Background: Cytoplasmic dynein-dynactin interactions involve IC and p150Glued.
Results: p150Glued binds to a bi-regional IC motif; intervening IC linker residues remain disordered.
Conclusion: p150Glued-bound IC is coiled-coil in region 1, partially disordered helix in region 2, and significantly disordered in the linkers.
Significance: Conserved region 1 is for recognition, variable region 2 is for regulation, and the longer disordered linker is for cargo recognition.
Keywords: Dynein, NMR, Protein Assembly, Protein Complexes, Protein Conformation, Protein Dynamics, Protein-Protein Interactions, Intrinsically Disordered Proteins
Abstract
Cytoplasmic dynein is a 1.2-MDa multisubunit motor protein complex that, together with its activator dynactin, is responsible for the majority of minus end microtubule-based motility. Dynactin targets dynein to specific cellular locations, links dynein to cargo, and increases dynein processivity. These two macromolecular complexes are connected by a direct interaction between dynactin's largest subunit, p150Glued, and dynein intermediate chain (IC) subunit. Here, we demonstrate using NMR spectroscopy and isothermal titration calorimetry that the binding footprint of p150Glued on IC involves two noncontiguous recognition regions, and both are required for full binding affinity. In apo-IC, the helical structure of region 1, the nascent helix of region 2, and the disorder in the rest of the chain are determined from coupling constants, amide-amide sequential NOEs, secondary chemical shifts, and various dynamics measurements. When bound to p150Glued, different patterns of spectral exchange broadening suggest that region 1 forms a coiled-coil and region 2 a packed stable helix, with the intervening residues remaining disordered. In the 150-kDa complex of p150Glued, IC, and two light chains, the noninterface segments remain disordered. The multiregion IC binding interface, the partial disorder of region 2 and its potential for post-translational modification, and the modulation of the length of the longer linker by alternative splicing may provide a basis for elegant and multifaceted regulation of binding between IC and p150Glued. The long disordered linker between the p150Glued binding segments and the dynein light chain consensus sequences could also provide an attractive recognition platform for diverse cargoes.
Introduction
Cytoplasmic dynein, the principal motor responsible for retrograde transport along microtubules, is involved in a variety of critical cellular processes, including orientation and assembly of the mitotic spindle and subsequent chromosome segregation, orientation and transport of organelles, and establishment of cell polarity (reviewed in Ref. 1). Dynein, a large ∼1.2-MDa multisubunit protein complex, is conceptually divided into two major functional domains. The motor domain is comprised of the dynein heavy chains and includes the sites for microtubule binding and the ATPase activity; the cargo attachment domain is comprised of the light intermediate chains, the intermediate chains (ICs),2 and the light chains Tctex1, LC8, and LC7 (2). In Drosophila melanogaster, IC (a 642-amino acid protein) consists of two structurally and functionally distinct subdomains as follows: a predominantly disordered N-terminal region (N-IC), which includes binding sites for the three light chains (3–7), and a C-terminal region, which includes seven WD40 repeats predicted to fold into a compact toroidal β-propeller structure. Molecular genetics analyses show that in D. melanogaster, IC mutations result in larval lethality, demonstrating the essential function of IC in vivo (8).
Commensurate with the numerous functions and activities of dynein within the cell, a multitude of adaptors and regulators has been identified (9), including dynactin, which is required for most types of cytoplasmic dynein activities (10). Dynactin, a heteromultimeric macromolecular complex, first identified through its role in activation of vesicular transport mediated by cytoplasmic dynein (11), also increases the processivity of transport by dynein along microtubules (12, 13). Dynactin's largest subunit, p150Glued, was discovered as a 150-kDa polypeptide copurified with cytoplasmic dynein (14), and it was subsequently determined to have extensive sequence and functional similarity to the D. melanogaster gene product Glued, a polypeptide of similar size (15).
Early in vitro studies show a direct interaction between dynactin p150Glued and dynein N-IC, implying that this interaction mediates mutual recognition between the two macromolecular complexes (11, 12). Subsequent molecular genetics studies in D. melanogaster demonstrate that additional copies of the dynein IC gene suppress the dominant rough eye phenotype of Glued (a mutation in the p150Glued) (16). In addition, overexpression of full-length p150Glued in COS-7 cells disrupts dynein-based motility (17), and conversely, overexpression of small IC fragments that bind p150Glued perturbs endomembrane and microtubule organization in COS-7 and HeLa cells. Both effects are proposed to arise from competitive inhibition (18).
Elucidating the structural basis for dynein-dynactin interactions and how their functional activities are regulated requires characterization of the constituent interacting subunits. N-IC is an intrinsically disordered protein (IDP) (3) and is predicted to form a coiled-coil in its N-terminal ∼31 residues. The term “IDP” collectively refers both to proteins that are completely disordered and to those consisting of a mixture of ordered and disordered residues, with “disordered” structure referring to a flexible ensemble of conformations that are, on average, aperiodic, extended, and not well packed by other protein atoms. IDPs play diverse roles in the promotion of supramolecular assembly and regulation of function in various binding partners and are themselves highly amenable to regulation through post-translational modification. Although the majority of IDPs in complex with binding partners have been structurally characterized in forms resulting from ligand binding-induced disorder-to-order transitions, a new class of IDPs has begun to emerge, in which these proteins do not fully fold even in the ligand-bound state, where they form partially disordered dynamic complexes (Ref 19 and references therein).
In contrast to the predominance of disorder in N-IC, p150Glued is predicted to contain extensive α-helical structures with two major coiled-coil regions (residues 214–567 and 982–1086 in D. melanogaster). Stoichiometric analysis of dynactin indicates two copies of p150Glued per dynactin complex (11), and coupled with images from deep-etch electron microscopy, these observations led to the widely accepted model of a homodimeric coiled-coil structure that constitutes the projecting arm of dynactin (10). Analyses of truncation mutants of both IC and p150Glued in various species have produced a coarse-grained approximation of the regions necessary for their binding interaction (18, 20–22). Although one study (23) suggests that the full-length IC includes a region that can interact with a segment (amino acids 600–811 in rat) of p150Glued, another study demonstrates binding between N-terminal residues of IC (residues 1–106 in Rattus norvegicus IC-2C isoform) and a region of p150Glued corresponding to the first predicted coiled-coil (p150Glued amino acids 214–567 in D. melanogaster) (18). To date, there is no report of a comparison of pre- versus post-complex structures of IC and p150Glued.
Although dynein is coupled with dynactin for many of its activities, dynein and dynactin are not constitutively colocalized in cells, which raises the question of how dynein-dynactin binding is regulated. Here, we report the multiregion recognition motif of the p150Glued binding interface in IC, and demonstrate that, in the assembled state, whether in binary combination with p150Glued or in a biologically relevant 150-kDa subcomplex with p150Glued and light chains, IC remains disordered in regions outside the interfaces with its binding partners. These results explain how alternative splicing and phosphorylation of IC might modulate its interaction with p150Glued.
