Background: ASCs are promising for skeletal regeneration, but their heterogeneity limits their use.
Results: Microfluidic analysis and FACS identified a cellular subset (CD105low) with enhanced osteogenic capacity.
Conclusion: CD105 depletion was found to enhance osteogenesis through reduction of TGF-β1 signaling.
Significance: We illuminate the functional relevance of hASC heterogeneity and enhance understanding of CD105 with respect to osteogenic differentiation.
Keywords: Adipose Tissue, Bone, Cell Sorting, SMAD Transcription Factor, Transforming Growth Factor Beta (TGFbeta), Adipose-derived Stromal Cells, Osteogenesis, Stem Cell Biology, Tissue Engineering
Abstract
Clinically available sources of bone for repair and reconstruction are limited by the accessibility of autologous grafts, infectious risks of cadaveric materials, and durability of synthetic substitutes. Cell-based approaches for skeletal regeneration can potentially fill this need, and adipose tissue represents a promising source for development of such therapies. Here, we enriched for an osteogenic subpopulation of cells derived from human subcutaneous adipose tissue utilizing microfluidic-based single cell transcriptional analysis and fluorescence-activated cell sorting (FACS). Statistical analysis of single cell transcriptional profiles demonstrated that low expression of endoglin (CD105) correlated with a subgroup of adipose-derived cells with increased osteogenic gene expression. FACS-sorted CD105low cells demonstrated significantly enhanced in vitro osteogenic differentiation and in vivo bone regeneration when compared with either CD105high or unsorted cells. Evaluation of the endoglin pathway suggested that enhanced osteogenesis among CD105low adipose-derived cells is likely due to identification of a subpopulation with lower TGF-β1/Smad2 signaling. These findings thus highlight a potential avenue to promote osteogenesis in adipose-derived mesenchymal cells for skeletal regeneration.
Introduction
Approximately 2 million bone grafting procedures are performed worldwide each year, procedures that are costly, painful, and often fail to provide a durable solution (1). Several recent studies, however, have shown that a cell-based approach using the stromal vascular fraction (SVF)6 from adipose tissue may represent an attractive alternative to traditional bone grafting (2–8). Although this strategy holds promise in the treatment of various bone defects, significant challenges must still be overcome. One such challenge is the marked heterogeneity present within the SVF (9, 10). Although cell surface antigen expression has been studied among the adipose SVF and culture-expanded cells (termed adipose-derived stromal cells or ASCs), no consensus exists on cell surface markers identifying adipose-derived mesenchymal progenitors or osteoprogenitor cells (9, 11–13). This difficulty is further compounded by the well documented, rapidly changing cell surface phenotype of culture-expanded ASCs (9, 13).
Several groups have reported expression of the cell surface receptor endoglin (CD105) to be correlated with stem cell capacity within mesenchymal cells of adipose tissue and bone marrow origin (10, 14–17). Although mesenchymal stromal cells have been traditionally defined by positive expression of CD105 (8, 18, 19), several groups have also observed considerable phenotypic drift within ASCs during in vitro expansion (10, 20–22). Specifically, CD105 expression has been found to rapidly increase from almost zero in freshly harvested cells to nearly ubiquitous expression after 4–7 days in culture (4, 16). This phenomenon may explain conflicting reports regarding CD105 as a marker for ASCs.
CD105 has also been found to be involved in a variety of other biological processes. Studies have linked endoglin to angiogenesis and neovascularization, the development of pre-eclampsia, scleroderma, and psoriasis (23–26). Furthermore, mutations in CD105 have been identified in patients with hereditary hemorrhagic telangiectasia, and recent reports have shown solubilized endoglin to bind with high affinity to bone morphogenetic proteins (BMPs)-9 and -10 (23).
In this study, using single cell transcriptional analysis, we found expression patterns for CD105 to be closely associated with the osteogenic potential of ASCs. Fluorescence-activated cell sorting based on CD105 was then employed to enrich ASCs for more highly osteogenic mesenchymal cells (10). By combining these approaches, we identified CD105low ASCs to be capable of enhanced osteogenic differentiation relative to CD105high and unsorted cells. Mechanistically, CD105 has been shown to function as a TGF-β1 co-receptor (23, 28). As TGF-β1 is a known inhibitor of mesenchymal cell osteogenic differentiation, cells with lower CD105 expression and thus diminished TGF-β1 signaling would be expected to more rapidly form bone (29, 30). This concept was further investigated in our study through RNA interference and analysis of multiple downstream signaling pathways.
Overall, from a clinical perspective, definition of the role CD105 plays in ASC osteogenic differentiation may enable development of improved strategies to treat various skeletal defects. Whether through isolation of an enriched population based on this cell surface marker or by targeted manipulation of CD105 levels, ASC-mediated bone regeneration may be potentially enhanced. Strategies in which harvested cells are re-implanted back into the same patient for the purposes of skeletal tissue engineering may ultimately prove to be an attractive alternative or adjuvant therapy to traditional bone grafting.
EXPERIMENTAL PROCEDURES
Lipoaspiration specimens were obtained after acquiring informed consent from patients, in accordance with Stanford University Institutional Review Board guidelines. ASCs were harvested from the adipose tissue of 15 female patients between the ages of 18 and 65 without major medical conditions who were undergoing elective lipoaspiration of the abdomen, flank, and/or thigh region as described previously (8, 31).
For gene expression studies, cells were serum-starved 24 h before analysis to synchronize cell cycle. Single cells were then sorted using a FACSAria (BD Biosciences) into the wells of 96-well plates containing Platinum Taq reverse transcriptase, polymerase master mix (Invitrogen), and TaqMan primers for each gene target (Applied Biosystems, Foster City, CA) (supplemental Table S1). Single cell lysates underwent target-specific 22-cycle preamplification, and quantitative PCR was performed in the BioMark Reader per the manufacturer's instructions (Fluidigm, San Francisco, CA). Data were normalized relative to the median expression of each gene and converted to base 2 logarithms. Single cell transcriptional profiles were clustered using an adaptive fuzzy c-means algorithm (32). This heuristic employs an objective optimization loop to determine the clustering parameters that best describe cells while minimizing theoretical information loss (supplemental Fig. S1A). Linear discriminant analysis was applied to determine those genes whose expression patterns maximally differed between groups, and the sensitivity and specificity of this partitioning scheme were assessed using traditional receiver operating characteristic (ROC) analysis of the true positive/negative and false positive/negative rates for multiple possible gene combinations (33).
Serum-starved and non-serum-starved ASCs were sorted following 36 h in culture using a fluorescein-conjugated anti-human CD105 antibody (R&D Systems, Minneapolis, MN). Unsorted cells and CD105high and CD105low cells were then seeded at 100,000 cells/well in a 6-well plate or 35,000 cells in a 12-well plate and treated with osteogenic differentiation medium (ODM) (31). For select experiments, ODM was supplemented with rhTGF-β1 (20 or 40 ng/ml), rhBMP-2 (200 ng/ml; R&D Systems), anti-BMP-2 neutralizing antibody (BMPi; 20 μg/ml), or the small molecule Alk-5 inhibitor SB-525334 (10 ng/ml) (Sigma-Aldrich). For other experiments, groups included unsorted hASCs transfected with either control short hairpin RNA (shRNA) or CD105 shRNA lentiviral particles (Santa Cruz Biotechnology, Santa Cruz, CA). Alkaline phosphatase staining and quantification were performed at 3 days, as described previously (34). Photometric quantification of Alizarin red stain was performed at 7 days to assay bone nodule formation (31). Adipogenic differentiation was assessed as described previously using Oil Red O staining and photometric quantification after 7 days of differentiation (31). RNA was harvested after 3 and 7 days of osteogenic and adipogenic differentiation, and gene expression was examined by quantitative RT-PCR using the Applied Biosystems Prism 7900HT sequence detection system (see supplemental Table S2 for a list of primers). Serum-free conditioned medium and cell lysate were prepared as described previously (35), and TGF-β1 protein was quantified by double sandwich ELISA according to the manufacturer's instructions (R&D Systems).
