Abstract
Silkworm hemolymph inhibits hemolysin production by Staphylococcus aureus. We purified a factor in the silkworm hemolymph responsible for this inhibitory activity. The final fraction with the greatest specific activity contained 220- and 74-kDa proteins. Determination of the N-terminal amino acid sequence revealed that the 220- and 74-kDa proteins were apolipophorin I and apolipophorin II, respectively, indicating that the factor was apolipophorin (ApoLp). The purified ApoLp fraction showed decreased expression of S. aureus hla encoding α-hemolysin, hlb encoding β-hemolysin, saeRS, and RNAIII, which activate the expression of these hemolysin genes. Injection of an anti-ApoLp antibody into the hemolymph increased the sensitivity of silkworms to the lethal effect of S. aureus. Hog gastric mucin, a mammalian homologue of ApoLp, decreased the expression of S. aureus hla and hlb. These findings suggest that ApoLp in the silkworm hemolymph inhibits S. aureus virulence and contributes to defense against S. aureus infection and that its activity is conserved in mammalian mucin.
Keywords: Apolipoproteins, Insect, Insect Immunity, Staphylococcus aureus, Toxins, Virulence Factors
Introduction
Studies of the interactions between bacteria and host animals at the molecular level are important for clarifying the pathogenetic mechanisms of infectious diseases. Host animals recognize bacterial components by Toll-like receptors and activate innate immune reactions. The innate immune system is conserved between mammalian and invertebrate animals. On the other hand, pathogenic bacteria recognize the host microenvironment by a two-component system and activate the expression of virulence genes.
Staphylococcus aureus is a pathogenic bacterium against humans. It exists on the skin, nasal cavity, and mucosa of 30% of healthy individuals. S. aureus causes various diseases, including toxic shock, necrotizing pneumonia, endocarditis, and impetigo. S. aureus produces various virulence factors such as adhesive factors, exotoxins, and immune disturbance factors. The expression of these virulence factors is regulated by a number of transcription factors, including SarA (1), Rot (2), SarZ (3), and the DNA-binding proteins of two-component systems (4). SaeRS, a two-component system, is required for the expression of exotoxins, including hemolysins, and is required for S. aureus virulence in mice (5). Expression of saeRS is activated by hydrogen peroxide, which kills bacteria in the phagosomes of macrophages, and an antimicrobial peptide, α-defensin (6–8). S. aureus secretes autoinducing peptide, which is encoded by the agrD gene in the agr locus and senses the amount of extracellular autoinducing peptide using the sensor protein AgrC, resulting in activation of the transcription of RNAIII from the P3 promoter (9). RNAIII regulates the expression of virulence genes according to S. aureus cell density (9, 10). Recently, Gresham and co-workers (11, 12) revealed that apolipoprotein B in mammalian blood and peroxides that are produced by macrophages inactivate the S. aureus quorum-sensing molecule autoinducing peptide and suppress S. aureus virulence. Invertebrate hemolymph contains antimicrobial peptides that inhibit bacterial growth (13, 14), although the factors that inhibit the bacterial gene expression necessary for virulence have not yet been identified.
We previously established an S. aureus infection model using silkworms and examined the interaction between host animal and pathogenic bacteria (15–22). Silkworms are larvae of the moth Bombyx mori, a lepidopteran species. S. aureus hemolysin kills silkworms (23), although deletion mutants of hemolysin genes of S. aureus do not show attenuated virulence against silkworms.2 These results led us to hypothesize that there is a factor in silkworm hemolymph that suppresses S. aureus hemolysin production. In the present study, we purified a factor that inhibited S. aureus production of hemolysin. The factor was apolipophorin (ApoLp),3 a lipid-carrying protein in the silkworm hemolymph. Furthermore, ApoLp inhibited the expression of the virulence regulatory genes saeRS and RNAIII and contributed to the defense systems of silkworms against S. aureus infection. The results serve as an example of a common defense system that suppresses bacterial virulence in both invertebrates and vertebrates.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
S. aureus strains were aerobically cultured in tryptic soy broth at 37 °C, and 12.5 μg of chloramphenicol/ml or 100 μg of kanamycin/ml was added to the medium if required. The JM109 strain of Escherichia coli was used as a host for pND50, pND50K, and their derivatives. E. coli strains transformed with the plasmids were cultured in Luria-Bertani broth containing 50 μg/ml kanamycin or 12.5 μg/ml chloramphenicol. Details of the bacterial strains and plasmids used in this study are shown in Table 1.
TABLE 1.
List of bacterial strains and plasmids used
| Strain or plasmid | Genotypes or characteristicsa | Source or Ref. |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | NCTC8325-4, restriction mutant | 30 |
| NCTC8325-4 | NCTC8325 cured of ϕ11, ϕ12, and ϕ13 | 40 |
| CK1844 | NCTC8325-4 Δagr::tetM (transduction from RN6911) | 20 |
| M1007-1 | NCTC8325-4 hla::pT1007, hlb::pT1811R; Cmr, Kmr | b |
| E. coli | ||
| JM109 | General purpose host strain for cloning | Takara Bio |
| Plasmids | ||
| pND50 | E. coli-S. aureus shuttle vector; Cmr | 41 |
| pluc | pND50 with luc+ with a ribosomal binding site | 20 |
| pluc-hla | pluc with hla promoter from RN4220 | 20 |
| pluc-sae P1 | pluc with sae P1 from NCTC8325-4 | This study |
| pluc-sae P3 | pluc with sae P3 from NCTC8325-4 | This study |
| pluc-sarA | pluc with sarA promoter from NCTC8325-4 | This study |
| pND50K | E. coli-S. aureus shuttle vector; Kmr | 42 |
| plucK | pND50K with luc+with a ribosomal binding site | This study |
| plucK-spa | plucK with spa promoter from RN4220 | This study |
| plucK-agr P2 | plucK with agr P2 from RN4220 | This study |
| plucK-agr P3 | plucK with agr P3 from RN4220 | This study |
a Cm, chloramphenicol; Km, kanamycin.
b S. Miyazaki, Y. Matsumoto, K. Sekimizu, and C. Kaito, unpublished data.
Measurement of Inhibitory Activity against S. aureus Hemolysin Production
An overnight culture of S. aureus NCTC8325-4 was inoculated into a 100-fold amount of fresh tryptic soy broth and cultured until the culture reached an A600 of 0.1–0.2. An 800-μl aliquot of the cultures was supplemented with 100 μl of silkworm hemolymph or protein solution obtained from each purification step and cultured at 37 °C for 3 h. The cultures were centrifuged, and hemolytic activity of the culture supernatant was measured. Two-fold serially diluted culture supernatants were mixed with an equal volume of 5% sheep erythrocytes and incubated at 37 °C for 1 h and at 4 °C overnight. Hemolytic activity was defined as the reciprocal of the dilution of supernatants that yielded 50% erythrocyte lysis. Hemolytic activities of the NCTC8325-4 strain, the hla-disrupted mutant, the hlb-disrupted mutant, and the hla/hlb-double disrupted mutant were 60, 40, 5, and 2.5 units, respectively, indicating that both α-hemolysin and β-hemolysin were observed in this assay.
