2007) J. Mol. Biol. 369, 413–418 [DOI] [PubMed] [Google Scholar]
- 25. Buzhynskyy N., Hite R. K., Walz T., Scheuring S. (2007) EMBO Rep. 8, 51–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fotiadis D., Liang Y., Filipek S., Saperstein D. A., Engel A., Palczewski K. (2003) Nature 421, 127–128 [DOI] [PubMed] [Google Scholar]
- 27. Kirchhoff H., Mukherjee U., Galla H. J. (2002) Biochemistry 41, 4872–4882 [DOI] [PubMed] [Google Scholar]
- 28. Kirchhoff H., Lenhert S., Büchel C., Chi L., Nield J. (2008) Biochemistry 47, 431–440 [DOI] [PubMed] [Google Scholar]
- 29. Kirchhoff H., Haferkamp S., Allen J. F., Epstein D. B., Mullineaux C. W. (2008) Plant Physiol. 146, 1571–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moreno-Herrero F., de Pablo P. J., Colchero J., Gomez-Herrero J., Baro A. M. (2000) Surface Sci. 453, 152–158 [Google Scholar]
- 31. Moreno-Herrero F., de Pablo P. J., Fernandez-Sanchez R., Colchero J., Gomez-Herrero J., Baro A. M. (2002) Appl. Phys. Lett. 81, 2620–2622 [Google Scholar]
- 32. Horcas I., Fernández R., Gómez-Rodríguez J. M., Colchero J., Gómez-Herrero J., Baro A. M. (2007) Rev. Sci. Instrum. 78, 013705 [DOI] [PubMed] [Google Scholar]
- 33. Semenova G. A. (1995) Can. J. Bot. 73, 1676–1682 [Google Scholar]
- 34. Nield J., Balsera M., De Las Rivas J., Barber J. (2002) J. Biol. Chem. 277, 15006–15012 [DOI] [PubMed] [Google Scholar]
- 35. Kouil R., Oostergetel G. T., Boekema E. J. (2011) Biochim. Biophys. Acta 1807, 368–374 [DOI] [PubMed] [Google Scholar]
- 36. Moreno-Herrero F., Colchero J., Gómez-Herrero J., Baró A. M. (2004) Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 69, 031915 [DOI] [PubMed] [Google Scholar]
- 37. Daum B., Nicastro D., Austin J., 2nd, McIntosh J. R., Kühlbrandt W. (2010) Plant Cell 22, 1299–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nield J., Barber J. (2006) Biochim. Biophys. Acta 1757, 353–361 [DOI] [PubMed] [Google Scholar]
- 39. Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S. (2004) Science 303, 1831–1838 [DOI] [PubMed] [Google Scholar]
- 40. Magis G. J., den Hollander M. J., Onderwaater W. G., Olsen J. D., Hunter C. N., Aartsma T. J., Frese R. N. (2010) Biochim. Biophys. Acta 1798, 637–645 [DOI] [PubMed] [Google Scholar]
- 41. Frese R. N., Pàmies J. C., Olsen J. D., Bahatyrova S., van der Weij-de Wit C. D., Aartsma T. J., Otto C., Hunter C. N., Frenkel D., van Grondelle R. (2008) Biophys. J. 94, 640–647 [DOI] [PMC free article] [PubMed] [Google Scholar]