Background: AKAP220 organizes the signaling enzymes PKA, GSK-3, and phosphoprotein phosphatase PP1.
Results: AKAP220 interacts with the scaffolding protein IQGAP1 to assimilate and process calcium and cAMP signals at leading edges of migrating cells.
Conclusion: AKAP220/IQGAP1 networks position calcium and cAMP-responsive signaling enzymes near substrates at the +TIPs of growing microtubules.
Significance: Anchored kinase/microtubule effector protein networks propagate cell motility.
Keywords: Akap, Cellular Regulation, Protein Kinases, Protein-Protein Interactions, Signal Transduction
Abstract
Cell movement requires the coordinated reception, integration, and processing of intracellular signals. We have discovered that the protein kinase A anchoring protein AKAP220 interacts with the cytoskeletal scaffolding protein IQGAP1 to influence cell motility. AKAP220/IQGAP1 networks receive and integrate calcium and cAMP second messenger signals and position signaling enzymes near their intended substrates at leading edges of migrating cells. IQGAP1 supports calcium/calmodulin-dependent association of factors that modulate microtubule dynamics. AKAP220 suppresses GSK-3β and positions this kinase to allow recruitment of the plus-end microtubule tracking protein CLASP2. Gene silencing of AKAP220 alters the rate of microtubule polymerization and the lateral tracking of growing microtubules and retards cell migration in metastatic human cancer cells. This reveals an unappreciated role for this anchored kinase/microtubule effector protein network in the propagation of cell motility.
Introduction
Signal transduction is the transfer of chemical, electrical, or mechanical signals across the plasma membrane or from one cellular location to another. Not surprisingly, cells have evolved an assortment of elegant mechanisms to fulfill this obligatory process. These include the diffusion of signals through the cytoplasm and nucleus and their processing by immobilized multiprotein complexes (1). Configuring enzymes such as protein kinases and phosphatases in this manner enhances the reception of this information and improves the fidelity of signal transfer. Consequently, an abundance of kinase and phosphatase-binding proteins tether their enzyme-binding partners to sites where they can respond to activating signals and be close to selected substrates (2).
A-kinase-anchoring proteins (AKAPs)3 are widely recognized for their role in directing the cAMP-dependent protein kinase (PKA) toward its substrates (1). This proceeds through stable interactions between a conserved region on the AKAP and a docking domain formed by the regulatory subunits of the tetrameric PKA holoenzyme (3–5). However, AKAPs also bind other classes of signaling molecules (6–8). Consequently, the selective recruitment of enzymes that respond to diverse second messengers into AKAP signaling complexes provide a means to fine-tune individual intracellular processes. AKAP220 is a ubiquitously expressed 220-kilodalton anchoring protein, encoded by the AKAP11 gene, which targets PKA to peroxisomes (9). Subsequent studies have implicated AKAP220 anchored PKA in the modulation of the small GTPase Rac (10) and identified other binding partners that include glycogen synthase kinase-3β (GSK-3β) (11) and protein phosphatase-1 (12). It was shown that AKAP220-mediated anchoring of PKA facilitates phosphorylation of GSK-3β on serine 9 to suppress the activity of this kinase (11). However, the physiological significance of this anchored regulatory mechanism has not yet been demonstrated.
In this report, we show that AKAP220 interacts with the cytoskeletal scaffold protein IQ domain GTPase-activating protein (IQGAP1). IQGAPs contain several protein interaction modules that allow them to bind a range of effector proteins that control changes in cell morphology and movement (13). These include MAPKs, actin, calmodulin, cadherin, and the small GTPases Rac and Cdc42. Accordingly, IQGAPs have been implicated in the control of cell growth and division, remodeling of the cytoskeleton, and cell-cell adhesion (14, 15). IQGAPs also interact with a variety of proteins that modulate microtubule dynamics at the cell cortex in migrating cells. These include the adenomatous polyposis coli (APC) tumor suppressor protein and microtubule plus-end tracking proteins (+TIPs), CLIP-170, and CLASP2 (16–19). In this context, specialized IQGAP sub-complexes are found at growing ends of microtubules just behind the leading edge of motile cells. These protein-protein interactions control cytoskeletal dynamics and establish cellular asymmetry in migrating cells (20). Our findings show that a union of AKAP220 with IQGAP1 forms a larger molecular network that can synchronize a succession of signal dependent protein-protein interactions to drive cell motility.
EXPERIMENTAL PROCEDURES
Mass Spectrometry
Proteins were identified as described (21).
Comet Tracking
HT1080 cells grown in glass-bottomed dishes coated with bovine collagen (Sigma) were transfected with siRNAs and EB3-GFP for 36 h prior to imaging. Sixteen hours before comet tracking, they were scratch wounded and maintained in Hank's balanced salt solution (Invitrogen) containing PDGF (Cell Signaling Technology). Comet tracking was performed on an Olympus DeltaVision microscope using a 63× differential interference contrast oil immersion lens (numerical aperture, 1.4) equipped with an environment chamber at 37 °C. Time lapse images were acquired at 2-s intervals over a 3-min time period. Four focal planes were acquired for each time point and deconvolved using DeltaVision software. These images were used to make maximum intensity projections in NIH ImageJ software. Velocity measurements and tracking diagrams were made using the Manual Tracking plugin for NIH ImageJ software and Adobe Photoshop. Statistical analyses were performed using an unpaired two-tailed Student's t test using GraphPad software. Quantitative measurements of the distribution of EB3-GFP tracks were done by plotting average pixel intensities along a thick line covering the cell edge using normalized projection images spanning 1 min in NIH ImageJ software.
Cell Migration Assays
MCF-7 or HT1080 human fibrosarcoma cells were grown on glass coverslips or glass-bottomed dishes coated with bovine collagen (Sigma). Cells were serum starved for 12 h in OptiMEM (Invitrogen) prior to wounding with a pipette tip and PDGF (Cell Signaling Tech.) stimulation. Phase contrast images of live HT1080 cells were taken on a Leica DMI6000B microscope using a 20× objective in a humidified chamber at 37 °C. Images were collected over 12 h at 20-min intervals. Multiple cells from each frame were used for quantitation. In each case, x and y coordinates were used to calculate distance traveled over time. Care was taken to avoid cells appearing unhealthy or that were perturbed by cellular debris. Statistical significance was determined by an unpaired two-tailed Student's t test using GraphPad software. Antibodies, Cells and Reagents, Cell Treatment and Lysis, and Immunofluorescence Staining are described in the supplemental Methods.
RESULTS
Identification of AKAP220-IQGAP1 Complex
We undertook a mass spectrometry based screen for additional AKAP220-binding partners. FLAG-tagged AKAP220 expressed in CVI origin SV-40 cells was isolated by affinity chromatography, and the resulting protein complex was separated on 4–12% SDS-PAGE gels (Fig. 1A). Tandem MS/MS identified several known binding partners of AKAP220 and 45 peptides derived from IQGAP 1, 2, and 3 isoforms (Fig. 1A and supplemental Fig. S1A). We have previously shown that only IQGAP2 is a PKA substrate (10). For this current study, we chose to focus on the functional ramifications of IQGAP1 interaction with the AKAP220 signaling complex.