Purification of S. aureus Hemolysin Inhibitor Produced from Silkworm Hemolymph
All procedures were performed at 4 °C. Hemolymph was collected from fifth instar larvae and supplemented with 1 mm N-phenylthiourea. The hemolymph was centrifuged at 8000 × g for 10 min at 4 °C, and the supernatant was stored at −80 °C and used in all experiments as silkworm hemolymph. The proteins from 50 ml of hemolymph were precipitated in 70% ammonium sulfate at 4 °C and centrifuged at 8000 × g for 30 min. The precipitate was dissolved and dialyzed in buffer A (50 mm MES (pH 6.2), 200 mm NaCl, 2 mm DTT, 5% glycerol). The sample was applied to a phosphocellulose column (bed volume, 47 ml). The proteins were eluted with a linear salt gradient (0.2–0.6 m NaCl). Fractions with inhibitory activity were pooled and dialyzed against 5 liters of buffer B (50 mm MES (pH 6.2), 100 mm NaCl, 2 mm DTT, 5% glycerol) followed by centrifugation at 8000 × g for 30 min to remove the insoluble materials. The supernatant was applied to a Mono S column (HR5/5; bed volume, 1 ml; GE Healthcare) pre-equilibrated with buffer C (50 mm MES (pH 6.2), 150 mm NaCl, 2 mm DTT, 5% glycerol). The proteins were eluted with a linear salt gradient (0.15–0.6 m NaCl) in a total volume of 30 ml using a fast protein liquid chromatography system. A 200-μl aliquot of the pooled fractions was applied to a SuperdexTM 200 (HR10/30; GE Healthcare) column pre-equilibrated with buffer A. The flow rate was 0.5 ml/min, and 0.5 ml was collected in each fraction. Gel filtration chromatography was calibrated using ferritin (395 kDa) and aldolase (191 kDa). Protein concentration was determined using the Bradford assay for all purification steps.
Determination of Amino Acid Sequence
The purified protein was subjected to SDS-PAGE and transferred to a PVDF membrane, which was stained with Coomassie Brilliant Blue, and the stained bands were excised. N-terminal sequencing was performed by Edman degradation (PPSQ-33A; Shimadzu Biotech).
Determination of cDNA Sequence of ApoLp by Rapid Amplification of 5′ cDNA End
We sequenced expressed sequence tag clones (24) of fbpv0376 and fbpv0284 containing bp 1720–2223 and 3973–4603 of apoLp-II/I, respectively. We obtained the bp 537–9954 DNA sequence of a hypothetical apoLp-II/I gene that was predicted by the Gene model of Scaffold Build2 (Kaikobase). To obtain a cDNA sequence encoding bp 1–536 of apoLp-II/I, we performed rapid amplification of 5′ cDNA ends. Total RNA was extracted from the fat body of fifth instar silkworms (Fu·Yo × Tsukuba·Ne) using the Qiagen RNeasy Mini kit, ligated with RNA oligonucleotides (5′-CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA-3′),and used for templates of reverse transcription using random primers (GeneRacerTM kit, Invitrogen). The ligated products were used as templates for PCR using GeneRacer 5′ primer and gene-specific primer (Table 2). The PCR products were cloned into vector plasmids and sequenced. Based on these sequences, we constructed five oligonucleotide primer pairs (Table 2) to amplify a cDNA encompassing bp 1–9954 of apoLp-II/I. Total RNA from the silkworm fat body was reverse transcribed into cDNA using a random hexamer and Superscript III reverse transcriptase (Invitrogen). Five DNA fragments encompassing bp 1–9954 of apoLp-II/I were amplified by PCR using five oligonucleotide primer pairs and cDNA as a template and sequenced using an ABI 3100 genetic analyzer.
TABLE 2.
Primers used in this study
RACE, rapid amplification of cDNA ends; F, forward; R, reverse.
| Target | Sequence (5′–3′) | Ref. |
|---|---|---|
| agrP2 | ||
| F | CGCGGTACCTTAAACAACTCATCAACTATT | 20 |
| R | GCGTCTAGAAAACTGGTCAATTTTATTATC | |
| agrP3 | ||
| F | CGCTCTAGATTAAACAACTCATCAACTATT | 20 |
| R | GCGGGTACCATGTCATTATACGATTTAGTA | |
| hlapromoter | ||
| F | CGCGGTACCTCCCGACGAAATTCCAAACAT | 20 |
| R | GCGTCTAGAACGATTTGAGGAAACAATAAT | |
| spapromoter | ||
| F | CGCGGTACCAGCACATTCAAAGCC | 20 |
| R | GCGTCTAGATGTATGTATTTGTAAAGTCAT | |
| sarApromoter | ||
| F | GAAGAATTCCTCATATGGGTGCAGCATT | This study |
| R | GGAGGATCCTGTCAGCATAAGTGACCATTGA | |
| saeP1 | ||
| F | GAAGAATTCTTATTGTGGCAAAAGGTT | 6 |
| R | GGTGGTACCTACCTTGATCTTGTGAAT | |
| saeP3 | ||
| F | GAAGAATTCACTGTTGAAGGTAAAGCTG | 6 |
| R | GGTGGTACCTCTGTTCTTACGACC | |
| RNAIII | ||
| F | TGGATTATCGACACAGTGAAC | 42 |
| R | CATGGTTATTAAGTTGGGATG | |
| 16 S rRNA | ||
| F | CAACGCGAAGAACCTTACCAA | 42 |
| R | GCGGGACTTAACCCAACATCT | |
| hla | ||
| F | GGTGCAAATGTTTCGATTGG | 42 |
| R | CGAAGTCTGGTGAAAACCCTG | |
| hlb | ||
| F | GCGGTTGTGGATTCGATAAT | This study |
| R | CAGCACCACAACGTGAATCT | |
| sarS | ||
| F | CGAGAGAAAATTGCAGAACGT | This study |
| R | TGTGATTCACTTTGATCTGCA | |
| saeQ | ||
| F | GAAAAATTAACGGGCGGATT | This study |
| R | ATTGCAATCTCTCCGAGTGG | |
| saeS | ||
| F | CGGCCATATGACACTAACTTTG | This study |
| R | TGCTTGCGTAATTTCCGTTA | |
| 5′-RACE | ||
| F | GGACACUGACAUGGACUGAAGGAGUA | Invitrogen |
| R | CGGCTGCGGTGCACAATAACTGCTC | This study |
| apoLp-II | ||
| apoLp-II-f | GGAATTCCATATGGGGACAATTAGTTTTAGTCTAAGTG | This study |
| apoLp-II-r | CCGCTCGAGTCTACGTCCTCTCGTTAGTCCATCTTC | |
| apoLp-I | ||
| apoLp-I-p1-f | CTAGCTAGCTCTGTCAAGACGGAAATTGATTC | This study |
| apoLp-I-p1-r | CCGCTCGAGTTCAATAAGTTTGATCTCCCG | |
| apoLp-I-p2-f | CTAGCTAGCGAACCCGTGTTTAATGCAAAC | This study |
| apoLp-I-p2-r | CCGCTCGAGATTGTCGGAGACCTCGTCAC | |
| apoLp-I-p3-f | CTAGCTAGCCTCGGTGATAGGAGCTACGC | This study |
| apoLp-I-p3-r | CCGCTCGAGCACAAGCTTCGTGCAAATC | |
| apoLp-I-p4f | CTAGCTAGCATCTGGCCGAAGCGATCCAC | This study |
| apoLp-I-p4r | CCGCTCGAGCTACTTCCGCCTGCGCGAAG | |
Preparation of Anti-ApoLp Antibody
A Japanese White rabbit was intracutaneously injected with 250 μg of ApoLp proteins with an equal volume of Freund's adjuvant four times at 2-week intervals. Immunoglobulins were purified from 10 ml of serum from the immunized rabbit using a protein G column (Prosep-G, Millipore).
