Background: P-glycoprotein plays a role in drug resistance and drug interactions.
Results: Surface plasmon resonance with P-glycoprotein in lipid nanodiscs indicates that inhibitory antibodies uncouple the ATP hydrolysis of P-gp from its transport function.
Conclusion: Inhibitory antibodies do not prevent P-gp flux through the catalytic cycle, but rather block drug efflux without inhibiting ATP hydrolysis.
Significance: The results clarify the mechanism of inhibitory antibodies to P-gp.
Keywords: ABC Transporter, ATPases, Drug Metabolism, Drug Resistance, Membrane Enzymes
Abstract
The human ATP-binding cassette (ABC) transporter, P-glycoprotein (P-gp; ABCB1), mediates the ATP-dependent efflux of a variety of drugs. As a result, P-gp plays a critical role in tumor cell drug resistance and the pharmacokinetic properties of most drugs. P-gp exhibits extraordinary substrate and inhibitor promiscuity, resulting in a wide range of possible drug-drug interactions. Inhibitory antibodies have long been considered as a possible strategy to modulate P-gp-dependent cancer cell drug resistance, and it is widely suggested that the antibodies MRK16 and UIC2 inhibit P-gp by capturing a single isoform and preventing flux through the catalytic cycle. Although the crystal structures of many bacterial whole transporters, as well as isolated nucleotide-binding domains, have been solved, high resolution structural data for mammalian ABC transporters are currently lacking. It has been extremely difficult to determine the detailed mechanism of transport of P-gp, in part because it is difficult to obtain purified protein in well defined lipid systems. Here we exploit surface plasmon resonance (SPR) to probe conformational changes associated with these intermediate states for P-gp in lipid bilayer nanodiscs. The results indicate that P-gp in nanodiscs undergoes functionally relevant ligand-dependent conformational changes and that previously described inhibitory antibodies bind to multiple nucleotide-bound states but not the ADP-VO4-trapped state, which mimics the post-hydrolysis state. The results also suggest that the substrate drug vinblastine is released at stages that precede or follow the post-hydrolysis ADP-PO4·P-gp complex.
Introduction
The ATP-binding cassette (ABC)2 superfamily of proteins serves a wide variety of functions in all higher organisms and prokaryotes (1–3). In humans, the isoform ABCB1, also known as P-glycoprotein (P-gp), is an efflux transporter that contributes to cancer cell drug resistance (4–6), and this has made it a target for therapeutics that modulate the effectiveness of many anti-cancer drugs. Early studies explored the utility of inhibitory antibodies, such as MRK16 and UIC2, to increase cancer drug exposure in target cells (7–10), but no antibodies have emerged in clinical practice. These antibodies remain as useful in vitro tools to study P-gp function (11, 12) and expression (13, 14), and it is widely asserted that they capture specific conformations of P-gp and therefore inhibit its flux through the catalytic cycle (12). The widespread observation that vinblastine efflux is prevented by MRK16 and UIC2 (7–10) suggests the hypothesis that either the drug is normally released from one of the P-gp states to which these antibodies bind or the antibodies prevent drug or nucleotide binding. However, these possibilities have not been clarified.
In addition to the long standing interest in P-gp as a target in cancer chemotherapy, there is great current interest in modulating P-gp function to improve the absorption, distribution, and elimination properties of many other drugs, and in particular, to improve CNS entry of drugs aimed at neurodegenerative diseases (15). High levels of expression of the human ABCB1 at the blood-brain barrier, intestine, and liver are consistent with its role in the protection of critical organs from chemical insult (3). In addition, P-gp is unusually substrate-promiscuous, as expected for a role in detoxification (2, 16). The x-ray crystal structure of the mouse ortholog, Abcb1a (17), together with numerous structures of bacterial ABC transporters (18–21), indicate that the ABC transporters are complex transmembrane proteins with several conserved structural elements. P-gp consists of four core domains: two transmembrane domains, containing multiple membrane-spanning α-helices (transmembrane helices (TMHs)), which form the drug-binding sites, and two nucleotide-binding domains (NBDs), which utilize the energy released in ATP binding and hydrolysis to power drug transport by an as yet undetermined mechanism (22). The NBDs of P-gp share a large degree of sequence identity with other ABC transporters, suggesting some commonality of mechanism. Conserved motifs that form the ATP-binding pocket within the NBDs include the Walker A and Walker B motifs, common to many nucleotide-binding proteins, as well as the ABC signature motif, the hallmark of the ABC superfamily (2). It is clear that large scale conformational changes transmitted between the NBDs and transmembrane domains contribute to the P-gp transport mechanism. Specifically, binding of ATP induces the engagement of the two NBDs, which likely transmits conformational change to the TMHs (23, 24). Conformational changes in the TMHs are also apparent in the quasi-stable ADP-VO4-trapped state, P-gp·ADP·VO4, which mimics the post-ATP hydrolysis state prior to the release of PO4 and ADP or the transition state leading to it (25, 26). The conformations of the TMHs during the progression of nucleotide-bound NBD states, in turn, control egress of drug from the drug-binding sites.