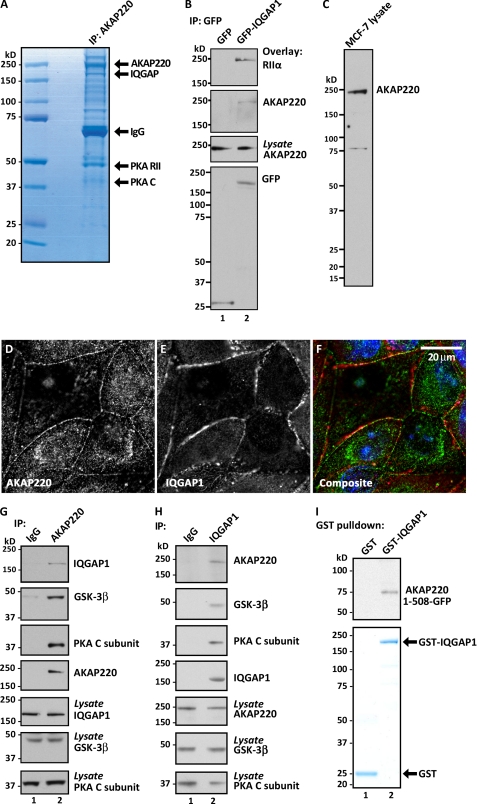
FIGURE 1.
AKAP220 assembles a complex that includes IQGAP1, GSK-3β, and PKA. A, FLAG-tagged AKAP220 immune complexes were isolated from HEK-293 cells and separated by SDS-PAGE. AKAP220 and selected binding partners were detected by Coomassie Blue stain. Molecular mass markers are indicated. B, immunoprecipitates of GFP-tagged IQGAP1 from COS cells were examined for co-purifying AKAPs by overlay of blots with digoxigenin-labeled type II PKA regulatory subunits (RIIα). C, immunoblot analysis of MCF-7 cell lysates using anti-AKAP220 antibody. D--F, immunofluorescent confocal analysis of endogenous AKAP220 (D and F, green) and IQGAP1 (E and F, red) in confluent MCF-7 cultures. Nuclei were identified with DRAQ5 dye (F, blue). Scale bar, 20 μm. G, co-IP of endogenous AKAP220, IQGAP1, GSK-3β, and the catalytic subunit of PKA (C subunit) from MCF-7 cells. H, reciprocal co-IP of endogenous IQGAP, AKAP220, GSK-3β, and PKAC subunit. I, recombinant GST-IQGAP1 was used to pull down in vitro-translated GFP-tagged AKAP220 (1–508 amino acids). Co-purification of AKAP220 was determined by immunoblotting for the GFP tag (top). GST tagged proteins were detected by Coomassie Blue stain (bottom). See also supplemental Fig. S1.
Our mass spectrometry results were verified initially by characterizing the AKAP220-IQGAP1 interaction in COS cells. GFP-tagged IQGAP1 was transfected into COS cells and immunoprecipitated from cell lysates. Immunocomplexes were probed for co-purification of AKAPs by overlay with digoxigenin-labeled type II PKA regulatory subunits. A single band at 220 kDa co-purified with IQGAP1 (Fig. 1B, top panel, lane 2). Immunoblot analysis confirmed that this was AKAP220 (Fig. 1B, upper middle panel, lane 2). A more detailed analyses of this protein-protein interaction were conducted in MCF-7 human breast cancer cells. Preliminary experiments confirmed that endogenous AKAP220 was present in MCF-7 cell lysates. A signal corresponding to an apparent molecular weight of 220 kDa was detected by immunoblot using affinity-purified antibodies raised against an N-terminal fragment of the recombinant protein (Fig. 1C). This antibody was subsequently used to detect AKAP220 in intact MCF-7 cells by immunofluorescence microscopy. These data revealed two locations for AKAP220, one that is consistent with our earlier report that this anchoring protein decorates peroxisomes (9) as evidenced by a punctate staining pattern in the cytoplasm. Additionally, a significant proportion of the anchoring protein was found near the cell cortex in confluent cultures (Fig. 1, D and F). The AKAP220 signal overlaps with IQGAP1 in this compartment (Fig. 1, D–F). Further control experiments confirmed that staining for AKAP220 was specific (supplemental Fig. S1B). When these results were considered along with our biochemical evidence, they suggested that AKAP220 and IQGAP1 interact. Support for their participation in a more elaborate protein network was provided by reciprocal studies that probed for binding partners in endogenous AKAP220 or IQGAP1 immunocomplexes (Fig. 1, G and H). Immunoblot analysis of AKAP220 immune complexes revealed co-purification of IQGAP1, GSK-3β, and PKA subunits (Fig. 1G, lane 2). Parallel studies detected AKAP220, GSK-3β, and PKA subunits in IQGAP1 complexes (Fig. 1H, lane 2). Furthermore, in vitro-translated protein products were used to detect a direct interaction between GST-IQGAP1 and a GFP-tagged fragment encompassing the first 508 amino acids of AKAP220 (Fig. 1I, top panels). Collectively, these data suggest that AKAP220 and IQGAP1 are direct binding partners and participate in a membrane localized signaling unit that includes GSK-3β and the PKA holoenzyme. Furthermore, an amino-terminal segment of AKAP220 (residues 1–508) is necessary and sufficient to bind IQGAP1.
IQ Domains of IQGAP1 Regulate Interaction with AKAP220
Calcium regulates the recruitment of several IQGAP1 binding partners. This involves the second messenger protein calmodulin (CaM), which reversibly associates with the IQ domains on this scaffolding protein (22). Mapping experiments implicated these IQ domains in the regulation of the AKAP220 interaction (Fig. 2, A–C). A family of internally deleted GFP-tagged IQGAP1 fragments (Fig. 2A) was expressed in COS cells and screened for interaction with AKAP220 (Fig. 2B, top panel, and Fig. 2C, densitometry analyses of AKAP220; Fig. 2B, top panel, normalized to IQGAP1 fragment expression, and Fig. 2B, bottom panel). Although each IQGAP1 fragment interacted with AKAP220 to some extent, the construct lacking IQ domains displayed a 3.6 (n = 4) fold enhancement in binding to AKAP220 (Fig. 2, B, lane 5, and C). Improved association with the anchoring protein correlated with a loss of binding to CaM (Fig. 2B, CaM panel, lane 5). This suggested that CaM binding to IQGAP1 could reduce interaction with AKAP220. Related experiments demonstrated Ca2+ inhibits the IQGAP1 interaction with CaM and the coincident recruitment of an activated form of the small GTPase Rac (Fig. 2, D and E, and supplemental Fig. S2A). These findings corroborate previous reports that IQGAPs serve as effectors of Rac and Cdc42 (15, 23).
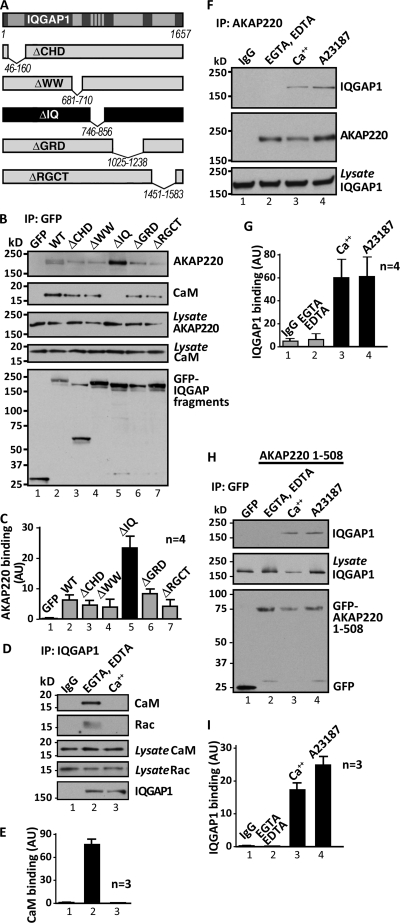
FIGURE 2.