For evaluation of in vivo osteogenesis, critical-sized (4-mm) calvarial defects were created in the right parietal bone of 60-day-old male CD-1 nude mice (Crl:CD-1-Foxn1nu; Charles River Laboratories, Wilmington, MA) (36). Apatite-coated (HA) PLGA scaffolds were fabricated, and for select experiments, these scaffolds were designed to release BMP-2 (200 μg/ml) (37). Each scaffold was implanted alone or was seeded with 150,000 cells (36). Animals were treated with HA PLGA scaffold, BMP-2 releasing scaffold, or anti-BMP-2 releasing scaffold seeded with the following cell types: 1) unsorted ASCs, 2) CD105high ASCs, 3) CD105low ASCs, 4) unsorted ASCs treated with a control shRNA, and 5) unsorted ASCs treated with a CD105 shRNA (Santa Cruz Biotechnology). As controls, no scaffold, as well as PLGA scaffold alone and BMP-2 releasing scaffold alone, were used.
Micro-CT was performed using a high-resolution MicroCAT IITM (ImTek Inc., Knoxville, TN) small animal imaging system as described previously (17). hASC viability in vivo was demonstrated weekly using an IVIS 200 (Caliper LifeSciences, Hopkinton, MA) after hASC transduction with a triple fusion lentivirus linked to luciferase and GFP (17). At 1 and 8 weeks postoperatively, calvaria were harvested, formalin-fixed, decalcified in 19% EDTA, and paraffin-embedded. Bone formation was assessed by aniline blue and pentachrome staining (38). In situ hybridization was performed at 1 week to detect expression of runx2, osteocalcin (Ocn), and TGF-β1, as described previously (34). FISH analysis for human sex chromosomes was also performed, as female hASCs were implanted in male mice (17).
Immunohistochemistry was performed using rabbit polyclonal anti-Smad2 or rabbit polyclonal anti-pSmad2 at 1:80 dilution (Santa Cruz Laboratories). VECTASTAIN ABC was used with the appropriate biotinylated secondary antibodies (Vector Laboratories, Burlingame, CA). Slides without primary antibody were used as a negative control. Visualization was performed with diaminobenzidine solution (Zymed Laboratories Inc., South San Francisco, CA).
Early passage hASCs were expanded in vitro to subconfluence before viral infection. Hexadimethrine bromide (5 g/ml) was added to augment infection efficiency of the CD105 shRNA lentiviral construct (Santa Cruz Biotechnology). Cells were infected four times, and puromycin (2 μg/ml) selection was performed for 3 days following the final infection. Flow cytometric analysis of GFP expression by the vehicle control was performed to assess transduction efficiency. Knockdown of endoglin was verified.
Means and standard deviations were calculated from numerical data. In figures, bar graphs represent means, and error bars represent one standard deviation. Unless otherwise stated, statistical analysis was performed using an ANOVA for multiple group comparisons and by a two-tailed Student's t test to directly compare two groups. Inequality of standard deviations was excluded by employing the Levene's test.
RESULTS
Osteogenic Gene Expression Is Heterogeneous across ASC Subpopulations
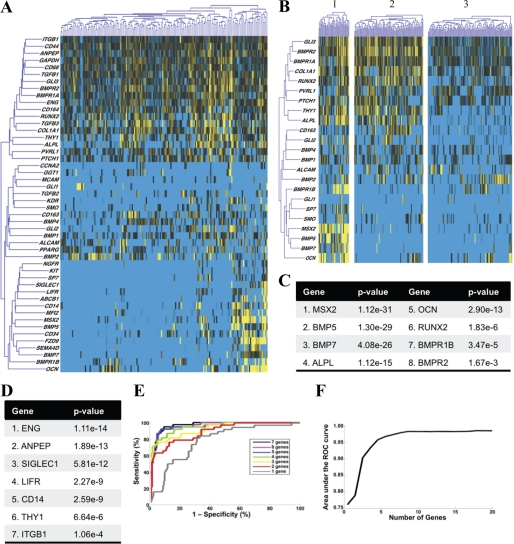
Using single cell transcriptional profiling of 48 genes, we observed considerable transcriptional heterogeneity among 200 human ASCs (Fig. 1A). Partitioning based on transcriptional profiles grouped cells into three clusters (Fig. 1B). Expression of osteogenic genes was highest in cluster 1, as determined qualitatively by the heat map visual representation of the single cell transcriptional data (Fig. 1B). To identify those genes best able to differentiate between cells with highly osteogenic transcriptional profiles (cluster 1) and those with lower expression of osteogenic genes (clusters 2 and 3), we compared the distribution of gene expression across all three clusters using a non-parametric, two-sample Kolmogorov-Smirnov analysis (39, 40). This analysis identified eight osteogenesis-related genes (Fig. 1C) whose distribution exhibited a statistically significant association with cluster membership (*, p < 0.01 following Bonferroni correction for multiple samples) (41). Expression levels of the genes MSX2, BMP-5, BMP-7, ALP, OCN, and RUNX2 were the most highly associated, with transcription of each differentially elevated in cluster 1 (i.e. expression of these genes predicted to which cluster a cell would be assigned). Thus, expression levels of these genes largely determine the osteogenic transcriptional profile of ASCs that were assigned membership to individual clusters. However, to isolate cells according to this osteogenic gene expression profile, it was necessary to correlate surface antigen expression patterns with the transcriptional patterns observed in our single cell analysis.
FIGURE 1.
Microfluidic-based single cell transcriptional analysis of human SVF. A, hierarchical clustering of single cell gene expression within human SVF cells. Genes and cells are organized in rows and columns, respectively. Gene expression is presented as -fold change from median. Considerable transcriptional heterogeneity is evident. B, clustering of gene expression data using an information theory-based algorithm that groups cells based on similarity of transcriptional profiles. The number of clusters is optimized objectively to minimize information loss, resulting in the partitioning of ASCs into three clusters. C, we observed eight genes that demonstrated a statistically significant association with the cluster membership of the ASCs using a strict cutoff of *, p < 0.01 (41). D, linear discriminant analysis was used to determine those surface antigen genes best able to differentiate between cluster 1 and clusters 2 and 3 (33). E, ROC analysis of the sensitivity and specificity of the clustering results. Groups of genes (i.e. best “1 gene” or best “3 gene” combination) were evaluated using ROC analysis. As expected, more accurate classification is achieved with a larger number of genes. F, ROC analysis of up to 20 genes for maximum achievable sensitivity and specificity, defined by the area under the associated ROC curves for each gene combination.