EXPERIMENTAL PROCEDURES
Protein Preparation
Constructs of the D. melanogaster IC (Fig. 1) were generated by PCRs using the cDNA of the Cdic2b gene (accession number AF 263371.1) as the template. An IC construct corresponding to residues 1–87 (IC:1–87) was generated with an N-terminal hexahistidine (His6) tag and a protease Factor Xa recognition sequence engineered immediately 5′ to the start codon. The PCR fragment was cloned into pCR2.1 TOPO (Invitrogen), followed by subcloning into a pET15d (Novagen) expression vector. For NMR studies, an IC construct containing two sequential LC8-binding sites (hereafter designated IC:1–143LL) was generated by PCR with a synthetic nucleotide template using the sequence LVYTKQTQTT in place of residues 111–120 (described in Ref. 24). The IC:1–143LL, as well as constructs corresponding to residues 1–143 (IC:1–143) and 1–40 (IC:1–40), were cloned into pET SUMO (Invitrogen). All sequences were verified by automated sequencing prior to transformation into Escherichia coli BL21(DE3) host cell lines for protein expression. The p150Glued construct (p150221–509Glued) from D. melanogaster (accession number AAF 49788, Dctn1 gene) was cloned into pET15d expression vector, containing an N-terminal His6 tag.
FIGURE 1.
IC constructs and isoforms. A, sequence alignment between fruit fly and rat IC. Top sequence is D. melanogaster (Dm, AF 263371.1), middle and bottom sequences are R. norvegicus IC-1A (IC-1A, NP 062107.1) and IC-2C (IC-2C, AAA 89165.1) isoforms, respectively. Identical residues (*), conserved residues (:), and semi-conservative substitutions (.) are designated below the aligned sequences. Predicted coiled-coil regions for all sequences are shown in boldface. The binding sites for Tctex1 and LC8 are highlighted in light and dark gray, respectively. Regions 1 and 2 of the p150Glued binding interface are designated above the sequence, as are linker 1 (abbreviated L1) and linker 2. B, IC constructs used in this study. Also highlighted are regions of predicted secondary structure (helices as cylinders and β-sheet as arrow) (44), coiled-coils (45) (double black bars, ICC), regions predicted to be disordered (raised black bars), and binding sites of light chains Tctex1 (light gray) and LC8 (dark gray).
Cells were grown in LB or minimal media at 37 °C to an A600 nm of ∼0.6. Isotopically labeled 15N and 13C,15N proteins for NMR studies were prepared using published protocols (25, 26). Protein expression and purification under native conditions were performed as described previously (27, 28). The His6 tag was cleaved from all IC proteins prepared for NMR and isothermal titration calorimetry (ITC) studies using Factor Xa protease (Novagen), and the SUMO tag was cleaved from all SUMO constructs using SUMO protease (Cornell University). Final purification of all proteins was performed via size-exclusion chromatography on a SuperdexTM 75 (or SuperdexTM 200 in the case of p150221–509Glued) (16:60) gel filtration column (GE Healthcare) with a running buffer of 50 mm sodium phosphate (pH 7.3) with 0.2 m sodium sulfate and 1 mm NaN3.
For NMR experiments, protein concentrations were determined from sequence-based calculated molar extinction coefficients at 280 nm (IC:1–143 and IC:1–143LL, 2980; LC8, 14,565; and p150221–509Glued, 1490 m−1 cm−1). For ITC experiments, protein concentrations were determined using reversed-phase HPLC with detection at 214 nm and calculation of peak intensity; sequence-based calculated molar extinction coefficients at 214 nm are: IC:1–143, 166,840; IC:1–87, 90,667; IC:1–40, 40,296; and p150221–509Glued, 392,888 m−1 cm−1 (29).
NMR Spectroscopy
Protein concentrations were 0.5–1 mm for IC:1–143, IC:1–40, IC:1–87, and IC:1–143LL and up to 2 mm for p150221–509Glued and LC8 in 10 mm sodium phosphate (pH 6.5) with 50 mm NaCl, 1 mm NaN3, 10% 2H2O, a mixture of protease inhibitors (Roche Applied Science), and 1 mm 2,2-dimethyl-2-silapentane-5-sulfonic acid. Chemical shifts were referenced with internal 2,2-dimethyl-2-silapentane-5-sulfonic acid (30). NMR spectra were collected on a 600 MHz Bruker DRX spectrometer. Triple resonance experiments CBCA(CO)NH and HNCACB (31), HNCA (32), and (H)CC(CO)NH (33) were recorded with 1024 (H) and at least 48(C) and 40(N) points. An HNN (34) experiment optimized for disordered proteins was also collected.
The three-dimensional 1H-15N NOESY-HSQC experiment was acquired with a mixing time of 0.20 s for IC:1–40 and IC:1–143 at 5 °C. 3J(HN-Hα) coupling constants for IC:1–40 and IC:1–143 at 5 °C were obtained from a three-dimensional HNHA (35) experiment with a dephasing/rephasing delay of 13.05 ms. T2 relaxation data were determined from experiments (36) recorded for IC:1–143 at 5 and 20 °C and on IC:1–143/p150Glued at 5 °C and IC:1–143LL/p150Glued/LC8 at 20 °C with a 1.7-s recycle delay and relaxation delay times of 15.44, 30.88, 46.32, 61.76, 77.20, 92.64, and 123.52 ms, and with at least one redundant data point to aid estimation of experimental error. Steady-state 1H-15N heteronuclear NOEs (36) were recorded at 20 °C for both apo- and p150Glued-bound IC:1–143, using a 3-s period of saturation and an additional delay of 1.5 s. Hydrogen/hydrogen exchange spectra were collected for apo- and p150Glued-bound IC:1–143 at 5 °C using the CLEANEX-PM-FHSQC pulse sequence (37) with a 20-ms mixing time.
Titration between 15N-labeled IC:1–143 and unlabeled p150221–509Glued was conducted at 5 °C with increasing IC:p150Glued molar ratios up to a 1:2.8 ratio and recording a series of 1H-15N HSQC spectra using echo-anti-echo phase discrimination of 256 increments with 1024 points. The spectrum collected for the 1:2.8 ratio evidenced no significant change from that of the 1:2 ratio, suggesting saturation of binding. 1H-15N TROSY-HSQC spectra were also collected in tandem during titration but did not improve the spectra. Sample quality was monitored with SDS-PAGE before, during, and after the titration process. The quaternary IC:1–143LL·p150Glued·LC8 complex was prepared with molar ratios of 2 (p150Glued) and 4 (LC8) relative to IC:1–143LL, and NMR data were collected at 20 °C.
