Background: Transcriptional activators interact with RNA polymerase to redefine gene expression.
Results: A phage activator engages a region of the specificity factor of E. coli RNA polymerase, which is normally bound by another portion of RNA polymerase.
Conclusion: Using an activator/co-activator system, the phage hijacks the host RNA polymerase.
Significance: Small transcriptional factors acting on defined local regions of RNA polymerase can fundamentally change gene expression.
Keywords: Bacteriophage, RNA Polymerase, Transcription, Transcription Initiation Factors, Transcription Regulation, MotA, Activation, β-Flap, σ70
Abstract
Sigma factors, the specificity subunits of RNA polymerase, are involved in interactions with promoter DNA, the core subunits of RNA polymerase, and transcription factors. The bacteriophage T4-encoded activator, MotA, is one such factor, which engages the C terminus of the Escherichia coli housekeeping sigma factor, σ70. MotA functions in concert with a phage-encoded co-activator, AsiA, as a molecular switch. This process, termed sigma appropriation, inhibits host transcription while activating transcription from a class of phage promoters. Previous work has demonstrated that MotA contacts the C terminus of σ70, H5, a region that is normally bound within RNA polymerase by its interaction with the β-flap tip. To identify the specific σ70 residues responsible for interacting with MotA and the β-flap tip, we generated single substitutions throughout the C terminus of σ70. We find that MotA targets H5 residues that are normally engaged by the β-flap. In two-hybrid assays, the interaction of σ70 with either the β-flap tip or MotA is impaired by alanine substitutions at residues Leu-607, Arg-608, Phe-610, Leu-611, and Asp-613. Transcription assays identify Phe-610 and Leu-611 as the key residues for MotA/AsiA-dependent transcription. Phe-610 is a crucial residue in the H5/β-flap tip interaction using promoter clearance assays with RNA polymerase alone. Our results show how the actions of small transcriptional factors on a defined local region of RNA polymerase can fundamentally change the specificity of polymerase.
Introduction
Gene expression is a highly orchestrated process involving dynamic interactions among proteins, DNA, RNA, small molecules, and sensors of the external environment. At the heart of this activity is RNA polymerase (RNAP),5 a multisubunit enzyme responsible for transcribing an RNA copy of DNA-encoded genes. Escherichia coli RNAP is composed of a core (subunits α1, α2, β, β′, and ω) and a specificity subunit, σ, which recognizes promoters, DNA sequences that signify the transcription start site. The primary σ in E. coli, σ70, is required for exponential growth. It is composed of four conserved domains, three of which (2, 3, and 4) are involved in recognition of conserved promoter DNA sequences: the −10 element, the −15 TG−14 (extended −10), and the −35 element, respectively (for review, see Ref. 1). The interface between σ and core RNAP is extensive (2–5). In particular, RNAP structures of Thermus aquaticus (3) and Thermus thermophilus (2) indicate that σ70 Region 2 binds to the coiled-coil domain of β′, whereas Region 4 and the C terminus of σ70, called H5, interact with the flap domain (β-flap) of the β subunit (Fig. 1A). These two interactions correctly position the DNA binding domains of σ70 to recognize the −10 and −35 promoter elements simultaneously (6) (Fig. 1B).
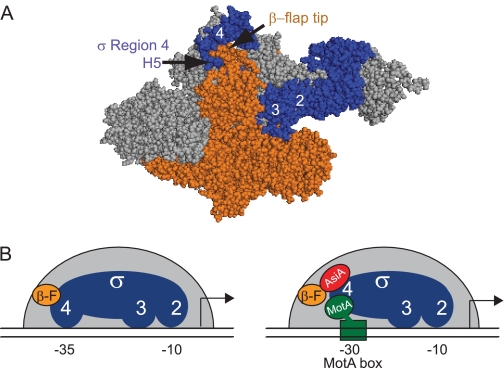
FIGURE 1.
The interaction of σ70 H5 with the β-flap tip is disrupted by AsiA. A, structure of T. thermophilus RNAP (2) (Protein Data Bank code 1IW7) shows the interaction between σ H5 and the β-flap tip. σ is shown in blue; β is shown in orange, and the remainder of core is shown in gray. B, schematic shows how the interaction of the β-flap (β-F; orange circle) with Region 4 of σ70 (blue) in RNAP (left panel) is remodeled by the T4 proteins AsiA (red) and MotA (green) (right panel). The remodeled σ Region 4 is no longer able to interact with the −35 element or the β-flap, and MotA interacts with the −30 region of the promoter DNA, the MotA box. Other interactions between σ70 Region 2 and the −10 element and Region 3 and the DNA are retained.
Given the primary role played by σ70 in promoter recognition, it is not surprising that secondary regulators have evolved to target this subunit. Class II activators use acidic residues to interact with basic amino acids between Arg-588 and Arg-603. These residues lie on a surface exposed helix within σ70 Region 4 (for review, see Refs. 7, 8). In contrast, the bacteriophage T4-encoded activator MotA uses a basic/hydrophobic cleft in its N-terminal domain (NTD) to engage H5 (9–11; for review, see Ref. 12).
MotA together with the T4 co-activator AsiA is required for the expression of prereplicative “middle” genes during infection. AsiA binds Region 4 of free σ70, and the AsiA-σ70 complex then associates with core to form an AsiA-associated RNAP (13). The binding of AsiA dramatically remodels the structure of Region 4 (14) so that Region 4 is now unable to bind to the −35 element DNA or to interact with the β-flap (Fig. 1B). It is thought that this remodeling makes H5 accessible for its interaction with MotANTD, a process termed sigma appropriation.