Reporter Assay
The RN4220 strain was transformed with reporter plasmids (Table 1). Plasmids were transferred from the RN4220 strain to the NCTC8325-4 strain by phage transduction using phage 80α. An 800-μl aliquot of S. aureus culture (A600 = 0.2) was supplemented with 100 μl of protein solution and cultured at 37 °C for 3 h. S. aureus cells were collected by centrifugation and lysed in a buffer (20 mm KH2PO4 (pH 7.8), 0.04% Triton X-100, 0.1 mm DTT, 10 μg/ml lysostaphin, one tablet of protease inhibitor (Complete, Roche Applied science)/50 ml). Cell lysate supernatant (100 μl) was incubated with an equal volume of luciferase substrate (Roche Applied Science), and luminescence was measured using a luminometer (Berthold Technologies, Bad Wildbad, Germany). The promoter activity was calculated as luminescence units per milligram of protein.
Quantitative Real Time PCR Analysis
An 800-μl aliquot of S. aureus culture (A600 = 0.2) was supplemented with 100 μl of protein solution and cultured at 37 °C for 4.5 h. S. aureus cells were collected by centrifugation, treated with RNAprotect Bacteria Reagent (Qiagen, Gaithersburg, MD), and lysed in a buffer (10 mm Tris-HCl (pH 8.0), 1 mm EDTA, 1 mg/ml lysostaphin). RNA was extracted using an RNeasy Mini kit (Qiagen). RNA was reverse transcribed to cDNA using Multiscribe Reverse Transcriptase (Applied Biosystems, Foster City, CA). Quantitative real time PCR was performed using cDNA as a template, FastStart Universal SYBR Green Master (Roche Applied Science), and primers (Table 2). The signals were detected using a StepOnePlusTM Real-Time PCR System (Applied Biosystems). The reaction mixture was incubated at 95 °C for 10 min and then for 40 cycles (95 °C, 15 s; 60 °C, 1 min). The data were normalized to 16 S rRNA.
Silkworm Infection Experiment
Fertilized eggs of B. mori were purchased from Ehime Sansyu (Ehime, Japan), and hatched larvae were raised to fifth instar larvae using an artificial diet in our laboratory (16). Fifth instar larvae were injected with S. aureus NCTC8325-4 cells or hla/hlb mutant cells mixed with anti-ApoLp immunoglobulins or control immunoglobulins. Silkworm survival was monitored. Statistical analyses of the survival curves were performed using one-sided rank log tests (Prism software package, GraphPad Software, San Diego, CA). LD50 values were determined by logistic regression analysis. Statistical analyses of the LD50 values were performed using likelihood ratio tests for logistic regression.
RESULTS
Silkworm Hemolymph Inhibits Hemolysin Production by S. aureus
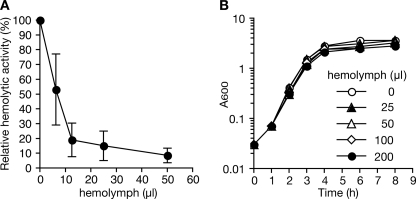
We examined whether silkworm hemolymph inhibits the production of hemolysin by S. aureus. Addition of the hemolymph to S. aureus culture medium decreased hemolysin production by S. aureus in a dose-dependent manner (Fig. 1A). In contrast, the hemolymph did not inhibit S. aureus growth (Fig. 1B). In addition, the hemolymph did not inhibit the activity of S. aureus hemolysins in the culture supernatant (data not shown). These results suggest that the silkworm hemolymph inhibits S. aureus hemolysin production without disturbing S. aureus growth.
FIGURE 1.
Silkworm hemolymph inhibits S. aureus hemolysin production. A, silkworm hemolymph was added to S. aureus NCTC8325-4 culture at an A600 of 0.1 and further incubated for 3 h. Hemolytic activity in the culture supernatant was measured using sheep erythrocytes. The horizontal axis indicates the amount of hemolymph added to the culture. The vertical axis indicates the relative hemolytic activity relative to control (no hemolymph). Data are presented as means ± S.D. from three independent experiments. B, growth curves of S. aureus cultured with various doses of silkworm hemolymph. Error bars indicate ± S.D.