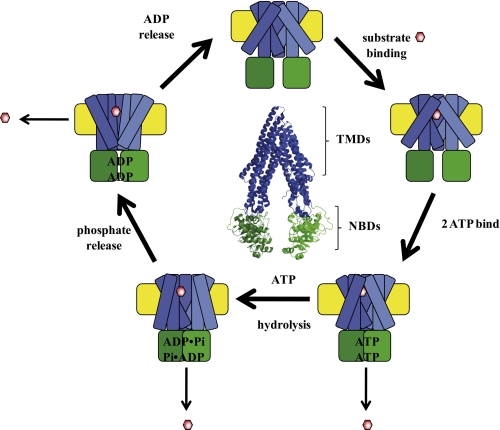
Despite the availability of these structural models (27, 28) and data from the biochemical studies (29, 30), the molecular mechanism of ABCB1 and the mechanism by which drugs and antibodies inhibit P-gp remain unknown. This is, in part, due to the difficulty in obtaining large quantities of purified P-gp in well defined membrane environments, and nearly all biochemical experiments are performed with crude membrane preparations (31, 32), detergent-solubilized species (33, 34), or proteoliposomes (29, 35, 36) or in whole cell assays (37–39) where multiple processes contribute to the net flux of drugs across the membrane. As a result, it has been difficult to study individual steps in the transport cycle or specific protein conformations that are populated. In fact, there are competing mechanistic models that differ in the point at which drug is released (Fig. 1) (40, 41). Although it is clear that at least one nucleotide-dependent conformational change takes place, the number of conformational states during the reaction cycle, their structural differences, and whether they precede or follow drug release have not been determined. Clearly, new methods are required to further define these mechanistic details.
FIGURE 1.
Schematized P-gp catalytic cycle. The structure of P-gp is schematized with the NBDs (green) undergoing nucleotide-dependent interactions. The TMHs are designated in blue and undergo nucleotide-dependent and drug (red hexagon)-dependent conformational changes. The model reflects the uncertainty about the point at which drug is released and the possibility that multiple states may be capable of releasing drug. At the center is the crystal structure of drug- and nucleotide-free mouse Abcb1a, Protein Data Bank (PDB) number 3G5U, with analogous color coding. TMDs, transmembrane domains.
Here purified P-gp was captured in lipid membrane nanodiscs, and the nucleotide-dependent conformations of P-gp were probed using surface plasmon resonance (SPR) and inhibitory antibodies. The results provide additional detail about the mechanism of inhibition by the MRK16 and UIC2 antibodies and the structure of intermediate conformations. Furthermore, the results suggest the possible conformations from which drug is released.
EXPERIMENTAL PROCEDURES
Chemicals
All chemicals were purchased from Sigma unless otherwise noted and were the highest quality available.
Protein Expression and Purification
Human dodeca-histidine-tagged P-gp was expressed using a baculovirus expression system in Trichoplusia ni cells and purified via nickel affinity chromatography as described (42). MSP1D1 was expressed in Escherichia coli and purified using the hepta-histidine tag via nickel affinity chromatography (42) as described.
Preparation of P-gp Nanodiscs
P-gp nanodisc preparations were made as described previously (42). Briefly, E. coli total lipid extract (Avanti Polar Lipids, Alabaster, AB) was dissolved in chloroform, dried to a lipid film under N2, and vacuum-desiccated overnight, prior to solubilization in disc-forming buffer (DFB: 20 mm Tris, 100 mm NaCl, pH 7.4) with the addition of 4-fold excess dodecyl β-maltopyranoside (Affymetrix, Fremont, CA). Purified P-gp, MSP1D1, and lipid were combined at a molar ratio of 0.02:1:35 with the addition of protease inhibitors, 1 mm benzamidine HCl, 40 μm leupeptin hemisulfate, and 1 μm pepstatin A (EMD Biosciences, Rockland, MA), and incubated at room temperature for 1 h with constant agitation. Prewashed Bio-Beads SM-2 media (Bio-Rad) were then added at 0.8 g/ml and incubated for 2 h at room temperature to remove the detergent. Resulting nanodiscs were recovered from the Bio-Beads using a 25-gauge needle.
Size Exclusion Chromatography
Nanodisc formation was analyzed using size exclusion chromatography, which was performed on a Superdex 200 10/300 column (GE Healthcare) at an isocratic flow rate of 0.5 ml/min, using DFB as the mobile phase. Gel filtration standards (Bio-Rad) were run on the column every 6 months to monitor column status.
ATPase Activity Assays
ATPase assays were performed as described previously (42), with the following modifications. The antibodies UIC2 (Millipore, Billerica, MA) and MRK16 (Kamiya Biomedical, Seattle, WA) were both purchased without BSA or azide. To determine the effect of UIC2 and MRK16 on ATPase activity of P-gp, the antibodies were first buffer-exchanged out of PBS and into DFB to minimize phosphate contamination. Antibody (5 μg, or DFB as a control) was added to 500 μl of P-gp nanodiscs at ∼80 nm (∼1 μg) and allowed to incubate for 30 min at 4 °C with constant agitation. Empty nanodiscs were also treated with antibody and DFB and used to determine the background amount of phosphate in the samples. Basal and drug (50 μm nicardipine or 10 μm nicardipine)-stimulated ATPase activity was determined using a colorimetric phosphate release assay (43). Drugs were made up as dimethyl sulfoxide (DMSO) stocks, and vehicle was added in the basal activity.