Analyses of AKAP220 and IQGAP1 interaction. A, diagram depicting domains of IQGAP1 wild-type and deletion mutants. CHD, calponin homology domain; WW, poly-proline protein-protein domain; IQ, 4 IQ motifs domain; GRD, rasGAP-related domain; RGCT, RasGAP carboxy terminal domain. B, IP of GFP-tagged IQGAP1 wild-type and indicated mutants from COS cells. Co-purification of AKAP220 and CaM is shown in blots overlaid with digoxigenin-labeled type II PKA regulatory subunits (RIIα) (top) or immunoblotted for endogenous CaM (middle). C, densitometry analysis of data in B. The AKAP220 signal in the top panel was normalized to IQGAP1 fragment expression shown in bottom panel (n = 4), mean ± S.E. D, IP of endogenous IQGAP1 from MCF-7 cells in the presence of chelators (EGTA and EDTA, 1 mm each), CaCl2 (1 mm), or from cells pretreated with the ionophore A23187 (5 μm) for 20 min. Co-precipitating, endogenous CaM (top) and Rac are shown. E, densitometry analysis of data in D. CaM in the top panel is normalized to CaM in lysates (middle panel, n = 3, mean ± S.E.). F, IP of endogenous AKAP220 from MCF-7 cells in the presence of chelators (1 mm each), CaCl2 (1 mm), or from cells pretreated for 20 min with ionophore A23187 (5 μm). Immunoblots show co-purification of endogenous IQGAP1 (top). Shown are AKAP220 (middle) and levels of IQGAP in the lysates (bottom). G, densitometry analysis of data in F. IQGAP1 in the top panel is normalized to IQGAP1 in lysates (bottom panel, n = 4, mean ± S.E.). H, IP of the GFP-tagged AKAP220 1–508-amino acid fragment from COS cells in the presence of chelators (1 mm each), CaCl2 (1 mm), or from cells pretreated for 20 min with ionophore A23187 (5 μm). Immunoblots show co-purification of endogenous IQGAP1. I, densitometry analysis. IQGAP1 in the top panel is normalized to IQGAP1 in lysates (middle panel, n = 3, mean ± S.E.). See also supplemental Fig. S2.
We next determined whether Ca2+/CaM regulates binding between IQGAP1 and AKAP220. Isolation of endogenous AKAP220 from MCF-7 cells pretreated with 5 μm of the calcium ionophore A23187 (20 min prior to lysis) increased binding to IQGAP1 (Fig. 2, F and G, lane 4). Similar results were obtained when cell lysates were prepared in the presence of 1 mm calcium (Fig. 2, F and G, lane 3). Furthermore, deletion of the IQ domains to create IQGAPΔIQ generated a protein that binds AKAP220 independent of calcium (supplemental Fig. S2B). Additional control experiments confirmed that association of AKAP220 with GSK-3β and PKA was unaffected by changes in calcium concentration (supplemental Fig. S2, C and D). In vitro mapping studies demonstrate that IQGAP directly interacts with a single region within the first 508 amino acids of the anchoring protein (Fig. 1I). This fragment, AKAP220-(1–508) is necessary and sufficient to bind IQGAP and does so in a calcium-dependent manner (Fig. 2, H, top panel, lanes 3 and 4, and I). Collectively, these results demonstrate that elevating calcium releases CaM (and Rac) from IQGAP1, while at the same time enhancing the interaction with AKAP220. Thus, fluctuations in intracellular calcium concentrations influence the association of IQGAP1 with AKAP220.
AKAP220 Anchors IQGAP and GSK-3 to Leading Edges of Migratory Cells
Growth factors such as PDGF promote cell migration via a mechanism that involves mobilization of calcium and elevation of intracellular cAMP (24). Recent evidence suggests that GSK-3β mediates regulation of IQGAP1 to control cell migration (19). As AKAP220 and IQGAP1 are found near the cell cortex (Fig. 1F), we wondered whether these proteins might be involved in this process. Initial experiments established that PDGF treatment favored formation of the AKAP220-IQGAP1 complex. Cells were exposed to PDGF (50 ng/ml for 20 min), and AKAP220 immune complexes were probed for IQGAP1 (Fig. 3A). PDGF stimulated the recruitment of IQGAP1 (Fig. 3, A, top panel, lane 3, and B). This effect was blocked upon the depletion of intracellular calcium with EGTA and EDTA (Fig. 3, A, top panel, lane 4, and B). Control experiments confirmed that AKAP220 interaction with GSK-3 was unaffected under these conditions (Fig. 3Aβ, GSK-3β panel, lanes 2–4). Related experiments confirmed that in PDGF-treated cells, the anchoring protein is asymmetrically distributed toward the leading edge (Fig. 3C). This was particularly evident when AKAP220 staining was compared with the distribution of a general membrane marker such as wheat germ agglutinin (Fig. 3, C–E). In contrast to what was observed in stationary epithelial cells, AKAP220 was enriched near the leading edge in migratory fibroblasts (compare Fig. 1, D–F, and 3, C–E and G–U).
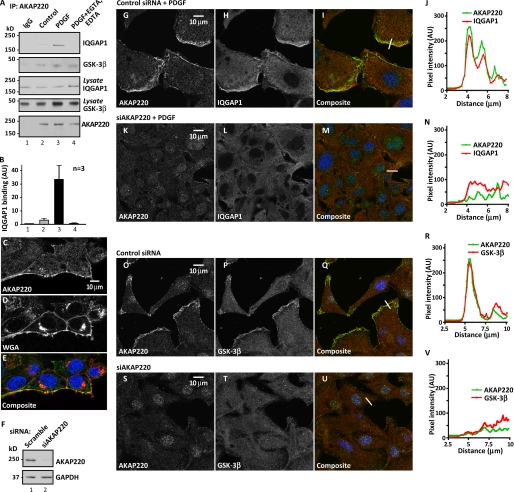
FIGURE 3.
AKAP220 organizes IQGAP1 and GSK-3β at the leading edge of motile cells in response to growth factors. A, formation of the AKAP220-IQGAP complex in response to growth factor stimulation (PDGF; 50 ng/ml for 20 min). AKAP220 immune complexes isolated from control (lane 2), PDGF-stimulated (lane 3), and PDGF+EGTA-treated (lane 4) cell lysates. Samples were probed for IQGAP1 (top), GSK-3β (top middle panel), and AKAP220 (bottom) by immunoblot. B, densitometry analysis. IQGAP1 in top panel (A) is normalized to IQGAP1 in lysates (middle panel, A) (n = 3), mean ± S.E. C–E, scratch-wounded and PDGF-stimulated cells were fixed and stained for fluorescence detection of AKAP220 (C) and Alexa Fluor 594-conjugated (D) wheat germ agglutinin (WGA) as a general membrane marker. E, nuclear staining with DRAQ5 is also shown in the composite image. F, gene silencing of AKAP220. Immunoblot detection of AKAP220 (top) and GAPDH (bottom) loading control. G–I and K–M, confocal immunofluorescence detection of AKAP220 (G) and IQGAP1 (H) in migrating HT1080 cells that had been scratch-wounded and PDGF-stimulated. Composite image (I) includes DRAQ5 staining as a nuclear marker (blue). K–M, gene silencing of AKAP220 prior to scratch wounding and PDGF stimulation altered the appearance of the AKAP220 and IQGAP1 immunofluorescent staining. J and N, measurement of pixel intensity (left to right) for AKAP220 (green) and IQGAP1 (red) along an arbitrary straight line (I and M) was plotted using NIH ImageJ software. O–Q and S–U, confocal immunofluorescence detection of AKAP220 (O) and GSK-3β (P) in scratch-wounded and PDGF-stimulated HT1080 cells. Composite image (Q) includes DRAQ5 staining as a nuclear marker. R and V, quantitative evaluation of overlap between the AKAP220 (green) and GSK-3β (red) signals at the indicated leading edge (white bar; Q and U) was plotted using NIH ImageJ software. S–U, the staining pattern for both proteins in cells treated with a siRNA against AKAP220 prior to scratch wounding and PDGF stimulation. All confocal images are taken from a region adjacent to the wound. AKAP220 siRNA-1 was used in knockdown experiments. See also supplemental Fig. S3. AU, arbitrary units.