Purification of Factor in Hemolymph of Silkworms That Inhibits Hemolysin Production by S. aureus
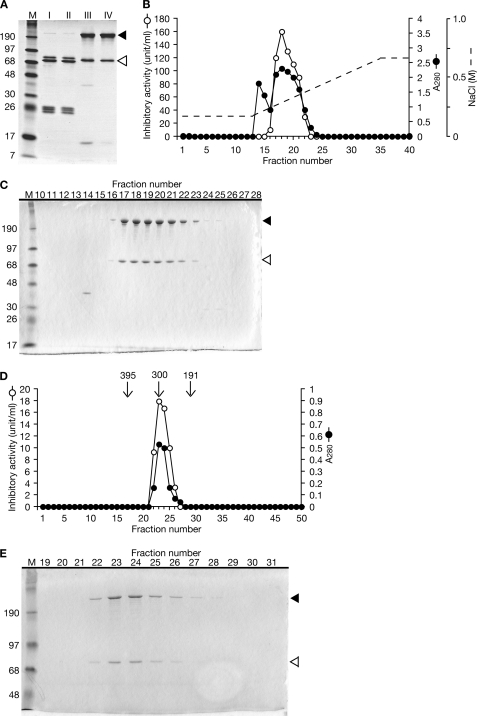
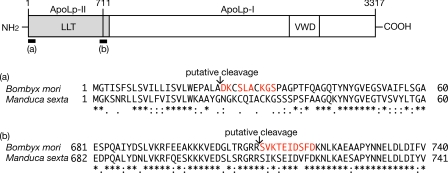
We defined the activity that decreases S. aureus hemolysin production by 50% as 1 unit and purified the inhibitory factor by monitoring the increase in specific activity. We obtained 90 ml of hemolymph from 210 fifth instar larvae of B. mori, and the proteins were precipitated by ammonium sulfate followed by fractionation using a phosphocellulose column and a Mono S column. The specific activity of the final fraction was 28-fold higher than that of the initial fraction. The recovery of activity was 59% (Table 3). Analysis by SDS-PAGE revealed that the final fraction (Fraction IV) contained 220- and 74-kDa proteins (Fig. 2A). The final step of Mono S chromatography revealed that the inhibitory activity against hemolysin production by S. aureus co-migrated with the 220- and 74-kDa proteins (Fig. 2, B and C). To confirm that these proteins were responsible for the inhibition of hemolysin production by S. aureus, we further fractionated the final fraction (Fraction IV) by gel column chromatography and confirmed that the inhibitory activity against S. aureus hemolysin production co-migrated with these proteins (Fig. 2, D and E). Using marker proteins, the molecular mass of native protein was calculated to be 300 kDa (Fig. 2D). These findings suggest that the inhibitory molecule against hemolysin production by S. aureus is a protein complex that comprises the 220- and 74-kDa proteins. The N-terminal amino acid sequences of the 220-kDa protein and the 74-kDa protein were matched with the sequences of ApoLp-I and ApoLp-II, respectively (Fig. 3). Insect ApoLp-I and ApoLp-II form a protein complex that is involved in transporting neutral lipids (25–27). Therefore, we concluded that ApoLp is the inhibitory factor against hemolysin production by S. aureus in the silkworm hemolymph. The concentration of ApoLp that effectively decreased S. aureus hemolysin production by 50% was calculated to be 17 μg/ml (60 nm) based on the specific activity (Table 3). The concentration of ApoLp in the silkworm hemolymph was calculated to be 1.4 mg/ml (4.8 μm) (Table 3), which is consistent with the amount of 2.0 mg/ml (6.9 μm) used in a previous report (28) and enough to decrease S. aureus hemolysin production.
TABLE 3.
Purification of inhibitor against S. aureus hemolysin production from silkworm hemolymph
| Fraction | Protein | Total activity | Specific activity | Yield | Purification |
|---|---|---|---|---|---|
| mg | units | units/mg protein | % | -fold | |
| I. Hemolymph | 3500 | 8500 | 2.4 | 100 | 1 |
| II. Ammonium sulfate | 3300 | 7900 | 2.4 | 93 | 1 |
| III. Phosphocellulose | 92 | 6000 | 65 | 71 | 27 |
| IV. Mono S | 75 | 5000 | 66 | 59 | 28 |
FIGURE 2.
Purification of factor in silkworm hemolymph that inhibits S. aureus hemolysin production. A, a 1-μg aliquot of proteins from each purification step was electrophoresed in a 12% SDS-polyacrylamide gel and stained with Coomassie Brilliant Blue. The black arrowhead indicates the 220-kDa band, and the white arrowhead indicates the 74-kDa band. Lane M, marker proteins. B, elution profile of ion exchange column chromatography (Mono S). Open circles indicate the activity that inhibits S. aureus hemolysin production. Closed circles indicate A280. The dotted line indicates the NaCl gradient. Fractions 16–22 were pooled as Fraction IV and further analyzed by gel filtration chromatography. C, SDS-PAGE analysis of Fractions 10–28 of Mono S column chromatography. The black arrowhead indicates the 220-kDa band, and the white arrowhead indicates the 74-kDa band. D, elution profile of gel filtration column chromatography. Open circles indicate the activity that inhibits S. aureus hemolysin production. Closed circles indicate A280. Arrows indicate the fractions that were eluted with marker proteins (ferritin, 395 kDa; aldolase, 191 kDa). Fraction 23 was estimated to be 300 kDa from the calibration curve based on the marker proteins. E, SDS-PAGE analysis of Fractions 19–31 of gel filtration column chromatography. The black arrowhead indicates the 220-kDa band, and the white arrowhead indicates the 74-kDa band.
FIGURE 3.
Determination of N-terminal amino acid sequence of inhibitory proteins against S. aureus hemolysin production. Because the amino acid sequence of ApoLp-II/I of B. mori was not identified in previous studies (43), we determined that the nucleotide sequence of cDNA encoding ApoLp-II/I (GenBankTM accession number AB640623). B, mori ApoLp-II (710 amino acids) and ApoLp-I (2607 amino acids) have 70.0 and 59.5% identity with that of Manduca sexta (GenBank accession number U57651 (44)), respectively. Sixty N-terminal residues of B. mori ApoLp-II and ApoLp-I were aligned with those of M. sexta by ClustalW (a and b). Identical residues are marked by an asterisk and similar residues are marked by dots below the alignment. Of 10 N-terminal amino acid residues of the 74-kDa protein in Fraction IV, eight (colored in red in a) were determined and matched with the internal sequence of B. mori ApoLp-II. Ten N-terminal amino acid residues of the 220-kDa protein in Fraction IV (colored in red in b) were matched with the internal sequence of B. mori ApoLp-I. LLT, large lipid transfer domain; VWD, von Willebrand factor module D domain (45).
ApoLp Decreases Expression of saeRS and RNAIII in S. aureus
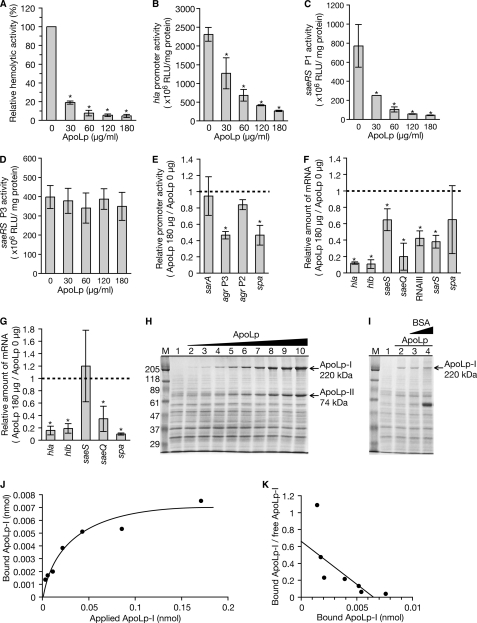
To elucidate the molecular mechanism by which ApoLp inhibits hemolysin production by S. aureus, we tested whether ApoLp decreases the expression of hla, which encodes α-hemolysin, and hlb, which encodes β-hemolysin. Addition of ApoLp to the S. aureus culture medium decreased hemolysin production by S. aureus (Fig. 4A) and decreased the promoter activity of the hla gene (Fig. 4B). Furthermore, addition of ApoLp to the S. aureus culture medium decreased the amount of hla mRNA and hlb mRNA (Fig. 4F). Therefore, ApoLp decreased the expression of the S. aureus hla and hlb genes.
