Abstract
Nonalcoholic steatohepatitis is characterized by hepatic steatosis, elevated levels of circulating free fatty acids (FFA), endoplasmic reticulum (ER) stress, and hepatocyte lipoapoptosis. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor 5 (DR5) is significantly elevated in patients with nonalcoholic steatohepatitis, and steatotic hepatocytes demonstrate increased sensitivity to TRAIL-mediated cell death. Nonetheless, a role for TRAIL and/or DR5 in mediating lipoapoptotic pathways is unexplored. Here, we examined the contribution of DR5 death signaling to lipoapoptosis by free fatty acids. The toxic saturated free fatty acid palmitate induces an increase in DR5 mRNA and protein expression in Huh-7 human hepatoma cells leading to DR5 localization into lipid rafts, cell surface receptor clustering with subsequent recruitment of the initiator caspase-8, and ultimately cellular demise. Lipoapoptosis by palmitate was not inhibited by a soluble human recombinant DR5-Fc chimera protein suggesting that DR5 cytotoxic signaling is ligand-independent. Hepatocytes from murine TRAIL receptor knock-out mice (DR−/−) displayed reduced palmitate-mediated lipotoxicity. Likewise, knockdown of DR5 or caspase-8 expression by shRNA technology attenuated palmitate-induced Bax activation and apoptosis in Huh-7 cells, without altering induction of ER stress markers. Similar observations were verified in other cell models. Finally, knockdown of CHOP, an ER stress-mediated transcription factor, reduced DR5 up-regulation and DR5-mediated caspase-8 activation upon palmitate treatment. Collectively, these results suggest that ER stress-induced CHOP activation by palmitate transcriptionally up-regulates DR5, likely resulting in ligand-independent cytotoxic signaling by this death receptor.
Keywords: Apoptosis, C/EBP Transcription Factor, Endoplasmic Reticulum Stress, Signal Transduction, TRAIL, Caspase-8, Free Fatty Acid, Nonalcoholic Fatty Liver Disease, Steatohepatitis
Introduction
Obesity and insulin resistance, cardinal features of the metabolic syndrome, are associated with enhanced lipolysis in adipose tissue (1, 2). This excessive lipolysis results in increased serum concentrations of free fatty acids (FFA),2 augmenting their delivery to non-adipose tissues such as liver, heart, and pancreatic β-cells (2). Inundation of these tissues with FFA overwhelms fatty acid oxidative pathways with toxic cellular consequences. An advanced response to toxic FFA is cellular demise by apoptosis, termed lipoapoptosis (3). Hepatocyte lipotoxicity is particularly germane to injury in the liver, where it contributes to the syndrome of nonalcoholic fatty liver disease (NAFLD) (4). For example, the magnitude of hepatocyte lipoapoptosis correlates with hepatic disease severity (4). Thus, the mechanisms initiating lipoapoptosis are of biomedical interest and human health relevance.
Lipotoxicity is likely multifactorial. Sustained endoplasmic reticulum (ER) stress (5–7), c-Jun N-terminal kinase (JNK) activation (8, 9), and oxidative stress (10) have all been implicated in lipotoxicity. Despite the fact that death receptors are potent mediators of cytotoxicity (11), especially in hepatic diseases, their contribution to lipotoxicity is incompletely defined. Fas, the prototype death receptor, has been implicated in adipocyte toxicity and inflammation (12), and its hepatic expression is increased in NAFLD (4). Tumor necrosis factor-α (TNF-α) and TNF receptor-1 (TNFR-1) have also been implicated in hepatic steatosis (13–15). In contrast, the role of TNF-related apoptosis-inducing ligand (TRAIL) and its cognate death receptors (DR)-4 and -5 in lipotoxicity is understudied. Yet, increasing data implicate a critical role for DR5 in lipotoxicity. For example, DR5 expression is increased in steatotic hepatocytes and sensitizes hepatocytes to exogenous TRAIL cytotoxicity (16). Thus, the role of DR5 in lipoapoptosis merits further investigation.
In this study, we explored the potential contribution of DR5 death signaling during lipoapoptosis by saturated FFA. The results implicate TRAIL death signaling by DR5 as a mediator of palmitate-induced hepatocyte lipoapoptosis downstream of ER stress-mediated CHOP induction of DR5 expression. These data further integrate and reconcile knowledge regarding the role of the DR5-mediated extrinsic pathway of apoptosis during hepatocyte lipotoxic processes.
EXPERIMENTAL PROCEDURES
Cells
Huh-7 and KMCH cells, human hepatoma and cholangiocarcinoma cell lines, respectively, were cultured in Dulbecco's modified Eagle's medium containing glucose (25 mm), 100,000 units/liter penicillin, 100 mg/liter streptomycin, and 10% fetal bovine serum. Jurkat cells, a human T lymphoblastoid cell line, wild-type (WT), and Jurkat variants I2.1 (lacking Fas-associated death domain protein or FADD) and I9.2 (lacking pro-caspase-8) were cultured at concentrations <1 × 106 cells/ml in RPMI 1640 medium supplemented with 15% FBS, 100,000 units/liter penicillin, 100 mg/liter streptomycin, and 2 mm glutamine. Huh-7 cells are a well validated model for the study of hepatocyte lipoapoptosis (8), whereas KMCH and Jurkat cells were used as model systems to investigate death receptor signaling in the context of lipoapoptosis.
C57BL/6 mice genetically deficient in the murine TRAIL receptor (DR−/−), the only pro-apoptotic death receptor for TRAIL in the mouse, have been described previously (17). Mouse hepatocytes were isolated from either C57BL/6 wild-type (The Jackson Laboratory, Bar Harbor, ME), DR−/−, or MRL/MpJ-Tnfrsf6lpr mice (Faslpr/lpl mice; The Jackson Laboratory) by collagenase perfusion, purified by Percoll (Sigma) gradient centrifugation, and plated as primary cultures. Human hepatocytes were prepared as described previously by us in detail (9).
Plasmid and Transfection
Short hairpin RNA (shRNA) silencing DR5, DR4, caspase-8, and CHOP were from Sigma and targeted nucleotides 1531–1551 of DR5 mRNA (NM_003842), 1499–1519 of DR4 mRNA (NM_003844), 1006–1026 of caspase-8 mRNA (NM_001228), and 550–570 of CHOP mRNA (NM_004083). shRNA silencing caspase-10 was from Open Biosystems (Thermo Fisher Scientific, Huntsville, AL) and targeted nucleotides 1834–1852 of caspase-10 mRNA (NM_032974). Huh-7 or KMCH cells were transfected with 1 μg/ml DNA plasmid using Lipofectamine (Invitrogen). Stably transfected clones were selected in medium containing 1200 mg/liter G418 and screened by immunoblot analysis.
Fatty Acid Treatment
Palmitic acid (PA) was prepared as described previously by us (9). The concentration of PA used in the main experiments varied between 400 and 800 μm and is similar to the fasting total FFA plasma concentrations observed in humans with nonalcoholic steatohepatitis (18, 19). The concentration of the vehicle, isopropyl alcohol, in the medium was 0.5%; this concentration was used as vehicle control.
Immunocytochemistry and TRAIL Receptor Cluster Analysis
Cells were cultured on glass coverslips. After palmitate treatment, cells were fixed with freshly prepared 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at 37 °C. Permeabilization was performed with 0.0125% (w/v) CHAPS in PBS at 37 °C, 10 min, for active Bax immunostaining, whereas cells were not permeabilized for DR5 or DR4 cell surface immunostaining. After incubation overnight at 4 °C with primary antibodies, cells were washed three times with PBS and incubated with secondary antibodies for 1 h at 37 °C. Primary antibodies were mouse anti-Bax antisera (1:500, clone 6A7; Santa Cruz Biotechnology, Santa Cruz, CA), goat anti-DR5 antisera (1:500, ALX-210–743-C200, Enzo Life Sciences, Plymouth Meeting, PA), or mouse anti-DR4 antisera (1:500, clone B9; Santa Cruz Biotechnology). Secondary antibodies were Alexa Fluor 488-conjugated anti-mouse IgG or Alexa Fluor 488-conjugated anti-goat IgG (Molecular Probes, Eugene, OR). To ensure that cell fixation with 4% paraformaldehyde does not permeabilize the plasma membrane, cellular immunofluorescence for α-tubulin, an abundant cytoplasmic protein, was performed on fixed and CHAPS-permeabilized or on fixed and nonpermeabilized Huh-7 cells. Rhodamine Red-X-conjugated anti-mouse IgG was used as a secondary antibody. ProLong antifade kit (Molecular Probes) was used as mounting medium, and images were acquired by confocal microscopy employing excitation and emission wavelengths of 488 and 507 nm for Alexa Fluor 488 and 570 and 590 nm for rhodamine Red-X, respectively. Fluorescence was quantified using the LSM210 imaging software (Carl Zeiss Microimaging Inc., Thornwood, NJ). 6A7-immunoreactive cells were quantified and expressed as a percentage of total cells counted as described previously by us (9). DR5 and DR4 fluorescent staining was assessed as relative fluorescent units per cell and expressed as fold change over the vehicle-treated cells.