Immunofluorescent confocal imaging also demonstrated a co-distribution of the AKAP220 and IQGAP1 signals at the leading edges of HT1080 human fibrosarcoma cells after scratch wounding (Fig. 3, G–I). A more detailed analysis using NIH ImageJ software to plot pixel intensities along a line (indicated in Fig. 3I) that bisects the leading edge of migrating cells emphasized the similar distribution of the green (AKAP220) and red (IQGAP1) channels (Fig. 3J). Additional experiments confirmed that IQGAP1 staining overlapped with actin, a marker for the leading edge in migrating cells (supplemental Fig. S3). Importantly, enrichment of IQGAP1 at the leading edge was lost upon siRNA-mediated gene silencing of the anchoring protein (Fig. 3, F and K–N). We also noted that upon gene silencing of AKAP220 the cells in the region adjacent to the wound appeared at a higher density perhaps owing to a decrease in the mobility of these cells (Fig. 3, K–M). (Note that all confocal images are taken from a region adjacent to the wound.)
We also confirmed that another AKAP220 binding partner GSK-3 was asymmetrically distributed in scratch-wounded PDGF-treated HT1080 cells (Fig. 3, O–R). Depletion of AKAP220 induced a redistribution of GSK-3β and an apparent decrease in the ability of these cells to move (Fig. 3, S–V). These observations led to a working hypothesis that AKAP220 and IQGAP1 act synergistically at the leading edges to control cell migration. This notion is substantiated by evidence that IQGAP and activated Rac or Cdc42 enable microtubules to be captured at the leading edge (25). We further reasoned that changes in cell polarity could be influenced if AKAP220 relayed signals to the plus-ends of microtubules.
Anchored GSK-3β Negatively Regulates IQGAP1-CLASP2 Interaction
IQGAP1 promotes cell migration by helping remodel the cytoskeleton (13). Recently, IQGAP1 was found to associate with CLIP-associating protein CLASP2 (19). Together, these proteins can control microtubule dynamics at the leading edge of motile cells. However, this complex can only be formed when the AKAP220-binding partner GSK-3β is inactive (19). Under resting conditions, this kinase tonically phosphorylates CLASP2 to prevent interaction with IQGAP1 (19). Thus, inhibition of GSK-3β at the leading edge would favor cell migration. This idea is substantiated by seminal work showing that GSK-3 supports microtubule dynamics at leading edges (26, 27). We postulated that these regulatory events could be mediated by AKAP220-anchored PKA, especially because PKA phosphorylation of serine 9 on GSK-3β inhibits the latter enzyme (28, 29). This was corroborated by immunofluorescence detection of phospho-Ser-9 GSK-3β at a number of leading edges in migrating cells after scratch wounding (Fig. 4, A–C). Antibody compatibility issues precluded dual labeling experiments for phospho-GSK-3β and AKAP220.
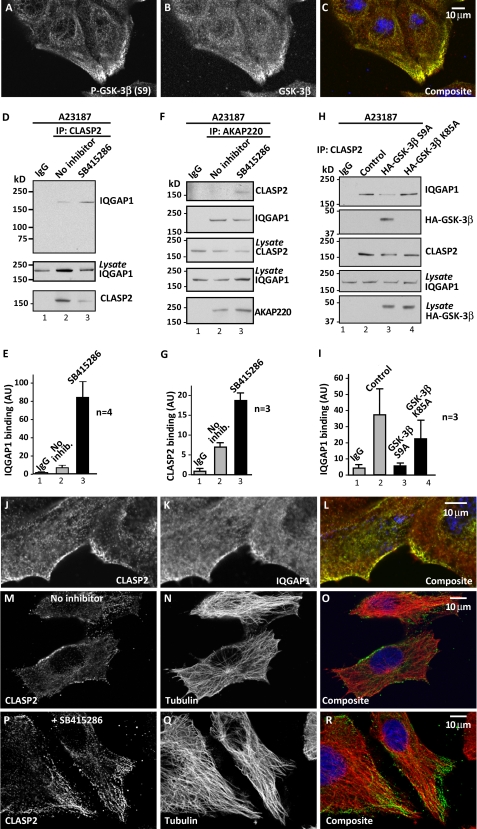
FIGURE 4.
CLASP2 interaction with IQGAP1. A–C, immunofluorescent confocal detection of phospho-Ser 9-GSK-3 (A), total GSK-3 (B), and composite images (C) in scratch-wounded and PDGF-stimulated (50 ng/ml) HT1080 cells. The image was taken from a region adjacent to the wound. D, endogenous CLASP2 was immunoprecipitated from cells pretreated with GSK-3 inhibitors (50 μm SB415286) for 1 h and subsequently treated with the ionophore A23187 (5 μm) for 20 min. Immunoblots show co-purification of endogenous IQGAP1. E, densitometry analysis. IQGAP1 in the top panel (C) is normalized to IQGAP1 in lysates (middle panel, C; n = 3), mean ± S.E. Analysis of lanes 2 and 3 using a two-tailed Student's t test revealed a p value = 0.022. F, endogenous AKAP220 complex was immunoprecipitated in the presence or absence of the GSK-3 inhibitor SB415286 (50 μm). Immunoprecipitates were probed for CLASP2 (top), IQGAP1 (second top panel), and AKAP220 (bottom). Levels of IQGAP1 and CLASP2 in the lysates are shown. G, densitometry analysis shows relative co-precipitation of CLASP2. Level of CLASP2 in the top panel of B normalized to CLASP2 in lysates (n = 3, mean ± S.E.). Analysis of lanes 2 and 3 using a two-tailed Student's t test revealed a p value = 0.007. H, endogenous CLASP2 was immunoprecipitated from cells transfected with GSK-3β mutants that are either constitutively active (S9A; lane 3) or inactive (K85A; lane 4). Immunoblots show co-purification of IQGAP1 (top) and GSK-3β (second top panel). Expression of HA-tagged GSK-3β is shown (bottom). I, densitometry analysis. IQGAP1 in the top panel (F) is normalized to IQGAP1 in lysates (middle bottom panel, F; n = 3, mean ± S.E.). J–L, immunofluorescent confocal detection of CLASP2 (J), IQGAP1 (K), and composite images (L) in scratch-wounded and PDGF-stimulated (50 ng/ml) HT1080 cells. Images are taken from a region of the culture dish adjacent to the wound. M–R, pharmacological inhibition of GSK-3 alters the distribution of CLASP2. Immunofluorescent confocal detection of CLASP2 (M), tubulin (N), and composite images (O) in HT1080 cells. Detection of CLASP2 (P), tubulin (Q), and composite images (R) HT1080 cells treated with SB415286 (50 μm). See also supplemental Fig. S4.