The C-terminal domain (CTD) of MotA, MotACTD, binds specifically to a T4 middle promoter element, the MotA box, positioned at −30 (10). Thus, MotA acts as a bridge between σ70 and T4 middle promoter DNA, replacing the recognition of the −35 element by σ70 Region 4 (Fig. 1B). Recent work suggests that there may be even more contacts among RNAP, MotA, and AsiA (15, 16).
The molecular details of the H5/MotA and H5/β-flap tip interactions in E. coli RNAP have not been determined. Furthermore, because of sequence differences between E. coli and the thermophilic bacterial RNAPs, deducing the details of the E. coli H5/β-flap tip contact from the structures of thermophilic RNAP is not straightforward. In this work, we have resolved at an amino acid level which residues within E. coli H5 are important for its interaction with MotA and with the β-flap tip. We show that MotA targets H5 residues that are normally engaged by the β-flap tip. Our results add further support to the model whereby AsiA remodeling of Region 4 is required to expose the MotA binding site on σ70. The MotA/AsiA system reveals how the actions of small transcriptional factors on a defined local region of RNAP can fundamentally change the specificity of polymerase.
EXPERIMENTAL PROCEDURES
Construction of Plasmids
Plasmids were constructed as described below using standard cloning procedures. Sequences of primers are available upon request. DNA sequence analyses (done by the Facility for Biotechnology Resources of the Food and Drug Administration) confirmed the sequences of the various constructs throughout the cloned regions.
Plasmids used in the two-hybrid assay were derived from pBRα-σ70 (17) and pACλcI32 (18). pBRα-σ contains an α-σ chimera gene, composed of RNAP α subunit residues 1–248 fused in-frame to σ70 Region 4 (residues 528–613). pBRα-σ70 D581G (19) is identical except for the substitution at Asp-581. In each case, the α-σ70 chimera gene is transcribed from tandem promoters, lpp and isopropyl-β-d-thiogalactopyranoside-inducible lacUV5. Mutant pBRα-σ70 and pBRα-σ70D581G constructs were created by two-step PCR. Two PCR products were first generated using pBRα-σ70 (or pBRα-σ70D581G), a primer that annealed within the σ70 DNA and contained the mutation, a primer that annealed either upstream or downstream of the σ70 DNA, and Pfu Turbo DNA polymerase (Stratagene). The resulting two PCR products were then mixed, and a PCR product was then obtained using this DNA, the upstream and downstream primers, and Pfu DNA polymerase. This product was then digested with SacI and AvrII, cloned into similarly digested pBRα-σ70, and transformed into E. coli XL1-Blue (Stratagene).
pACλcI32 encodes residues 1–236 of the bacteriophage λ cI protein under the control of the isopropyl-β-d-thiogalactopyranoside-inducible lacUV5 promoter. pcI-AsiA (10) and pMWT (9) contain the asiA gene or the NTD of motA (encoding residues 1–98), respectively, fused in-frame to cI in pACλcI32. pACλcI-β-flap (6) contains the rpoB gene encoding residues 858–946 of the β subunit of RNAP (the β-flap moiety) fused in-frame to cI in pACλcI32.
For purification of wild-type (WT) σ70 and σ70 mutant proteins, the plasmid pETσFL-CFI was created. pETσFL-CFI is identical to pETσFL (11) except that the XhoI site downstream of the rpoD gene was destroyed by a linker comprising unique AvrII and SalI sites. The removal of the downstream XhoI site left only one XhoI site in the plasmid, at the start of σ70 Region 4. Thus, pETσFL-CFI facilitates cloning of σ70 Region 4 mutants into the pETσFL background through unique 5′ XhoI and 3′ AvrII or SalI sites. A PCR product was generated using the pBRα-σ70 derivative containing the desired mutation, the primers that annealed upstream and downstream of the Region 4 insert in pBRα-σ70, and Pfu Turbo DNA polymerase (Stratagene). The product was digested with XhoI and AvrII and then ligated with pETσFL-CFI that had been previously digested with XhoI and AvrII.
Transcription Templates
pDKT90 (20) contains the T4 middle promoter PuvsX and the σ70-dependent promoter Pminor, a cryptic −35/−15TG−14 promoter located downstream of PuvsX (21). pPRE# (22) contains the extended −10 promoter PRE# (23). Both plasmids were digested with BsaAI to generate linear templates for transcription. The PuvsX/σ* and galP1/cons (24) templates were generated by annealing single-stranded oligonucleotides (Operon, purified by reversed-phase/ion exchange chromatography), containing template and nontemplate sequences from −66 to +34 relative to the start of transcription. PuvsX/σ* is identical to PuvsX/σ (13) except that the +1A was replaced with a G. This change prevents slippage of the RNAP at the A tract that surrounds the start site (25).
Buffers
Protein buffer contained 20 mm Tris-Cl (pH 7.9), 28 mm Tris acetate (pH 7.9), 45 mm NaCl, 23% glycerol, 0.5 mm EDTA, 0.06 mm EGTA, 0.2 mm DTT, 0.003% Triton X-100, 3 mm imidazole, 100 mm potassium glutamate, 2.8 mm magnesium acetate, 69 μg/ml BSA, and 13 mm potassium phosphate (pH 6.5). DNA/ribonucleoside triphosphate (NTP) buffer contained 0.5 mm Tris-Cl (pH 7.9), 57 mm Tris acetate (pH 7.9), 48 mm potassium phosphate (pH 6.5), 12% glycerol, 0.43 mm EDTA, 0.24 mm EGTA, 0.38 mm DTT, 214 mm potassium acetate, 5.7 mm magnesium acetate, 140 μg/ml BSA, 0.48 mm each ATP, GTP, and CTP, and 0.012 mm [α-32P]UTP (1 × 105 dpm/pmol).