TRAIL receptor clustering was examined by total internal reflection microscopy (TIRF) (20). Huh-7 cells cultured on coverslips were transfected with respective DR4-EGFP or DR5-EGFP plasmid (21) using FuGENE HD transfection reagent (Roche Applied Science) 36 h prior to treatment. Cells were treated with palmitate or M2 antibody-aggregated FLAG-tagged TRAIL (21) at the indicated time and fixed with double distilled H2O containing 2.5% formaldehyde, 0.1 m PIPES, 1.0 mm EGTA, and 3.0 mm MgSO4 for 20 min at 37 °C. Cells were then washed three times in PBS, one time in water and mounted using Prolong Antifade (Invitrogen). The slides were analyzed with a TIRF microscope (Zeiss AxioObserver.Z1, Munich, Germany). Cells with EGFP clusters were quantified in 100 cells randomly selected for each condition. EGFP fluorescence at the plasma membrane was also quantified using image analysis software (Carl Zeiss Vision GmbH, Munich, Germany), and data were expressed as the average fluorescence intensity in the cell multiplied by the number of pixels above the background.
To assess individual cell transfection efficiency for DR4-EGFP and DR5-EGFP plasmids, we employed digitized fluorescent microscopy (22). Briefly, cells were cultured on glass coverslips in 35-mm dishes (Mattek, Ashland, MA). Thirty six hours following transfection, individual cellular fluorescence was visualized using excitation and emission filters of 490 and 520 nm, respectively. EGFP fluorescence per cell was quantified as the average fluorescence intensity in the cell multiplied by the number of pixels above background using MetaFluor software (Molecular Devices, Sunnyvale, CA).
Quantitation of Apoptosis
Huh-7 and KMCH cells were stained with 5 μg/ml 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI) for 30 min at room temperature and visualized under fluorescence microscopy (Nikon Eclipse TE200, Nikon Corporation, Japan). Apoptotic cells were quantified by counting 300 random cells per study and expressed as a percent of total cells counted. Cells with the characteristic nuclear changes of chromatin condensation and nuclear fragmentation were considered apoptotic. For caspase-3/-7 activity, cells were plated in 96-well plates. Caspase-3/-7 activation in cell lines was measured using Apo-ONE homogeneous caspase-3/-7 kit (Promega, Madison, WI) according to the manufacturer's instructions (9).
Jurkat cells were harvested and stained with propidium iodide to determine DNA content by flow cytometry (23). Cells were collected, washed with PBS, fixed, and permeabilized with ice-cold 70% ethanol overnight, followed by incubation with 0.1% Triton X-100, 0.1 mg/ml RNase A (Sigma) and 5 μg/ml propidium iodide for 30 min at 37 °C in the dark. Data acquisition and cell cycle analysis were performed on a FACScan flow cytometer with the accompanying CellQuest software (BD Biosciences).
Quantitative Real Time-PCR
Five micrograms of TRIzol-extracted total RNA was reverse-transcribed with Moloney leukemia virus reverse transcriptase and random primers (both from Invitrogen). Real time PCR was performed as described previously by us (9) using SYBR Green fluorescence technology. Primers are listed in Table 1.
TABLE 1.
Primers used for quantitative real time PCR
| Gene name | Accession no. | Primer sequence | Product size |
|---|---|---|---|
| Human and mouse 18 S | X03205 | Forward, 5′-CGTTCTTAGTTGGTGGAGCG-3′ | 212 bp |
| Reverse, 5′-CGCTGAGCCAGTCAGTGTAG-3′ | |||
| Human DR5 | NM_003842 | Forward, 5′-GGGAAGAAGATTCTCCTGAGATGTG-3′ | 292 bp |
| Reverse, 5′-ACATTGTCCTCAGCCCCAGGTCG-3′ | |||
| Human DR4 | NM_003844 | Forward, 5′-CAGAACGTCCTGGAGCCTGTAAC-3′ | 299 bp |
| Reverse, 5′-ATGTCCATTGCCTGATTCTTTGTG-3′ | |||
| Human TRAIL | NM_003810 | Forward, 5′-GTCCTCAGAGAGTAGCAGC-3′ | 157 bp |
| Reverse, 5′-CTCAAGTGCAAGTTGCTCAG-3′ | |||
| Human CHOP | NM_004083 | Forward, 5′-ATGGCAGCTGAGTCATTGCCTTTC-3′ | 177 bp |
| Reverse, 5′-AGAAGCAGGGTCAAGAGTGGTGAA-3′ | |||
| Mouse DR | NM_020275 | Forward, 5′-TGACGGGGAAGAGGAACTGA-3′ | 466 bp |
| Reverse, 5′-GGCTTTGACCATTTGGATTTGA-3′ |
Immunoblot Analysis
Whole cell lysates were prepared and subjected to immunoblot analysis as described previously (9). Samples containing 80 μg of protein were resolved by 12.5% SDS-PAGE, transferred to nitrocellulose membranes, and incubated with primary antibodies overnight at 4 °C. Membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (BIOSOURCE) for 1 h at room temperature. Bound antibody was visualized using chemiluminescent substrate (ECL, Amersham Biosciences) and exposed to Kodak X-OMAT film (Eastman Kodak Co.).
Immunoprecipitation
20 × 106 cells treated with either vehicle or palmitate were solubilized in death-inducing signaling complex (DISC) buffer containing 1% Triton X-100, 150 mm NaCl, 10% glycerol, 20 mm Tris-HCl, pH 7.5, 2 mm EDTA, 1 mm PMSF, 50 mm NaF, 1 mm Na3VO4 and a commercial protease inhibitor mixture (Roche Applied Science). After samples were incubated on ice for 30 min, insoluble debris was removed by centrifugation for 15 min at 14,000 × g at 4 °C. The protein concentration of the supernatant was determined by the Bradford assay and adjusted to 7 mg of protein in 1 ml of lysis buffer. Supernatants were pre-cleared using a mixture of protein A-agarose (30 μl) and protein G-Sepharose beads (50 μl) for 1 h at 4 °C and then incubated for 1 h at 4 °C with either 2 μg of goat anti-caspase-8 antibody (SC-6136, Santa Cruz Biotechnology) or 2 μg of goat anti-DR5 antibody (ALX-210-743-C200, Enzo Life Sciences). Eighty microliters of the mixture of protein A and G beads were next added to each sample and incubated under agitation overnight at 4 °C. Immune complexes were then pelleted by centrifugation for 5 min at 14,000 × g, washed seven times with lysis buffer, released from the beads by boiling for 5 min in SDS sample buffer, and examined by immunoblotting as described above.
Membrane Lipid Raft and Non-raft Fractionation
Lipid raft and non-raft soluble fractions were separated by discontinuous sucrose density gradients as described previously (24). Briefly, 20 × 108 cells treated with vehicle or palmitate were lysed on ice for 30 min in 2 ml of a Triton buffer containing 1% Triton X-100, 25 mm MES, 150 mm NaCl, pH 6.5, supplemented with 50 mm NaF, 1 mm Na3VO4 and a commercial protease inhibitor mixture (Roche Applied Science), and then homogenized. The homogenates were mixed with 2 ml of 90% sucrose solution prepared with Triton buffer and placed at the bottom of a centrifuge tube. The samples were then overlaid with an equal volume (4 ml) of 35% sucrose and 5% sucrose solutions (both prepared with Triton-buffer) and centrifuged at 39,000 rpm in SW41Ti rotor (Beckman Instruments) for 18 h at 4 °C. Twelve fractions of 1 ml were collected from the top to the bottom of the gradient and examined by Western blotting. Caveolin-1 antibody was used to identify lipid raft/caveolae fractions (fractions 3–7) from the non-raft fractions (fractions 8–12). The lipid raft/caveolae fractions or the non-raft fractions were mixed together, and proteins were precipitated by the addition of trichloroacetic acid (TCA) and then examined by immunoblot analysis.