Immunoprecipitation experiments revealed that CLASP2 interacts with IQGAP1 preferentially when calcium levels are increased (supplemental Fig. S4). Pharmacological inhibition of GSK-3 activity with the compound SB41586 augmented this protein-protein interaction (Fig. 4, D, top panel, lane 3, and E). These binding events were also examined in the context of the AKAP220-signaling complex. Pharmacological inhibition of GSK-3 during the isolation of AKAP220 complexes allowed the detection of CLASP2 (Fig. 4, F, top panel, lane 3, and G). This inferred that AKAP220 serves as a platform for the assembly of a CLASP2-IQGAP sub-complex. Expression of a constitutively active GSK-3β mutant (GSK-3β-S9A) in HT1080 cells reduced the co-purification of this sub-complex (Fig. 4, H, top panel, lane 3, and I). Control experiments using a catalytically inactive (kinase-dead) mutant had no effect (Fig. 4, H, top panel, lane 4, and I). Immunoblotting confirmed that the constitutively active GSK-3β-S9A mutant co-precipitated with CLASP2 (Fig. 4H, top middle panel, lane 3). These data are consistent with our hypothesis that AKAP220 organizes a GSK-3-CLASP2 sub-complex. Incidental support for this model was provided by immunofluorescence detection of IQGAP1 and CLASP2 at leading edges of scratch wounded cultures (Fig. 4, J–L). Furthermore, immunofluorescence data presented in Fig. 4, M–R, suggest that subcellular distribution of CLASP2 becomes more polarized and organized toward leading edges of HT1080 cells that have been treated with the GSK-3 small molecule inhibitor SB415286 (Fig. 4, M–Q and P--R). Tubulin was used as a marker of microtubules (Fig. 4, N and Q). Collectively, the observations in Fig. 4 are consistent with the notion that inhibition of GSK-3 favors formation of the IQGAP1-CLASP2 sub-complex.
AKAP220 Contributes to Microtubule Dynamics and Cell Motility
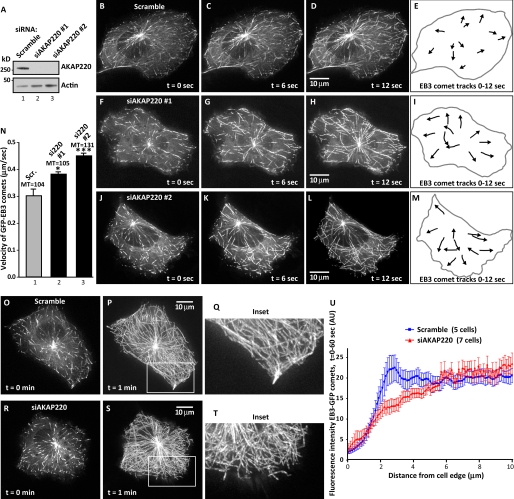
Microtubules undergo alternating phases of growth and shortening in a process known as “dynamic instability” (30, 31). This intrinsic property of microtubules is modulated by the synergistic action of cellular enzymes and effector proteins that associate with the plus ends of microtubules. Our cumulative data suggest that stimulation of AKAP220-associated PKA, phosphorylation of GSK-3β, and elevations in calcium cooperatively drive the formation of an IQGAP1-CLASP2 sub-complex. Because association of CLASP2 with plus-end tips is correlated with microtubule stabilization and suppression of microtubule dynamics (32, 33), we reasoned that removal of the anchoring protein could prevent the formation of this sub-complex and impact microtubule growth. Two independent siRNAs suppressed AKAP220 in HT1080 cells as assessed by immunoblot (Fig. 5A, top panel, lanes 2 and 3). Control experiments confirmed that siRNA encoding a scrambled sequence had no effect on AKAP220 expression (Fig. 5A, top panel, lane 1). Equivalent levels of total protein were present in all samples (Fig. 5A, bottom panel). Time-lapse fluorescent imaging evaluated the impact of AKAP220 knockdown on microtubule polymerization in HT1080 cells (supplemental Movies S1–S3). GFP-tagged EB3 (end binding protein 3) served as a marker for the plus-end tips of growing microtubules (34). Velocity measurements of EB3-GFP comets and tracking patterns of individual +TIPs were generated using the NIH ImageJ software and the Manual Tracking plug-in. Control cells transfected with scrambled siRNA exhibited microtubule tip velocities of 0.31 ± 0.02 μm/sec (Fig. 5, B–E, and N, column 1, microtubules= 104). The velocity of EB3-GFP tracking increased by 21% (0.39 ± 0.01 μm/sec; microtubules = 105) and by 33% (0.46 ± 0.01 μm/sec; MT = 131) in cells treated with siAKAP220-1 and siAKAP220-2, respectively (Fig. 5, F–M, and N, columns 2 and 3). Representative EB3-GFP comet tracks are presented for each siRNA (0–12 s, Fig. 5, E, I, and M). This shows that gene silencing of AKAP220 leads to increased rates of microtubule polymerization. This is consistent with loss of CLASP-selective microtubule stabilization (16–19). However, in the case of AKAP220 knockdown, we believe that the IQGAP1-CLASP2 sub-complex is mislocalized rather than depleted.
FIGURE 5.
AKAP220 impacts microtubule dynamics. A, top, immunoblot showing siRNA knockdown of AKAP220 in HT1080 human fibrosarcoma cells. Actin (bottom) is used as loading control. B–D, EB3-GFP comet tracks recorded in scrambled siRNA-treated HT1080 cells that were scratch-wounded and treated with PDGF (50 ng/ml). Images of projections at 0 s (B) and spanning 0–6 s (C) and 0–12 s (D) are shown. E, representative EB3 comet tracks spanning 12 s traced over an outline of the cell. Comet tracks were prepared using the Manual Tracking plug-in for NIH ImageJ software. F–M, EB3-GFP comets were tracked as above in cells depleted of the anchoring protein with siAKAP220-1 or siAKAP220-2 as indicated. Images of projections (F–H) and (J–L) and representative comet tracks (I and M) are shown. N, quantitative analysis of EB3-GFP comet velocities in live siRNA-treated HT1080 cells (as in B–M) with S.E. and statistical significance (unpaired two-tailed Student's t test; *, p ≤ 0.05 and ***, p ≤ 0.001). The number of microtubules (MT) used in analyses is indicated above each column. O–T, time 0 and 1 min projections of EB3-GFP comet tracks near the periphery in cells treated with scramble (Scr.) siRNA (O–Q) and siAKAP220 (R–T). U, pixel intensities along a line from the cell edge to the interior were measured using the 1-min projections of EB3-GFP comets in scramble siRNA (5 cells, blue line) and siAKAP220-treated cells (7 cells, red line). Error bars, S.E. See also supplemental Movies S1–S3.