Proteins
pETσFL-CFI and the mutant derivatives were used for the expression and subsequent purification of WT σ70 and σ70 Region 4 mutants, all His6-tagged at the N terminus. Proteins were expressed and purified as described for His6-tagged σ70 (11). In the absence of the T4 proteins, all of the mutants, except S609A and D613K, saturated core polymerase with a ratio of 7:1 or less. E. coli RNAP core was purchased from Epicenter Technologies. MotA (9) and His6-AsiA (26) were purified as described.
Two-hybrid Assays
β-Galactosidase assays were performed as described (26) using cultures of E. coli KS1 (27) containing the indicated plasmids except that cultures were grown for 3 h in LB medium supplemented with the appropriate antibiotics at the following concentrations: 50 μg/ml carbenicillin, 25 μg/ml chloramphenicol, and 50 μg/ml kanamycin. β-Galactosidase activity was determined in Miller units (28). For the assays using MotANTD or the β-flap, relative β-galactosidase activities at a 100 μm isopropyl-β-d-thiogalactopyranoside concentration were calculated as follows: (Miller units with the Region 4 mutant − Miller units with the pBRα control)/(Miller units with pBRα-σ70 − Miller units with the pBRα control). For the assays with AsiA, the relative values were calculated as (Miller units with the Region 4 mutant/Miller units with the WT Region 4) because the strain with pBRα shows a reduction in viability when AsiA is induced and thus cannot be used as a background. Values represent the average of three or more assays (MotA and β-flap assays in Fig. 2) or the average of two (AsiA assays in Fig. 2; all assays in supplemental Fig. S1).
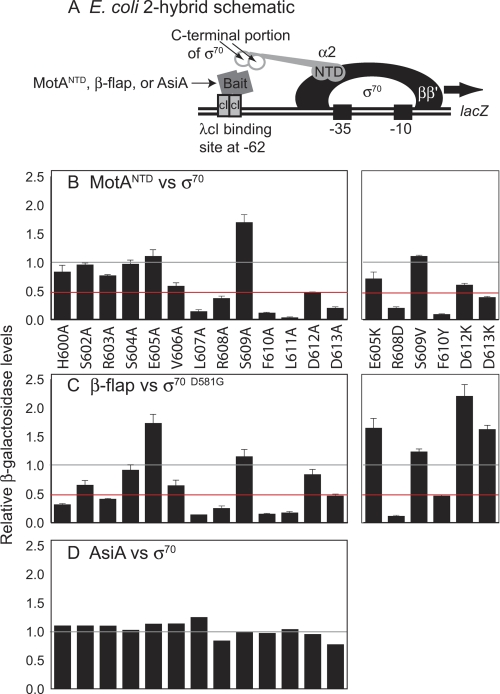
FIGURE 2.
Detection of protein interactions using the E. coli two-hybrid assay. A, schematic shows the E. coli two-hybrid system used to detect interactions between the C-terminal region of σ70 and other proteins. Positions of RNA polymerase subunits β, β′, and σ70 and the α-σ70 chimera are located upstream of the reporter gene (lacZ). The cI-bait fusion protein is indicated. The α-σ70 chimera consists of the NTD of α fused to the C terminus of σ70. B–D, relative β-galactosidase activity is shown for assays using α-σ70 chimera with σ70 mutations and cI-bait fusion proteins containing MotANTD (B), E. coli β-flap (C), or AsiA (D). Gray and red lines indicate normalized WT level and a 2-fold reduction, respectively. S.D. values (error bars) in B and C were determined from three or more independent experiments.
MotA/AsiA Activation Assays
Transcription reactions were assembled as follows. A solution (2.9 μl) containing 0.38 pmol of σ70 (WT or mutant), 0.05 pmol of core, and 4 pmol of His6-AsiA, when indicated, in protein buffer was incubated for 15 min at 37 °C. (In reactions containing AsiA, the His6-AsiA was preincubated with the σ70 at 37 °C for 10 min to ensure formation of the σ70-His6-AsiA complex.) A second solution (2.1 μl), comprising 0.1 pmol of DNA, 1.9 pmol of MotA (when indicated), and NTPs in DNA/NTP buffer, was then added to the first solution and incubated at 37 °C for 20 s. Rifampicin (0.5 μl, 300 ng) was then added. Thus, these are stringent conditions that only allow 20 s for the formation of open complex. Reactions were then incubated for an additional 7 min and collected on dry ice.
Gel load solution (25 μl of ½ × TBE, 7 m urea, 0.1% bromphenol blue, 0.1% xylene cyanol) was added, and the solution was heated at 95 °C for 2 min before aliquots were subjected to electrophoresis on 4% polyacrylamide, 7 m urea denaturing gels run in ½ × TBE. Gels were imaged using a Fujifilm FLA-5100 imaging system or by autoradiography followed by scanning with a Powerlook 2100XL densitometer. Quantification was performed using Quantity One software from Bio-Rad.
Promoter Clearance Assays
Transcription reactions were assembled as detailed above, except that the reactions lacked AsiA, MotA, or NTPs and contained 1 pmol of σ, 0.4 pmol of core, and 0.4 pmol of either PuvsX/σ* or galP1/cons DNA. The solution was incubated at 37 °C for 15 min to form stable complexes; NTPs (1 μl; final concentration of 200 μm for the unlabeled NTPs and 50 μm (∼104 dpm/pmol) for the α-32P labeled NTP (indicated in the figure legends)) and 0.5 μg of heparin were then added to initiate a single round of transcription. Reactions were incubated at 37 °C for 7.5 min and as indicated treated with 5 units of calf intestine phosphatase at 37 °C for 15 min to lower the background of unincorporated labeled NTP after electrophoresis. Samples were treated as detailed above except that the gel load solution was 13 μl of 10 mm EDTA (pH 7), 0.1% bromphenol blue, and 0.1% xylene cyanol in formamide, and samples were electrophoresed on 23% polyacrylamide, 7 m urea denaturing gels.