Antibodies and Reagents
Rabbit anti-DR5, rabbit anti-DR4, and rabbit anti-cFLIPL were from ProSci (1:1000, Poway, CA). Rabbit anti-JNK, rabbit anti-phospho-JNK (Thr-183/Tyr-185), and mouse anti-caspase-8 were from Cell Signaling Technology (1:1000, Beverly, MA). Mouse anti-caspase-10 (clone 4C1) was purchased from MBL (1:1000, MBL International, Woburn, MA). Goat anti-DR5 (sc-7192, 1:100), mouse anti-Bax antisera (1:500, clone 6A7), mouse anti-DR4 antisera (1:500, clone B9), goat anti-caspase-8 antibody (SC-6136), and goat anti-β-actin (1:100) were from Santa Cruz Biotechnology (Santa Cruz, CA). Goat anti-DR5 antisera (1:500, ALX-210-743-C200) and mouse anti-FLIP (1:200, ALX-804-428) were from Alexis (Enzo Life Sciences, Plymouth Meeting, PA). Rabbit anti-caveolin-1 (1:1000) and mouse anti-tubulin (1:500) were a gift from Dr. Mark McNiven (Mayo Clinic, Rochester, MN). The pan-caspase inhibitor Z-VAD-fmk and the caspase-8 inhibitor Z-IETD-fmk were from MP Biomedicals (Aurora, OH). Recombinant human TRAIL (375-TEC) was from R&D Systems (Minneapolis, MN), and human DR5-Fc chimera (ALX-522-005-C050) and FLAG-TRAIL (ALX-522-003) were from Enzo Life Sciences. Thapsigargin and tunicamycin were from EMD Biosciences (Calbiochem). Human recombinant TNFα, actinomycin D, and M2 monoclonal anti-FLAG antibody were from Sigma.
Xbp-1 Splicing
Total RNA was isolated, and spliced Xbp-1 (Xbp-1s) and unspliced Xbp-1 (Xbp-1u) were detected by RT-PCR followed by digestion by the restriction enzyme Pst-I, as described previously by us (25).
Statistical Analysis
All data represent at least three independent experiments and are expressed as means ± S.E. Differences between groups were compared using Student's t tests and one-way analysis of variance with post hoc Dunnett test, and p values <0.05 were considered statistically significant.
RESULTS
DR5 Expression Is Increased by Palmitate Treatment and Contributes to Hepatocyte Lipoapoptosis
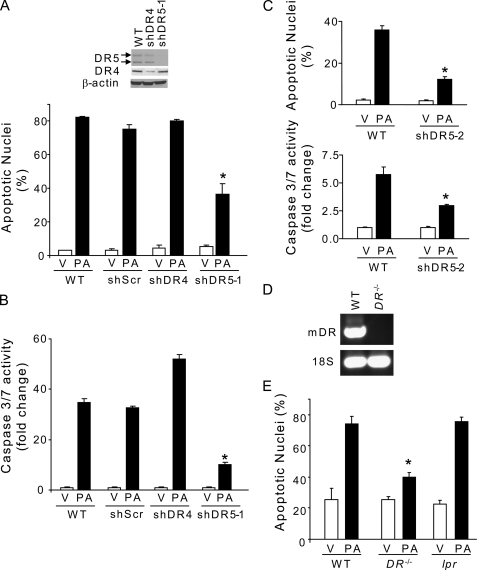
Consistent with prior observations (16), treatment of Huh-7 cells for 8 h with the saturated FFA palmitate resulted in a 2–3-fold increase in DR5 mRNA expression as compared with vehicle-treated cells (Fig. 1A). Similarly, an increase in DR5 cellular protein expression was observed in palmitate-treated Huh-7 cells (Fig. 1B), which was also associated with an increase in DR5 on the cell surface (Fig. 1, C and E). Palmitate-induced DR5 up-regulation was also confirmed in primary human hepatocytes (Fig. 1B). In contrast, DR4 expression was essentially unchanged by palmitate treatment (Fig. 1, A, B, D, and E). To ascertain whether DR5, or perhaps DR4, directly contributes to saturated FFA-induced apoptosis, Huh-7 cells were stably transfected with a DR5 targeting shRNA (shDR5) or a DR4 targeting shRNA (shDR4). Knockdown of DR5 expression attenuated palmitate-induced apoptosis in Huh-7 cells in multiple clones and concentrations of palmitate (400 and 800 μm) as assessed by both morphological and biochemical criteria (Fig. 2, A–C). In contrast, shRNA-targeted knockdown of DR4 did not protect against palmitate-induced apoptosis (Fig. 2, A and B). Further evidence for TRAIL DR contribution to saturated FFA-mediated toxicity was obtained by demonstrating that hepatocytes from DR−/− mice (Fig. 2D) were more resistant to lipoapoptosis by palmitate than wild-type (WT) hepatocytes (Fig. 2E); these hepatocytes genetically deficient in murine DR remained sensitive to apoptosis induced by TNFα (28 ng/ml) plus actinomycin D (0.2 μg/ml) for 24 h (37 ± 2 apoptotic nuclei) as compared with WT hepatocytes (43 ± 3 apoptotic nuclei). Death receptor specificity for these observations was confirmed as hepatocytes from lpr mice, which contain a Fas receptor that is unable to signal for death, and readily underwent lipoapoptosis by palmitate (Fig. 2E). Thus, TRAIL death receptor DR5 is up-regulated by saturated FFA and contributes to hepatocyte lipotoxicity in vitro.
FIGURE 1.
Palmitate increases DR5 cell surface expression. A, total RNA was isolated from Huh-7 cells treated for 8 h with PA at 400 and 800 μm. Vehicle (V) was used as control. DR5 and DR4 mRNA were quantified by real time PCR. Fold induction is expressed in relation to the housekeeping rRNA, 18 S. Data represent the mean ± S.E. of three independent experiments, *, p < 0.05. B, whole cell lysate was prepared 8 h after treatment with PA or vehicle. The concentration of PA was 400 and 800 μm for Huh-7 cells and 100, 200, and 400 μm for human primary hepatocytes. Immunoblot analyses were performed for DR5 and DR4, and β-actin was used as a control for protein loading. C and D, Huh-7 cells were treated for 6 h with PA at 800 μm or vehicle (V). Next cells were fixed with 4% paraformaldehyde, and DR5 and DR4 cell surface expressions (green) were assessed by confocal microscopy using specific antibodies recognizing the N-terminal region of the receptors. Nuclei were stained with DAPI (blue). α-Tubulin immunofluorescence (red) was observed only in Huh-7 cells fixed and permeabilized with 0.0125% CHAPS, ensuring that fluorescent staining for DR5 and DR4 observed in nonpermeabilized cells was at the cell surface. Representative images of three independent experiments are depicted. E, fluorescence intensity for the green channel was quantified in 10 random nonpermeabilized cells for each condition with automated software. Data represent mean ± S.E. of three experiments, *, p < 0.05; RFU: relative fluorescent units.
FIGURE 2.
DR5 knockdown attenuates palmitate-induced apoptosis in hepatocytes. A, WT Huh-7 cells or Huh-7 cells stably expressing short hairpin RNA targeting DR4 (shDR4) or DR5 (shDR5–1) were treated for 16 h with PA at 800 μm or vehicle (V). Scrambled shRNA-transfected Huh-7 cells (shScr) were also used in these experiments to discount any changes to the gene expression profile that may result from the shRNA delivery method or from clonal selection. Effective and selective down-regulation of DR4 or DR5 protein levels in shDR4 or shDR5 Huh-7 cells was verified by immunoblot analysis on whole cell lysates. DR5/β-actin and DR4/β-actin ratios were calculated from three immunoblots by densitometry. DR5/β-actin ratio values in WT Huh-7 cells were 1.03 ± 0.03 as compared with 0.12 ± 0.01 in shDR5 Huh-7 cells. DR4/β-actin ratio values in WT Huh-7 cells were 0.85 ± 0.01 as compared with 0.23 ± 0.04 in shDR4 Huh-7 cells. Apoptosis was assessed by morphological criteria after DAPI staining. B, cells treated as in A were assayed for caspase-3/7 activity. C, DR5 contribution to palmitate-induced apoptosis was further confirmed using WT Huh-7 cells and a second clone of HuH-7 cells stably transfected with shRNA against DR5 (shDR5-2) treated for 16 h with PA at 400 μm or vehicle. Apoptosis was assessed by morphological criteria after DAPI staining and biochemically by measuring caspase-3/7 activity. D, total RNA was isolated from WT or DR knock-out (DR−/−) primary mouse hepatocytes. DR mRNA expression was assessed by real time PCR. In WT primary mouse hepatocytes, the cycle threshold (ct) values for murine DR ranged from 30 to 31 cycles, whereas no detectable signal was recorded in DR−/− primary mouse hepatocytes over the course of 40 cycles. PCR products were then run on a 1.5% agarose gel and visualized by ethidium bromide staining. 18 S was used as an internal control. E, isolated WT, DR−/− or lpr primary mouse hepatocytes were treated for 16 h with PA at 400 μm or vehicle, and apoptotic nuclei were counted after DAPI staining. All data are expressed as mean ± S.E. for three experiments; *, p < 0.05.