Nucleotide and Drug Binding to P-gp
Sodium orthovanadate was prepared by boiling and adjusting the pH of the solution to 10 until the solution became colorless and the pH stabilized. Nucleotides were made up in 10 mm Tris, pH 7.4. For the ADP/VO4 samples, ADP was added to the ATPase buffer and mixed thoroughly prior to the addition of VO4 to prevent a local pH change, which would cause VO4 aggregation. All reactions were carried out in ATPase buffer (50 mm Tris, 150 mm NH4Cl, 5 mm MgSO4, 0.02% NaN3, pH 7.4). P-gp nanodiscs (50 nm) were incubated in ATPase buffer with in ATPase buffer with or without nucleotide (5 mm ADP, 5 mm ADP, and 5 mm Vi, or 5 mm AMPPNP) in the presence and absence of 10 μm vinblastine for 30 min at 37 °C. Dilutions of each conformation were made using the same buffer to maintain the drug, nucleotide, and/or VO4 concentrations to ensure that the drug or nucleotide was bound for the duration of the experiment.
SPR and Antibody Immobilization
SPR experiments were performed at the University of Washington Bioanalytical Pharmacy Core on a Biacore T100 (GE Healthcare). CM5 chips and amine-coupling reagents were purchased from GE Healthcare. The antibodies were diluted into 10 mm sodium acetate buffer at the appropriate pH at a concentration of 30 μg/ml and immobilized using amine-coupling chemistry. The level of the immobilized antibody was 3000 response units, which was reached using the immobilization wizard of the Biacore T100 controller software. An activated and capped surface without antibody was used as the reference, which was subtracted from each of the flow cells containing the antibodies. For all experiments, the running buffer used was DFB, and the surface was regenerated by two 20-s injections of 10 mm glycine, pH 1.5, at a flow rate of 30 μl/min.
Antibody Capture Kinetics
P-gp nanodiscs (0–12.5 nm) in ATPase buffer with and without drug (10 μm vinblastine) and nucleotide (5 mm AMPPNP, 5 mm ADP, or 5 mm ADP/VO4) were flowed over the immobilized antibody at a flow rate of 75 μl/min and allowed to associate for 60 s followed by a 120- or 300-s dissociation phase. Due to the lack of dissociation, only association data were fit using Prism 5 (GraphPad). The temperature of the sample chamber was maintained at 10 °C to ensure sample stability, and the chip surface temperature was kept at 25 °C.
Nucleotide Binding to Captured P-gp Nanodiscs
P-gp nanodiscs (30 nm) in ATPase buffer without nucleotide were captured by both antibodies by flowing over the surface at a flow rate of 75 μl/min for 60 s. After a 120-s stabilization phase, nucleotides at the same concentrations as before or ATPase buffer were flowed over the captured P-gp nanodiscs at 75 μl/min for 120 s followed by a 120-s dissociation phase.
RESULTS
ATPase Activity
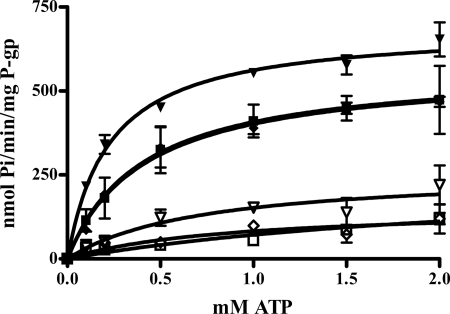
Human ABCB1 was incorporated into lipid bilayer nanodiscs as described previously using the engineered apolipoprotein scaffold MSP1D1 and E. coli total lipid extract. Although the inhibitor effects of MRK16 and UIC2 on substrate efflux from whole cells are well established, no data concerning their effects on the ATPase function have been published. It is reasonable to presume that these antibodies would also inhibit ATPase function if they capture specific P-gp conformations and prevent flux through the catalytic cycle, as widely suggested. ATPase activity was therefore determined for P-gp in nanodiscs in the presence or absence of UIC2 or MRK16. In contrast to expectations, neither antibody inhibited basal or drug-stimulated ATPase activity (Fig. 2). In fact, MRK16 slightly activated the basal and nicardipine-stimulated ATPase activity, with the major impact on the Vmax term. These data indicate that the NBDs are capable of binding and hydrolyzing ATP even in the presence of antibody and that nicardipine (or vinblastine, Km, ATP = 0.4077 ± 0.2076 mm, data not shown) binds and stimulates ATPase activity even with antibody present. MRK16 and UIC2 clearly do not prevent the NBDs from sampling conformations required for drug-stimulated ATP hydrolysis, and MRK16 actually stimulates this activity. The data are summarized in Table 1.
FIGURE 2.
ATPase activity of P-gp in nanodiscs in the presence of antibodies MRK16 and UIC2. Open squares, buffer control, no drug, no antibody. Open inverted triangles, with MRK16, no drug. Filled inverted triangles, with MRK16, + 50 μm nicardipine. Open diamonds, with UIC2, no drug. Filled squares, no antibody, + 50 μm nicardipine. Filled diamonds, with UIC2, + 50 μm nicardipine. The solid line represents the fit to the Michaelis-Menten equation, the values of which are reported in Table 1. Error bars indicate S.D.