The sequences of the abortive RNAs were assigned from transcriptions in which one or more of the NTPs was missing (supplemental Fig. S2, lanes 1–3 versus lane 4). As reported previously (29), the migration of the phosphatase-treated, short RNAs is dependent on base composition as well as length, resulting in a complex pattern (supplemental Fig. S2, lane 8). The total amount of abortives sized +2 to +10 and the amount of full-length RNA were quantified using Quantity One software (Bio-Rad) after autoradiography and scanning using a Powerlook 2100XL densitometer. In the case of galP1/cons, the amount of short RNAs made using RNAP lacking σ was also subtracted as background before determining the relative ratio.
RESULTS
Mutations at Specific σ70 H5 Residues Impair the Interaction of σ70 with MotANTD
Sequence analyses of the C-terminal ∼90 amino acids of various σs identify a highly conserved portion (Region 4, residues 529–599 in σ70) and a less conserved portion at the C terminus (H5, residues 600–613 in σ70) (30). MotANTD interacts with H5, and this association is needed for MotA/AsiA activation (9–11). In particular, deletion of residues 604–613 essentially eliminates MotA activation (10).
To determine which H5 residues are important for this interaction, we assayed the effect of single H5 substitutions on the interaction of the C-terminal portion of σ70 (Region 4 plus H5) with MotANTD in an E. coli two-hybrid assay. In this assay, the level of β-galactosidase activity is an indirect read-out of the strength of the protein-protein interaction (Fig. 2A). Alanine substitutions at Leu-607, Arg-608, Phe-610, Leu-611, Asp-612, and Asp-613 reduced the relative level of β-galactosidase 2-fold or more, suggesting that these residues are important for the interaction of σ70 H5 with MotANTD (Fig. 2B). Of these, Phe-610 and Leu-611 are critical residues; alanine substitutions here nearly eliminated β-galactosidase activity.
To investigate the σ70 H5 residues in more detail, we assayed additional substitutions. Single “charge swap” mutants, E605K, R608D, D612K, and D613K, showed minimal or no reduction in β-galactosidase activity relative to the alanine substitutions. Thus, the presence of charge at these positions appears unimportant (Fig. 2B). To test the importance of the aromatic side chain at position Phe-610, we replaced this phenylalanine with tyrosine, which differs only by the addition of a single hydroxyl moiety on the aromatic ring. F610Y reduced the β-galactosidase activity relative to WT to the same extent as F610A (Fig. 2B), suggesting that the phenylalanine side chain is crucial for the interaction in this assay. Finally, we substituted Ser-609 with valine, a large hydrophobic amino acid. S609V showed only a marginal increase compared with WT activity. We conclude that the side chain at position 609 is not significant for the MotA/H5 interaction. How the S609A substitution increases the MotANTD/σ70 Region 4 interaction is unclear.
H5/β-Flap Tip and H5/MotA Interactions Involve a Similar Set of H5 Residues
Structural and biochemical analyses have indicated that the interface between the C-terminal portion of σ70 and the β-flap is extensive (2, 3, 11, 31, 32) (Fig. 1A) and like the σ70/MotANTD interface, includes residues within σ70 H5 (2, 32, 33). Consequently, we repeated the two-hybrid assays using the C-terminal portion of σ70 and a construct containing the β-flap. In this case, σ70 also contained a D581G substitution; previous work has demonstrated that this mutation is required to observe the interaction between σ70 and the β-flap in this assay (6).
Although there were subtle differences between the pattern of H5 mutations that impair the interaction with MotA or the β-flap, what is striking is the set of residues (Leu-607, Arg-608, Phe-610, Leu-611, and Asp-613) that are important for both (Fig. 2, B versus C). In addition, the σ70/β-flap interaction is also affected by mutations at residues His-600 and Arg-603, positions that have little importance for the interaction with MotA (Fig. 2B).
To ensure that this result was not affected by the D581G mutation, we repeated the two-hybrid assay using MotANTD and the series of σ70 mutants that also contain D581G. Results were essentially the same with (supplemental Fig. S1) or without (Fig. 2B) the mutation at 581. To ensure that the effects of the mutations were specific for MotA and the β-flap, we repeated the assays using AsiA because AsiA interacts with multiple residues within Region 4, but not with H5 (11, 14, 33, 34). As expected, none of the introduced mutations negatively impacted the σ70 interaction with AsiA either in the WT (Fig. 2D) or D581G background (supplemental Fig. S1).
Phe-610 and Leu-611 Are Crucial for MotA/AsiA Activation
To investigate how the H5 mutations affect MotA/AsiA activation, we performed single round in vitro transcriptions with a template containing the T4 middle promoter PuvsX and a control promoter, Pminor (Fig. 3A). E. coli RNAP alone generates a basal level of transcription from PuvsX, (35) (Fig. 3B). We used this level to determine the intrinsic activity of σ mutants and then the extent of activation by AsiA and MotA. Single alanine mutants from Leu-607 to Asp-613 and the mutants F610Y, D612K, and D613K were tested.