Palmitate-mediated Apoptosis by DR5 Is Independent of TRAIL
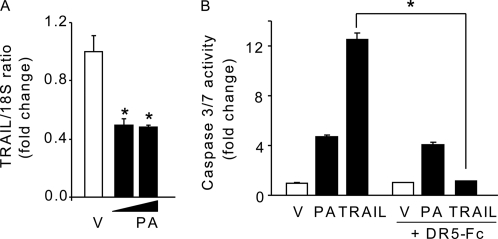
Because palmitate-treated Huh-7 cells display increased sensitivity to exogenous TRAIL (data not shown), we examined TRAIL mRNA expression in Huh-7 cells. Interestingly, TRAIL mRNA was not increased but rather decreased by palmitate treatment (Fig. 3A), suggesting that palmitate-induced lipotoxicity is unlikely due to increased TRAIL expression. Consistent with these observations, incubation of Huh-7 cells with a soluble human recombinant DR5-Fc chimera protein, which inhibits TRAIL-mediated death by competing with cell surface DR4 and DR5 for TRAIL, did not inhibit palmitate-induced apoptosis (Fig. 3B). As a control, DR5-Fc completely blocked Huh-7 cell apoptosis by exogenous TRAIL (Fig. 3B). These results suggest that palmitate-induced apoptosis by DR5 in liver cells does not involve TRAIL binding to its receptor.
FIGURE 3.
Palmitate-induced apoptosis by DR5 is TRAIL-independent. A, total RNA was isolated from Huh-7 cells treated for 8 h with PA at 400 and 800 μm or vehicle (V). TRAIL mRNA was quantified by real time PCR. Fold induction is expressed in relation to internal control 18 S. B, Huh-7 were treated with PA at 800 μm for 16 h or with TRAIL at 10 ng/ml for 6 h in the presence or absence of DR5-Fc protein chimera (250 ng/ml). Vehicle was used as a control, and caspase-3/7 catalytic activity was measured by the fluorogenic assay. All data are expressed as mean ± S.E. for three experiments; *, p < 0.05.
Palmitate Promotes DR5 Receptor Clustering and Redistribution to Lipid Rafts Resulting in Pro-apoptotic Receptor Signaling
TRAIL death receptor clustering is required for the initiation of death signaling and ultimately the induction of apoptosis (26, 27). Therefore, we used Huh-7 cells expressing EGFP-tagged DR5 or DR4 (Fig. 4A). We examined TRAIL receptor clustering upon treatment with palmitate for up to 120 min, and TIRF microscopy was used to image fluorescent clusters. TIRF microscopy visualizes fluorescence within 100 nm of the cell surface, and therefore changes in fluorescence largely reflect events at the plasma membrane. Under basal conditions (time 0), DR5-EGFP fluorescence was dim and was largely randomly distributed on the cell surface (Fig. 4B). Within 60 min of palmitate addition, the amount of DR5-EGFP at the cell surface markedly increased leading to the formation of bright macro-aggregates, consistent with receptor trafficking to and clustering within the plasma membrane; indeed, these macroaggregates are highly analogous to the signaling protein oligomer transduction structures previously reported for Fas on the plasma membrane (28). By 120 min of palmitate stimulation, DR5-EGFP fluorescence decreased to basal values (Fig. 4B), consistent with an internalization of the receptor complex (21). This pattern of DR5-EGFP cell surface dynamics was observed whether total cellular fluorescence or the percentage of cells with fluorescent punctae were quantified. Conversely, palmitate failed to induce the formation of DR4-EGFP macro-aggregates on the cell surface (Fig. 4C). As a control, clustering of DR4, as well as DR5, was readily observed when incubating cells with exogenous M2 antibody-aggregated FLAG-tagged TRAIL (Fig. 4D) for 30 min. Differences in DR4- or DR5-EGFP clustering during palmitate stimulation could not be attributed to different levels of EGFP-tagged receptor expression as Huh-7 cells are efficiently transfected by both DR5- and DR4-EGFP (Fig. 4A).
FIGURE 4.
Palmitate induces DR5 receptor clustering within the plasma membrane. Huh-7 cells were transfected with DR5-EGFP (A, B, and D) or DR4-EGFP plasmid (A, C, and D) for 36 h. A, total cellular EGFP fluorescence was visualized by digitized fluorescent microscopy. Both individual cellular EGFP fluorescence and EGFP-positive cells were quantified using imaging software to demonstrate that DR5-EGFP or DR4-EGFP plasmids transfected into the cell with the same efficiency. Data are expressed as means ± S.E. for three experiments. B and C, cells transfected with DR5-EGFP or DR4-EGFP for 36 h were treated with PA at 800 μm for 0, 30, 60, and 120 min. D, cells transfected with DR5-EGFP or DR4-EGFP for 36 h were treated with FLAG-tagged TRAIL (400 ng/ml) for 0 and 30 min. FLAG-tagged TRAIL was pre-oligomerized using the M2 monoclonal anti-FLAG antibody (2 μg/ml) as described previously (21). Cells were then fixed and EGFP fluorescence localized at the plasma membrane was analyzed by TIRF microscopy. Representative images are depicted for each condition. The percentage of cells with EGFP clusters was quantified and is expressed as means ± S.E. for three experiments. EGFP fluorescent intensity per cell was also quantified using image analysis software and is expressed as mean ± S.E. for three experiments; *, p < 0.05, RFU, relative fluorescent units.
Because lipid rafts serve as a platform to aggregate DR5 receptors (26, 27), we next examined whether TRAIL receptors co-localize with lipid rafts of the plasma membrane; detergent-insoluble membrane fractions contains caveolin-1 (Fig. 5A) (29). In vehicle-treated Huh-7 cells, DR5 predominantly localized in the detergent-soluble fractions, and little to no DR5 protein was observed by immunoblot analysis in the fraction that contains the lipid rafts/caveolae. By 8 h of treatment with palmitate, substantial accumulation of DR5 protein was found in the lipid raft fraction (Fig. 5A). In contrast, a minimal amount of DR4 was already present in the lipid rafts of vehicle-treated cells with no further accumulation in this fraction following palmitate stimulation (Fig. 5A). Taken together, these data are consistent with palmitate-induced accumulation and aggregation of DR5 within the plasma membrane, a required step for receptor signaling.
FIGURE 5.
Palmitate induces DR5 receptor recruitment to lipid rafts and caspase-8 activation. A, Huh-7 cells were treated with PA at 800 μm for 0, 4, or 8 h and subjected to discontinuous sucrose density gradients for separation of lipid rafts/caveolae and non-rafts fractions as described under “Experimental Procedures.” Immunoblot analysis was performed for DR5, DR4. and caveolin-1 (a lipid rafts marker). B and C, cell lysates were prepared from Huh-7 cells treated with vehicle or PA at 800 μm for 8 h. Immunoprecipitations (IP) were performed using a goat anti-DR5 antibody (B) or a goat anti-caspase-8 (C8) antibody that recognizes the p55/53 full-length and the p43/41-cleaved intermediate forms of caspase-8 (C). Whole cell lysates and immunoprecipitates were analyzed by immunoblotting for caspase-8, DR5, DR4, and the long and short isoforms of c-FLIP (c-FLIPL and c-FLIPs). To avoid interference with the heavy chain of goat immunoglobulins, rabbit anti-DR5 was used to detect DR5 in immunoprecipitates, whereas goat anti-DR5 was used in whole cell lysates. β-actin was assessed in whole cell lysates and was used as a control for protein loading.