TABLE 1.
Kinetic constants from Michaelis-Menten fit to ATPase assay in the presence and absence of antibodies
|
Vmax |
Km |
|||
|---|---|---|---|---|
| Basal | Nicardipine-stimulated | Basal | Nicardipine-stimulated | |
| nmol Pi/min/mg of P-gp | mm | |||
| DFB | 140.0 ± 42.6 | 571.5 ± 64.1 | 0.964 ± 0.632 | 0.401 ± 0.143 |
| MRK16 | 262.2 ± 64.5 | 688.9 ± 24.0 | 0.723 ± 0.449 | 0.226 ± 0.030 |
| UIC2 | 153.8 ± 62.6 | 568.1 ± 35.8 | 0.860 ± 0.824 | 0.413 ± 0.086 |
Antibody Binding and SPR
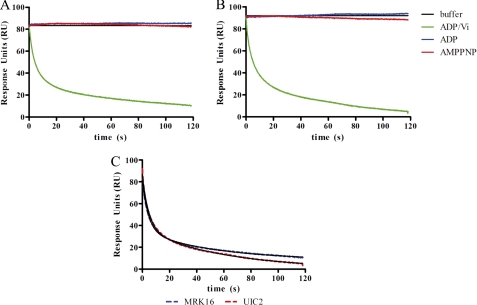
SPR was used to probe the conformations of P-gp that bind to the antibodies. Here we use the term conformation as it relates to the protein structure, which is presumably linked to varying states of occupancy by drug or nucleotide. We presume that the antibodies would distinguish between different states of ligand occupancy only if they were accompanied by changes in P-gp structure. Conversely, all ligand-bound states of P-gp that bind to the antibody are not necessarily identical. The P-gp nanodiscs were incubated, in the presence or absence of the transportable substrate vinblastine, with saturating concentrations of the nonhydrolyzable ATP analog AMPPNP, ADP, or ADP and VO4 to obtain the well characterized and quasi-irreversible trapped state that mimics the post-hydrolysis P-gp·ADP·PO4 complex or the transition state leading to it. These P-gp nanodiscs complexes were then analyzed for binding to surface-immobilized antibodies MRK16 or UIC2 by SPR. The overall experimental design is summarized in Fig. 3, where the binding (association) of a single concentration of P-gp nanodiscs, as analyte, to immobilized antibody is shown. Both antibodies have epitopes on the extracellular surface of P-gp. Fig. 4 and the corresponding parameters in Table 2 demonstrate that the P-gp is clearly capable of binding to both antibodies in the absence of drug and in the presence of AMPPNP or ADP. The affinity and on-rates of the P-gp nanodisc particles were unaffected by the presence of vinblastine. The off-rates of the particles were too slow to measure in each case, so only the association rates are reported in Table 2. Although the off-rates were not experimentally recovered because they are too slow, the limit of the instrument, 1 × 10−5 s−1, can be used to approximate the upper limit of the Kd for each antibody to each state in Fig. 4. Attempts to fit the association data to a more complex two-component model (two binding sites with different kinetic parameters) improved the fit slightly. However, although one of the recovered on-rates from the two-component model was consistently nearly identical to the on-rate from the one-component fit, the second on-rate was dramatically slower (∼100-fold). Such slow on-rates are difficult to interpret and likely represent some nonspecific binding. The likelihood that this slow rate was due to nonspecific binding is supported by the observation that it was nearly constant across each of the nucleotide-bound states. Because the fits to the simple binding model adequately demonstrate the unique properties of the vanadate-trapped state, the results of the simpler model are reported in Table 2. The most striking result is that neither antibody binds to the ADP-VO4-trapped state, in marked contrast to the other nucleotide-bound states for which the antibodies have extremely high affinity. Apparently, in the trapped state, the conformation of the extracellular P-gp epitopes is very different from any of the other states of the catalytic cycle that are modeled here.
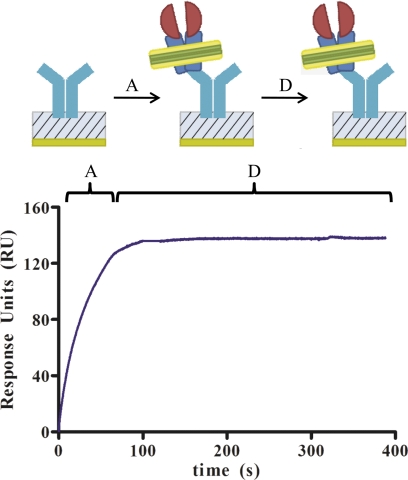
FIGURE 3.
Experimental design and example of SPR data. The association phase (indicated by A) and dissociation phase (indicated by D) are shown for a single concentration of P-gp nanodiscs, 12.5 nm, binding to MRK16 that is immobilized on the surface. The association phase has not reached equilibrium at the time the P-gp nanodiscs are removed from the analyte buffer and the dissociation phase begins. However, because the dissociation is extremely slow, even undetectable, the response units (RU) remain nearly constant during the dissociation phase. In subsequent experiments, only the association phase was studied.