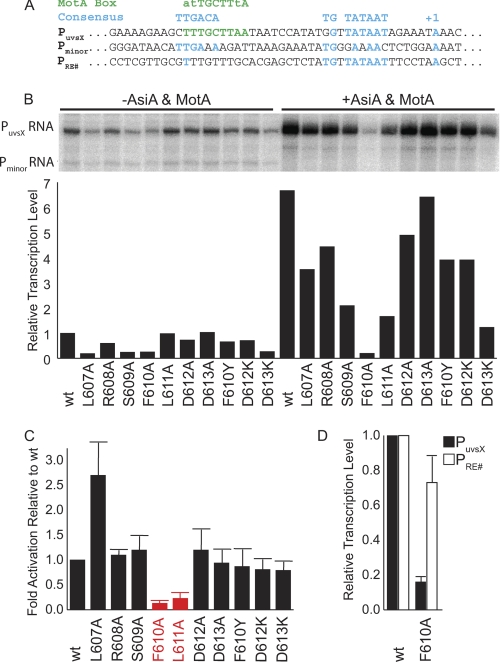
FIGURE 3.
In vitro transcription analysis of σ70 mutants. A, sequences of a MotA/AsiA activated, T4 middle promoter (PuvsX) and promoters not activated by MotA (Pminor and PRE#). B, single round in vitro transcription using WT and mutant σ70. The presence or absence of T4 transcription factors AsiA and MotA, which gives rise to activated or basal levels of transcription, respectively, is indicated at the top of the figure. Transcripts originating from PuvsX or Pminor are indicated on the left. Histogram showing the transcription level normalized to WT basal level is shown below each lane. C, histogram showing the -fold activation (activated level/basal level relative to transcriptions performed with WT σ70) for each σ. The alanine substitutions having the most pronounced effect, F610A and L611A, are shown in red. S.D. values (error bars) were determined from three or more independent experiments. D, histogram showing the relative transcription level from PuvsX or PRE# using RNAP reconstituted with either WT σ70 or σ70F610A. S.D. values were determined from four independent experiments.
Transcription from PuvsX is activated ∼7-fold over the basal level when using RNAP with WT σ70, AsiA, and MotA (Fig. 3B). Only two mutants, F610A and L611A, significantly impaired MotA/AsiA activation (colored red in Fig. 3C), indicating that these are the crucial residues for MotA/AsiA activation.
The level of basal transcription for several of the H5 mutants was less than that observed with WT (Fig. 3B, left). This result is consistent with the idea that the H5/β-flap tip helps to position Region 4 for its interaction with the −35 DNA. However, none of the H5 mutants affected the ability of AsiA alone to inhibit transcription, either from Pminor, which cannot be activated by MotA (Fig. 3B), or from PuvsX in the absence of MotA (data not shown). This result is expected because the AsiA interaction with Region 4 and its inhibition of transcription do not require H5 (10, 11). Although L607A showed an increase over the WT level of activation (Fig. 3C), this may reflect the low basal level of transcription observed with this mutant (Fig. 3B). Surprisingly, F610Y, which showed a dramatic reduction of activity in the two-hybrid assay (Fig. 2B), had only a minimal effect on MotA activation. We speculate that conformational differences between the complete RNAP-DNA-MotA-AsiA complex and how MotANTD interacts with H5 in the two-bybrid assay allow the additional hydroxyl moiety to be accommodated.
Gross Misfolding of σ70 F610A Does Not Account for Its Impairment in MotA Activation
In the MotA/AsiA activation assay, the F610A mutation greatly reduced both basal and activated transcription. Thus, it was possible that the inability of σ70 F610A to function in MotA activation was due to a general misfolding of the protein. We tested this possibility using the extended −10 promoter PRE# because transcription from this extended −10 promoter does not require Region 4 (36).
Whereas the F610A mutant displayed only 25% MotA-independent transcriptional activity compared with WT σ70 when using PuvsX, it retained ∼75% activity with PRE# (Fig. 3D), indicating that this purified σ mutant is functional. In addition, in transcriptions performed under other conditions detailed below (Fig. 4), RNAP reconstituted with the F610A mutant gave levels of PuvsX RNA similar to those seen with WT RNAP. Thus, the F610A mutant is not simply inactive.
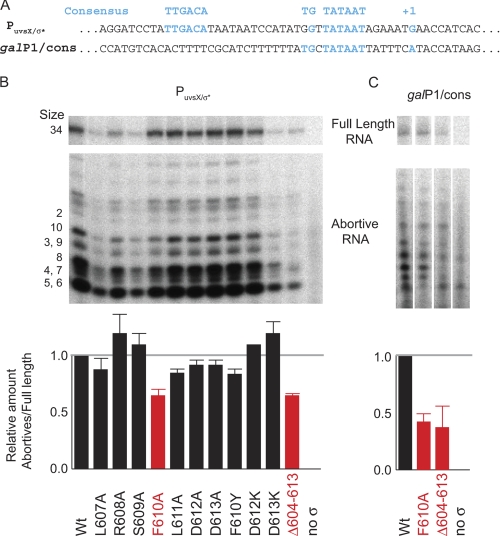
FIGURE 4.
RNAP containing σ70F610A or σ70Δ604–613 is more efficient at promoter clearance. A, sequences of the −35/−10 promoter PuvsX/σ* and the extended −10 promoter galP1/cons. B and C, single round in vitro transcription using WT and the indicated mutant σ and either PuvsX/σ* (B) or galP1/cons (C). The labeled nucleotide in the reaction was [α-32P]ATP. Each template produces a full-length product of 34 nucleotides as well as abortive RNAs from 2 to ∼10 nucleotides. Because the RNA products were treated with phosphatase before electrophoresis, the abortive RNAs migrate according to size and base composition (see supplemental Fig. S2 for details). Histograms below show the amount of abortive RNA relative to the amount of full-length RNA for each lane. In the case of galP1/cons, the amount of short RNAs made using RNAP lacking σ was also subtracted as background before determining the relative ratio. The mutations having the most significant effect, F610A and Δ604–613, are shown in red. S.D. values (error bars) were determined from three or more independent experiments. The error bars with RNAP containing σ70D612A are too small to be seen.