NMR Data Analysis
NMR spectra were processed with NMRPipe (38) and analyzed using NMRView (39). Sequential assignments for IC:1–143 were based on HNCA, HNCACB, and CBCA(CO)NH spectra using Burrow-Owl (40) with additional analysis of the (H)CC(CO)NH-TOCSY and HNN spectra. Secondary chemical shifts (ΔδCα and ΔδCβ) were calculated relative to random coil shifts corrected for temperature, pH, and for primary sequence (41). For all dynamics experiments, peak intensities were measured as the peak height at the highest point and the associated error taken to be the spectral base-line noise. T2 and NOE values were calculated as described previously (42). For titration experiments, peak intensities were measured as peak volumes. To account for differences in concentration across the titration series, a normalization factor was determined from the peak volume of residue 143, which is presumably not altered by p150Glued binding. Changes in peak intensities Ibound/Ifree were calculated as the ratio between the peak volumes in spectra of the complex and apo-IC:1–143. For CLEANEX-PM data, peak intensities were normalized as the ratio to the peak intensity of residue 137.
Isothermal Titration Calorimetry
IC:1–40, IC:1–87, IC:1–143, and p150221–509Glued were prepared in a buffer containing 50 mm sodium phosphate (pH 7.5) with 50 mm sodium chloride and 0.5 mm NaN3. Binding thermodynamics were determined using a VP-ITC isothermal titration calorimeter (MicroCal, Northampton, MA) at 25 °C with p150Glued in the cell and the IC constructs in the syringe, using cell/syringe concentrations of 23.6 μm (31.7 μm for titration with IC:1–40), 0.40 mm for p150Glued and each IC construct. Data were processed using Origin 7.0 (OriginLab Corp., Northampton, MA). The average of the last 2–4 enthalpies of injection were subtracted from the binding data prior to fitting. The IC:1–87 and IC:1–143 constructs bind p150Glued with a stoichiometry (n) of 0.99 ± 0.02. Data were fit to a single-site binding model, with small deviations from the best fit. Data reported are for experiments performed in duplicates. Error estimates are based upon deviations from the theoretical best fit, with duplicates yielding similar parameter values and associated uncertainties.
Sequence Analysis
Sequence alignment between the D. melanogaster IC isoform 2 (accession number AF 263371.1) and R. norvegicus IC-1A and IC-2C isoforms (accession numbers NP 062107.1 and AAA 89165.1, respectively) was performed with manual adjustment to achieve the alignment in the “serine-rich” region (43). The Jpred 3 (44) and Paircoil2 (45) programs were used to predict secondary structure and coiled-coil propensities, respectively. The DISOPRED (46) program was used for disorder prediction.
RESULTS
Resonance Assignments and Secondary Structure of IC:1–143
D. melanogaster dynein intermediate chain IC:1–143 containing residues 1–143 includes the LC8 (residues 126–138) and Tctex1 (residues 110–122) recognition sequences (Fig. 1). IC:1–143 is predicted to contain two segments of α-helical secondary structure, encompassing residues 3–36 and 49–59 (the second segment corresponding to residues 53–64 and 54–65 in R. norvegicus IC-1A and IC-2C isoforms, respectively), a β-strand in the light chains binding site, and disorder in the remainder of the sequence (Fig. 1B). IC:1–143 also includes a serine-rich region (43), corresponding to residues 71–89 and 97–114 in D. melanogaster and the R. norvegicus IC-1A isoform, respectively. The ICs in both species contain a predicted coiled-coil at the N terminus, spanning residues 2–35 in D. melanogaster and 1–44 and 1–52 in R. norvegicus IC-1A and IC-2C, respectively (Fig. 1A).
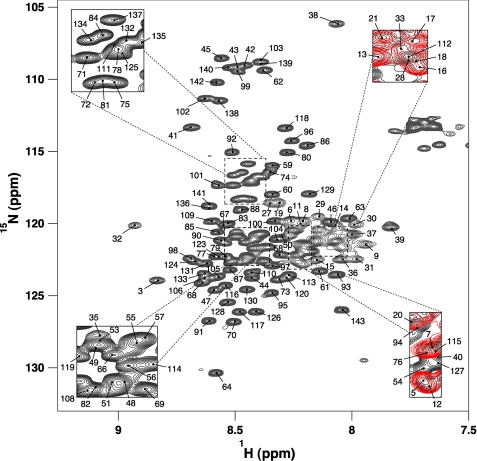
The 1H-15N HSQC spectrum of IC:1–143 at 5 °C (Fig. 2) exhibits a narrow 7.7 to 8.9 ppm amide chemical shift range, suggesting a predominantly disordered or helical structure. Because of the high sequence redundancy, a “divide-and-conquer” approach to chemical shift assignments was necessary, whereby smaller (and sequentially overlapping) constructs, IC:1–40 and IC:1–87, were assigned to identify and verify residue assignments in IC:1–143. Assignments were further aided by comparison with chemical shifts for IC:84–143 collected at 20 °C (42). Backbone amide assignments at 5 °C were completed for 131 of the 138 nonproline residues.
FIGURE 2.
Resonance assignments of IC:1–143. Two-dimensional 1H-15N HSQC spectrum of 0.6 mm 15N IC:1–143 in 10 mm sodium phosphate buffer (pH 6.5), 50 mm NaCl, 1 mm NaN3 recorded at 5 °C. Peaks are labeled according to their backbone resonance assignments. Several peaks corresponding to the N-terminal 40 residues are indicated; the rest are demonstrated through an overlay of the IC:1–40 HSQC (red) on that of IC:1–143 (black).
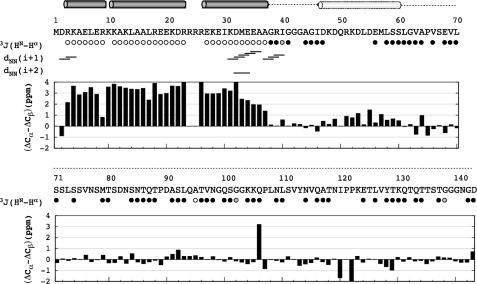
Structural characterization of IC:1–143 by NMR spectroscopy included determination of 3J(HN-Hα) scalar coupling constants, analysis of sequential amide-amide NOEs, and measurement of secondary chemical shifts (Fig. 3). Excluded residues correspond to overlapped peaks or to peaks too weak for reliable measurement. The majority of residues included in the analysis exhibit 3J-coupling values in the 6–8 Hz range, indicative of disordered conformations; however, there also exist contiguous stretches of residues (3–9, 11–23, and 27–37) near the N terminus with 3J scalar coupling values of less than 5 Hz, indicative of helical structure. Further suggestive of helical structure in this region is the presence of several dNN(i + 1) NOEs spanning residues 2–4, 31–36, 37–40, and dNN(i + 2) NOEs between residues 32 and 34 (Fig. 3). Several additional dNN NOEs were present but not unambiguously assigned due to the extreme spectral overlap. No amide-amide NOEs were observed for resolved peaks outside of residues 1–40.
FIGURE 3.