A more detailed examination of EB3-GFP comet projections (1 min) revealed that lateral tracking of +TIPs along the cell periphery was reduced upon depletion of AKAP220 (Fig. 5, O–T, and supplemental Movies S1–S3). Quantification of this effect was provided by measuring the density of EB3-GFP comets in relation to their distance from the leading edge (Fig. 5U). Analysis showed that gene silencing of AKAP220 reduced the cortical distribution of EB3-GFP comets (Fig. 5U, red line) when compared with controls (Fig. 5U, blue line). This observation is in keeping with experiments showing that depletion of IQGAP1 uncouples microtubule growth to the cortical actin cytoskeleton (16, 17). Additional experiments using antibodies toward tyrosinated tubulin, a post-translational mark of newly formed microtubules (35, 36), confirmed that loss of AKAP220 reduces the enrichment of newly formed microtubules near the cell periphery in migrating cells (supplemental Fig. S5). Taken together, these results suggest that AKAP220 supports the spatial and temporal synchronization of signaling machinery with +TIP proteins to regulate aspects of microtubule dynamic instability.
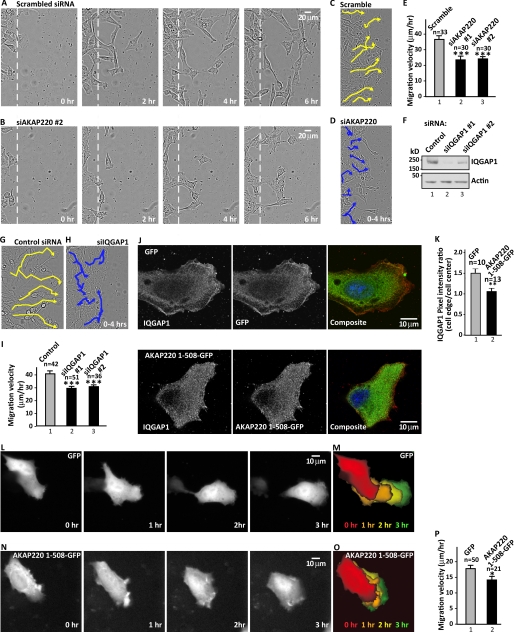
Cell movement requires a coordinated restructuring of microtubules and remodeling of actin. Because AKAP220 spatially restricts protein kinases and microtubule effectors near the leading edge, we reasoned that this anchoring protein might contribute to the control of cell movement. PDGF-stimulated HT1080 cell migration velocities were measured by imaging at 20-min intervals over a 12-h period following scratch wounding. The migration rate of control cells transfected with scrambled siRNA was 35.9 ± 2.4 μm/h (n = 33 cells) (Fig. 6, A, C, and E, column 1, and supplemental Movie S4). In contrast, cells depleted of AKAP220 with siAKAP220-2 exhibited a 34% decrease in average migration velocity (23.8 ± 1.5 μm/h (n = 30); Fig. 6, B, D, and E). Cells treated with siAKAP220-1 exhibited a 36% reduction in average migration velocity (23.1 ± 2.4 μm/h (n = 30); Fig. 6E and supplemental Movie S5). Related experiments evaluated cell migration upon gene silencing of IQGAP1 (Fig. 6, F–I). Depletion of IQGAP1 reduced the migration velocity by 27% (siIQGAP1-1, 30 ± 1.5 μm/h (n = 51), Fig. 6I) when compared with control cells (41. 1 ± 2.6 μm/h (n = 42), Fig. 6I). In cells transfected with a different siRNA targeting IQGAP1, the average migration velocity was reduced by 30% (siIQGAP1-2, 31.6 ± 1.8 μm/h (n = 36), Fig. 6I). Depletion of IQGAP1 reduced persistent migration and enhanced the lateral movement of migrating cells (Fig. 6, G and H). Thus, knockdown of IQGAP1 has a similar effect as gene silencing of AKAP220 in HT1080 cells.
FIGURE 6.
AKAP220 promotes cell migration. A and B, representative time course images of cell migration across wound edge of scratch-wounded and PDGF-stimulated (50 ng/ml) HT1080 cells with either scrambled siRNA (A) or siAKAP220-2 (B). Images shown are from 120-min intervals over 6 h. The dashed line indicates initial wound edge. See also supplemental Movies S4 and S5. C and D, nuclei from individual cells were tracked every 20 min for 4 h using NIH Image J software and the Manual Tracking plug-in for either scramble (C) or siAKAP220 (D). E, quantitation of migration velocity (μm/h) from live-cell imaging of scratch-wounded HT1080 cells with scrambled siRNA (n = 33), siAKAP220-1 (n = 30), and siAKAP220-2 (n = 30) with statistical significance (unpaired two-tailed Student's t test; *** p ≤ 0.001) and S.E. F, immunoblot of IQGAP1 level in HT1080 cells treated with either scramble, siIQGAP1-1, or siIQGAP1-2. Actin serves as loading control. G and H, nuclei from individual cells treated with either control siRNA (G) or siIQGAP1 (H) were tracked every 20 min for 4 h as above. I, quantitation of migration velocity (μm/h) for cells treated with control siRNA (n = 42), siIQGAP1-1 (n = 51), and siIQGAP1-2 (n = 36) with statistical significance (unpaired two-tailed Student's t test; ***, p ≤ 0.001) and S.E. J, immunofluorescent confocal detection of IQGAP1 subcellular location (left panels, gray) in cells expressing GFP control or AKAP220-(1–508)-GFP (middle panels, gray). Composite images are shown for IQGAP1 (red), GFP (green), and nuclei (blue). K, quantitative analysis of IQGAP1 distribution from defined regions at the cell edge and cell center. Ratio of pixel intensity at the cell edge versus the cell interior from cells expressing GFP (n = 10) and AKAP220-(1–508)-GFP (n = 13) is presented with statistical significance (unpaired two-tailed Student's t test; **, p ≤ 0.01) and S.E. L–O, representative migration path (beginning upper left) of a cell expressing either GFP control (L and M) or AKAP220-(1–508)-GFP (N and O). Images shown in L and N are from time 0 through 3 h at 1-h intervals. Images M and O show progressive positions of each cell at the color-coded time points as indicated. P, quantitation of average migration velocity in L and N. Data are shown from GFP (n = 50) and AKAP220-(1–508)-GFP (n = 21) with statistical significance (unpaired two-tailed Student's t test; *, p ≤ 0.05) and S.E. See also supplemental Movies S4–S7.
Finally, we evaluated the effects of interrupting IQGAP1 association with AKAP220 on the migration patterns of HT1080 cells. Expression of a GFP-tagged AKAP220 fragment encompassing the IQGAP1 binding domain (AKAP220-(1–508)-GFP) served as a dominant interfering agent that prevented accumulation of IQGAP1 at leading edges (Fig. 6, J and K). Furthermore, the GFP moiety provided a fluorescent marker to monitor cell migration (Fig. 6, L–O). Fluorescent time-lapse microscopy revealed that expression of AKAP220-(1–508)-GFP reduced the migration velocity by 21% (14 ± 1.2 μm/h (n = 21 cells), Fig. 6, N–P, and supplemental Movie S7) compared with control cells expressing GFP alone (17.7 ± 1.1 μm/h (n = 50 cells), Fig. 6, L–N, and supplemental Movie S6). A montage of images at 1-h intervals best illustrates the migration paths of single cells (Fig. 6, M and O). Collectively, the data presented in Fig. 6 indicate that AKAP220 anchored IQGAP1 supports cell motility.