σ70 H5 Residue Phe-610 Is Also Important in Promoter Clearance
During transcription initiation, short abortive RNAs are generated as the RNAP-promoter complex proceeds from an initiating to an elongating complex (for review, see Ref. 37). Mutations within Region 4 or H5 that are predicted to reduce its interaction with the −35 DNA element and/or the β-flap facilitate promoter clearance, decreasing the level of abortive products relative to full-length RNA (32, 37–39). This result supports a model wherein the interaction of σ70 H5 with the β-flap impedes the progression from initiation to elongation.
To investigate the effect of each H5 residue on promoter clearance, we performed single round in vitro transcription reactions using short templates containing either a consensus −35/−10 promoter (PuvsX/σ*) or an extended −10 promoter (galP1/cons) (Fig. 4A). Each template produces a full-length product of 34 nucleotides plus abortive products of 2 to ∼10 nucleotides (Fig. 4B and C, and supplemental Fig. S2). To ensure that we were assaying promoter clearance only, we incubated RNAP with the template for 15 min at 37 °C before the addition of NTPs to maximize the formation of stable open complexes. The amount of abortive RNA relative to the amount of full-length RNA is a measure of promoter clearance.
Using PuvsX/σ*, either σ70 F610A or a σ70 missing residues 604–613 (Δ604–613) yielded ∼60% of the abortive/full-length RNA ratio of that seen with WT (shown in red in Fig. 4B). The effect of other mutations was modest, although the L607A, L611A, and F610Y mutants resulted in small decreases. Because the F610A mutant and Δ604–613 produced similar results, we conclude that the σ70 phenylalanine at 610 is the most crucial residue within the H5/β-flap tip interaction when RNAP is transcribing from a −35/−10 promoter.
Previous work has shown that a L607P substitution, which we expect would disrupt the local structure of H5, decreases the amount of abortives relative to full-length transcript when using galP1/cons (32). Thus, even though an extended −10 promoter, like galP1/cons, does not require Region 4 for recognition, the loss of the H5/β-flap tip interaction still affects promoter clearance (Fig. 4C). Transcription using either σ70F610A or σ70Δ604–613 resulted in a decrease of this ratio to ∼40% of that seen using WT, suggesting that Phe-610 is also the crucial residue for promoter clearance from an extended −10 promoter.
DISCUSSION
The C-terminal portion of σ70 (Region 4 plus H5) is involved both in the basic function of RNAP as well as multiple mechanisms that regulate transcription (10, 36, 40, 41) (for review, see Refs. 8, 42). Within RNAP, its interaction with the β-flap positions Region 4 so that σ70 can interact with both the −10 and −35 elements of promoter DNA (6). This contact also affects promoter clearance, because the newly synthesized RNA travels along a channel composed in part by the β-flap (43). Thus, the extruding RNA must abrogate the contact between the β-flap tip and H5, as the RNA exits RNAP.
The C-terminal portion of σ70 also functions as a target for multiple factors that affect the regulation and distribution of RNAP at promoters throughout the cell. Two such factors, AsiA and MotA, are encoded by bacteriophage T4. Previous work has indicated that MotA, like the β-flap tip, engages H5 (9–11). Although hundreds of activators of E. coli RNAP have been identified, T4 MotA is the only regulator known to function through an interaction with H5.
Our work here resolves the contributions of individual H5 residues in the interaction of E. coli σ70 with MotA and with the β-flap tip of RNAP. Our results indicate that both MotA and the β-flap tip engage the same C-terminal residues of σ70. Leu-607, Arg-608, Phe-610, Leu-611, and Asp-613 are important for either interaction in the two-hybrid assay. The hydrophobic residue Phe-610 is the key element for promoter clearance and is one of two residues, along with Leu-611, that is crucial for MotA/AsiA activation.
It is not surprising that only a subset of the residues identified in the two-hybrid assay have a significant effect on transcription. The two-hybrid assay detects interactions between small domains. However, the transcription complex contains the full complement of contacts that could compensate for the loss of a single interaction.
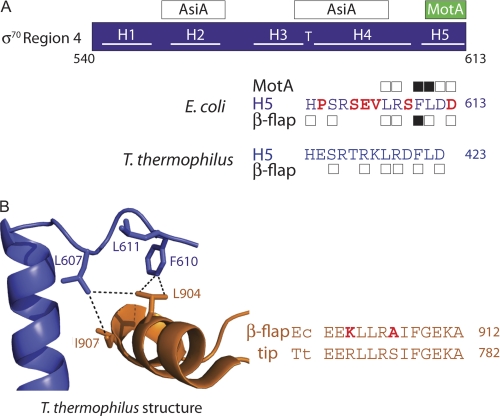
Our findings are important because structures of E. coli H5 with MotA or with the β-flap tip are not yet available. Consequently, molecular details must be deduced from biochemical analyses and from the predictions of the available structures of thermophilic RNAPs (2, 3) or the C-terminal portion of E. coli σ70 with AsiA (14) or the host anti-sigma factor Rsd (44). It is particularly important to test whether the H5/β-flap tip contacts seen in the thermophilic RNAP structures apply to E. coli RNAP because the sequence of H5 deviates between these bacteria (Fig. 5A).
FIGURE 5.