Secondary structure of IC:1–143. 3J-coupling constants are grouped as <5 Hz (open circles) for helical structure, between 6 and 8 Hz (filled black circles) for random coil, and >8 Hz (filled gray circles). Sequential amide-amide connectivities are shown short and medium range NOEs. Secondary chemical shifts for Cα and Cβ atoms are presented in bar plot format below the inter-residue NOE connectivities. The secondary structure estimated from 3J-coupling values and secondary chemical shifts is plotted above the sequence and includes the following: helices (solid cylinders), nascent helix (outlined cylinder), and random coil conformation (dotted line).
Secondary chemical shifts, sensitive indicators of protein secondary structure, were calculated for the 13Cα and 13Cβ nuclei from the difference between the observed and sequence-corrected random coil chemical shifts (41). Their difference (ΔCα–ΔCβ) is presented for all assigned resonances (Fig. 3). The majority of residues show only small deviations from zero, suggestive of random coil conformations. A large positive difference, indicative of helical conformations (47), was observed in stretches spanning residues 3–23 and 26–37 (the apparent punctuation arises from a lack of chemical shift assignments for residues Arg-24 and Arg-25). An additional stretch of lower average positive values was observed for residues 47–60. The predominance of disordered segments with regions of helicity localized to the N terminus matches the sequence-based structure prediction (Fig. 1) and is consistent with the CD-detected (27) structural content.
Dynamics of IC:1–143
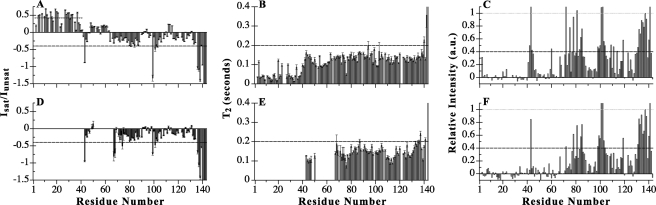
Significant deviation from random coil behavior in the nanosecond-picosecond time scale is reflected in the 1H-15N steady-state heteronuclear NOEs recorded at 20 °C (Fig. 4A); this temperature affords the best contrast in dynamic behavior between different regions of the protein. As expected for a primarily disordered protein, the majority of residues exhibit negative heteronuclear NOE values with larger negative values at the C terminus. Region 1–41, however, has an average heteronuclear NOE value of 0.42, whereas region 48–61 has an average value of 0.16 (Fig. 4A). The sign and magnitude of the NOE values within these two regions are indicative of some ordered structure.
FIGURE 4.
Dynamics of 15N-labeled IC:1–143 in its apo bound (A–C) and p150Glued-bound states (D–F). Steady-state 1H-15N heteronuclear NOE values shown as Isat/Iunsat for 15N IC:1–143 in its apo bound (A) and p150221–509Glued-bound (D) states at pH 6.5 and 20 °C. Average steady-state heteronuclear NOE values calculated for the assigned N-terminal 41 residues and for residues 48–61 are plotted as dotted lines for those regions (A). NOE values lower than negative 1.5 corresponding to the C-terminal residues were truncated in these plots. T2 relaxation rates and CLEANEX-PM relative cross-peak intensities were measured at pH 6.5 and 5 °C for apo bound (B and C) and p150221–509Glued-bound 15N IC:1–143 (E and F), respectively. T2 values greater than 0.4 s corresponding to the C termini were truncated in these plots (B and E). CLEANEX-PM peak intensities (C and F) were recorded with a 20-ms mixing time. Dashed lines are drawn through the plots (A–F) to facilitate visual comparison.
Backbone dynamics, also inferred from T2 experiments collected at 5 °C (Fig. 4B), allow motions on the microsecond-millisecond time scale to be probed. Observed T2 values range from 0.013 to 0.57 s with an overall average value of 0.114 s. The broad distribution of T2 values is indicative of nonrandom structure in intermediate exchange on the NMR time scale, particularly so for residues evincing the lowest T2 values, such as residues 1–40 (average T2 value of 0.046 s). The lower T2 values in this region are consistent with the increased peak broadness and nebulosity apparent for residues 1–40 in relation to the other peaks (Fig. 2 and right insets therein). Overall broader peaks are observed for NMR spectra of the IC:1–40 construct alone (data not shown), indicating that exchange broadening is characteristic of this region, independent of the size of the protein. Further information on the conformational ensemble of IC:1–143 was garnered from amide hydrogen/hydrogen exchange experiments (Fig. 4C). For a 20-ms mixing time, numerous residues in IC:1–143 exhibit strong peak intensities, indicating efficient exchange with the solvent and a lack of ordered structure on this time scale. There are also numerous residues with peaks apparently missing from the CLEANEX-PM spectrum, including residues 4–40 and 48–58, 61–66, and 69–70, indicating a relatively more protected local environment within these regions.
Mapping the IC Binding Interface with p150Glued
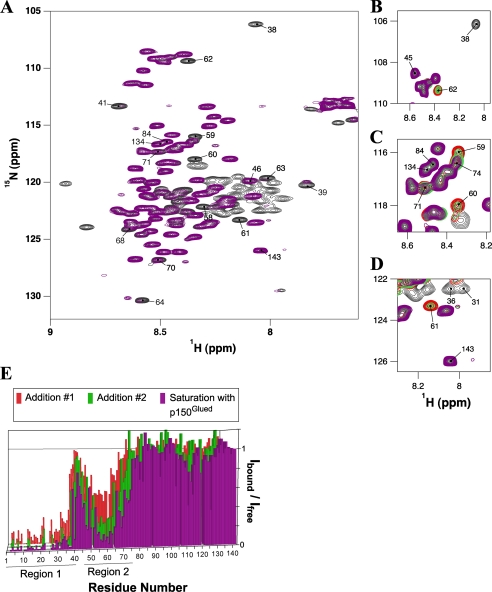
To identify the IC:1–143 interface with p150Glued, an NMR titration experiment was performed wherein 1H-15N HSQC spectra of 15N-labeled IC:1–143 were recorded with stepwise addition of unlabeled p150221–509Glued (Fig. 5). A superimposition of 1H-15N HSQC spectra of 15N-labeled IC:1–143, collected in the absence and presence of a saturating amount of unlabeled p150221–509Glued (Fig. 5A), reveals no novel peaks for IC:1–143 residues in its p150Glued-bound conformation. Rather than moving in chemical shift space, numerous peaks for IC:1–143 were significantly attenuated, presumably due to intermediate chemical exchange effected by the binding association with p150Glued, leading to significant peak broadening and apparent disappearance of the peaks from the spectrum. Of particular interest is the pattern of peak disappearances over the course of the titration. The first set of peaks to disappear are for residues 1–41 (black peaks in Fig. 5, B–D), followed by a gradual disappearance of peaks for residues 46–75 with increasing p150Glued concentrations (Fig. 5, B–D, e.g. red, green, and attenuated purple peaks). Collectively, the 1–41 peaks (region 1, Fig. 5E) are the most strongly attenuated (an average of greater than 90%). Overall, the 46–75 peaks (region 2, Fig. 5E) are attenuated by an average of 72%, with the 48–66 peaks more strongly attenuated (80% on average) than the remaining peaks (residues 67–75, 57% on average), suggesting that binding behavior is different for these two segments of region 2.