FIGURE 4.
ApoLp decreases expression of S. aureus hla, saeRS, and RNAIII. A, various doses of ApoLp were added to S. aureus NCTC8325-4 culture and further cultured at 37 °C for 3 h. Hemolytic activity of the culture supernatant was measured using sheep erythrocytes. Data shown are means ± S.D. from three independent experiments. B, C, and D, S. aureus NCTC8325-4 strains transformed with reporter plasmids carrying the hla promoter (B) and saeRS promoters (P1, C; P3, D) were cultured in the presence of various doses of ApoLp at 37 °C for 3 h. Luciferase activities of the cell lysates were measured. Means ± S.D. of three independent experiments are shown. Asterisks indicate Student's t test p values of less than 0.05 between the presence and absence of ApoLp. E, S. aureus NCTC8325-4 strains that were transformed with reporter plasmids carrying promoters of spa, sarA, agr P2, and agr P3 were cultured in the presence or absence of 180 μg/ml ApoLp. Luciferase activities of the cell lysates were measured. Data are presented as the relative values against the luciferase activity in the absence of ApoLp. Means ± S.D. of three independent experiments are shown. Asterisks indicate Student's t test p values of less than 0.05 between the presence and absence of ApoLp. F, total RNA was extracted from S. aureus NCTC8325-4 cells cultured in the presence or absence of 180 μg/ml ApoLp. The amounts of hla mRNA, hlb mRNA, saeS mRNA, saeQ mRNA, RNAIII, and sarS mRNA were measured by quantitative RT-PCR. The horizontal axis shows the relative value against the amount of RNAs in the absence of ApoLp. Means ± S.D. of three independent experiments are shown. Asterisks indicate Student's t test p values of less than 0.05 between the presence and absence of ApoLp. G, total RNA was extracted from the agr deletion mutant cells of the NCTC8325-4 strain cultured in the presence or absence of 180 μg/ml ApoLp. RNA amounts of hla, hlb, saeS, saeQ, and spa were measured by quantitative RT-PCR. The horizontal axis shows the relative value against the amount of RNAs in the absence of ApoLp. Means ± S.D. of three independent experiments are shown. Asterisks indicate Student's t test p values of less than 0.05 between the presence and absence of ApoLp. H, S. aureus NCTC8325-4 cells (7.5 × 108 cfu) were incubated with an increasing amount of ApoLp in 100 μl of tryptic soy broth at 25 °C for 1 h. The samples were centrifuged, and the pellets were analyzed by SDS-PAGE. Lane 1, absence of ApoLp; lane 2, 0.67 pmol of ApoLp; lane 3, 1.3 pmol of ApoLp; lane 4, 2.7 pmol of ApoLp; lane 5, 5.3 pmol of ApoLp; lane 6, 11 pmol of ApoLp; lane 7, 21 pmol of ApoLp; lane 8, 43 pmol of ApoLp; lane 9, 85 pmol of ApoLp; lane 10, 170 pmol (37.5 μg) of ApoLp. Lane M, marker proteins. The data are representative of two independent experiments. I, S. aureus NCTC8325-4 cells (7.5 × 108 cfu) were incubated with 5 pmol of ApoLp in the presence of an increasing amount of BSA in 100 μl of tryptic soy broth at 25 °C for 1 h. The samples were centrifuged, and the pellets were analyzed by SDS-PAGE. Lane 1, absence of ApoLp and BSA; lane 2, 5 pmol of ApoLp and absence of BSA; lane 3, 5 pmol of ApoLp and 170 pmol of BSA; lane 4, 5 pmol of ApoLp and 1.7 nmol of BSA. The data are representative of two independent experiments. J and K, the amount of ApoLp-I in the S. aureus pellet in H was measured and plotted on a linear plot (J) and a Scatchard plot (K). The band intensity of ApoLp-I in lanes 2 and 3 in H was beyond the calibration curve of densitometric scanning and was thus removed from the graphs. RLU, relative luciferase units.
We then examined whether ApoLp decreases the expression of genes that regulate the expression of hla and hlb. The two-component system saeRS (29); a quorum-sensing system, agr (30); and a transcription factor, sarA (31), positively regulate the expression of hla and hlb. A polycistronic transcript, saeP-saeQ-saeR-saeS, is transcribed from the P1 promoter of saeRS, whereas saeQ-saeR-saeS is transcribed from the P3 promoter (32, 33). The saeQ has a role in the saeRS function (33). The addition of ApoLp did not alter the P3 promoter activity of saeRS (Fig. 4D), whereas it decreased the P1 promoter activity of saeRS to less than one-tenth (Fig. 4C). Furthermore, the addition of ApoLp decreased the amount of saeQ and saeS mRNAs (Fig. 4F). In addition, ApoLp deceased the P3 promoter activity of the agr locus and decreased the amount of RNAIII transcribed from the P3 promoter (Fig. 4, E and F). In contrast, ApoLp did not decrease the P2 promoter activity of agr or the sarA promoter activity (Fig. 4E). These findings suggest that ApoLp decreases the expression of genes that positively regulate the expression of hla and hlb.
To determine whether ApoLp affects the expression of other virulence genes, we examined the expression of the spa gene encoding protein A. The addition of ApoLp decreased spa promoter activity (Fig. 4E), although it did not significantly decrease the amount of spa mRNA (Fig. 4F). The sarS gene positively regulates the expression of spa (34). The addition of ApoLp decreased the amount of the sarS mRNA (Fig. 4F). Therefore, ApoLp might decrease the promoter activity of spa by suppressing sarS.
To verify whether the effects of ApoLp on virulence genes require the expression of the agr locus, we examined the effect of ApoLp against an agr deletion mutant of S. aureus. ApoLp decreased the expression of hla, hlb, spa, and saeQ genes in the agr deletion mutant, although it did not decrease the expression of saeS (Fig. 4G). Therefore, ApoLp decreases the expression of hla, hlb, spa, and saeQ genes via an agr-independent pathway.
To clarify the mechanism by which ApoLp decreases the expression of various virulence genes in S. aureus, we tested whether ApoLp binds S. aureus cells. ApoLp was mixed with S. aureus cells, and the amount of ApoLp in the S. aureus pellet was measured. The amount of ApoLp in the S. aureus pellet increased with an increase in the amount of ApoLp applied to S. aureus cells (Fig. 4, H and J). The amount of ApoLp in the S. aureus pellet was not affected by the presence of a 34- or 340-fold amount of BSA (Fig. 4I). Scatchard plot analysis revealed that the dissociation constant (Kd) of ApoLp with S. aureus cells was 130 nm (Fig. 4K). These results suggest that ApoLp specifically binds to surface molecules of S. aureus and decreases the expression of virulence genes in S. aureus.