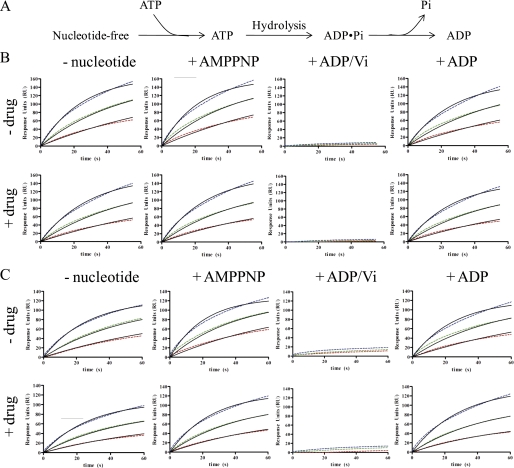
FIGURE 4.
Association of P-gp nanodiscs, at various nucleotide-bound NBD states to MRK16 and UIC2. A, schematic of ATP hydrolysis reaction with discreet states probed in the SPR experiments. B, binding to MRK16 in the presence of the indicated nucleotide, with and without 10 μm VO4 (Vi) (drug). C, binding to UIC2 in the presence of the indicated nucleotide, with and without 10 μm vinblastine. Red, 3.1 nm P-gp nanodiscs. Green, 6.2 nm P-gp nanodiscs. Blue, 12.5 nm P-gp nanodiscs. Dashed lines are raw data. Solid lines are best fits to the standard SPR binding model. Recovered parameters are summarized in Table 2.
TABLE 2.
On-rates and upper limit of Kd values for SPR data
Vi, VO4; ND, not determined; NA, not applicable.
| Drug-free |
10 μm vinblastine |
|||
|---|---|---|---|---|
| kon | Kda | kon | Kda | |
| mm−1s−1 | pm | m−1s−1 | pm | |
| MRK16 | ||||
| Nucleotide-free | 3.1 × 106 ± 1.7 × 104 | 3.22 | 2.6 × 106 ± 1.7 × 104 | 3.85 |
| ADP | 3.0 × 106 ± 2.2 × 104 | 3.33 | 2.7 × 106 ± 1.9 × 104 | 3.71 |
| AMPPNP | 3.6 × 106 ± 2.4 × 104 | 2.78 | 2.4 × 106 ± 1.6 × 104 | 4.17 |
| ADP/Vi | ND | NA | ND | NA |
| UIC2 | ||||
| Nucleotide-free | 2.8 × 106 ± 1.5 × 104 | 3.57 | 2.4 × 106 ± 1.3 × 104 | 4.17 |
| ADP | 3.1 × 106 ± 2.3 × 104 | 3.23 | 1.9 × 106 ± 1.2 × 104 | 5.26 |
| AMPPNP | 3.8 × 106 ± 2.3 × 104 | 2.63 | 2.6 × 106 ± 1.4 × 104 | 3.85 |
| ADP/Vi | ND | NA | ND | NA |
a The Kdvalues are estimates based on the recovered on-rates and assuming the slowest off-rate, 1 ×10−5 s−1, detectable by the instrument.
To confirm this result, a second experimental design was used wherein P-gp nanodiscs were captured by the antibodies and ligands were then flowed across the biosensor chip surface (Fig. 5). In this case, ADP alone or AMPPNP had no effect on the response units and the affinity of the P-gp nanodiscs was not decreased. Again in marked contrast, when both ADP and VO4 were flowed across the surface, the P-gp nanodiscs rapidly dissociated from the immobilized antibody. In effect, formation of the trapped state released the P-gp nanodiscs from the antibodies. For both antibodies, the data fit best to two dissociation rates. For UIC2, the rates and fractional contributions were 0.2097 ± 0.0017 s−1 (57.6%) and 0.0178 ± 0.002 (42.4%). For MRK16, the rates and fractional contributions were 0.2247 ± 0.0014 (63.4%) and 0.0193 ± 0.0002 (46.6%). These experiments do not directly reveal the source of heterogeneous kinetics, which is likely due to heterogeneity of the nucleotide-binding sites. The results further demonstrate that the conformation of the trapped state is significantly different from the conformation in the presence of the other ligands, and neither antibody retains its affinity for this state. The antibodies are unable to prevent formation of the trapped conformation.
FIGURE 5.
Dissociation of P-gp nanodiscs from UIC2 or MRK16 upon formation of the ADP·VO4-trapped complex. After the P-gp nanodiscs were captured, ADP and VO4 (green) were flowed across the sensor chip, leading to rapid dissociation from both antibodies. ADP alone (blue) or AMPPNP (red) did not induce dissociation. Solid lines are best fits of a two-component dissociation model. The recovered off-rates are included in the text. RU, response units; Vi, VO4. A, dissociation of P-gp nanodiscs from UIC2. B, dissociation of P-gp nanodiscs from MRK16. C, the dissociation curves induced by ADP/VO4 for UIC2 (blue) and MRK16 (red) are shown with best fits to a two-component model.