σ70 H5 residues important for interaction with the β-flap tip and MotANTD. A, schematic of σ70 Region 4, showing locations of interactions with AsiA and MotA. The C-terminal sequence (residues 600–613) of E. coli σ70 is shown. Boxes above/below the sequence denote residues where alanine substitutions decrease the interaction of Region 4 with MotANTD or the β-flap by 2-fold or more in the two-hybrid assays (Fig. 2); black boxes denote residues that also affect MotA/AsiA activation (Fig. 3) or promoter clearance (Fig. 4). The corresponding H5 sequence of T. thermophilus is shown below; σ residues that are within 5 Å of the β-flap in the T. thermophilus structure (2) are denoted with a box. B, left, portion of the structure of the T. thermophilus RNAP depicting interactions between the C terminus of σ in blue and the β-flap tip in orange (Protein Data Bank code 1IW7) (2). Stick renderings of residues discussed in the text, which are identical between E. coli and T. thermophilus RNAP, are shown and numbered according to E. coli RNAP (E. coli/T. thermophilus numbering: σLeu-607/Leu-418, σPhe-610/Phe-421, σLeu-611/Leu-422, βLeu-904/Leu-774, βIle-907/Ile-777). Dashed lines indicate distances <5 Å. B, right, alignment of H5 sequences and β-flap tip sequences in E. coli (Ec) and T. thermophilus (Tt). E. coli residues that differ from those in T. thermophilus are colored in red.
In addition, the β-flap of E. coli contains a large insertion, βi9, not found in the thermophilic RNAPs. A recent structure of the E. coli β-flap, which includes βi9, has been reported (45). However, this structure was obtained from a complex of E. coli β-flap with T4 gp33, a component of the T4 late σ factor, and the position of the β-flap tip is not suitable for its needed interaction with σ70 H5, presumably because gp33 reorients the β-flap tip. Furthermore, the results of the two-hybrid assays using the E. coli σ Region 4 and the E. coli β-flap suggest that there are interactions that differ from those predicted by the T. thermophilus structure (Fig. 5). This is not surprising because a comparison of the T. thermophilus H5 structure with the structure of E. coli H5 in a complex with the anti-sigma factor Rsd (44) reveals significant structural differences (supplemental Fig. S3). In fact, within the σ70-Rsd complex, E. coli H5 is reoriented relative to the position of the thermophilic H5 (supplemental Fig. S3). This result suggests that this region of σ may be highly malleable and capable of adopting various structures.
Despite these differences, it is clear that hydrophobic contacts play a significant role in the interaction of either the thermophilic or E. coli H5 with the β-flap and in the interaction of E. coli H5 with MotA. These hydrophobic contacts are seen directly within the T. thermophilus RNAP structure (2) (Fig. 5B), and conserved hydrophobic residues within the β-flap tip of E. coli (Leu-901, Leu-902, Ile-905, and Phe-906) are important for growth, for transcription from a −35/−10 promoter, and for an interaction with σ70 (31). A hydrophobic cleft within MotANTD also serves as the contact site for H5 (9). Thus, the intimate association of Phe-610 with these hydrophobic surfaces appears to be a salient feature of the interaction of H5 with either the β-flap or MotA.
Taken together, our results indicate that in sigma appropriation, MotA and the β-flap tip indirectly compete for a common target in σ70 H5. However, the overall σ70/β-flap interface is composed of many contacts throughout Region 4 (2, 3, 32, 45, 46) whereas the interaction of MotA with H5 is composed of a few contacts. Therefore, it is not surprising that MotA cannot significantly activate transcription on its own; it is unlikely to compete adequately for σ70 directly by displacing or precluding the σ70/β-flap interaction. It is reasonable then that MotA relies on the action of AsiA, which binds tightly to free σ70 and remodels Region 4 before σ70 associates with core (13, 47) (for review, see Ref. 12). In doing so, AsiA both prevents the interaction of σ70 with the β-flap and disrupts the −35 promoter element recognition motif, ultimately reducing host transcription. As a consequence, competition with the β-flap is eliminated, H5 is now free to be bound by MotA, polymerase is redirected to T4 middle promoters, and the T4 viral cycle is achieved.
Supplementary Material
Acknowledgments
We thank Alice Boulanger-Castaing, Kim Decker, Tamara James, and Meng Hsieh for helpful discussions.
This work was supported, in whole or in part, by the Intramural Research Program of the NIDDK, National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- RNAP
- RNA polymerase
- CTD
- C-terminal domain
- NTD
- N-terminal domain
- NTP
- ribonucleoside triphosphate.