Caspase-8 is recruited to and activated at the TRAIL receptor DISC and plays a crucial role in mediating TRAIL-induced apoptosis (30, 31). Therefore, to determine whether the observed plasma membrane receptor clustering was capable of pro-apoptotic signaling, we sought to ascertain if caspase-8 was recruited to and active within a DR5 receptor complex. Cleavage and activation of pro-caspase-8 into p43/p41 intermediate products were readily identified in palmitate-treated Huh-7 cells (Figs. 5, B and C, and 6A). Caspase-8 activation by palmitate was independent of a decrease in cellular FLICE-like inhibitory protein (c-FLIP) expression (Figs. 5B and 6A), a protein that may be recruited to the DISC and acts to inhibit TRAIL pro-apoptotic signaling (32, 33). To examine the ability of palmitate to induce a DR5-mediated processing of caspase-8, immunoprecipitation of DR5 was performed followed by immunoblot analysis (Fig. 5B). Indeed, palmitate treatment resulted in the appearance of the p43 and p41 cleaved forms of caspase-8 in DR5 immune complexes. In contrast, no recruitment of cFLIPL or the short isoform of c-FLIP (c-FLIPS) within a DR5 receptor complex was observed (Fig. 5B). Because DR5 expression is up-regulated by palmitate, larger amounts of DR5 were immunoprecipitated from palmitate-treated cells as compared with control (Fig. 5B). The reciprocal immunoprecipitation paradigm was performed using an anti-caspase-8 antibody that recognizes both the full-length and p43 and p41 forms of caspase-8; DR5 was more efficiently immunoprecipitated as pro-caspase-8 was processed into cleaved products upon palmitate treatment (Fig. 5C). In contrast, no binding of caspase-8 products to DR4 was observed (Fig. 5C). To further confirm a role for DR5-activated caspase-8 during lipoapoptosis processes, we generated Huh-7 cells with a stable knockdown of caspase-8 (shC8) (Fig. 6A). Consistent with prior studies (30, 31), these shC8 cells are resistant to TRAIL-induced apoptosis (data not shown). More importantly, shC8 cells were resistant to palmitate-induced lipoapoptosis as compared with wild-type (WT) Huh-7 cells (Fig. 6, B and C). Besides caspase-8, caspase-10 may also be recruited to the TRAIL DISC and contribute to TRAIL killing (34); however, silencing of caspase-10 by targeted shRNA (Fig. 6D) did not alter palmitate-induced toxicity (Fig. 6, E and F), indicating caspase-8 specificity in this process.
FIGURE 6.
DR5-activated caspase-8 contributes to Bax activation and apoptosis by palmitate. A, WT Huh-7 cells or Huh-7 cells stably expressing short hairpin RNA complementary to caspase-8 (shC8) were treated with PA at 400 or 800 μm or vehicle (V) for 8 h. Immunoblot analysis was performed for c-FLIPL and caspase-8 (C8) displaying cleaved caspase-8 products p43/p41. Efficient knockdown of caspase-8 in shC8 Huh-7 cells was observed. Images were cut and combined from the same radiograph. B and C, WT and shC8 Huh-7 cells treated as in A for 16 h. Apoptosis was assessed by morphological criteria after DAPI staining (B) and biochemically by measuring caspase-3/7 activity (C). D, efficient knockdown of caspase-10 in Huh-7 cells stably expressing short hairpin RNA complementary to caspase-10 (shC10) was observed by immunoblot analysis. β-Actin was used as a control for protein loading. E and F, WT and shC10 Huh-7 cells were treated with PA at 800 μm or vehicle (V) for 16 h, and apoptosis was assessed by morphological criteria after DAPI staining (E) and biochemically by measuring caspase-3/7 activity (F). G, WT, shDR5, shDR4, and shC8 Huh-7 cells were treated with PA at 600 μm or vehicle for 16 h. Bax activation was assessed using conformation-specific antisera (6A7) and immunofluorescence microscopy. Representative images of three independent experiments are depicted. Bax activation (green) was quantified in five random high power fields for each condition with automated software. Nuclei were stained with DAPI (blue). All data are expressed as mean ± S.E. for three experiments; *, p < 0.05.
Activation of the pro-apoptotic protein Bax, a known mediator of mitochondrial dysfunction, represents a critical molecular event during lipoapoptosis (8, 9). Because active caspase-8 promotes Bax activation (35), we examined the potential relationship between DR5 downstream death signaling and Bax activation upon palmitate treatment. Bax activation in response to palmitate was readily detected in WT Huh-7 cells using the active conformation-specific 6A7 monoclonal antibody (Fig. 6G). In contrast, both knockdown of DR5 or caspase-8 expression efficiently reduced palmitate-activated Bax (Fig. 6G). Taken together, these results implicate DR5-activated caspase-8 as a key step in mediating hepatocyte lipotoxicity by palmitate.
DR5-mediated Toxicity Is Downstream of ER Stress-mediated CHOP Induction
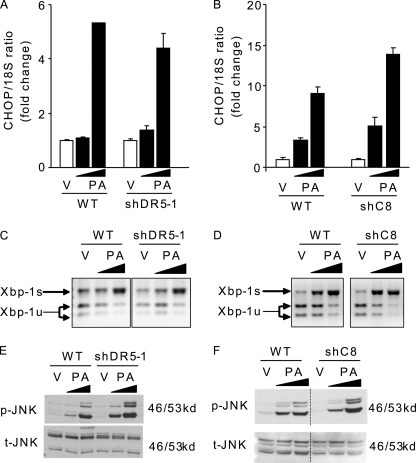
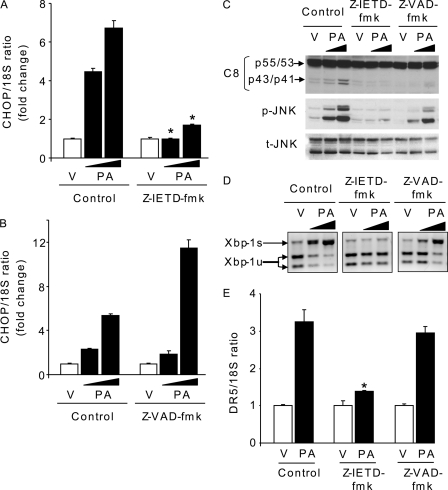
Saturated FFA activate ER stress-signaling pathways (7, 25), and a recent study suggests that DR5-activated caspase-8 may directly induce ER stress (36). Therefore, in an attempt to relate DR5 activation to ER stress in the context of saturated FFA cytotoxicity, we examined several ER stress markers, including CHOP, XBP-1, and JNK, in shDR5 and shC8 Huh-7 cells during treatment with palmitate. However, selective shRNA-targeted knockdown of DR5 or caspase-8 did not prevent CHOP induction (Fig. 7, A and B), XPB-1 splicing (Fig. 7, C and D), or JNK-activating phosphorylation (Fig. 7, E and F). These data suggest that palmitate-activated DR5 death signaling is not upstream of ER stress. To reconcile our data with those implicating DR5 signaling as a cause of ER stress, we used the pharmacological approach employed in the prior study to inhibit caspase-8 (36). Caspase-8 inhibition using the pharmacological inhibitor Z-IETD-fmk completely prevented CHOP induction, XBP-1 splicing, and JNK activation (Fig. 8, A, C, and D), an observation consistent with the prior studies by Tiwary et al. (36). However, this protective effect was only partial when using the pan-caspase inhibitor Z-VAD-fmk, which also efficiently prevents caspase-8 activation by palmitate (Fig. 8, B–D). Because neither shRNA knockdown of caspase-8 nor its inhibition with Z-VAD-fmk completely blocks ER stress by palmitate, the ability of Z-IETD-fmk to fully inhibit ER stress is potentially an off-target effect of this drug. Interestingly, Z-IETD-fmk did not inhibit CHOP up-regulation by thapsigargin or tunicamycin (data not shown), two potent pharmacological ER stress inducers, indicating that Z-IETD-fmk is not a general inhibitor of ER stress responses but rather antagonizes ER stress induction by palmitate; the mechanisms by which Z-IETD-fmk blocks palmitate-induced ER stress upstream of the ER require further delineation.
FIGURE 7.
DR5 or caspase-8 knockdown does not modify palmitate-induced ER stress. WT, shDR5, and shC8 Huh-7 cells were treated with PA at 400 or 800 μm or vehicle (V) for 8 h. A and B, total RNA was isolated, and CHOP mRNA was quantified by real time PCR. Fold induction is relative to internal control 18 S. Data represent mean ± S.E. of three experiments. C and D, XBP-1 cDNA was amplified by PCR, followed by incubation with PstI. In vehicle-treated cells, most Xbp-1 PCR products were cut by PstI, producing 290- and 180-bp amplification products, indicative of the native and unspliced form of Xbp-1 mRNA (Xbp-1u). A 471-bp amplification product, indicative of spliced Xbp-1 mRNA (Xbp-1s), was observed in PA-treated cells. E and F, whole cell lysates were prepared, and immunoblot analysis was performed for phosphorylated JNK (p-JNK) protein expression and total JNK (t-JNK), a control for protein loading. Images were cut and combined from the same radiograph.
FIGURE 8.