ApoLp Contributes to Silkworm Resistance to Lethal Effects of S. aureus
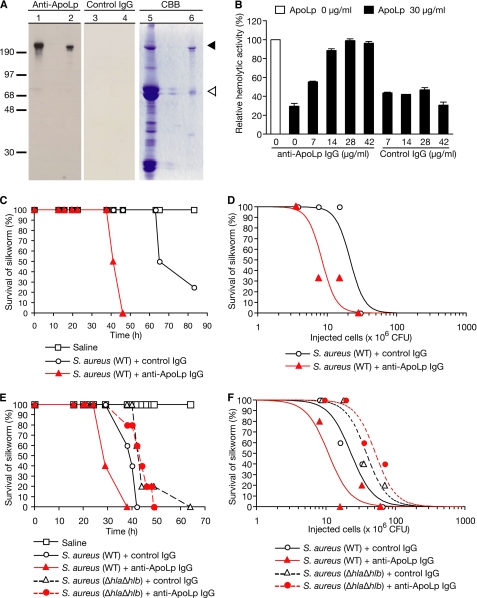
To determine whether ApoLp contributes to silkworm resistance against the lethal effect of S. aureus, we evaluated the impact of injecting anti-ApoLp antibodies into the silkworm hemolymph on the killing effect by S. aureus. Anti-ApoLp antibodies were prepared by affinity purification of IgG from the serum of a rabbit immunized with ApoLp. Western blot analysis using the resulting anti-ApoLp revealed the presence of the 220-kDa subunit of ApoLp (ApoLp-I; Fig. 5A). In contrast, IgG from non-immunized rabbit (control IgG) did not react with the protein (Fig. 5A), indicating that the 220-kDa protein detected by the antibody was ApoLp-I. We then tested whether the anti-ApoLp antibody inhibits the activity of ApoLp, which inhibits hemolysin production by S. aureus. The addition of the anti-ApoLp antibody blocked the inhibitory effect of ApoLp against hemolysin production by S. aureus (Fig. 5B). In contrast, control IgG did not block the inhibitory effect of ApoLp against hemolysin production by S. aureus (Fig. 5B). Therefore, the anti-ApoLp antibody inactivated ApoLp, which inhibited hemolysin production by S. aureus. We then examined whether administration of the anti-ApoLp antibody to silkworms increases the susceptibility to S. aureus infection. Silkworms injected with anti-ApoLp IgG died faster than silkworms injected with control IgG after injection of S. aureus (Fig. 5C). In addition, the LD50, the bacterial cell number that causes the death of half of the silkworms, was decreased in silkworms injected with anti-ApoLp IgG compared with silkworms that were injected with control IgG (Fig. 5D). These results suggest that ApoLp contributes to the silkworm resistance to S. aureus infection. To further verify whether the defensive role of ApoLp against S. aureus virulence in silkworms is due to the inhibition of S. aureus hemolysin production, we examined whether administration of anti-ApoLp IgG to silkworms increases the susceptibility to the hla/hlb-double disrupted mutant of S. aureus. Silkworms injected with anti-ApoLp IgG showed survival curves similar to those of silkworms injected with control IgG after injection of the hla/hlb-double disrupted mutant of S. aureus (Fig. 5E). In contrast, anti-ApoLp IgG-injected silkworms died faster than control IgG-injected silkworms after injection of wild-type S. aureus (Fig. 5E). Furthermore, the LD50 value was not decreased in silkworms injected with anti-ApoLp IgG and the hla/hlb mutant compared with silkworms that were injected with control IgG and the hla/hlb mutant (Fig. 5F). The LD50 was decreased in silkworms injected with anti-ApoLp IgG and wild-type S. aureus compared with silkworms that were injected with control IgG and wild-type S. aureus (Fig. 5F). Therefore, the defensive effect of ApoLp against the lethal effect of S. aureus is due to inhibition of the expression of hla and hlb genes in S. aureus.
FIGURE 5.
Administration of anti-ApoLp increases sensitivity of silkworms against S. aureus. A, detection of ApoLp by Western blot analysis using anti-ApoLp IgG. The left panel shows the membrane detected with anti-ApoLp IgG. The middle panel shows the membrane detected with control IgG. The right panel shows the gel stained with Coomassie Brilliant Blue (CBB). Fifty micrograms of hemolymph proteins of silkworms were electrophoresed in lanes 1, 3, and 5. One microgram of Fraction III (Table 3) was electrophoresed in lanes 2, 4, and 6. B, effect of anti-ApoLp antibodies against the activity of ApoLp, which inhibits S. aureus hemolysin production. Thirty micrograms of ApoLp were mixed with various doses of anti-ApoLp antibodies or control IgG, added to S. aureus culture, and further cultured at 37 °C for 3 h. Hemolytic activity of the culture supernatants was measured. The vertical axis shows the relative hemolytic activity against the value in the absence of ApoLp and antibodies. C, infection experiments in silkworms administered anti-ApoLp antibodies. Silkworms (four larvae per group) were injected with 2.1 × 106 cfu of S. aureus NCTC8325-4 (WT) and 200 μg of anti-ApoLp IgG or control IgG. Silkworm survival was monitored after the injection. The one-sided rank log test p value between the survival curves of anti-ApoLp IgG-injected silkworms and control IgG-injected silkworms is 0.0047. D, dose-response relationship between various doses of S. aureus (WT) and silkworms administered 200 μg of anti-ApoLp IgG or control IgG (three larvae per group). Survival was measured at 40 h after the injection. To determine LD50 values, logistic regression analysis was used to fit smooth curves to the survival proportions. The p value determined using likelihood ratio tests between the LD50 of anti-ApoLp IgG-injected silkworms and that of control IgG-injected silkworms is 0.0122. E, silkworms (five larvae per group) were injected with 6.5 × 107 cfu of S. aureus NCTC8325-4 (WT) or 7.0 × 107 cfu of the hla/hlb-double disrupted mutant M1007-1 (ΔhlaΔhlb) and 200 μg of anti-ApoLp IgG or control IgG. Silkworm survival was monitored after the injection. The one-sided rank log test indicated that the survival curve of silkworms injected with anti-ApoLp IgG and S. aureus (WT) significantly differed from that of silkworms injected with control IgG and S. aureus (WT) (p = 0.0003). The survival curves did not differ significantly between silkworms injected with anti-ApoLp IgG and the hla/hlb mutant and silkworms injected with control IgG and the hla/hlb mutant (p = 0.8899). F, dose-response relationship between various doses of S. aureus (WT) or the hla/hlb mutant (ΔhlaΔhlb) and silkworms administered 200 μg of anti-ApoLp IgG or control IgG (five larvae per group). Survival was measured at 44 h after the injection. To determine LD50 values, logistic regression analysis was used to fit smooth curves to the survival proportions. Likelihood ratio tests indicated that the LD50 of silkworms injected with anti-ApoLp IgG and S. aureus (WT) significantly differed from that of silkworms injected with control IgG and S. aureus (WT) (p = 0.0294). The LD50 values did not differ significantly between silkworms injected with anti-ApoLp IgG and the hla/hlb mutant and silkworms injected with control IgG and the hla/hlb mutant (p = 0.3504).