CD105 Allows for Optimal Clustering Based on Osteogenic Gene Expression
We applied linear discriminant analysis (42) to determine which surface antigen genes were expressed in patterns that most closely mirrored expression of the osteogenic genes identified above (supplemental Fig. S1B). This identified seven surface antigen genes (Fig. 1D) whose expression patterns were predictive of the cluster-defining osteogenic genes (Fig. 1C). We found the expression pattern of the surface antigen gene endoglin to exhibit the highest single association, which suggests that this marker may be used to separate cells according to differences in the transcriptional state of these osteogenic genes. Given the complexity of transcriptional networks, however, characterizing the exact nature of this relationship would require considerably more rigorous analysis. Nevertheless, these data indicate the expression pattern of the surface-accessible marker CD105 to be predictive for expression of other osteogenic genes. As such, CD105 represents a good candidate marker to use for separating cells based on osteogenic gene expression (Fig. 1, D and E). A second cell surface receptor that also appeared to correlate with expression of osteogenic genes independent from CD105 was Thy-1 (Fig. 1D). Known as CD90, this surface marker has previously been shown to be associated with osteoprogenitor cells and may thus be used to potentially isolate osteogenic cells (55).
Partitions outlined by linear discriminant analysis (and therefore the surface antigens identified as candidates for prospective isolation) were highly sensitive and specific to produce robust clustering of ASCs (Fig. 1, E and F). This therefore confirmed that the three clusters identified in Fig. 1B represent transcriptionally defined subpopulations that can be reliably identified based on single cell transcriptional data. Accordingly, we would expect that prospective isolation of these subpopulations would separate ASCs with differences in osteogenic capacity. Although we cannot necessarily predict the nature of these differences, with respect to CD105 and its function as a co-receptor for TGF-β1, we would expect that CD105low cells would exhibit reduced TGF-β1 signaling.
CD105low Sorted Cells Demonstrate Increased Osteogenic Potential in Vitro
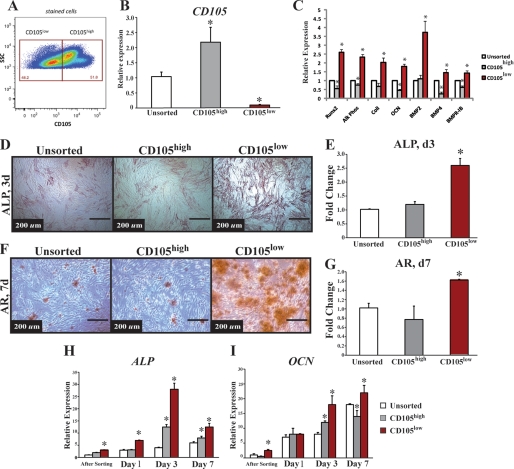
A strategy using FACS sorting on as few markers as possible would allow for the largest capture of cells in the shortest amount of time. Because gene expression of ENG was best predictive of osteogenic gene expression, we sorted non-serum-starved ASCs into CD105high, CD105low, and unenriched (sorted for live cells only) groups using FACS (data not shown and Fig. 2A). Sorting at 36 h yielded a CD105high population comprising 51.8% of the cells and a CD105low population comprising 48.2%. Analysis following FACS revealed over 98.3% enrichment for each population. Importantly, CD105high cells demonstrated significantly higher gene expression of CD105 (Fig. 2B, *, p < 0.05), thus further confirming the fidelity of the sort.
FIGURE 2.
FACS analysis and gene expression of CD105 and osteogenic differentiation of CD105high, CD105low, and unsorted ASCs. A, FACS analysis of CD105 expression 36 h after ASC harvest. B, to confirm that cell surface marker sorting is indicative of transcriptional profile, we demonstrated that cells sorted with high CD105 do indeed have significantly higher expression of CD105 (*, p < 0.05). C, gene expression of early (RUNX-2, ALP, and COL1) and late (OCN) osteogenesis as well as genes involved in the BMP pathway (BMP-2, BMP-4, and BMPR1B). Across all genes, CD105low cells had greater expression (*, p < 0.05). D, alkaline phosphatase stain. E, quantification of unsorted, CD105high, and CD105low ASCs (*, p < 0.05). F, Alizarin red stain. G, quantification comparing unsorted, CD105high, and CD105low hASCs. (*, p < 0.05). H, gene expression of ALP (*, p < 0.05). I, OCN during osteogenesis over time starting immediately after the sort and following for 7 days. The CD105low cells appear to maintain a higher expression profile of ALP and OCN over time (*, p < 0.05). Statistical analysis was performed with either a one-way ANOVA (cell population) or a two-way ANOVA (cell population and time) followed by post hoc individual comparisons.
Transcriptional analysis immediately after sorting revealed that the CD105low subpopulation was enriched for a more osteogenic subtype as demonstrated by increased expression of runx2 ALP, collagen1 (COL1), and OCN (Fig. 2C, *, p < 0.05). In addition, we found that the CD105low subpopulation exhibited significantly greater expression of the BMP pathway genes BMP-2, BMP-4, and BMPR-1B, all of which have been shown to play a key role in osteogenic differentiation (Fig. 2C, *, p < 0.05) (43). These findings, however, represent relative gene expression and do not necessarily reflect how these cells would respond to osteogenic differentiation. Furthermore, CD105high and CD105low populations represented cell populations after removal of non-SVF cells based on size gates, as well as removal of doublets (supplemental Fig. S2). This purity, we believe, accounted for some of the large differences in gene expression.
To determine whether this increase in osteogenic gene expression correlated with an increase in osteogenic differentiation, we exposed the two sorted ASC subpopulations (CD105high and CD105low) as well as the unsorted population to in vitro osteogenic differentiation conditions. Staining at day 3 for alkaline phosphatase (ALP), an early marker of bone formation, was increased in the CD105low population (Fig. 2, D and E, *, p < 0.05). We assessed terminal osteogenic differentiation using Alizarin red staining for extracellular matrix mineralization at day 7 and found that CD105low ASCs created a significantly greater amount of mineralized extracellular matrix when compared with CD105high or unsorted cells (Fig. 2, F and G, *, p < 0.05). In addition, the CD105low population demonstrated persistent up-regulation of the osteogenic genes ALP (Fig. 2H, *, p < 0.05) and OCN (Fig. 2I, *, p < 0.05) during osteogenic differentiation.
To eliminate any potential for the introduction of artifact through changes in cell cycle profile following FACS isolation, serum-starved ASCs were also sorted for CD105 prior to evaluation of in vitro osteogenic differentiation. No differences were appreciated in CD105 expression between serum-starved and non-serum-starved ASCs (data not shown). Also, despite synchronization of cell cycle, similar differences were noted in both alkaline phosphatase and Alizarin red staining, with serum-starved CD105low ASCs demonstrating enhanced osteogenesis relative to similarly treated CD105high and unsorted cells (supplemental Fig. S3, *, p < 0.05). Collectively, single cell transcriptional analysis thus allowed for correlation of cell surface markers with an osteogenic transcriptional profile that persisted over time in vitro. These data strongly suggest that cell sorting based on expression of CD105 effectively enriches for a more osteogenic ASC subpopulation.
Cells primed to undergo osteogenic differentiation are theoretically less likely to undergo adipogenic differentiation as an inverse relationship has been repeatedly observed between these differentiation pathways (44). Indeed, we observed the CD105high population to have marked adipogenic potential by Oil Red O staining and gene expression analysis (supplemental Fig. S4, A and B). With respect to chondrogenic differentiation, pentachrome staining after cell implantation demonstrated an absence of cartilage (blue), indicating that bone formation occurred by intramembranous ossification for unsorted, CD105high, and CD105low ASCs (supplemental Fig. S4C).