DISCUSSION
In this report, we show that AKAP220 participates in the integration and processing of calcium and cAMP signals that underlie directional mobilization of a microtubule-associated network at the leading edge of cells. This helps to establish cellular asymmetry, a prerequisite for cell migration. A role for AKAP220 in the control of microtubule dynamics at the plus ends ascribes a biological function for this poorly understood anchored signaling complex.
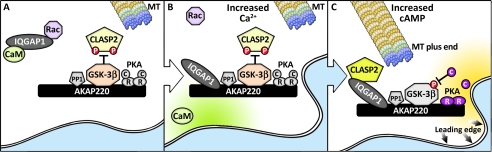
A hallmark of cell movement and growth cones is mobilization of actin polymerization followed by the recruitment of microtubules (37, 38). Cell migration involves a preferential reduction in microtubule dynamics at the leading edge relative to the trailing edge of cells (39). We propose that regulated AKAP220 and IQGAP1 association brings together the correct combination of signaling enzymes and microtubule effectors that are required for the polarization of microtubule dynamics at the leading edge. This model is depicted in Fig. 7. AKAP220 targets PKA, GSK-3β, and protein phosphatase-1, whereas IQGAP1 sequesters calmodulin and the Rac GTPase and is prevented from interacting with the anchoring protein (Fig. 7A). We propose that calcium loading liberates calmodulin and the Rac GTPase from IQGAP1 (Fig. 7B). Rac is now free to influence actin polymerization, whereas IQGAP1 can organize a sub-complex of AKAP220 binding partners for the modulation of microtubules at the leading edge. It is within this cellular context that the actions of IQGAP1 come under the influence of cAMP (Fig. 7C). This two-stage process involves (a) PKA-mediated suppression of GSK-3β activity and (b) the dephosphorylation of serines 533 and 537 on CLASP2 (Fig. 7C). This dephosphorylated form of CLASP2 can then interact with microtubule +TIPs and the AKAP220-anchored IQGAP1. We speculate that protein phosphatase-1 within the AKAP220 complex can have some bearing on the localized dephosphorylation of CLASP2 (12). The net effect is persistent cell motility (Fig. 7C).
FIGURE 7.
Model depicting the proposed role of AKAP220 in the control of cell migration. A, AKAP220 and IQGAP1 complexes in unstimulated cells. IQGAP1 is in a ternary complex with calmodulin CaM and Rac near the cell cortex. Anchored GSK-3β phosphorylates CLASP2 to prevent interaction with IQGAP1. B, as intracellular calcium increases, CaM and active Rac are released from IQGAP1, which can now bind to AKAP220. C, as cAMP levels rise, PKA (r, regulatory subunit; C, catalytic subunit) phosphorylates and inhibits GSK-3β, augmenting the CLASP2 interaction with anchored IQGAP1. This promotes microtubule rescue and cell migration.
Data presented in Fig. 4 support our theory that suppression of GSK-3β enables CLASP2 recruitment to the AKAP220 complex. This configuration is reminiscent of the APC tumor suppressor complex where GSK-3β activity can be attenuated upon phosphorylation on serine 9 by the lipid-dependent enzyme protein kinase B/Akt (29). Another parallel worthy of note is that CLASPs and IQGAPs interact with this APC complex where localized dephosphorylation of CLASP2 proceeds through associated protein phosphatase-2A (16, 17). In fact, the complementary functions of AKAP220, APC, and their overlapping repertoire of binding partners illustrate the degree of redundancy that is built into essential biological systems such as the cell migration machinery. One possible advantage of these related signaling complexes co-habiting the same subcellular environment would be to permit the utilization of alternate second messengers. AKAP220 transduces cAMP signals to activate PKA, whereas APC receives phospholipid signals downstream of Akt. In either case, the net result is the inhibition of GSK-3β activity and the subsequent recruitment of the +TIP protein CLASP2 to the leading edges of motile cells. Our findings also provide further molecular context for recent elegant observations showing that GSK-3β activity affects microtubules through control of the microtubule-actin cross-linking protein ACF7 (40, 41). This raises the intriguing possibility that the AKAP220/PKA/GSK-3β signaling consortium might play a broader role in microtubule behavior by impacting the function of this spectraplakin.
Dynamic instability of microtubules is characterized by stochastic transition between persistent growth and disassembly (30). A host of modulators have evolved to regulate microtubule dynamics in living cells. For example, growing tubulin polymers that transition to rapid disassembly (called “catastrophe”) can be rescued upon binding of CLASP2 (42). The imaging studies presented in Fig. 5 suggest that knockdown of AKAP220 influences the rate of microtubule polymerization in a manner analogous to the gene silencing of CLASP2. This +TIP binding protein promotes polymerization and stabilization of microtubules by selectively promoting microtubule rescue at the cell periphery (16–19, 32, 42). Paradoxically, gene silencing of CLASP2 increases the overall rate of microtubule polymerization by increasing the availability of tubulin throughout the cytoplasm because previously rescued/stabilized microtubules are now free to depolymerize. We postulate that the mislocalization of CLASP2 and the other signaling enzymes that occur as a consequence of AKAP220 knockdown has a similar effect. Close inspection of microtubule tracking patterns of >1 min revealed that the lateral movement of EB3 comets along the cell periphery occurs less frequently in cells lacking AKAP220. Thus, it would appear that removal of AKAP220 impacts microtubule dynamics in a manner that is consistent with the displacement of both IQGAP1 and CLASP2.
Our cell-based studies presented in Figs. 5 and 6 argue that AKAP220 provides a molecular foundation for the management and spatial patterning of microtubule effectors in response to signaling events. Live-cell imaging studies confirm that siRNA depletion of AKAP220 increases global microtubule polymerization rates consistent with a loss of polarized microtubule stabilization. This retards the migratory behavior of invasive human fibrosarcoma cells. We propose that IQGAP1/CLASP2 function is compromised upon gene silencing of AKAP220. This could occur in a variety of ways. Elimination of AKAP220 and the concomitant mislocalization of PKA could locally derepress GSK-3β activity leading to enhanced phosphorylation of CLASP2, destabilization of microtubules, and retarded cell migration. Alternatively, displacement of the IQGAP1-CLASP2 complex from its site of action at the leading edge hampers cell motility. Disruption of the actin cytoskeleton and effects on cell-cell adhesion also remain possible mechanisms contributing to the change in cell migration. Regardless of which prospective regulatory event predominates, our identification of AKAP220 as a modulator of cell motility and the discovery of an anchored GSK-3β/IQGAP1/CLASP2 axis provides an important mechanistic glimpse as to how AKAP220 could play an essential role in cytoskeletal polarization during metastasis and invasion of cancer cells.
Supplementary Material
Acknowledgments
We thank members of the Scott lab for critical evaluation of this manuscript and T. Pawson (Mount Sinai Hospital Research Institute, Toronto) for mass spectrometry.
This work was supported, in whole or in part, by National Institutes of Health Grants DK54441 (to J. D. S.), GM69429 (to L. W.), and CA93645 (to D. B. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, Figs. S1–S5, and Movies S1–S7.
- AKAP
- A-kinase-anchoring protein
- PKA
- cAMP-dependent protein kinase
- GSK-3β
- glycogen synthase kinase-3β
- APC
- adenomatous polyposis coli
- CaM
- calmodulin
- +TIP
- plus-end tracking protein
- COS
- CVI origin SV-40.