REFERENCES
- 1. Hook-Barnard I. G., Hinton D. M. (2007) Gene Regulation and Systems Biology http://www.la-press.com/transcription-initiation-by-mix-and-match-elements-flexibility-for-pol-article-a481 [PMC free article] [PubMed]
- 2. Vassylyev D. G., Sekine S., Laptenko O., Lee J., Vassylyeva M. N., Borukhov S., Yokoyama S. (2002) Nature 417, 712–719 [DOI] [PubMed] [Google Scholar]
- 3. Murakami K. S., Masuda S., Darst S. A. (2002) Science 296, 1280–1284 [DOI] [PubMed] [Google Scholar]
- 4. Burgess R. R., Anthony L. (2001) Curr. Opin. Microbiol. 4, 126–131 [DOI] [PubMed] [Google Scholar]
- 5. Arthur T. M., Anthony L. C., Burgess R. R. (2000) J. Biol. Chem. 275, 23113–23119 [DOI] [PubMed] [Google Scholar]
- 6. Kuznedelov K., Minakhin L., Niedziela-Majka A., Dove S. L., Rogulja D., Nickels B. E., Hochschild A., Heyduk T., Severinov K. (2002) Science 295, 855–857 [DOI] [PubMed] [Google Scholar]
- 7. Rhodius V. A., Busby S. J. (2000) J. Mol. Biol. 299, 311–324 [DOI] [PubMed] [Google Scholar]
- 8. Decker K. B., Hinton D. M. (2009) Mol. Microbiol. 73, 137–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bonocora R. P., Caignan G., Woodrell C., Werner M. H., Hinton D. M. (2008) Mol. Microbiol. 69, 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pande S., Makela A., Dove S. L., Nickels B. E., Hochschild A., Hinton D. M. (2002) J. Bacteriol. 184, 3957–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baxter K., Lee J., Minakhin L., Severinov K., Hinton D. M. (2006) J. Mol. Biol. 363, 931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hinton D. M. (2010) Virol. J. 7, 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hinton D. M., Vuthoori S. (2000) J. Mol. Biol. 304, 731–739 [DOI] [PubMed] [Google Scholar]
- 14. Lambert L. J., Wei Y., Schirf V., Demeler B., Werner M. H. (2004) EMBO J. 23, 2952–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuan A. H., Hochschild A. (2009) Mol. Microbiol. 74, 1018–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuan A. H., Nickels B. E., Hochschild A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6597–6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dove S. L., Huang F. W., Hochschild A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13215–13220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu J. C., Kornacker M. G., Hochschild A. (2000) Methods 20, 80–94 [DOI] [PubMed] [Google Scholar]
- 19. Nickels B. E., Dove S. L., Murakami K. S., Darst S. A., Hochschild A. (2002) J. Mol. Biol. 324, 17–34 [DOI] [PubMed] [Google Scholar]
- 20. March-Amegadzie R., Hinton D. M. (1995) Mol. Microbiol. 15, 649–660 [DOI] [PubMed] [Google Scholar]
- 21. Hook-Barnard I., Johnson X. B., Hinton D. M. (2006) J. Bacteriol. 188, 8352–8359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Q., Decker K. B., Boucher P. E., Hinton D., Stibitz S. (2010) Mol. Microbiol. 77, 1326–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keilty S., Rosenberg M. (1987) J. Biol. Chem. 262, 6389–6395 [PubMed] [Google Scholar]
- 24. Burns H. D., Belyaeva T. A., Busby S. J., Minchin S. D. (1996) Biochem. J. 317, 305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hinton D. M. (1991) J. Biol. Chem. 266, 18034–18044 [PubMed] [Google Scholar]
- 26. Pal D., Vuthoori M., Pande S., Wheeler D., Hinton D. M. (2003) J. Mol. Biol. 325, 827–841 [DOI] [PubMed] [Google Scholar]
- 27. Dove S. L., Joung J. K., Hochschild A. (1997) Nature 386, 627–630 [DOI] [PubMed] [Google Scholar]
- 28. Miller J. (1972) Experiments in Molecular Genetics, pp. 352–355, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 29. Scherzinger E., Lanka E., Hillenbrand G. (1977) Nucleic Acids Res. 4, 4151–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gruber T. M., Gross C. A. (2003) Annu. Rev. Microbiol. 57, 441–466 [DOI] [PubMed] [Google Scholar]
- 31. Geszvain K., Gruber T. M., Mooney R. A., Gross C. A., Landick R. (2004) J. Mol. Biol. 343, 569–587 [DOI] [PubMed] [Google Scholar]
- 32. Nickels B. E., Garrity S. J., Mekler V., Minakhin L., Severinov K., Ebright R. H., Hochschild A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4488–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gregory B. D., Nickels B. E., Garrity S. J., Severinova E., Minakhin L., Urbauer R. J., Urbauer J. L., Heyduk T., Severinov K., Hochschild A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4554–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Minakhin L., Camarero J. A., Holford M., Parker C., Muir T. W., Severinov K. (2001) J. Mol. Biol. 306, 631–642 [DOI] [PubMed] [Google Scholar]
- 35. Hinton D. M., March-Amegadzie R., Gerber J. S., Sharma M. (1996) Methods Enzymol. 274, 43–57 [DOI] [PubMed] [Google Scholar]
- 36. Kumar A., Malloch R. A., Fujita N., Smillie D. A., Ishihama A., Hayward R. S. (1993) J. Mol. Biol. 232, 406–418 [DOI] [PubMed] [Google Scholar]
- 37. Hsu L. M. (2009) Methods 47, 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carpousis A. J., Stefano J. E., Gralla J. D. (1982) J. Mol. Biol. 157, 619–633 [DOI] [PubMed] [Google Scholar]
- 39. Vo N. V., Hsu L. M., Kane C. M., Chamberlin M. J. (2003) Biochemistry 42, 3798–3811 [DOI] [PubMed] [Google Scholar]
- 40. Kumar A., Grimes B., Fujita N., Makino K., Malloch R. A., Hayward R. S., Ishihama A. (1994) J. Mol. Biol. 235, 405–413 [DOI] [PubMed] [Google Scholar]
- 41. Klocko A. D., Wassarman K. M. (2009) Mol. Microbiol. 73, 152–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Browning D. F., Busby S. J. (2004) Nat. Rev. Microbiol. 2, 57–65 [DOI] [PubMed] [Google Scholar]
- 43. Vassylyev D. G., Vassylyeva M. N., Perederina A., Tahirov T. H., Artsimovitch I. (2007) Nature 448, 157–162 [DOI] [PubMed] [Google Scholar]
- 44. Patikoglou G. A., Westblade L. F., Campbell E. A., Lamour V., Lane W. J., Darst S. A. (2007) J. Mol. Biol. 372, 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Opalka N., Brown J., Lane W. J., Twist K. A., Landick R., Asturias F. J., Darst S. A. (2010) PLoS Biol. 8, e1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mekler V., Kortkhonjia E., Mukhopadhyay J., Knight J., Revyakin A., Kapanidis A. N., Niu W., Ebright Y. W., Levy R., Ebright R. H. (2002) Cell 108, 599–614 [DOI] [PubMed] [Google Scholar]
- 47. Stevens A. (1976) in RNA Polymerase (Losick R., Chamberlin M. eds) pp. 617–627, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.