CD105low CD90high Sorted Cells Demonstrate Increased Osteogenic Potential in Vitro
As CD90 expression was similarly identified by transcriptional analysis to be associated with osteogenic cells, FACS isolation of ASCs was also performed using this surface marker (Fig. 1D and supplemental Fig. S5). Like CD105, CD90 sorted cells were heterogeneous and appeared to cluster based on osteogenic gene expression (supplemental Fig. S5B). We then sorted and clustered cells on both CD105 and CD90 and demonstrated that CD105low CD90high cells appeared to identify ASCs with high osteogenic gene expression (supplemental Fig. S5, C and D). Therefore, we would expect cells with different combinations of CD105 and CD90 expression to exhibit different osteogenic transcriptional profiles and capacity for osteogenic differentiation.
Evaluation of in vitro osteogenesis revealed CD105low CD90high cells to be more osteogenic when compared with CD105low CD90lowcells (supplemental Fig. S6). Importantly, however, the CD105low CD90high population represents just 16.0% of ASCs and less than 3% of all harvested cells. Because of these small numbers, from a clinical perspective, we do not believe that the incremental difference in osteogenesis was significant enough to warrant sorting on two markers over just one.
CD105low Enriched Cells Demonstrate High Osteogenic Gene Expression in Vivo and Accelerate Healing of Critical-sized Calvarial Defects
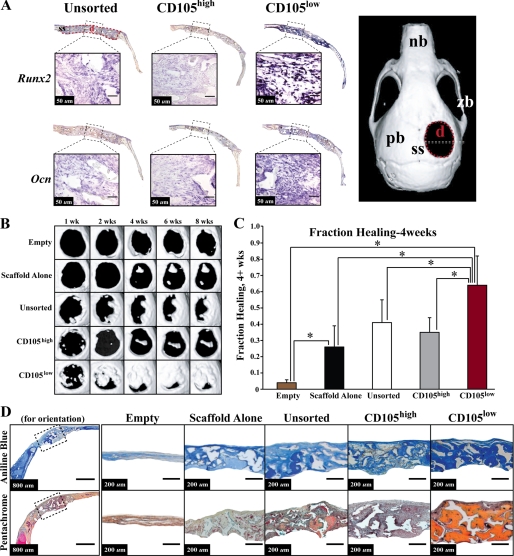
To evaluate the in vivo osteogenic capacity of these ASC populations, we created 4-mm critical-sized parietal bone defects in skeletally mature athymic CD-1 mice (17, 45). Repair was performed using CD105high, CD105low, or unsorted cells seeded onto apatite-coated PLGA scaffolds (17). Controls included defects treated with an empty scaffold and untreated defects without scaffold or cells. We performed in situ hybridization at day 5 following implantation of cells and found an increase in Runx2 (Fig. 3A, top row) and Ocn (Fig. 3A, bottom row) expression within defects containing CD105low ASCs relative to defects receiving CD105high or unsorted cells (Fig. 3A). Control sense probes of Runx2 and Ocn showed minimal background staining (data not shown).
FIGURE 3.
Osteogenic gene expression in vivo and healing of a critical-sized 4-mm calvarial defect. A, our analysis demonstrated an increase in early osteogenic marker Runx-2 (top row) and late osteogenic marker Ocn (bottom row) transcript expression via in situ hybridization within defects containing CD105low ASCs at day 5. The full defect is shown above each image with a scale bar shown within each close up of the defect site. Right side, nb = nasal bone, zb = zygomatic bone, pb = parietal bone, ss = sagittal suture, d = defect. B, micro-CT of mouse calvaria from 1 week until 8 weeks after injury. At each time point, the CD105low-treated defects demonstrated improved bone healing. C, quantification of osseous healing by micro-CT revealed significantly more healing with CD105low cells relative to both CD105high cells and unsorted cells as well as empty and scaffold alone control at the 4-week time point (*, p < 0.05). D, calvarial defects 4 mm in size were allowed to heal for 8 weeks before histological analysis by aniline blue and pentachrome stains. Five treatment groups included those defects left empty (Empty), those treated with a scaffold but without hASCs (Scaffold Alone), and those treated with unsorted, CD105high, or CD105low hASCs. Pictures were taken in the middle of the defect site. In aniline blue stains, bone appears dark blue. In pentachrome stains, bone appears yellow.
Radiographic analysis was performed from 1 to 8 weeks using micro-CT (n = 4 animals per group). Defects treated with CD105low ASCs demonstrated significantly increased de novo bone regeneration when compared with defects treated with CD105high ASCs or empty scaffold (Fig. 3B). The unsorted population demonstrated a moderate healing capacity (Fig. 3B, third row). Quantification of healing at 4 weeks postoperatively demonstrated increased bone formation in defects treated with CD105low cells when compared with all other groups (Fig. 3C, *, p < 0.05).
Histological analysis with aniline blue (in which osteoid appears dark blue) and pentachrome staining (in which mature bone stains yellow) of calvarial defects at 8 weeks correlated with micro-CT findings. Defects treated with scaffold alone showed some bony healing, particularly at the edges of the defect (Fig. 3D, third column). In contrast, robust and thick bony regeneration was observed throughout defects treated with CD105low cells (Fig. 3D, far right). Defects treated with CD105high ASCs demonstrated some bone formation, but to a lesser degree when compared with the CD105low groups (Fig. 3D, fifth column).
Persistence and viability of human cells within defects during the time of bone regeneration were demonstrated with in vivo imaging systems after infecting ASCs with a GFP/luciferase lentivirus before implantation (supplemental Fig. S7A). Persistence was verified in the region of the newly generated bone by FISH analysis for the human female X chromosome as all xenografts were sex mismatched (supplemental Fig. S7B).
Improved Osteogenesis in the CD105low Population Is Associated with Decreased TGF-β1 Signaling
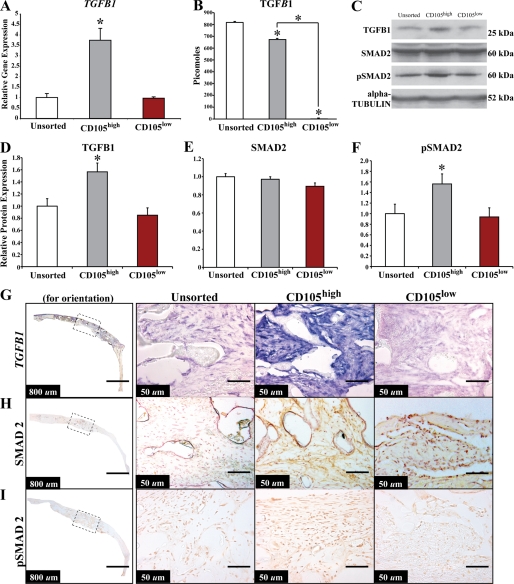
CD105 is a known co-receptor for TGF-β1 (46). Although the effects of TGF-β1 signaling on osteogenesis are variable, overall continuous TGF-β1 treatment reduces late osteogenic differentiation. Our laboratory and others have previously demonstrated that TGF-β1 inhibits osteogenesis in osteocompetent cell lines (29, 30). In aggregate, the CD105high cells had increased TGF-β1 gene expression (Fig. 4A, *, p < 0.05). These findings were consistent with the single cell gene expression findings (Fig. 1A), which demonstrated a strong pairwise linear correlation of 0.755 between TGF-β1 and ENG gene expression, suggesting a positive relationship of expression between these two genes. Moreover, CD105high cells showed a statistically significant increase in secretion of TGF-β1 relative to CD105low ASCs, as measured by an ELISA assay of serum-free cell-conditioned medium when normalized to cell number (Fig. 4B, *, p < 0.05). Finally, by Western blot analysis, CD105high cells demonstrated higher levels of TGF-β1 protein (Fig. 4, C and D).