REFERENCES
- 1. Scott J. D., Pawson T. (2009) Science 326, 1220–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pidoux G., Taskén K. (2010) J. Mol. Endocrinol. 44, 271–284 [DOI] [PubMed] [Google Scholar]
- 3. Carr D. W., Stofko-Hahn R. E., Fraser I. D., Bishop S. M., Acott T. S., Brennan R. G., Scott J. D. (1991) J. Biol. Chem. 266, 14188–14192 [PubMed] [Google Scholar]
- 4. Gold M. G., Lygren B., Dokurno P., Hoshi N., McConnachie G., Taskén K., Carlson C. R., Scott J. D., Barford D. (2006) Mol. Cell 24, 383–395 [DOI] [PubMed] [Google Scholar]
- 5. Kinderman F. S., Kim C., von Daake S., Ma Y., Pham B. Q., Spraggon G., Xuong N. H., Jennings P. A., Taylor S. S. (2006) Mol. Cell 24, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coghlan V. M., Perrino B. A., Howard M., Langeberg L. K., Hicks J. B., Gallatin W. M., Scott J. D. (1995) Science 267, 108–111 [DOI] [PubMed] [Google Scholar]
- 7. Wong W., Goehring A. S., Kapiloff M. S., Langeberg L. K., Scott J. D. (2008) Sci. Signal 1, ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kapiloff M. S., Piggott L. A., Sadana R., Li J., Heredia L. A., Henson E., Efendiev R., Dessauer C. W. (2009) J. Biol. Chem. 284, 23540–23546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lester L. B., Coghlan V. M., Nauert B., Scott J. D. (1996) J. Biol. Chem. 271, 9460–9465 [DOI] [PubMed] [Google Scholar]
- 10. Logue J. S., Whiting J. L., Tunquist B., Langeberg L. K., Scott J. D. (2011) J. Biol. Chem. 286, 22113–22121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tanji C., Yamamoto H., Yorioka N., Kohno N., Kikuchi K., Kikuchi A. (2002) J. Biol. Chem. 277, 36955–36961 [DOI] [PubMed] [Google Scholar]
- 12. Schillace R. V., Scott J. D. (1999) Curr. Biol. 9, 321–324 [DOI] [PubMed] [Google Scholar]
- 13. Brown M. D., Sacks D. B. (2006) Trends Cell Biol. 16, 242–249 [DOI] [PubMed] [Google Scholar]
- 14. Noritake J., Watanabe T., Sato K., Wang S., Kaibuchi K. (2005) J. Cell Sci. 118, 2085–2092 [DOI] [PubMed] [Google Scholar]
- 15. Johnson M., Sharma M., Henderson B. R. (2009) Cell Signal. 21, 1471–1478 [DOI] [PubMed] [Google Scholar]
- 16. Watanabe T., Wang S., Noritake J., Sato K., Fukata M., Takefuji M., Nakagawa M., Izumi N., Akiyama T., Kaibuchi K. (2004) Dev. Cell 7, 871–883 [DOI] [PubMed] [Google Scholar]
- 17. Barth A. I., Caro-Gonzalez H. Y., Nelson W. J. (2008) Semin. Cell Dev. Biol. 19, 245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S., Akhmanova A. (2005) J. Cell Biol. 168, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Watanabe T., Noritake J., Kakeno M., Matsui T., Harada T., Wang S., Itoh N., Sato K., Matsuzawa K., Iwamatsu A., Galjart N., Kaibuchi K. (2009) J. Cell Sci. 122, 2969–2979 [DOI] [PubMed] [Google Scholar]
- 20. Galjart N. (2005) Nat. Rev. Mol. Cell Biol. 6, 487–498 [DOI] [PubMed] [Google Scholar]
- 21. Jin J., Smith F. D., Stark C., Wells C. D., Fawcett J. P., Kulkarni S., Metalnikov P., O'Donnell P., Taylor P., Taylor L., Zougman A., Woodgett J. R., Langeberg L. K., Scott J. D., Pawson T. (2004) Curr. Biol. 14, 1436–1450 [DOI] [PubMed] [Google Scholar]
- 22. Briggs M. W., Sacks D. B. (2003) FEBS Lett. 542, 7–11 [DOI] [PubMed] [Google Scholar]
- 23. Kuroda S., Fukata M., Nakagawa M., Fujii K., Nakamura T., Ookubo T., Izawa I., Nagase T., Nomura N., Tani H., Shoji I., Matsuura Y., Yonehara S., Kaibuchi K. (1998) Science 281, 832–835 [DOI] [PubMed] [Google Scholar]
- 24. Graves L. M., Bornfeldt K. E., Sidhu J. S., Argast G. M., Raines E. W., Ross R., Leslie C. C., Krebs E. G. (1996) J. Biol. Chem. 271, 505–511 [DOI] [PubMed] [Google Scholar]
- 25. Kaibuchi K., Kuroda S., Amano M. (1999) Annu. Rev. Biochem. 68, 459–486 [DOI] [PubMed] [Google Scholar]
- 26. Kumar P., Lyle K. S., Gierke S., Matov A., Danuser G., Wittmann T. (2009) J. Cell Biol. 184, 895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wittmann T., Waterman-Storer C. M. (2005) J. Cell Biol. 169, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. (1995) Nature 378, 785–789 [DOI] [PubMed] [Google Scholar]
- 29. Fang X., Yu S. X., Lu Y., Bast R. C., Jr., Woodgett J. R., Mills G. B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11960–11965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mitchison T., Kirschner M. (1984) Nature 312, 237–242 [DOI] [PubMed] [Google Scholar]
- 31. Rodionov V. I., Borisy G. G. (1997) Science 275, 215–218 [DOI] [PubMed] [Google Scholar]
- 32. Akhmanova A., Hoogenraad C. C., Drabek K., Stepanova T., Dortland B., Verkerk T., Vermeulen W., Burgering B. M., De Zeeuw C. I., Grosveld F., Galjart N. (2001) Cell 104, 923–935 [DOI] [PubMed] [Google Scholar]
- 33. Drabek K., van Ham M., Stepanova T., Draegestein K., van Horssen R., Sayas C. L., Akhmanova A., Ten Hagen T., Smits R., Fodde R., Grosveld F., Galjart N. (2006) Curr. Biol. 16, 2259–2264 [DOI] [PubMed] [Google Scholar]
- 34. Komarova Y., Lansbergen G., Galjart N., Grosveld F., Borisy G. G., Akhmanova A. (2005) Mol. Biol. Cell 16, 5334–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gundersen G. G., Kalnoski M. H., Bulinski J. C. (1984) Cell 38, 779–789 [DOI] [PubMed] [Google Scholar]
- 36. Webster D. R., Gundersen G. G., Bulinski J. C., Borisy G. G. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 9040–9044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Waterman-Storer C. M., Salmon E. D. (1997) J. Cell Biol. 139, 417–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suter D. M., Forscher P. (1998) Curr. Opin Neurobiol. 8, 106–116 [DOI] [PubMed] [Google Scholar]
- 39. Salaycik K. J., Fagerstrom C. J., Murthy K., Tulu U. S., Wadsworth P. (2005) J. Cell Sci. 118, 4113–4122 [DOI] [PubMed] [Google Scholar]
- 40. Zaoui K., Benseddik K., Daou P., Salaün D., Badache A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 18517–18522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu X., Shen Q. T., Oristian D. S., Lu C. P., Zheng Q., Wang H. W., Fuchs E. (2011) Cell 144, 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Al-Bassam J., Kim H., Brouhard G., van Oijen A., Harrison S. C., Chang F. (2010) Dev. Cell 19, 245–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.