DISCUSSION
The data summarized here demonstrate the utility of lipid nanodiscs as a platform to study conformational aspects of P-gp, which have eluded analysis by many other methods due to the difficulty in obtaining adequate quantities in well defined membrane environments. These results also clarify the mechanism of inhibition of P-gp by MRK16 and UIC2, which have been used extensively to modulate P-gp in cell-based assays and which have provided proof of principle for many years that modulation of P-gp is a reasonable strategy to increase cancer cell exposure to numerous anti-cancer drugs. Based on the lack of inhibition of the ATPase function, it is clear that these antibodies do not capture a specific conformation and prevent flux through the P-gp catalytic cycle. Their effect on the cell efflux of various drugs, combined with the results here, indicates that these antibodies uncouple the ATP hydrolysis from transport. Inasmuch as many small molecule drugs also uncouple ATP hydrolysis from transport, the antibodies MRK16 and UIC2 may inhibit by a similar mechanism. Drugs that uncouple P-gp may be trapped within the TMHs without being effluxed and prevent the efflux of other drugs, without preventing ATP hydrolysis in the NBDs.
The results further suggest that the species from which vinblastine is released to the extracellular milieu is not the post-hydrolysis P-gp·ADP·PO4 state, to the extent that this state is mimicked by the vanadate-trapped species. Although some ATP-hydrolytic enzymes are covalently modified by vanadate (44), the available data indicate that this does not occur with P-gp and that the interaction is reversible (25, 45) Here we presume that the protein conformations of both ligand states P-gp·ADP·PO4 and P-gp·ADP·VO4 are nearly identical. The fast dissociation of both antibodies from the vanadate-trapped species would suggest the possibility of dissociation of vinblastine or other drugs from the structurally similar P-gp·ADP·PO4 state. In contrast, antibody bound to other states in the catalytic cycle at the extracellular epitopes would sterically block drug dissociation, although the NBDs apparently cycle through the conformations required for efficient ATP hydrolysis. An alternative possibility is that drugs do dissociate from the post-hydrolysis state, but their off-rate is sufficiently slow to not compete with the dissociation and reassociation of the antibodies at the extracellular surface.
It is interesting to consider the energetic aspects of the conformational change associated with forming the trapped state. From the concentration dependence of the association of antibodies to the nucleotide-bound state other than the ADP-VO4-trapped state, a crude estimate of Kd for the antibody is 1–10 pm, which in turn corresponds to a ΔG for binding of ∼14–16 kcal/mol at 20 °C. Inasmuch as the ADP-VO4-trapped state has no measurable affinity for these antibodies, the ΔΔG between the nucleotide-bound states and the trapped state must be at least ∼14 kcal/mol. This is a remarkable energy differential, suggestive of a very large conformational change upon reaching the post-hydrolysis state prior to the release of PO4. This could reflect the transfer of exposed extracellular epitopes to a partially membrane-buried state.
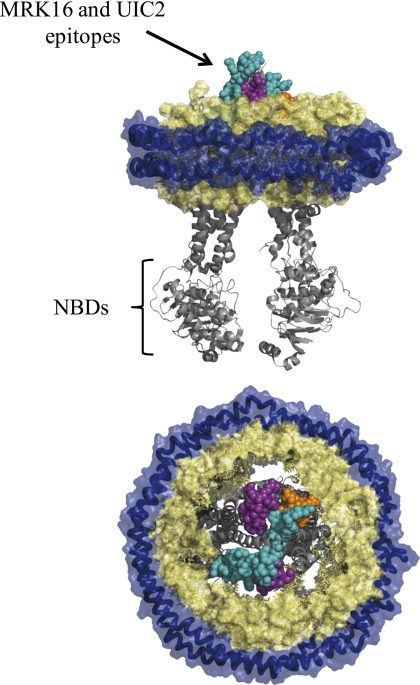
The conformational analysis is strikingly analogous to the results based on two-dimensional crystal analysis wherein the low resolution electron density suggests a difference in the accessibility to solvent of the drug-binding site in the trapped state, which is inaccessible, as compared with the ATP analog-bound and ADP-bound states, for which the channel is more open (46). The epitopes for UIC2 and MRK16 are both discontinuous and made up of multiple extracellular loops (Fig. 6) (13, 47, 48). Presumably, these loops must undergo conformational changes when the TMHs rearrange, as reflected in the two-dimensional structures, resulting in abolition of both epitopes at the ADP·VO4-trapped state. The electron density suggests that the channel is closed in this state. Thus the two-dimensional crystal data are consistent with the current SPR data and suggest that drug is not released from the trapped state.
FIGURE 6.
Epitopes of MRK16 and UIC2 mapped onto a model of P-gp in nanodiscs. The mouse Abcb1a ligand-free structure is shown (PDB number 3G5U). The royal blue helices are the MSP proteins of the nanodiscs. The space-filled wheat-colored structures are lipids. Some lipids have been removed to visualize the P-gp structure. Teal, MRK16 epitope. Purple, UIC2 epitope. Orange, overlapping epitope. Gray structures are the remainder of P-gp, not included in the antibody epitopes. Top, edge-on view. Bottom, top-down view, from the extracellular side.