Specific caspase-8 inhibitor, Z-IETD-fmk, but not the pan-caspase inhibitor, Z-VAD-fmk, inhibits palmitate-induced ER stress. Huh-7 cells were treated with PA at 400 or 800 μm or vehicle (V) for 8 h. Caspase-8 activity was inhibited using the specific inhibitor Z-IETD-fmk (10 μm) or the pan-caspase inhibitor Z-VAD-fmk (10 μm). A and B, total RNA was isolated, and CHOP mRNA was quantified by real time PCR. Fold induction is relative to internal control 18 S. Data represent mean ± S.E. of three experiments; *, p < 0.05. C, whole cell lysates were prepared, and immunoblot analysis was performed for caspase-8 (C8), phosphorylated JNK (p-JNK), total JNK (t-JNK), and β-actin, a control for protein loading. D, spliced Xbp-1 (Xbp-1s) was detected by RT-PCR followed by digestion by Pst-I. E, cells were treated with PA at 800 μm or vehicle (V) for 8 h. DR5 mRNA was quantified by real time PCR relative to internal control 18 S. Data represent mean ± S.E. of three experiments; *, p < 0.05.
Further evidence that ER stress pathways precede DR5 activation during hepatocyte lipoapoptosis was obtained by demonstrating that inhibition of CHOP induction, either by targeted shRNA (shCHOP) (Fig. 9A) (7) or by the drug Z-IETD-fmk, prevented DR5 mRNA up-regulation by palmitate (Figs. 9B and. 8E). Consistent with transcriptional regulation of DR5 expression by CHOP (37), CHOP knockdown also reduced caspase-8 activation upon palmitate treatment (Fig. 9C). Thus, the results indicate that DR5 up-regulation and activation follows ER stress-induced CHOP induction during hepatocyte lipoapoptosis.
FIGURE 9.
CHOP knockdown reduces DR5 up-regulation and caspase-8 activation by palmitate. WT Huh-7 cells or Huh-7 cells stably expressing short hairpin RNA targeting CHOP (shCHOP) were treated for 8 h with PA at 800 μm or vehicle (V). A and B, total RNA was isolated, and CHOP and DR5 mRNA were quantified by real time PCR. Fold induction is relative to internal control 18 S. Data represent mean ± S.E. of three experiments; *, p < 0.05. C, immunoblot analysis for caspase-8 (C8) was performed on whole cell lysate. β-Actin was used as a control for protein loading. Images were cut and combined from the same radiograph.
Palmitate-activated DR5 Death Signaling also Mediates the Lipotoxic Insult in Other Cell Models
To further implicate DR5 death signaling in mediating the lipotoxic insult, we used the human T lymphoblastoid cell line, Jurkat cells. These cells express DR5 but little to no DR4 (38) and therefore represent a useful tool to study specifically DR5-mediated death signaling. Similarly to Huh-7 cells, treatment with palmitate up-regulates DR5 protein expression and activates caspase-8 in WT Jurkat cells (Fig. 10A), which results in cellular demise (Fig. 10, B and C). FADD is also essential for death receptor pro-apoptotic signaling (39). Consistent with the above studies, Jurkat variants deficient in FADD (ΔFADD, I2.1) or caspase-8 (ΔC8, I9.2) were also resistant to palmitate-induced lipotoxicity and displayed reduced caspase-8 activation following palmitate treatment (Fig. 10, B and C). Like Huh-7 cells, these caspase-8- and FADD-deficient Jurkat cells also underwent ER stress upon exposure to palmitate (Fig. 10, D and E).
FIGURE 10.
FADD and caspase-8 deficiency protects Jurkat cells against palmitate-induced toxicity without altering ER stress responses. A, whole cell lysate was prepared from WT Jurkat cells treated for 8 h with vehicle (V) or PA at the dose of 800 μm. Immunoblot analyses were performed for DR5 and caspase-8. β-Actin was used as a control for protein loading. B and C, WT, ΔFADD, and ΔC8 Jurkat cells were treated with vehicle or PA at 800 μm for 16 h. Apoptosis was assessed by sub-G1 analysis by flow cytometry after propidium iodide staining (B) and biochemically by measuring caspase-3/7 activity (C). D and E, Jurkat cells were treated as in B and C. Total RNA was isolated, and CHOP mRNA was quantified by real time PCR. Fold induction is relative to internal control 18 S (D). Whole cell lysates were prepared, and immunoblot analysis was performed for phosphorylated JNK (p-JNK) and total JNK (t-JNK), a control for protein loading (E). All data represent mean ± S.E. of three experiments; *, p < 0.05.
We also employed the cholangiocarcinoma cell line KMCH as another cell model. Again, suppression of DR5 expression by targeted shRNA (Fig. 11A) protected the cells from palmitate-induced toxicity in several clones (Fig. 11, B and C). Despite increased level in DR4 protein expression, shDR5 KMCH cells demonstrated a reduction in caspase-8 activation upon palmitate treatment (Fig. 11D). Also, like hepatocytes, the knockdown of DR5 did not confer resistance to palmitate-mediated ER stress (Fig. 11E). Thus, these results in additional cell lines further establish DR5 death signaling as a potent contributor to lipoapoptosis.
FIGURE 11.
DR5 knockdown protects KMCH cells against palmitate toxicity. A, efficient and selective knockdown of DR5 in KMCH cells stably expressing short hairpin RNA complementary to DR5 (shDR5) was observed by immunoblot analysis. β-Actin was used as a control for protein loading. B and C, WT or two shDR5 KMCH clones (shDR5-1 and shDR5-2) were treated for 16 h with vehicle (V) or PA at the dose of 800 μm. Scrambled shRNA-transfected KMCH cells (shScr) were also used in these experiments to discount any changes to the gene expression profile that may result from the shRNA delivery method or from clonal selection. Apoptosis was assessed by morphological criteria after DAPI staining (B) and biochemically by measuring caspase-3/7 activity (C). All data represent means ± S.E. of three experiments; *, p < 0.05. D and E, WT or shDR5 KMCH cells were treated for 8 h with vehicle (V) or PA at the dose of 800 μm. Whole cell lysates were prepared, and immunoblot analysis was performed for caspase-8 and β-actin, a control for protein loading (D). Total RNA was isolated, and spliced Xbp-1 (Xbp-1s) was detected by RT-PCR followed by digestion by PstI (E).
DISCUSSION
FFA-induced hepatic injury plays a major role in NAFLD. The principal findings of this study provide mechanistic insights regarding the contribution of DR5 death signaling to hepatocyte lipoapoptosis. Our results indicate the following: (i) the toxic saturated FFA palmitate directly up-regulates DR5 by a CHOP-dependent mechanism downstream of ER stress; (ii) genetic deletion or shRNA targeted knockdown of DR5 expression attenuates lipoapoptosis by palmitate; and (iii) palmitate directly induces plasma membrane DR5 association with lipid rafts and receptor clustering independent of its cognate ligand TRAIL resulting in caspase-8 activation and cellular demise. These observations are more thoroughly discussed below.
DR5 mRNA expression is significantly elevated in patient with nonalcoholic steatohepatitis (16, 40) and in experimental murine models of steatohepatitis (41). Although we had previously demonstrated that DR5 cellular expression is increased in steatotic hepatocytes (16), here we describe for the first time the direct contribution of DR5 death signaling to hepatocyte lipoapoptosis. For example, short hairpin RNA-targeted knockdown of DR5 renders Huh-7 cells partially resistant to palmitate cytotoxicity. Similarly, genetic deficiency of mouse DR, homologous to human DR5, protected mouse primary hepatocytes from lipoapoptosis. Consistent with DR5-mediated cytotoxicity, palmitate-induced cytotoxicity was caspase-8-dependent. Activated caspase-8 further activates the pro-apoptotic members of the Bcl-2 family, Bax (35), resulting in Bax translocation in the outer mitochondrial membrane with subsequent mitochondrial dysfunction, downstream activation of the effector caspase-3 and -7, and ultimately cellular demise. Prior studies have reported that the broad inhibition of caspases can reduce hepatocyte apoptosis in animal models of nonalcoholic steatohepatitis (42, 43), and caspase-8 inhibition in this model could account, in part, for the protection of hepatocyte against steatosis-induced apoptosis. Thus, this implicates a functional role for DR5 death signaling in lipotoxicity suggesting that its enhanced expression is not an epi-phenomenon in this cellular process.