FIGURE 4.
Analysis of the TGF-β1 pathway in vitro and in vivo of unsorted, CD105high, and CD105low cells. A, TGF-β1 quantitative RT-PCR of unsorted, CD105high, and CD105low cells (*, p < 0.05). B, TGF-β1 ELISA demonstrating the amount of TGF-β1 in unsorted, CD105high, and CD105low cells (*, p < 0.05). C, Western blot images of TGF-β1, Smad2, pSmad2, and α-tubulin. D, TGF-β1 protein levels in the cytosolic fraction of unsorted, CD105high, and CD105low cells (*, p < 0.05). E, Smad2 levels in the cytosolic portion of unsorted, CD105high, and CD105low cells. F, pSmad2 levels in the nuclear fraction of unsorted, CD105high, and CD105low cells (*, p < 0.05). G, in situ hybridization for TGF-β1 gene expression in calvarial defects treated with unsorted, CD105high, and CD105low cells demonstrating a greater signal in the CD105high-treated defects. H, immunohistochemistry for Smad2 of unsorted, CD105high, and CD105low cells demonstrating similar signal in all groups. I, immunohistochemistry for pSmad2 of unsorted, CD105high, and CD105low cells demonstrating greater nuclear signal in the CD105high-treated defects. The left column shows parietal bone for orientation with the dotted line indicating the center of the defect and the region that is magnified in subsequent images on the right.
We examined whether this difference in TGF-β1 expression affected downstream Smad2 signaling. Overall, CD105high ASCs showed increased pSmad2 levels relative to other cells, as demonstrated by Western blot (Fig. 4, C, E, and F). These data indicate that the difference in osteogenic differentiation seen between the two cell populations may be associated with differences in TGF-β1/Smad2 signaling and suggest that this pathway may be manipulated to augment osteogenesis. In vivo analysis using in situ hybridization demonstrated significantly increased TGF-β1 transcript abundance, as well as increased pSmad2 activation in the CD105high population when compared with defects treated with CD105low or unsorted cells (Fig. 4, G–I).
TGF-β1 Inhibits Osteogenic Differentiation and Inhibition of Alk-5 Signaling Enhances Osteogenic Differentiation
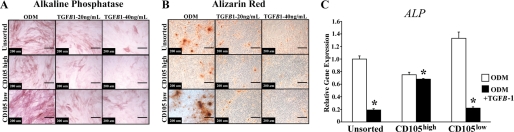
We next supplemented unsorted, CD105high, and CD105low cells with TGF-β1 protein in higher doses than previously investigated (29). At these high doses, TGF-β1 inhibited osteogenesis in all three populations (Fig. 5, A and B). TGF-β1 treatment also resulted in similar inhibition of ALP gene expression. (Fig. 5C, *, p < 0.05).
FIGURE 5.
Osteogenic differentiation of unsorted, CD105high-, and CD105low-treated cells with TGF-β. A, alkaline phosphatase staining after 3 days of differentiation with ODM alone or in combination with TGF-β1 (20 and 40 ng/ml). B, Alizarin red staining after 7 days of differentiation with ODM alone or in combination with TGF-β1 (20 and 40 ng/ml). C, ALP expression after TGF-β1 (40 ng/ml) addition to ODM after 3 days of differentiation (*, p < 0.05).
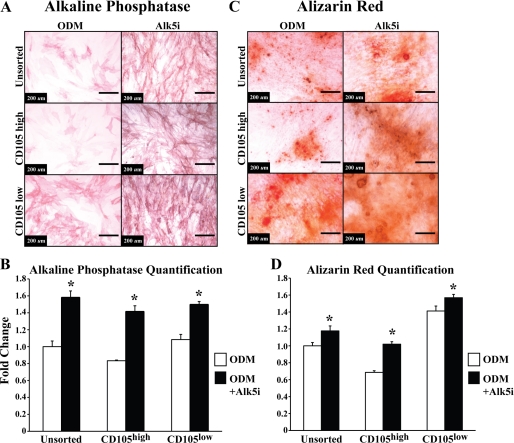
Having demonstrated that an increase in TGF-β1 signaling decreased osteogenic differentiation, we next determined whether blocking this pathway through a small molecule inhibitor would do the opposite and make CD105high cells more CD105low-like. Because Alk-5 exists downstream from TGF-β1 and CD105, inhibition of Alk-5 might promote osteogenesis. As expected, treatment of sorted cells with SB-525334, a small molecule Alk-5 inhibitor, resulted in increased osteogenic differentiation of all ASCs at both early and late time points (Figs. 6, A–D, *, p < 0.05), in effect making CD105high cells more CD105low-like.
FIGURE 6.
Osteogenic differentiation of unsorted, CD105high, and CD105low cells treated with Alk-5 inhibitor. A, alkaline phosphatase staining after 3 days of differentiation with ODM alone or in combination with Alk-5 inhibitor (10 ng/ml). B, quantification of alkaline phosphatase activity normalized to total protein content after 3 days of differentiation with ODM alone or in combination with Alk-5 inhibitor (10 ng/ml) (*, p < 0.05). C, Alizarin red staining after 7 days of differentiation with ODM alone or in combination with Alk-5 inhibitor (10 ng/ml). D, photometric quantification of Alizarin red staining. Pictures are of 10× magnification, n = 3 (*, p < 0.05). Statistical analysis was performed with either a one-way ANOVA (cell population) or a two-way ANOVA (cell population and treatment) followed by post hoc individual comparisons.
Differences in Osteogenic Differentiation between the CD105high, CD105low and Unsorted Groups Are Independent of the Smad1/5 Pathway
ASCs have been shown to signal through the BMP-2 pathway, and we noted that BMP-2 gene expression was elevated in the CD105low population. We thus set out to explore whether there were differences in downstream BMP signaling through the Smad1/5 pathway. We noted that at baseline, CD105high, CD105low, and unsorted cells had similar Smad5 levels (supplemental Fig. S8, A and B). After stimulation with BMP-2, both CD105high and CD105low cells showed up-regulated levels of pSmad1/5 (supplemental Fig. S8, A and C). Likewise, both CD105high and CD105low ASCs behaved similarly to exogenous BMP-2, with enhanced Runx2 gene expression and in vitro differentiation (supplemental Fig. S8, D–F). Finally, in response to BMP-2 released from a scaffold, both CD105high and CD105low ASCs demonstrated increased pSmad1/5 levels and accelerated bone regeneration in vivo (supplemental Fig. S8, G and H). Therefore, CD105 does not appear to alter ASC responsiveness to BMP.