FIGURE 5.
Titration of 15N-labeled IC:1–143 with p150221–509Glued. A, overlay of 1H-15N HSQC spectra of IC:1–143 (black) and p150221–509Glued bound IC:1–143 (purple). Several peaks are labeled whose relative peak volumes are attenuated by more than 70% in the bound state; peaks corresponding to residues 84, 134, and 143 (undiminished relative peak volumes) are also indicated for comparison. The spectra were recorded at pH 6.5 and 5 °C with ∼0.6 mm 15N IC:1–143 in 10 mm sodium phosphate buffer and greater than 2-fold excess of p150221–509Glued in the case of the fully bound state (purple). Portions of HSQC spectra (B–D) show selected sets of peaks during the titration process. Peaks in these overlaid spectra are colored corresponding to 15N IC:1–143 with 0 eq of p150221–509Glued (black), two subsequent additions of p150221–509Glued (red and green, in that order), and greater than 2 eq (purple) of p150221–509Glued. E, numerical plot of relative integrated peak intensity (Ibound/Ifree) versus residue number. Relative peak intensity is defined as the ratio of the integrated peak volume in the spectrum of the complex to the integrated peak volume in the spectrum of the free IC:1–143 protein. Two distinct series of peaks (labeled region 1and region 2) in IC:1–143 exhibited significant attenuation upon interaction with p150221–509Glued.
Dynamics of IC:1–143 Bound to p150Glued
To determine the effect of p150Glued binding on the dynamics of IC:1–143, the same NMR dynamics experiments collected for the apo-state were performed for the bound state (Fig. 4, D--F). Peak disappearance essentially rendered all residues of region 1 and a major portion of region 2 spectroscopically invisible, thus preventing determination of their dynamic properties in the bound state. In 1H-15N steady-state heteronuclear NOE and T2 spectra, peaks are visible for residues 43–47, 50, 67–69, 71–75, and the span of residues 76–143. Peaks for residues 67–69 and 71–75 (the latter part of region 2) are somewhat weaker but of sufficient intensity for dynamics characterization. For residues 76–143, the NOE and T2 values are essentially unchanged from their values in the unbound state, indicating little change in the degree of their disorder. For residues 43–47 (linking region 1 and region 2), as well as residues 67–69 and 71–75, the NOE and T2 values are also largely unchanged and are indicative of conformational disorder on the picosecond-nanosecond time scale in the bound complex. For a 20-ms mixing time, CLEANEX-PM experiments show that the majority of residues exhibiting strong, visible peak intensities in the apo-state do so for the bound state as well. Notable exceptions include the following: residues 41–47 (excluding residue 43), which roughly correspond to termini of regions 1 and 2; residues 59, 60, 68, and 72–75, all included within region 2; and residues 78–81, which are located just C-terminal to region 2. The diminished intensities are indicative of slower exchange with the solvent, suggesting that the termini and residues surrounding the binding interface become more ordered in the bound state.
Thermodynamics of p150Glued-IC Interactions
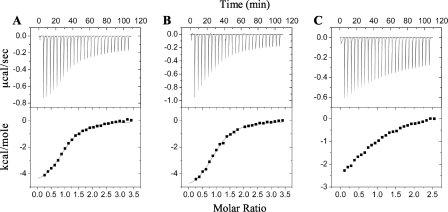
Isothermal titration calorimetry measurements confirm the newly determined multiregion binding footprint of p150Glued upon IC. Data were collected for titration of p150221–509Glued with three IC constructs (Fig. 1) as follows: IC:1–143, containing both regions of the p150Glued recognition motif determined here, as well as the smallest previously reported (18) p150Glued-binding domain of IC (residues 1–106 of isoform IC-2C in rat corresponding to residues 1–97 in D. melanogaster); IC:1–87, containing both regions of the recognition motif for p150Glued; and IC:1–40, containing only region 1 of the p150Glued binding footprint. Representative data from each titration are shown in Fig. 6. p150221–509Glued binds to both IC:1–143 and IC:1–87 with moderate affinity and similar Kd values of 3.5 ± 0.3 μm (with ΔH0 = −5.0 ± 0.2 kcal/mol) and 3.6 ± 0.3 μm (with ΔH0 = −5.5 ± 0.2 kcal/mol), respectively. The titration between IC:1–40 and p150221–509Glued does not reach saturation, and the data were not fit. The weaker binding affinity for IC:1–40, inferred from the failure to reach saturation, suggests that the first 40 residues of IC are involved in only a fraction of the interactions with p150Glued, consistent with the smaller binding enthalpies of the initial injections (Fig. 6, lower panels). The initial normalized enthalpies of injection for titrations of p150221–509Glued with IC:1–87 and IC:1–143 are similar, although the value for IC:1–40 is approximately half that.
FIGURE 6.
Representative ITC data for IC constructs binding to p150Glued. Isothermal titration calorimetry data with thermograms (top panels) and isotherms (bottom panels) shown for titration of p150221–509Glued with IC:1–143 (A), IC:1–87 (B), and IC:1–40 (C). Data were collected at 25 °C in 50 mm sodium phosphate, 50 mm sodium chloride (pH 7.5). For the data collected with titrants IC:1–143 (A) and IC:1–87 (B), solid lines correspond to the nonlinear least squares fit for an A + B AB binding model.
Dynamics of IC:1–143 Bound to p150Glued and Light Chains
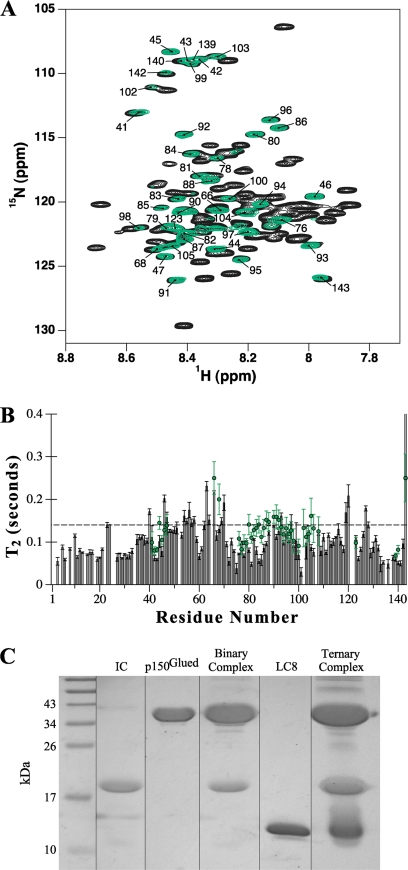
To investigate the dynamics of IC:1–143 in a more biologically relevant context, we conducted NMR studies on IC:1–143 bound to p150221–509Glued and to two light chains. To simplify NMR spectra, we used a construct of IC:1–143 in which the Tctex1 binding region is replaced with an LC8 recognition sequence (e.g. IC:1–143LL). IC:1–143LL binds 2 eq of LC8 with higher affinity than IC:1–143 binds Tctex1 and LC8, and the structure of IC at the light chains interface is similar in both cases (24). Noticeably, peaks corresponding to residues in the LC8-binding sites (residues 108–120 and 124–138) are absent from the spectrum of the quaternary complex (Fig. 7A) as observed with titration of IC:84–143 with Tctex1 and LC8 (42). As in the case of the binary complex between IC:1–143 and p150221–509Glued, a similar disappearance of peaks due to binding with p150Glued also occurs in the spectra of the quaternary complex. Although peaks corresponding to IC residues in the binding interfaces with p150Glued and the light chains are absent, numerous peaks remain with similar intensity as in the apo-form (Fig. 7A). The mere presence of these intense peaks in the spectrum of a 150-kilodalton complex suggests that the segment connecting the p150Glued-binding site to the LC8 sites retains a considerable degree of conformational flexibility. The T2 values of the remaining peaks (Fig. 7B) are largely unchanged in the quaternary complex, indicating that residues that are not directly at the binding interfaces with p150Glued and the light chains remain disordered on the nanosecond-picosecond time scale.