Death receptor activation is a complex multifactorial process requiring translocation of receptors to the plasma membrane, receptor association often within lipid rafts, and subsequent recruitment of platforms and effector molecules to the receptor complex (11). Although cognate ligands are usually required for these activation processes, ligand-independent receptor activation has been well documented. Indeed, bile acids stimulate ligand-independent activation of Fas (44, 45) and DR5 (46). Palmitate also appears to initiate ligand-independent activation of DR5. Several observations support this interpretation of our data. Death receptor-mediated apoptosis is regulated at the cell surface by the density of death receptors (45), and palmitate promotes increased cell surface expression of DR5 with downstream initiation of a death-signaling pathway. Exogenous addition of soluble human recombinant DR5-Fc chimera protein, which binds soluble TRAIL and prevents its access to the receptor, also failed to prevent palmitate cytotoxicity. In addition, palmitate actually reduced, rather than increased, TRAIL mRNA expression by hepatocytes. Like Fas receptor (28), the formation of cell surface receptor oligomeric structures, referred to as signaling protein oligomer transduction structures, were also observed for DR5-EGFP by TIRF microscopy following palmitate stimulation. An additional report suggests that formation of ceramide-enriched membrane platforms serves to trap and cluster DR5, a prerequisite for the induction of apoptosis by DR5 (26, 27); and plasma membrane DR5 associates with lipid raft upon palmitate treatment. Interestingly, palmitate is a precursor of de novo ceramide synthesis, which may contribute to the observed DR5 recruitment in the lipid rafts and receptor clustering. Although it was also reported that post-translational lipid modification of death receptors by palmitoylation leads to their redistribution into lipid rafts and oligomerization (47), palmitoylation of TRAIL receptors was observed only for DR4 but not DR5 (47). Thus, the precise mechanisms by which palmitate induces DR5 clustering and pro-apoptotic signaling independent of TRAIL remains to be delineated.
Accumulating studies have demonstrated the triggering of ER stress responses during saturated FFA-induced apoptosis (7, 25, 48); and ER-stress pathways are observed in both rodent and human steatohepatitis (5, 6). ER stress responses are sensed and transduced by ER-resident proteins and can result in the activation/induction of pro-apoptotic proteins, such as CHOP (7, 25, 48). The mechanisms by which saturated FFA trigger ER stress responses are unclear but appear to be due to the release of calcium from the ER stores (49, 50). More recently, a study has reported that ER stress can be triggered by the extrinsic pathway of apoptosis induced by DR5 (36). However, in our studies, we observed that palmitate-induced ER stress was upstream and not downstream of DR5-activated caspase-8. DR5 cellular levels are transcriptionally up-regulated by p53-dependent and -independent mechanisms, including CHOP-dependent mechanisms (37). p53 is mutated and functionally inactive in Huh-7 cells making it unlikely that p53 mediates DR5 expression by palmitate (51). Consistent with a transcriptional regulation of DR5 by CHOP (37), shRNA-targeted knockdown of CHOP reduced DR5 up-regulation by palmitate, a mechanism that could account into the resistance of these cells to palmitate-induced apoptosis (7). We have previously implicated the BH3-only proteins Bim and PUMA in mediating mitochondrial dysfunctions and hepatocyte apoptosis during saturated FFA insult (9, 52). Both Bim and PUMA are regulated downstream of ER stress responses (9, 53) and are required for TRAIL cytotoxicity (54, 55). Likely, the BH3-only protein stress exerted by Bim and PUMA up-regulation serves to sensitize the cell to TRAIL cytotoxicity by the mitochondrial pathway. Likely, caspase-8 generation of truncated Bid by DR5 activation (11), also a pro-apoptotic BH3-only protein, in the context of PUMA and Bim up-regulation, overwhelms the anti-apoptotic defenses of the cell, culminating in cell death. Thus, our studies help assimilate multiple injurious effects of FFA on the cell by linking ER stress and alterations in Bcl-2 protein family members to a death receptor pathway of apoptosis (Fig. 12).
FIGURE 12.
DR5 death signaling contributes to palmitate-induced apoptosis. ER stress-induced CHOP expression by saturated FFA increases cellular DR5 expression thereby augmenting its cell surface density. FFA also promote DR5 clustering that serves to recruit and activate caspase-8, which ultimately promotes Bax activation. Cytotoxic FFA also induce Bim and PUMA expression causing cellular BH3-only stress. This BH3-only stress sensitizes the cell to DR5 cytotoxic signaling by enhancing Bax-mediated mitochondrial dysfunction with subsequent caspase activation and apoptosis.
Lipotoxicity is almost certainly a pleiotropic process with multiple causes of cellular dysfunction and death. This study notably provides further insights regarding the specific role of the death receptor-mediated extrinsic pathway in FFA-induced hepatocyte lipotoxicity but also integrates these observations with prior studies highlighting a pathogenetic role for ER stress and alterations in Bcl-2 protein family members in this process. The results implicate DR5 up-regulation with subsequent receptor clustering and activation of caspase-8 as critical steps in mediating saturated FFA-mediated apoptosis downstream of ER stress. Thus, clinical trials of caspase inhibitors in human steatohepatitis or other approaches to reduce DR5 activation (56) hold promise to ameliorate hepatocyte lipoapoptosis in human fatty liver disease.
Acknowledgments
We thank Dr. Anuradha Krishnan and Dr. Michael R. Charlton for providing isolated human primary hepatocytes. We thank Courtney Riddle for excellent secretarial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DK41876 (to G. J. G.), DK069757 (to M. C. R.), DK079875 (to J. L. M.), CA69008 (to S. H. K.), CA123258 (to W. S. E.-D.), and P30DK084567 (to Optical Microscopy Core of the Mayo Clinic Center for Cell Signaling in Gastroenterology and the Mayo Foundation).
- FFA
- free fatty acid
- c-FLIP
- cellular FLICE-like inhibitory protein
- CHOP
- C/EBP homologous protein
- DISC
- death-induced signaling complex
- DR
- death receptor
- ER
- endoplasmic reticulum
- FADD
- Fas-associated death domain protein
- NAFLD
- nonalcoholic fatty liver disease
- PA
- palmitic acid
- scr
- scrambled
- sh
- short hairpin
- TIRF
- total internal reflection microscopy
- TRAIL
- tumor necrosis factor-related apoptosis-inducing ligand
- V
- vehicle
- EGFP
- enhanced GFP
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone.
REFERENCES
- 1. Angulo P. (2002) N. Engl. J. Med. 346, 1221–1231 [DOI] [PubMed] [Google Scholar]
- 2. Parekh S., Anania F. A. (2007) Gastroenterology 132, 2191–2207 [DOI] [PubMed] [Google Scholar]
- 3. Unger R. H., Orci L. (2002) Biochim. Biophys. Acta 1585, 202–212 [DOI] [PubMed] [Google Scholar]
- 4. Feldstein A. E., Canbay A., Angulo P., Taniai M., Burgart L. J., Lindor K. D., Gores G. J. (2003) Gastroenterology 125, 437–443 [DOI] [PubMed] [Google Scholar]
- 5. Wang D., Wei Y., Pagliassotti M. J. (2006) Endocrinology 147, 943–951 [DOI] [PubMed] [Google Scholar]
- 6. Puri P., Mirshahi F., Cheung O., Natarajan R., Maher J. W., Kellum J. M., Sanyal A. J. (2008) Gastroenterology 134, 568–576 [DOI] [PubMed] [Google Scholar]
- 7. Cazanave S. C., Elmi N. A., Akazawa Y., Bronk S. F., Mott J. L., Gores G. J. (2010) Am. J. Physiol. Gastroentest. Liver Physiol. 299, G236–G243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malhi H., Bronk S. F., Werneburg N. W., Gores G. J. (2006) J. Biol. Chem. 281, 12093–12101 [DOI] [PubMed] [Google Scholar]