Reduced Endoglin Expression Promotes hASC Osteogenic Differentiation
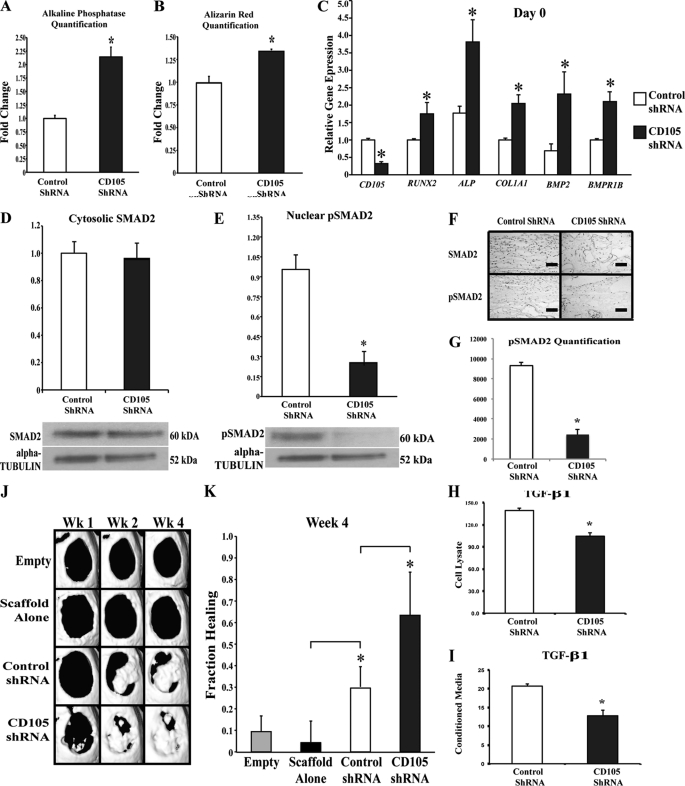
To further test the biology of the endoglin/TGF-β1 pathway, we knocked down endoglin transcript using a lentiviral vector containing an shRNA construct against CD105. Transduction efficiency was over 98% by FACS for GFP and immunofluorescence (data not shown). In vitro shRNA knockdown of CD105 enhanced osteogenic differentiation in a manner consistent with CD105low sorting (Fig. 7, A and B, *, p < 0.05). In addition, early osteogenic gene expression was up-regulated in CD105 shRNA-treated cells, including RUNX-2, ALP, and COL1 (Fig. 7C, *, p < 0.05). As with the sorted CD105low population, those cells with CD105 knockdown demonstrated decreased levels of pSmad2 despite similar levels of non-phosphorylated Smad2 in vitro and in vivo (Fig. 7, D–G, *, p < 0.05). Also, like the sorted populations, CD105 shRNA-transfected cells had a decrease in TGF-β1 ligand levels as measured by an ELISA of cell lysate and conditioned medium (Fig. 7, H and I, *, p < 0.05). Finally, ASCs transfected with CD105 shRNA lentiviral particles allowed for more rapid bone regeneration in vivo (Fig. 7, J and K, *, p < 0.05).
FIGURE 7.
In vitro analysis of osteogenic activity in CD105 knockdown ASCs. A, alkaline phosphatase quantification after 3 days of differentiation of control shRNA ASCs (left) and CD105 knockdown ASCs (right) (*, p < 0.05). B, Alizarin red quantification after 7 days of differentiation (*, p < 0.05). C, CD105 gene expression levels on day 0, confirming successful knockdown of CD105. Osteogenic gene expression levels of Runx2, ALP, Col1, Bmp2, and Bmpr1b at day 0 were determined (*, p < 0.05). D, Western blot analysis of control shRNA-transfected versus CD105 knockdown cells analyzing Smad2 expression. E, Western blot analysis of control shRNA-transfected versus CD105 knockdown cells analyzing pSmad2 expression (*, p < 0.05). F, immunohistochemical analysis of Smad2 and pSmad2 in defects treated with control shRNA or CD105 shRNA-transfected cells. G, quantification of immunohistochemical staining showed significantly less pSmad2 with CD105 knockdown cells (*, p < 0.05). H, TGF-β1 ELISA of cell lysate for control and CD105 shRNA-transfected cells (*, p < 0.05). I, TGF-β1 ELISA of conditioned medium for control and CD105 shRNA-transfected cells (*, p < 0.05). J, representative three-dimensional micro-CT reconstructions of 4-mm calvarial defects treated with no scaffold or cells (Empty), a scaffold alone, a scaffold seeded with ASCs transfected with control shRNA, or a scaffold seeded with CD105 shRNA-transfected ASCs. K, fraction healing of 4-mm calvarial defects. (n = 4 for each group, *, p < 0.05). Statistical analysis was performed with a one-way ANOVA (cell population) followed by post hoc individual comparisons.
DISCUSSION
At present, prospective enrichment of the stromal vascular fraction of human adipose tissue to isolate homogeneous stem cell subpopulations remains elusive. In this study, single cell transcriptional analysis utilizing a microfluidic-based process was employed to determine correlations between expression of various surface markers and clusters of osteogenic transcriptional activity. With this approach, both endoglin (CD105) and Thy-1 (CD90) were found to be associated with differences in expression of osteogenic genes among ASCs.
Identification of these candidate cell surface markers allowed for subsequent isolation of subpopulations to evaluate osteogenic differentiation. Immediately after ASC harvest, however, the CD105low population represents over 90% of cells. Although an early sort would appear to offer only 10% enrichment, isolation of cells based on size gates, singlet gates, and propidium iodide staining would still remove a large unwanted fraction composed of non-SVF and non-viable cells. In our study, we used ASCs 36 h after harvest and isolated an extremely pure population By sorting at 36 h, we were able to obtain equivalent sized fractions of CD105high and CD105low ASCs. This allowed for controlled experiments in large numbers and more accurate analysis of the mechanism behind the observed cellular differences.
Studies have shown endoglin to be a transmembrane co-receptor for TGF-β1 receptors Alk-1 and Alk-5 (47). The effect of endoglin on vascular endothelial cells and hemostasis has been widely studied (48, 49); however, a detailed analysis of its relevance to hASC osteogenic differentiation has not been performed. Previous studies have used expression of this receptor to sort various cells for osteogenic and chondrogenic enrichment (10, 14, 46). Although these studies have found varying outcomes when using the CD105+ population, they did not use ASCs from subcutaneous adipose tissue. Gazit and co-workers (14) found that bone marrow-derived cells demonstrated enhanced in vitro osteogenic differentiation in the CD105+ fraction; however, a comparison of these cells with the CD105low and unsorted populations was not provided. In our study, we similarly found CD105high cells to have osteogenic potential in vitro and in vivo; however, the CD105low cells appeared to be more osteogenic. In addition to examining a different cell type, the study by Gazit and co-workers (14) utilized different methodologies than in the current work and failed to show any differences in vivo. During their cell isolation, they used magnetic beads for enrichment, and they noted that only 80% of the CD105+ cells were positive for CD105. In contrast, following our sort, the CD105high population showed significantly greater expression of CD105, and FACS analysis revealed over 98.3% enrichment. Also lacking in the study by Gazit and co-workers (14) was an unsorted control, which is of key importance. Furthermore, the high magnification images of defects treated with the CD105low population did not demonstrate bone matrix mineralization, which is contrary to the nature of mesenchymal stem cells. Lastly, their in vivo model utilized a collagen sponge with rhBMP-2 and CD105+ cells, and they failed to compare this with CD105low cells or the sponge alone. Importantly, we have previously demonstrated that BMP-2 alone can stimulate bone formation in vivo (50).