A surprising result of our studies is that MRK16 and UIC2 do not show differential binding to the P-gp states modeled here, in contrast to the literature that suggests that UIC2 is “conformation-specific,” whereas MRK16 is not. This difference is based on studies in whole cells where UIC2 displays increased immunoreactivity in the presence of P-gp substrates (13), if the ATP levels within the cell have been depleted (49), or in mutant P-gps containing a lysine-to-methionine mutation in either of the NBD Walker A motifs, which are able to bind nucleotide but are deficient in ATP hydrolysis activity (12). MRK16 shows no such differential immunoreactivity for any of these cases. Possibly, in steady-state efflux experiments in whole cells, P-gp states are populated that are not accessed in our studies targeted to the discrete states that mimic ATP-bound, ADP-bound, and ADP·PO4-bound states. If states other than these are differentially bound by MRK16 and UIC2, then they could elicit different effects in transport assays without detectable differences in our studies.
In summary, lipid nanodiscs provide a powerful platform to probe conformation and mechanism of P-glycoprotein by methods that have been previously intractable, such as SPR. The results clarify the mechanism of inhibition of the inhibitory antibodies MRK16 and UIC2, and they suggest that some drugs are not released from P-gp into the extracellular space from the immediate post-hydrolysis state.
This work was supported, in whole or in part, by National Institutes of Health Grants GM 32165 and 62284 (to W. M. A.).
- ABC
- ATP-binding cassette
- P-gp
- P-glycoprotein
- TMH
- transmembrane helix
- NBD
- nucleotide-binding domain
- AMPPNP
- adenosine 5′-(β,γ-imido) triphosphate tetralithium salt hydrate
- DFB
- disc-forming buffer
- MSP
- membrane scaffold protein.
REFERENCES
- 1. Higgins C. F. (1992) Annu. Rev. Cell Biol. 8, 67–113 [DOI] [PubMed] [Google Scholar]
- 2. Jones P. M., O'Mara M. L., George A. M. (2009) Trends Biochem. Sci. 34, 520–531 [DOI] [PubMed] [Google Scholar]
- 3. Leonard G. D., Fojo T., Bates S. E. (2003) Oncologist 8, 411–424 [DOI] [PubMed] [Google Scholar]
- 4. Juliano R. L., Ling V. (1976) Biochim. Biophys. Acta 455, 152–162 [DOI] [PubMed] [Google Scholar]
- 5. Gottesman M. M. (2002) Annu. Rev. Med. 53, 615–627 [DOI] [PubMed] [Google Scholar]
- 6. Gillet J. P., Gottesman M. M. (2010) Methods Mol. Biol. 596, 47–76 [DOI] [PubMed] [Google Scholar]
- 7. Hunter J., Hirst B. H., Simmons N. L. (1993) Pharm. Res. 10, 743–749 [DOI] [PubMed] [Google Scholar]
- 8. Druley T. E., Stein W. D., Roninson I. B. (2001) Biochemistry 40, 4312–4322 [DOI] [PubMed] [Google Scholar]
- 9. Ruth A., Stein W. D., Rose E., Roninson I. B. (2001) Biochemistry 40, 4332–4339 [DOI] [PubMed] [Google Scholar]
- 10. Hunter J., Jepson M. A., Tsuruo T., Simmons N. L., Hirst B. H. (1993) J. Biol. Chem. 268, 14991–14997 [PubMed] [Google Scholar]
- 11. Mechetner E. B., Roninson I. B. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 5824–5828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mechetner E. B., Schott B., Morse B. S., Stein W. D., Druley T., Davis K. A., Tsuruo T., Roninson I. B. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12908–12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schinkel A. H., Arceci R. J., Smit J. J., Wagenaar E., Baas F., Dollé M., Tsuruo T., Mechetner E. B., Roninson I. B., Borst P. (1993) Int. J. Cancer 55, 478–484 [DOI] [PubMed] [Google Scholar]
- 14. Kobayashi Y., Yamashiro T., Nagatake H., Yamamoto T., Watanabe N., Tanaka H., Shigenobu K., Tsuruo T. (1994) Biochem. Pharmacol. 48, 1641–1646 [DOI] [PubMed] [Google Scholar]
- 15. Jeynes B., Provias J. (2011) Neurosci. Lett. 487, 389–393 [DOI] [PubMed] [Google Scholar]
- 16. Kimura Y., Morita S. Y., Matsuo M., Ueda K. (2007) Cancer Sci. 98, 1303–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aller S. G., Yu J., Ward A., Weng Y., Chittaboina S., Zhuo R., Harrell P. M., Trinh Y. T., Zhang Q., Urbatsch I. L., Chang G. (2009) Science 323, 1718–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dawson R. J., Locher K. P. (2007) FEBS Lett. 581, 935–938 [DOI] [PubMed] [Google Scholar]