FIGURE 7.
Dynamics of 15N-labeled IC:1–143 in a quaternary complex with p150Glued and light chains. A, overlay of 1H-15N HSQC spectra of apo-IC:1–143 (black) and IC:1–143LL in complex with excess p150221–509Glued and LC8 (green); spectra were recorded at pH 6.5 and 20 °C in 10 mm sodium phosphate buffer. Labeled peaks correspond to those whose relative peak volumes remain at 60% or greater intensity for IC:1–143LL in the quaternary complex. B, T2 relaxation times collected at 20 °C for apo-IC:1–143 (bars) and the remaining visible peaks of IC:1–143LL in complex with excess p150Glued and LC8 (green circles). A dashed line is drawn through the plot to facilitate visual comparison. C, strips from SDS-polyacrylamide gel analysis of components and of the assembled IC quaternary complex used for NMR experiments; IC:1–143LL (lane 2), p150221–509Glued (lane 3), a binary complex of IC:1–143LL saturated with p150221–509Glued (lane 4), LC8 (lane 5), and a quaternary complex of IC:1–143LL combined with saturating amounts of both p150221–509Glued and LC8 (lane 6).
DISCUSSION
In our ongoing efforts to characterize the structure, function, and regulation of the cargo-attachment domain of dynein, we have mapped the shortest IC segment necessary for binding dynactin p150Glued to IC residues 1–75 (1–101 in rat DIC1A). Mapping is based on NMR analyses, which also reveal three important structural features of IC residues involved in dynein-dynactin binding. First, the binding footprint on IC is multiregional, i.e. it involves two noncontiguous IC recognition sequences. Second, the IC regions bound to p150Glued are helical in nature; the more N-terminal region 1 is likely coiled-coil as inferred from spectral exchange broadening and sequence-based structure prediction of the apoprotein. Third, the intervening residues between IC regions interacting with dynactin remain disordered in the complex. Furthermore, the IC linker residues connecting the p150Glued binding regions and the light chains recognition sequences remain disordered in the quaternary complex. We propose that the multiregion binding footprint of p150Glued on IC, along with the interplay of order and disorder in the resulting complex, function in regulation of dynein-dynactin interactions and in cargo recognition.
Residual Structure in IC:1–143
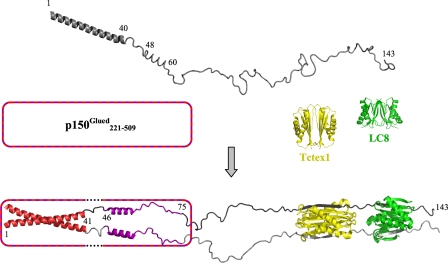
NMR experiments show that predominantly disordered apo-IC:1–143 contains two segments of α-helical structure, namely residues 1–40 and 48–60, largely in agreement with sequence-based prediction (Fig. 1B). Stretches of helical structure are inferred from 3J-coupling constants, dNN sequential NOEs, and deviation of 13Cα and 13Cβ secondary chemical shifts from random coil values. These structure-based analyses are further bolstered by complementary dynamics measurements (1H-15N steady-state heteronuclear NOE, T2, and CLEANEX-PM) that indicate greater order in the regions of inferred α-helical structure relative to the rest of the protein. Although structural and dynamics data from NMR suggest a well formed helix for residues 1–40, residues 48–60 apparently form a nascent helix as indicated by the small positive values of secondary chemical shift differences and of steady-state heteronuclear NOEs. A model depicting these features of IC in its unbound state is presented in Fig. 8, top.
FIGURE 8.
Model for assembly of dynein intermediate chain with p150Glued and light chains. The model depicts apo-IC:1–143 (top) as a primarily disordered and monomeric ensemble of conformations with one defined helical region encompassing the N-terminal 40 residues and another region of nascent helicity encompassing residues ∼48–60, as determined from NMR measurements. Segments of IC for which NMR spectral characteristics are altered by p150Glued binding are indicated in red (region 1, residues 1–41) and purple (region 2, residues 46–75). The helical nature of regions 1 and 2 in bound IC (bottom) is inferred as described in the text, whereas all other regions in the bound complex are directly measured by NMR spectroscopy or x-ray crystallography. In accordance with sequence-based prediction of coiled-coil conformations in both the N-terminal α-helical region of IC and the p150Glued construct used in this study, the N-terminal 40 residues of IC in the bound state are depicted in a generic coiled-coil configuration. The p150Glued·IC complex is a tetramer that could be formed either from two p150Glued/IC heterodimeric coiled-coils (i.e. one chain from IC and one chain of p150Glued) or from two homodimeric coiled-coils (i.e. two chains of IC/IC coiled-coil packed against two chains of p150Glued coiled-coil). The p150Glued construct is represented as a box with a striped border. In the IC-bound state, dotted lines in the box indicate that p150Glued does not interact with IC residues 42–45 (linker 1). Residues in linker 1 and linker 2 (residues 76–106) are disordered, as indicated by NMR (Fig. 7). The IC·Tctex1·LC8 subcomplex portion of the model is from a crystal structure (Protein Data Bank entry 3FM7) (24), with Tctex1 (yellow) and LC8 dimers (green), and the corresponding IC segments in gray (one subunit light gray and the other dark gray). All structures, including apo-Tctex1 (Protein Data Bank entry 1YGT) and apo-LC8 (Protein Data Bank entry 3BRI) were generated using PyMOL (51).