Other studies analyzing cells based on CD105 expression have also offered little detail about the mechanism behind the differences noted in their enriched populations (10). Endoglin has been shown to bind to TGF-β1 and TGF-β3 but not TGF-β2 (47). TGF-β signaling has been shown to function through both Smad-dependent and Smad-independent pathways, including MAPK signaling (51–53). We chose to focus on the Smad-dependent pathway as other studies have failed to demonstrate the functional relevance of MAPK signaling to TGF-β-mediated responses in vivo (54). Furthermore, other groups have previously shown that endoglin enhances Alk-5/Smad2 signaling (27, 46). In this study, we demonstrated that the CD105high subpopulation of ASCs has higher levels of pSmad2 relative to CD105low cells. Because TGF-β1 has been shown to inhibit osteogenesis in mesenchymal cells, we hypothesized that lower TGF-β1 signaling, specifically through Alk-5, would lead to enhanced osteogenesis (30). This was what we observed as we demonstrated that our CD105low population had decreased pSmad2 levels and enhanced osteogenesis in vitro and in vivo. Similarly, RNAi-mediated suppression of CD105 resulted in a parallel acceleration of bone differentiation by ASCs. Finally, if a decrease in CD105 causes an increase in osteogenic differentiation due to a reduction in TGF-β1 signaling, we hypothesized that TGF-β1 administration should mitigate the osteogenic differentiation advantage of the CD105low cells. Indeed, this was precisely what was observed in our results.
Regardless of CD105 expression level, however, all cells responded to BMP-2 with an up-regulation of pSmad1/5 and an increase in osteogenic differentiation in vitro. Furthermore, in vivo BMP-2 treatment increased pSmad1/5 levels and enhanced healing of calvarial defects with both CD105high and CD105low cells. Interestingly, other studies have shown endoglin to bind BMP-9 and -10, inhibiting interaction with type II receptors (23). As both CD105high and CD105low ASCs appear to respond well to BMP in vitro and in vivo, however, direct interaction between CD105 and the pro-osteogenic BMP-2 appears less likely. Instead, the data suggest that the osteogenic effects of CD105 may be secondary to alterations in the signaling pathway of TGF-β1. Furthermore, the BMP-Smad1/5 pathway may be independent to the TGF-β signaling pathway in ASCs. Future studies may thus look to decrease TGF-β1 signaling while simultaneously increasing BMP-2 signaling to optimize healing of skeletal defects.
We anticipate that findings from our approach will illuminate the functional relevance of hASC transcriptional heterogeneity and enhance our understanding of CD105 with respect to osteogenic differentiation. Our findings also have potential translational ramifications as the feasibility of cutting through stem cell heterogeneity to enrich for specific subpopulations has wide significance. It is thus foreseeable that cell-based strategies for bone tissue engineering may one day incorporate enrichment strategies or targeted suppression of CD105 to enhance skeletal regeneration. Also, given the inverse relationship between bone and fat, a better understanding of CD105 and its mechanism in regulating ASC differentiation may similarly lead to development of novel treatment strategies aimed at adipogenic regeneration.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants 1 R21 DE019274-01 and 1 RC2 DE020771-01 from the NIDCR (to M. T. L.) and 2 RO1 DK074095-07 from the NIDDK (to G. C. G). This work was also supported by grants from the National Endowment for Plastic Surgery, the Oak Foundation, and the Hagey Laboratory for Pediatric Regenerative Medicine (to M. T. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. S1–S8.
- SVF
- stromal vascular fraction
- ASC
- adipose-derived stromal cell
- hASC
- human ASC
- BMP
- bone morphogenetic protein
- ALP
- alkaline phosphatase
- pSmad
- phosphorylated Smad
- ROC
- receiver operating characteristic
- ODM
- osteogenic differentiation medium
- PLGA
- poly(lactic-co-glycolic acid)
- ANOVA
- analysis of variance
- micro-CT
- micro-computed tomography
- rh
- recombinant human.
REFERENCES
- 1. Levit K., Wier L., Stranges E., Ryan K., Elixhauser A. (2007) HCUP Facts and Figures: Statistics on Hospital-based Care in the United States, United States Agency for Healthcare Research and Quality, Rockville, MD [Google Scholar]
- 2. Lendeckel S., Jödicke A., Christophis P., Heidinger K., Wolff J., Fraser J. K., Hedrick M. H., Berthold L., Howaldt H. P. (2004) J. Craniomaxillofac. Surg. 32, 370–373 [DOI] [PubMed] [Google Scholar]
- 3. Yoon S. H., Shim Y. S., Park Y. H., Chung J. K., Nam J. H., Kim M. O., Park H. C., Park S. R., Min B. H., Kim E. Y., Choi B. H., Park H., Ha Y. (2007) Stem Cells 25, 2066–2073 [DOI] [PubMed] [Google Scholar]
- 4. Mitchell J. B., McIntosh K., Zvonic S., Garrett S., Floyd Z. E., Kloster A., Di Halvorsen Y., Storms R. W., Goh B., Kilroy G., Wu X., Gimble J. M. (2006) Stem Cells 24, 376–385 [DOI] [PubMed] [Google Scholar]
- 5. Peterson B., Zhang J., Iglesias R., Kabo M., Hedrick M., Benhaim P., Lieberman J. R. (2005) Tissue Eng. 11, 120–129 [DOI] [PubMed] [Google Scholar]
- 6. Hattori H., Sato M., Masuoka K., Ishihara M., Kikuchi T., Matsui T., Takase B., Ishizuka T., Kikuchi M., Fujikawa K., Ishihara M. (2004) Cells Tissues Organs 178, 2–12 [DOI] [PubMed] [Google Scholar]
- 7. Aust L., Devlin B., Foster S. J., Halvorsen Y. D., Hicok K., du Laney T., Sen A., Willingmyre G. D., Gimble J. M. (2004) Cytotherapy 6, 7–14 [DOI] [PubMed] [Google Scholar]
- 8. Zuk P. A., Zhu M., Ashjian P., De Ugarte D. A., Huang J. I., Mizuno H., Alfonso Z. C., Fraser J. K., Benhaim P., Hedrick M. H. (2002) Mol. Biol. Cell 13, 4279–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoshimura K., Shigeura T., Matsumoto D., Sato T., Takaki Y., Aiba-Kojima E., Sato K., Inoue K., Nagase T., Koshima I., Gonda K. (2006) J. Cell Physiol. 208, 64–76 [DOI] [PubMed] [Google Scholar]
- 10. Rada T., Reis R. L., Gomes M. E. (2011) Stem Cell Rev. 7, 64–76 [DOI] [PubMed] [Google Scholar]
- 11. Lin C. S., Xin Z. C., Deng C. H., Ning H., Lin G., Lue T. F. (2010) Histol. Histopathol. 25, 807–815 [DOI] [PubMed] [Google Scholar]
- 12. Zimmerlin L., Donnenberg V. S., Pfeifer M. E., Meyer E. M., Péault B., Rubin J. P., Donnenberg A. D. (2010) Cytometry A 77, 22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zannettino A. C., Paton S., Arthur A., Khor F., Itescu S., Gimble J. M., Gronthos S. (2008) J. Cell Physiol. 214, 413–421 [DOI] [PubMed] [Google Scholar]