- 19. Diederichs K., Diez J., Greller G., Müller C., Breed J., Schnell C., Vonrhein C., Boos W., Welte W. (2000) EMBO J. 19, 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karpowich N., Martsinkevich O., Millen L., Yuan Y. R., Dai P. L., MacVey K., Thomas P. J., Hunt J. F. (2001) Structure 9, 571–586 [DOI] [PubMed] [Google Scholar]
- 21. Locher K. P., Lee A. T., Rees D. C. (2002) Science 296, 1091–1098 [DOI] [PubMed] [Google Scholar]
- 22. Callaghan R., Ford R. C., Kerr I. D. (2006) FEBS Lett. 580, 1056–1063 [DOI] [PubMed] [Google Scholar]
- 23. Crowley E., O'Mara M. L., Kerr I. D., Callaghan R. (2010) FEBS J. 277, 3974–3985 [DOI] [PubMed] [Google Scholar]
- 24. Martin C., Berridge G., Mistry P., Higgins C., Charlton P., Callaghan R. (2000) Biochemistry 39, 11901–11906 [DOI] [PubMed] [Google Scholar]
- 25. Urbatsch I. L., Sankaran B., Weber J., Senior A. E. (1995) J. Biol. Chem. 270, 19383–19390 [DOI] [PubMed] [Google Scholar]
- 26. Urbatsch I. L., Tyndall G. A., Tombline G., Senior A. E. (2003) J. Biol. Chem. 278, 23171–23179 [DOI] [PubMed] [Google Scholar]
- 27. O'Mara M. L., Tieleman D. P. (2007) FEBS Lett. 581, 4217–4222 [DOI] [PubMed] [Google Scholar]
- 28. Rosenberg M. F., Callaghan R., Modok S., Higgins C. F., Ford R. C. (2005) J. Biol. Chem. 280, 2857–2862 [DOI] [PubMed] [Google Scholar]
- 29. Sauna Z. E., Kim I. W., Nandigama K., Kopp S., Chiba P., Ambudkar S. V. (2007) Biochemistry 46, 13787–13799 [DOI] [PubMed] [Google Scholar]
- 30. Loo T. W., Bartlett M. C., Clarke D. M. (2003) J. Biol. Chem. 278, 1575–1578 [DOI] [PubMed] [Google Scholar]
- 31. Loo T. W., Clarke D. M. (2005) Biochem. Biophys. Res. Commun. 329, 419–422 [DOI] [PubMed] [Google Scholar]
- 32. Zolnerciks J. K., Wooding C., Linton K. J. (2007) FASEB J. 21, 3937–3948 [DOI] [PubMed] [Google Scholar]
- 33. Qu Q., Russell P. L., Sharom F. J. (2003) Biochemistry 42, 1170–1177 [DOI] [PubMed] [Google Scholar]
- 34. Liu R., Siemiarczuk A., Sharom F. J. (2000) Biochemistry 39, 14927–14938 [DOI] [PubMed] [Google Scholar]
- 35. Ambudkar S. V., Lelong I. H., Zhang J., Cardarelli C. (1998) Methods Enzymol. 292, 492–504 [DOI] [PubMed] [Google Scholar]
- 36. Bucher K., Belli S., Wunderli-Allenspach H., Krämer S. D. (2007) Pharm. Res. 24, 1993–2004 [DOI] [PubMed] [Google Scholar]
- 37. Takeuchi T., Yoshitomi S., Higuchi T., Ikemoto K., Niwa S., Ebihara T., Katoh M., Yokoi T., Asahi S. (2006) Pharm. Res. 23, 1460–1472 [DOI] [PubMed] [Google Scholar]
- 38. Hsiao P., Bui T., Ho R. J., Unadkat J. D. (2008) Drug Metab. Dispos. 36, 481–484 [DOI] [PubMed] [Google Scholar]
- 39. Taub M. E., Podila L., Ely D., Almeida I. (2005) Drug Metab. Dispos. 33, 1679–1687 [DOI] [PubMed] [Google Scholar]
- 40. Higgins C. F., Linton K. J. (2004) Nat. Struct. Mol. Biol. 11, 918–926 [DOI] [PubMed] [Google Scholar]
- 41. Ambudkar S. V., Kim I. W., Sauna Z. E. (2006) Eur. J. Pharm. Sci. 27, 392–400 [DOI] [PubMed] [Google Scholar]
- 42. Ritchie T. K., Grinkova Y. V., Bayburt T. H., Denisov I. G., Zolnerciks J. K., Atkins W. M., Sligar S. G. (2009) Methods Enzymol. 464, 211–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chifflet S., Torriglia A., Chiesa R., Tolosa S. (1988) Anal. Biochem. 168, 1–4 [DOI] [PubMed] [Google Scholar]
- 44. Davies D. R., Hol W. G. (2004) FEBS Lett. 577, 315–321 [DOI] [PubMed] [Google Scholar]
- 45. Russell P. L., Sharom F. J. (2006) Biochem. J. 399, 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee J. Y., Urbatsch I. L., Senior A. E., Wilkens S. (2008) J. Biol. Chem. 283, 5769–5779 [DOI] [PubMed] [Google Scholar]
- 47. Georges E., Tsuruo T., Ling V. (1993) J. Biol. Chem. 268, 1792–1798 [PubMed] [Google Scholar]
- 48. Zhou Y., Gottesman M. M., Pastan I. (1999) Arch Biochem. Biophys. 367, 74–80 [DOI] [PubMed] [Google Scholar]
- 49. Goda K., Nagy H., Mechetner E., Cianfriglia M., Szabó G., Jr. (2002) Eur. J. Biochem. 269, 2672–2677 [DOI] [PubMed] [Google Scholar]