Multiregion Binding Footprint for p150221–509Glued on IC:1–143
NMR titration of 15N-labeled IC:1–143 with unlabeled p150Glued identifies a new binding motif, namely a two-region binding footprint involving IC residues 1–41 (region 1) and residues 46–75 (region 2), with no binding to the intervening linker residues (Fig. 8, bottom). Intriguingly, both regions correspond to or contain the α-helical structures identified in apo-IC:1–143, although their structure in the p150Glued-bound state cannot be directly ascertained through NMR spectroscopy as the corresponding peaks essentially disappear from the spectra. ITC analysis demonstrates that both regions are required for full binding affinity. The requirement of both regions for significant IC-p150Glued interaction explains the results of Vaughan and Vallee (20), in which binding was observed between a fragment containing the first 123 residues of rat IC-1A and p150150–811Glued, whereas binding was not observed between p150Glued and a fragment of rat IC-1A containing only the first 61 residues (lacking region 2) nor with a truncation mutant of IC in which the first 61 residues had been deleted (lacking region 1).
Structure and Dynamics of Assembled IC
Our interpretive model for IC in its assembled state with p150Glued and light chains is based upon a combination of x-ray crystallography data, structural and dynamic information from NMR spectroscopy, as well as sequence-based prediction of structural propensity. Both helices of unbound IC (Fig. 8, top) are included in the multiregion binding footprint of p150Glued, with the first consisting of the entirety of region 1 whose associated peaks are first to disappear during NMR titration. The significant peak broadening upon binding to p150Glued is attributed to multiple exchange processes possibly including intermediate exchange between bound and free states of IC, as well as exchange between helical and coiled-coil conformations in an oligomerization process that results in a tetrameric p150Glued·IC complex. Evidence from NMR spectroscopy (analysis of identical HSQC spectra for increasingly dilute samples of IC:1–40) as well as analytical ultracentrifugation studies on IC:1–289 (3) indicate a predominantly monomeric state for apo-IC. The proposed coiled-coil conformation for region 1 in the bound state derives from the complex exchange processes and sequence-based prediction of a coiled-coil in both IC (residues 1–35, Fig. 1B) and the p150Glued construct used in this study; the coiled-coil assemblage depicted in the model could represent either an IC/IC coiled-coil packed on p150Glued/p150Glued coiled-coil (the latter not shown in the model) or two p150Glued/IC coiled-coils, each formed from one chain of IC and one chain of p150Glued (only one coiled-coil is shown in the model).
The second nascent helix in apo-IC is contained within the p150Glued recognition motif in region 2, with associated peaks diminishing more gradually and exhibiting less attenuation of intensity than those of region 1. The less dramatic disappearance of region 2 peaks is attributed to less complex exchange broadening processes, likely chemical exchange between the free and p150Glued-bound states of IC as well as structural fluctuation between nascent and fully formed helix within IC. Thus, the nascent helical structure depicted in the model for apo-IC is proposed to persist and perhaps stabilize in the bound state. For residues ∼67–75 of region 2, less attenuation of peak intensity suggests even simpler exchange processes, likely between free and bound IC with minimal structural changes. The retained disorder in this part of region 2 in bound p150Glued-IC is confirmed by dynamics experiments. The attribution of peak disappearance primarily to exchange broadening (rather than to the larger size and increased rotational correlation time of the complex with p150Glued) is supported by the observation that titration of IC with 2H-labeled p150221–509Glued (data not shown) does not improve spectra of p150Glued-bound IC.
Between regions 1 and 2 of the p150Glued-binding motif on IC is linker 1, a short intervening segment (residues 42–45) that remains disordered in the bound state. It is possible that linker 1 turns to allow region 2 to pack against region 1, a structure that is an alternative to the model presented in Fig. 8. In this scenario, the disappearance of peaks in region 2 could be attributed to its packing against region 1 and not to direct interaction with p150Glued; our data cannot differentiate between the two possibilities. Linker 2 (residues 76–107), connecting the C-terminal end of the p150Glued recognition sequences with the N-terminal end of the light chain binding domain, also remains disordered in assembled IC. IC residues at the light chain binding interfaces are absent from spectra of the quaternary complex (Fig. 7A) and so cannot be probed in this study, but the structure of IC bound to the light chains is known to assume β-strands (24, 48, 49), depicted accordingly in the model (Fig. 8).
Functional Implications of Disorder in Assembled IC
Assembled IC is a dynamic complex, integrating both order and disorder, with IC remaining largely disordered and flexible in regions outside the segments that form an interface with any of the binding partners. At the IC-p150Glued interface, a multiregion motif suggests distinct regional functions with three-dimensionally packed and conserved region 1 primarily for recognition and partially disordered and variable region 2 primarily for regulation by post-translational modification and/or alternative splicing (Fig. 1A). In this vein, phosphorylation of residues in the serine-rich cluster at the C-terminal end of region 2 (22) has been proposed as a potential regulator of IC activity, including its interaction with p150Glued (20, 43). Partial disorder in region 2 would allow access for regulatory chemical modifications of the serine-rich zone.
The disorder in linker 2 may present accessible binding interfaces for other IC-binding partners, possibly including NudC/CL (50), NudE/EL, Spindly, or zw10 (9). Linker 2 is also subject to alternative splicing (Fig. 1A), which produces variability in linker length that could modulate IC binding affinity for any of these binding partners. Furthermore, the IC-p150Glued interaction may be enhanced by light chains binding through the “multivalency effect” (24). Association with a bivalent ligand such as Tctex1 or LC8 essentially increases the local effective concentrations of proximate IC regions, with the extent of binding enhancement for another ligand such as p150Glued depending upon the length of linker 2; a shorter linker is expected to result in greater binding enhancement than a longer one. The potential modulation of IC-p150Glued interaction by processes related to IC linker 2 disorder is consistent with the emerging theme of assembled IC as a flexible polybivalent scaffold whose affinities for its multitude of binding partners are finely tuned to provide versatility and reversibility in response to changing conditions in the cellular environment (24, 27, 42).
In summary we propose that a multiregion IC binding interface and disorder of the linkers between binding interfaces together provide a basis for elegant and multifaceted regulation of binding between IC and p150Glued, and thus of the association of dynein with one of its key regulators. The linker disorder retained throughout assembled polybivalent IC may also maintain accessibility of segments involved in cargo recognition.
Acknowledgments
We thank Professor Clare Woodward for critical reading of the manuscript and Dr. Gregory C. Benison for assistance with NMR experiment setup.
This work was supported by National Science Foundation Grant MCB 0818896.
- IC
- 74-kDa dynein intermediate chain corresponding to gene Cdic2b
- N-IC
- IC residues 1–289
- HSQC
- heteronuclear single-quantum coherence
- ITC
- isothermal titration calorimetry
- IDP
- intrinsically disordered protein
- SUMO
- small ubiquitin-related modifier.