- 14. Aslan H., Zilberman Y., Kandel L., Liebergall M., Oskouian R. J., Gazit D., Gazit Z. (2006) Stem Cells 24, 1728–1737 [DOI] [PubMed] [Google Scholar]
- 15. Yu G., Wu X., Dietrich M. A., Polk P., Scott L. K., Ptitsyn A. A., Gimble J. M. (2010) Cytotherapy 12, 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang T., Liu W., Lv X., Sun H., Zhang L., Liu Y., Zhang W. J., Cao Y., Zhou G. (2010) Biomaterials 31, 3564–3571 [DOI] [PubMed] [Google Scholar]
- 17. Levi B., James A. W., Nelson E. R., Vistnes D., Wu B., Lee M., Gupta A., Longaker M. T. (2010) PLoS ONE 5, e11177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop Dj., Horwitz E. (2006) Cytotherapy 8, 315–317 [DOI] [PubMed] [Google Scholar]
- 19. Gronthos S., Franklin D. M., Leddy H. A., Robey P. G., Storms R. W., Gimble J. M. (2001) J. Cell Physiol. 189, 54–63 [DOI] [PubMed] [Google Scholar]
- 20. Rada T., Gomes M. E., Reis R. L. (2011) J. Tissue Eng. Regen. Med. 5, 655–664 [DOI] [PubMed] [Google Scholar]
- 21. Katz A. J., Tholpady A., Tholpady S. S., Shang H., Ogle R. C. (2005) Stem Cells 23, 412–423 [DOI] [PubMed] [Google Scholar]
- 22. McIntosh K., Zvonic S., Garrett S., Mitchell J. B., Floyd Z. E., Hammill L., Kloster A., Di Halvorsen Y., Ting J. P., Storms R. W., Goh B., Kilroy G., Wu X., Gimble J. M. (2006) Stem Cells 24, 1246–1253 [DOI] [PubMed] [Google Scholar]
- 23. Castonguay R., Werner E. D., Matthews R. G., Presman E., Mulivor A. W., Solban N., Sako D., Pearsall R. S., Underwood K. W., Seehra J., Kumar R., Grinberg A. V. (2011) J. Biol. Chem. 286, 30034–30046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pohl D., Andrýs C., Borská L., Fiala Z., Hamaková K., Ettler K., Krejsek J. (2011) Acta Medica 54, 59–62 [DOI] [PubMed] [Google Scholar]
- 25. Holmes A. M., Ponticos M., Shi-Wen X., Denton C. P., Abraham D. J. (2011) J. Cell Commun. Signal. 5, 173–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nassiri F., Cusimano M. D., Scheithauer B. W., Rotondo F., Fazio A., Yousef G. M., Syro L. V., Kovacs K., Lloyd R. V. (2011) Anticancer Res. 31, 2283–2290 [PubMed] [Google Scholar]
- 27. Guerrero-Esteo M., Sanchez-Elsner T., Letamendia A., Bernabeu C. (2002) J. Biol. Chem. 277, 29197–29209 [DOI] [PubMed] [Google Scholar]
- 28. Zhang L., Magli A., Catanese J., Xu Z., Kyba M., Perlingeiro R. C. (2011) Blood 118, 88–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levi B., James A. W., Xu Y., Commons G. W., Longaker M. T. (2010) Plast. Reconstr. Surg. 126, 412–425 [DOI] [PubMed] [Google Scholar]
- 30. Maeda S., Hayashi M., Komiya S., Imamura T., Miyazono K. (2004) EMBO J. 23, 552–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Levi B., James A. W., Glotzbach J. P., Wan D. C., Commons G. W., Longaker M. T. (2010) Plast. Reconstr. Surg. 126, 822–834 [DOI] [PubMed] [Google Scholar]
- 32. Glotzbach J. P., Januszyk M., Vial I. N., Wong V. W., Gelbard A., Kalisky T., Thangarajah H., Longaker M. T., Quake S. R., Chu G., Gurtner G. C. (2011) PLoS One 6, e21211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peterson L. E., Coleman M. A. (2008) Int. J. Approx. Reason. 47, 17–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. James A. W., Theologis A. A., Brugmann S. A., Xu Y., Carre A. L., Leucht P., Hamilton K., Korach K. S., Longaker M. T. (2009) PLoS One 4, e7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li S., Quarto N., Longaker M. T. (2007) Am. J. Physiol. Cell Physiol. 293, C1834–C1842 [DOI] [PubMed] [Google Scholar]
- 36. Cowan C. M., Shi Y. Y., Aalami O. O., Chou Y. F., Mari C., Thomas R., Quarto N., Contag C. H., Wu B., Longaker M. T. (2004) Nat. Biotechnol. 22, 560–567 [DOI] [PubMed] [Google Scholar]
- 37. Lee M., Chen T. T., Iruela-Arispe M. L., Wu B. M., Dunn J. C. (2007) Biomaterials 28, 1862–1870 [DOI] [PubMed] [Google Scholar]
- 38. Xu Y., Hammerick K. E., James A. W., Carre A. L., Leucht P., Giaccia A. J., Longaker M. T. (2009) Tissue Eng. Part A 15, 3697–3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ong L. D., LeClare P. C. (1968) Health Phys. 14, 376. [PubMed] [Google Scholar]
- 40. Eadie W., Drijard F., Roos M., Sadoulel B. (1971) Statistical Methods in Experimental Physics, pp. 313–317, North-Holland Publishing Co., Amsterdam [Google Scholar]
- 41. Devlin B., Roeder K., Wasserman L. (2003) Genet. Epidemiol. 25, 36–47 [DOI] [PubMed] [Google Scholar]
- 42. Wallace S. A., Anderson D. I., Anderson D., Mayo A. M., Nguyen K. T., Ventre M. A. (1999) Percept. Mot. Skills 88, 759–764 [DOI] [PubMed] [Google Scholar]
- 43. Wan D. C., Shi Y. Y., Nacamuli R. P., Quarto N., Lyons K. M., Longaker M. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12335–12340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gimble J. M., Nuttall M. E. (2004) Endocrine 23, 183–188 [DOI] [PubMed] [Google Scholar]
- 45. Gupta D. M., Kwan M. D., Slater B. J., Wan D. C., Longaker M. T. (2008) J. Craniofac. Surg. 19, 192–197 [DOI] [PubMed] [Google Scholar]
- 46. Bernabeu C., Conley B. A., Vary C. P. (2007) J. Cell. Biochem. 102, 1375–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cheifetz S., Bellón T., Calés C., Vera S., Bernabeu C., Massagué J., Letarte M. (1992) J. Biol. Chem. 267, 19027–19030 [PubMed] [Google Scholar]
- 48. Li C., Issa R., Kumar P., Hampson I. N., Lopez-Novoa J. M., Bernabeu C., Kumar S. (2003) J. Cell Sci. 116, 2677–2685 [DOI] [PubMed] [Google Scholar]
- 49. Kumar P., Wang J. M., Bernabeu C. (1996) J. Pathol. 178, 363–366 [DOI] [PubMed] [Google Scholar]
- 50. Levi B., Nelson E. R., Li S., James A. W., Hyun J. S., Montoro D. T., Lee M., Glotzbach J. P., Commons G. W., Longaker M. T. (2011) Stem Cells 29, 1241–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yu L., Hébert M. C., Zhang Y. E. (2002) EMBO J. 21, 3749–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mulder K. M. (2000) Cytokine Growth Factor Rev. 11, 23–35 [DOI] [PubMed] [Google Scholar]
- 53. Yue J., Mulder K. M. (2000) J. Biol. Chem. 275, 30765–30773 [DOI] [PubMed] [Google Scholar]
- 54. Janssens K., ten Dijke P., Janssens S., Van Hul W. (2005) Endocr. Rev. 26, 743–774 [DOI] [PubMed] [Google Scholar]
- 55. Chan C. K., Chen C. C., Luppen C. A., Kim J. B., DeBoer A. T., Wei K., Helms J. A., Kuo C. J., Kraft D. L., Weissman I. L. (2009) Nature 457, 490–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.