Inhibitory Mechanism by ApoLp against Expression of Bacterial Virulence Genes Is Conserved in Mammals
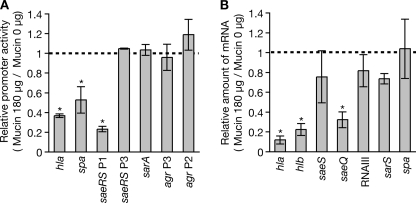
C-terminal amino acid residues of ApoLp are similar to mammalian mucin, which is a component of mucus (26, 35, 36). We hypothesized that the mechanism by which ApoLp inhibits S. aureus virulence is conserved in mammals. The addition of hog gastric mucin to S. aureus culture medium decreased the promoter activities of hla, spa, and saeRS P1 (Fig. 6A) and decreased the amount of hla, hlb, and saeQ mRNAs (Fig. 6B). In contrast, the addition of hog gastric mucin did not alter the promoter activities of saeRS P3, sarA, or agr (Fig. 6A) or the amounts of saeS mRNA, RNAIII, sarS mRNA, and spa mRNA (Fig. 6B). Therefore, mammalian mucin shows activities that repress the expression of hla, hlb, and saeQ, whereas it does not have the inhibitory effect of ApoLp against the expression of RNAIII.
FIGURE 6.
Hog gastric mucin deceases expression of hla and hlb encoding hemolysins. A, S. aureus strains transformed with reporter plasmids carrying promoters of hla, spa, saeRS P1, saeRS P3, sarA, agr P3, and agr P2 were cultured for 3 h in the presence or absence of 180 μg of hog gastric mucin (Wako, Tokyo, Japan)/ml. Luciferase activities of the cell lysates were measured. Data shown are the relative values against luciferase activity in the absence of mucin. Means ± S.D. of three independent experiments are shown. Asterisks indicate Student's t test p values of less than 0.05 between the presence and absence of mucin. B, total RNA was extracted from S. aureus cells cultured in the presence or absence of 180 μg of hog gastric mucin/ml. RNA amounts of hla, hlb, saeS, saeQ, RNAIII, sarS, and spa were measured by quantitative RT-PCR. The horizontal axis shows the relative value against the amount of RNAs in the absence of mucin. Means ± S.D. of three independent experiments are shown. Asterisks indicate Student's t test p values of less than 0.05 between the presence and absence of mucin.
DISCUSSION
The findings of the present study demonstrated that ApoLp in the silkworm hemolymph contributes to resistance against S. aureus infection by suppressing the expression of virulence genes. ApoLp is the first molecule identified in a multicellular organism other than mammals that inhibits the expression of bacterial virulence genes. These findings also revealed that the mechanism of ApoLp to inhibit S. aureus virulence is conserved in mammalian mucin. Thus, a mechanism by which host factors inhibit bacterial virulence might have been conserved during the long battle between host and pathogenic bacteria.
Peroxides produced by macrophages and blood apolipoprotein B directly inactivate the extracellular quorum-sensing molecule of S. aureus that is encoded in the agr locus (11, 12). In contrast, the present study revealed that ApoLp binds to the S. aureus cell surface and suppresses the expression of hemolysins and the virulence regulators saeRS and RNAIII. Furthermore, ApoLp decreased the expression of hemolysin genes via the agr-independent pathway. Therefore, the mechanism of virulence suppression by ApoLp differs from that by peroxides and apolipoprotein B. Thus, ApoLp might bind a receptor molecule of a two-component system, resulting in the transfer of the signal into the cells followed by the suppression of some virulence genes. S. aureus possesses 16 two-component systems, and not all their ligands have been identified (4). Additional studies are needed to identify the surface molecule to which ApoLp binds and reveal the molecular mechanism by which ApoLp decreases the expression of virulence genes.
ApoLp-III, which interacts with ApoLp-II/I, is involved in the pattern recognition of pathogenic bacteria and subsequent encapsulation by interacting with β-glucan and LPS (37–39). To our knowledge, there are no reports that ApoLp-II/I contributes to defense against pathogens. The present study suggests that ApoLp-II/I suppresses virulence. ApoLp-II/I and ApoLp-III may function together in a coordinated defense mechanism against pathogens.
Acknowledgments
We thank Kazuei Mita for providing the expressed sequence tag clone. We thank Timothy J. Foster, Keiichi Hiramatsu, and Richard P. Novick for providing the bacterial strains.
This work was supported by grants-in-aid for scientific research and in part by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO) and the Genome Pharmaceutical Institute.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) AB640623.
S. Miyazaki, Y. Matsumoto, K. Sekimizu, and C. Kaito, unpublished data.
- ApoLp
- apolipophorin.