REFERENCES
- 1. Vallee R. B., Williams J. C., Varma D., Barnhart L. E. (2004) J. Neurobiol. 58, 189–200 [DOI] [PubMed] [Google Scholar]
- 2. Pfister K. K., Shah P. R., Hummerich H., Russ A., Cotton J., Annuar A. A., King S. M., Fisher E. M. (2006) PloS Genet. 2, 11–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Makokha M., Hare M., Li M., Hays T., Barbar E. (2002) Biochemistry 41, 4302–4311 [DOI] [PubMed] [Google Scholar]
- 4. Lo K. W., Naisbitt S., Fan J. S., Sheng M., Zhang M. J. (2001) J. Biol. Chem. 276, 14059–14066 [DOI] [PubMed] [Google Scholar]
- 5. Mok Y. K., Lo K. W., Zhang M. J. (2001) J. Biol. Chem. 276, 14067–14074 [DOI] [PubMed] [Google Scholar]
- 6. Susalka S. J., Nikulina K., Salata M. W., Vaughan P. S., King S. M., Vaughan K. T., Pfister K. K. (2002) J. Biol. Chem. 277, 32939–32946 [DOI] [PubMed] [Google Scholar]
- 7. Hall J., Song Y., Karplus P. A., Barbar E. (2010) J. Biol. Chem. 285, 22566–22575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boylan K. L., Hays T. S. (2002) Genetics 162, 1211–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kardon J. R., Vale R. D. (2009) Nat. Rev. Mol. Cell Biol. 10, 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schroer T. A. (2004) Annu. Rev. Cell Dev. Biol. 20, 759–779 [DOI] [PubMed] [Google Scholar]
- 11. Gill S. R., Schroer T. A., Szilak I., Steuer E. R., Sheetz M. P., Cleveland D. W. (1991) J. Cell Biol. 115, 1639–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kardon J. R., Reck-Peterson S. L., Vale R. D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. King S. J., Schroer T. A. (2000) Nat. Cell Biol. 2, 20–24 [DOI] [PubMed] [Google Scholar]
- 14. Collins C. A., Vallee R. B. (1989) Cell Motil. Cytoskeleton 14, 491–500 [DOI] [PubMed] [Google Scholar]
- 15. Holzbaur E. L., Hammarback J. A., Paschal B. M., Kravit N. G., Pfister K. K., Vallee R. B. (1991) Nature 351, 579–583 [DOI] [PubMed] [Google Scholar]
- 16. Boylan K., Serr M., Hays T. (2000) Mol. Biol. Cell 11, 3791–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Quintyne N. J., Gill S. R., Eckley D. M., Crego C. L., Compton D. A., Schroer T. A. (1999) J. Cell Biol. 147, 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. King S. J., Brown C. L., Maier K. C., Quintyne N. J., Schroer T. A. (2003) Mol. Biol. Cell 14, 5089–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uversky V. N. (2011) Chem. Soc. Rev. 40, 1623–1634 [DOI] [PubMed] [Google Scholar]
- 20. Vaughan K. T., Vallee R. B. (1995) J. Cell Biol. 131, 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karki S., Holzbaur E. L. (1995) J. Biol. Chem. 270, 28806–28811 [DOI] [PubMed] [Google Scholar]
- 22. Vaughan P. S, Leszyk J. D., Vaughan K. T. (2001) J. Biol. Chem. 276, 26171–26179 [DOI] [PubMed] [Google Scholar]
- 23. Vaughan P. S., Miura P., Henderson M., Byrne B., Vaughan K. T. (2002) J. Cell Biol. 158, 305–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hall J., Karplus P. A., Barbar E. (2009) J. Biol. Chem. 284, 33115–33121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nyarko A., Barbar E. (2011) J. Biol. Chem. 286, 1556–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Makokha M., Huang Y. J., Montelione G., Edison A. S., Barbar E. (2004) Protein Sci. 13, 727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nyarko A., Hare M., Hays T. S., Barbar E. (2004) Biochemistry 43, 15595–15603 [DOI] [PubMed] [Google Scholar]
- 28. Barbar E., Kleinman B., Imhoff D., Li M., Hays T. S., Hare M. (2001) Biochemistry 40, 1596–1605 [DOI] [PubMed] [Google Scholar]
- 29. Kuipers B. J., Gruppen H. (2007) J. Agric. Food Chem. 55, 5445–5451 [DOI] [PubMed] [Google Scholar]
- 30. Markley J., Bax A., Arata Y., Hilbers C., Kaptein R., Sykes B., Wright P., Wuthrich K. (1998) Pure Appl. Chem. 70, 117–142 [Google Scholar]
- 31. Grzesiek S., Bax A. (1992) J. Magn. Reson. 96, 432–440 [Google Scholar]
- 32. Kay L., Ikura M., Tschudin R., Bax A. (1990) J. Magn. Reson. 89, 496–514 [DOI] [PubMed] [Google Scholar]
- 33. Grzesiek S., Anglister J., Bax A. (1993) J. Magn. Reson. B 101, 114–119 [Google Scholar]
- 34. Panchal S. C., Bhavesh N. S., Hosur R. V. (2001) J. Biomol. NMR 20, 135–147 [DOI] [PubMed] [Google Scholar]
- 35. Vuister G., Bax A. (1993) J. Am. Chem. Soc. 115, 7772–7777 [Google Scholar]
- 36. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 37. Hwang T. L., van Zijl P. C., Mori S. (1998) J. Biomol. NMR 11, 221–226 [DOI] [PubMed] [Google Scholar]
- 38. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 39. Johnson B. A. (2004) Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 40. Benison G., Berkholz D. S., Barbar E. (2007) J. Magn. Reson. 189, 173–181 [DOI] [PubMed] [Google Scholar]
- 41. Kjaergaard M., Brander S., Poulsen F. M. (2011) J. Biomol. NMR 49, 139–149 [DOI] [PubMed] [Google Scholar]
- 42. Benison G., Nyarko A., Barbar E. (2006) J. Mol. Biol. 362, 1082–1093 [DOI] [PubMed] [Google Scholar]
- 43. Nurminsky D. I., Nurminskaya M. V., Benevolenskaya E. V., Shevelyov Y. Y., Hartl D. L., Gvozdev V. A. (1998) Mol. Cell. Biol. 18, 6816–6825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cole C., Barber J. D., Barton G. J. (2008) Nucleic Acids Res. 36, W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McDonnell A. V., Jiang T., Keating A. E., Berger B. (2006) Bioinformatics 22, 356–358 [DOI] [PubMed] [Google Scholar]
- 46. Ward J. J., McGuffin L. J., Bryson K., Buxton B. F., Jones D. T. (2004) Bioinformatics 20, 2138–2139 [DOI] [PubMed] [Google Scholar]
- 47. Wishart D. S., Sykes B. D., Richards F. M. (1991) J. Mol. Biol. 222, 311–333 [DOI] [PubMed] [Google Scholar]
- 48. Williams J. C., Roulhac P. L., Roy A. G., Vallee R. B., Fitzgerald M. C., Hendrickson W. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10028–10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Benison G., Karplus P. A., Barbar E. (2007) J. Mol. Biol. 371, 457–468 [DOI] [PubMed] [Google Scholar]
- 50. Zhou T., Zimmerman W., Liu X., Erikson R. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9039–9044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]