- 9. Cazanave S. C., Mott J. L., Elmi N. A., Bronk S. F., Werneburg N. W., Akazawa Y., Kahraman A., Garrison S. P., Zambetti G. P., Charlton M. R., Gores G. J. (2009) J. Biol. Chem. 284, 26591–26602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li N., Frigerio F., Maechler P. (2008) Biochem. Soc. Trans. 36, 930–934 [DOI] [PubMed] [Google Scholar]
- 11. Guicciardi M. E., Gores G. J. (2009) FASEB J. 23, 1625–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wueest S., Rapold R. A., Schumann D. M., Rytka J. M., Schildknecht A., Nov O., Chervonsky A. V., Rudich A., Schoenle E. J., Donath M. Y., Konrad D. (2010) J. Clin. Invest. 120, 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin H. Z., Yang S. Q., Chuckaree C., Kuhajda F., Ronnet G., Diehl A. M. (2000) Nat. Med. 6, 998–1003 [DOI] [PubMed] [Google Scholar]
- 14. Feldstein A. E., Werneburg N. W., Canbay A., Guicciardi M. E., Bronk S. F., Rydzewski R., Burgart L. J., Gores G. J. (2004) Hepatology 40, 185–194 [DOI] [PubMed] [Google Scholar]
- 15. Romanatto T., Roman E. A., Arruda A. P., Denis R. G., Solon C., Milanski M., Moraes J. C., Bonfleur M. L., Degasperi G. R., Picardi P. K., Hirabara S., Boschero A. C., Curi R., Velloso L. A. (2009) J. Biol. Chem. 284, 36213–36222 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16. Malhi H., Barreyro F. J., Isomoto H., Bronk S. F., Gores G. J. (2007) Gut 56, 1124–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Finnberg N., Gruber J. J., Fei P., Rudolph D., Bric A., Kim S. H., Burns T. F., Ajuha H., Page R., Wu G. S., Chen Y., McKenna W. G., Bernhard E., Lowe S., Mak T., El-Deiry W. S. (2005) Mol. Cell. Biol. 25, 2000–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Belfort R., Harrison S. A., Brown K., Darland C., Finch J., Hardies J., Balas B., Gastaldelli A., Tio F., Pulcini J., Berria R., Ma J. Z., Dwivedi S., Havranek R., Fincke C., DeFronzo R., Bannayan G. A., Schenker S., Cusi K. (2006) N. Engl. J. Med. 355, 2297–2307 [DOI] [PubMed] [Google Scholar]
- 19. Sanyal A. J., Campbell-Sargent C., Mirshahi F., Rizzo W. B., Contos M. J., Sterling R. K., Luketic V. A., Shiffman M. L., Clore J. N. (2001) Gastroenterology 120, 1183–1192 [DOI] [PubMed] [Google Scholar]
- 20. Steyer J. A., Almers W. (2001) Nat. Rev. Mol. Cell Biol. 2, 268–275 [DOI] [PubMed] [Google Scholar]
- 21. Akazawa Y., Mott J. L., Bronk S. F., Werneburg N. W., Kahraman A., Guicciardi M. E., Meng X. W., Kohno S., Shah V. H., Kaufmann S. H., McNiven M. A., Gores G. J. (2009) Gastroenterology 136, 2365–2376.e1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ribeiro A., Bronk S. F., Roberts P. J., Urrutia R., Gores G. J. (1999) Hepatology 30, 1490–1497 [DOI] [PubMed] [Google Scholar]
- 23. Meng X. W., Heldebrant M. P., Kaufmann S. H. (2002) J. Biol. Chem. 277, 3776–3783 [DOI] [PubMed] [Google Scholar]
- 24. Ostrom R. S., Insel P. A. (2006) Methods Mol. Biol. 332, 181–191 [DOI] [PubMed] [Google Scholar]
- 25. Akazawa Y., Cazanave S., Mott J. L., Elmi N., Bronk S. F., Kohno S., Charlton M. R., Gores G. J. (2010) J. Hepatol. 52, 586–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Song J. H., Tse M. C., Bellail A., Phuphanich S., Khuri F., Kneteman N. M., Hao C. (2007) Cancer Res. 67, 6946–6955 [DOI] [PubMed] [Google Scholar]
- 27. Dumitru C. A., Gulbins E. (2006) Oncogene 25, 5612–5625 [DOI] [PubMed] [Google Scholar]
- 28. Siegel R. M., Muppidi J. R., Sarker M., Lobito A., Jen M., Martin D., Straus S. E., Lenardo M. J. (2004) J. Cell Biol. 167, 735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Michel V., Bakovic M. (2007) Biol. Cell 99, 129–140 [DOI] [PubMed] [Google Scholar]
- 30. Jin Z., El-Deiry W. S. (2006) Mol. Cell. Biol. 26, 8136–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Suliman A., Lam A., Datta R., Srivastava R. K. (2001) Oncogene 20, 2122–2133 [DOI] [PubMed] [Google Scholar]
- 32. Kataoka T., Tschopp J. (2004) Mol. Cell. Biol. 24, 2627–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sharp D. A., Lawrence D. A., Ashkenazi A. (2005) J. Biol. Chem. 280, 19401–19409 [DOI] [PubMed] [Google Scholar]
- 34. Engels I. H., Totzke G., Fischer U., Schulze-Osthoff K., Jänicke R. U. (2005) Mol. Cell. Biol. 25, 2808–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deng Y., Lin Y., Wu X. (2002) Genes Dev. 16, 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tiwary R., Yu W., Li J., Park S. K., Sanders B. G., Kline K. (2010) Plos One 5, e11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamaguchi H., Wang H. G. (2004) J. Biol. Chem. 279, 45495–45502 [DOI] [PubMed] [Google Scholar]
- 38. Mühlenbeck F., Schneider P., Bodmer J. L., Schwenzer R., Hauser A., Schubert G., Scheurich P., Moosmayer D., Tschopp J., Wajant H. (2000) J. Biol. Chem. 275, 32208–32213 [DOI] [PubMed] [Google Scholar]
- 39. Juo P., Woo M. S., Kuo C. J., Signorelli P., Biemann H. P., Hannun Y. A., Blenis J. (1999) Cell Growth Differ. 10, 797–804 [PubMed] [Google Scholar]
- 40. Kahraman A., Schlattjan M., Kocabayoglu P., Yildiz-Meziletoglu S., Schlensak M., Fingas C. D., Wedemeyer I., Marquitan G., Gieseler R. K., Baba H. A., Gerken G., Canbay A. (2010) Hepatology 51, 92–102 [DOI] [PubMed] [Google Scholar]
- 41. Farrell G. C., Larter C. Z., Hou J. Y., Zhang R. H., Yeh M. M., Williams J., dela Pena A., Francisco R., Osvath S. R., Brooling J., Teoh N., Sedger L. M. (2009) J. Gastroenterol. Hepatol. 24, 443–452 [DOI] [PubMed] [Google Scholar]
- 42. Witek R. P., Stone W. C., Karaca F. G., Syn W. K., Pereira T. A., Agboola K. M., Omenetti A., Jung Y., Teaberry V., Choi S. S., Guy C. D., Pollard J., Charlton P., Diehl A. M. (2009) Hepatology 50, 1421–1430 [DOI] [PubMed] [Google Scholar]
- 43. Anstee Q. M., Concas D., Kudo H., Levene A., Pollard J., Charlton P., Thomas H. C., Thursz M. R., Goldin R. D. (2010) J. Hepatol. 53, 542–550 [DOI] [PubMed] [Google Scholar]
- 44. Graf D., Kurz A. K., Fischer R., Reinehr R., Häussinger D. (2002) Gastroenterology 122, 1411–1427 [DOI] [PubMed] [Google Scholar]
- 45. Higuchi H., Gores G. J. (2003) Am. J. Physiol. Gastroentest. Liver Physiol. 284, G734–G738 [DOI] [PubMed] [Google Scholar]
- 46. Lim S. C., Duong H. Q., Choi J. E., Lee T. B., Kang J. H., Oh S. H., Han S. I. (2011) Carcinogenesis 32, 723–731 [DOI] [PubMed] [Google Scholar]
- 47. Rossin A., Derouet M., Abdel-Sater F., Hueber A. O. (2009) Biochem. J. 419, 185–192 [DOI] [PubMed] [Google Scholar]
- 48. Wei Y., Wang D., Topczewski F., Pagliassotti M. J. (2006) Am. J. Physiol. Endocrinol. Metab. 291, E275–E281 [DOI] [PubMed] [Google Scholar]
- 49. Wei Y., Wang D., Gentile C. L., Pagliassotti M. J. (2009) Mol. Cell. Biochem. 331, 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fu S., Yang L., Li P., Hofmann O., Dicker L., Hide W., Lin X., Watkins S. M., Ivanov A. R., Hotamisligil G. S. (2011) Nature 473, 528–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen J. J., Chou C. W., Chang Y. F., Chen C. C. (2008) J. Immunol. 180, 8030–8039 [DOI] [PubMed] [Google Scholar]
- 52. Barreyro F. J., Kobayashi S., Bronk S. F., Werneburg N. W., Malhi H., Gores G. J. (2007) J. Biol. Chem. 282, 27141–27154 [DOI] [PubMed] [Google Scholar]
- 53. Puthalakath H., O'Reilly L. A., Gunn P., Lee L., Kelly P. N., Huntington N. D., Hughes P. D., Michalak E. M., McKimm-Breschkin J., Motoyama N., Gotoh T., Akira S., Bouillet P., Strasser A. (2007) Cell 129, 1337–1349 [DOI] [PubMed] [Google Scholar]
- 54. Werneburg N. W., Guicciardi M. E., Bronk S. F., Kaufmann S. H., Gores G. J. (2007) J. Biol. Chem. 282, 28960–28970 [DOI] [PubMed] [Google Scholar]
- 55. Lee D. H., Rhee J. G., Lee Y. J. (2009) Br. J. Pharmacol. 157, 1189–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pockros P. J., Schiff E. R., Shiffman M. L., McHutchison J. G., Gish R. G., Afdhal N. H., Makhviladze M., Huyghe M., Hecht D., Oltersdorf T., Shapiro D. A. (2007) Hepatology 46, 324–329 [DOI] [PubMed] [Google Scholar]