REFERENCES
- 1. Chien Y., Manna A. C., Projan S. J., Cheung A. L. (1999) J. Biol. Chem. 274, 37169–37176 [DOI] [PubMed] [Google Scholar]
- 2. McNamara P. J., Milligan-Monroe K. C., Khalili S., Proctor R. A. (2000) J. Bacteriol. 182, 3197–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kaito C., Morishita D., Matsumoto Y., Kurokawa K., Sekimizu K. (2006) Mol. Microbiol. 62, 1601–1617 [DOI] [PubMed] [Google Scholar]
- 4. Cheung A. L., Bayer A. S., Zhang G., Gresham H., Xiong Y. Q. (2004) FEMS Immunol. Med. Microbiol. 40, 1–9 [DOI] [PubMed] [Google Scholar]
- 5. Liang X., Yu C., Sun J., Liu H., Landwehr C., Holmes D., Ji Y. (2006) Infect. Immun. 74, 4655–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geiger T., Goerke C., Mainiero M., Kraus D., Wolz C. (2008) J. Bacteriol. 190, 3419–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuroda H., Kuroda M., Cui L., Hiramatsu K. (2007) FEMS Microbiol. Lett. 268, 98–105 [DOI] [PubMed] [Google Scholar]
- 8. Palazzolo-Ballance A. M., Reniere M. L., Braughton K. R., Sturdevant D. E., Otto M., Kreiswirth B. N., Skaar E. P., DeLeo F. R. (2008) J. Immunol. 180, 500–509 [DOI] [PubMed] [Google Scholar]
- 9. Novick R. P. (2003) Mol. Microbiol. 48, 1429–1449 [DOI] [PubMed] [Google Scholar]
- 10. Novick R. P., Ross H. F., Projan S. J., Kornblum J., Kreiswirth B., Moghazeh S. (1993) EMBO J. 12, 3967–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rothfork J. M., Timmins G. S., Harris M. N., Chen X., Lusis A. J., Otto M., Cheung A. L., Gresham H. D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13867–13872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peterson M. M., Mack J. L., Hall P. R., Alsup A. A., Alexander S. M., Sully E. K., Sawires Y. S., Cheung A. L., Otto M., Gresham H. D. (2008) Cell Host Microbe 4, 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Okada M., Natori S. (1983) Biochem. J. 211, 727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoffmann J. A. (1995) Curr. Opin. Immunol. 7, 4–10 [DOI] [PubMed] [Google Scholar]
- 15. Kaito C., Akimitsu N., Watanabe H., Sekimizu K. (2002) Microb. Pathog. 32, 183–190 [DOI] [PubMed] [Google Scholar]
- 16. Kaito C., Kurokawa K., Matsumoto Y., Terao Y., Kawabata S., Hamada S., Sekimizu K. (2005) Mol. Microbiol. 56, 934–944 [DOI] [PubMed] [Google Scholar]
- 17. Kaito C., Sekimizu K. (2007) Drug Discov. Ther. 1, 89–93 [PubMed] [Google Scholar]
- 18. Ishii K., Hamamoto H., Kamimura M., Sekimizu K. (2008) J. Biol. Chem. 283, 2185–2191 [DOI] [PubMed] [Google Scholar]
- 19. Ishii K., Hamamoto H., Kamimura M., Nakamura Y., Noda H., Imamura K., Mita K., Sekimizu K. (2010) J. Biol. Chem. 285, 28635–28642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsumoto Y., Kaito C., Morishita D., Kurokawa K., Sekimizu K. (2007) Infect. Immun. 75, 1964–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nagata M., Kaito C., Sekimizu K. (2008) J. Biol. Chem. 283, 2176–2184 [DOI] [PubMed] [Google Scholar]
- 22. Matsumoto Y., Xu Q., Miyazaki S., Kaito C., Farr C. L., Axelrod H. L., Chiu H. J., Klock H. E., Knuth M. W., Miller M. D., Elsliger M. A., Deacon A. M., Godzik A., Lesley S. A., Sekimizu K., Wilson I. A. (2010) Structure 18, 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hossain M. S., Hamamoto H., Matsumoto Y., Razanajatovo I. M., Larranaga J., Kaito C., Kasuga H., Sekimizu K. (2006) J. Biochem. 140, 439–444 [DOI] [PubMed] [Google Scholar]
- 24. Mita K., Morimyo M., Okano K., Koike Y., Nohata J., Kawasaki H., Kadono-Okuda K., Yamamoto K., Suzuki M. G., Shimada T., Goldsmith M. R., Maeda S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14121–14126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weers P. M., Van Marrewijk W. J., Beenakkers A. M., Van der Horst D. J. (1993) J. Biol. Chem. 268, 4300–4303 [PubMed] [Google Scholar]
- 26. Babin P. J., Bogerd J., Kooiman F. P., Van Marrewijk W. J., Van der Horst D. J. (1999) J. Mol. Evol. 49, 150–160 [DOI] [PubMed] [Google Scholar]
- 27. Chino H., Murakami S., Harashima K. (1969) Biochim. Biophys. Acta 176, 1–26 [PubMed] [Google Scholar]
- 28. Tsuchida K., Arai M., Tanaka Y., Ishihara R., Ryan R. O., Maekawa H. (1998) Insect Biochem. Mol. Biol. 28, 927–934 [DOI] [PubMed] [Google Scholar]
- 29. Mainiero M., Goerke C., Geiger T., Gonser C., Herbert S., Wolz C. (2010) J. Bacteriol. 192, 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peng H. L., Novick R. P., Kreiswirth B., Kornblum J., Schlievert P. (1988) J. Bacteriol. 170, 4365–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ziebandt A. K., Weber H., Rudolph J., Schmid R., Höper D., Engelmann S., Hecker M. (2001) Proteomics 1, 480–493 [DOI] [PubMed] [Google Scholar]
- 32. Steinhuber A., Goerke C., Bayer M. G., Döring G., Wolz C. (2003) J. Bacteriol. 185, 6278–6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Adhikari R. P., Novick R. P. (2008) Microbiology 154, 949–959 [DOI] [PubMed] [Google Scholar]
- 34. Cheung A. L., Schmidt K., Bateman B., Manna A. C. (2001) Infect. Immun. 69, 2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kutty R. K., Kutty G., Kambadur R., Duncan T., Koonin E. V., Rodriguez I. R., Odenwald W. F., Wiggert B. (1996) J. Biol. Chem. 271, 20641–20649 [DOI] [PubMed] [Google Scholar]
- 36. Avarre J. C., Lubzens E., Babin P. J. (2007) BMC Evol. Biol. 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Whitten M. M., Tew I. F., Lee B. L., Ratcliffe N. A. (2004) J. Immunol. 172, 2177–2185 [DOI] [PubMed] [Google Scholar]
- 38. Pratt C. C., Weers P. M. (2004) Biol. Chem. 385, 1113–1119 [DOI] [PubMed] [Google Scholar]
- 39. Niere M., Dettloff M., Maier T., Ziegler M., Wiesner A. (2001) Biochemistry 40, 11502–11508 [DOI] [PubMed] [Google Scholar]
- 40. Novick R. (1967) Virology 33, 155–166 [DOI] [PubMed] [Google Scholar]
- 41. Matsuo M., Kurokawa K., Nishida S., Li Y., Takimura H., Kaito C., Fukuhara N., Maki H., Miura K., Murakami K., Sekimizu K. (2003) FEMS Microbiol. Lett. 222, 107–113 [DOI] [PubMed] [Google Scholar]
- 42. Kaito C., Saito Y., Nagano G., Ikuo M., Omae Y., Hanada Y., Han X., Kuwahara-Arai K., Hishinuma T., Baba T., Ito T., Hiramatsu K., Sekimizu K. (2011) PLoS Pathog. 7, e1001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sakashita K., Tatsuke T., Masaki Y., Lee J. M., Kawaguchi Y., Kusakabe T. (2009) J. Fac. Agric. Kyusyu Univ. 54, 401–406 [Google Scholar]
- 44. Sundermeyer K., Hendricks J. K., Prasad S. V., Wells M. A. (1996) Insect Biochem. Mol. Biol. 26, 735–738 [DOI] [PubMed] [Google Scholar]
- 45. Smolenaars M. M., Kasperaitis M. A., Richardson P. E., Rodenburg K. W., Van der Horst D. J. (2005) J. Lipid Res. 46, 412–421 [DOI] [PubMed] [Google Scholar]