Abstract
NALP3 inflammasome, composed of the three proteins NALP3, ASC, and Caspase-1, is a macromolecular complex responsible for the innate immune response against infection with bacterial and viral pathogens. Formation of the inflammasome can lead to the activation of inflammatory caspases, such as Caspase-1, which then activate pro-inflammatory cytokines by proteolytic cleavage. The assembly of the NALP3 inflammasome depends on the protein-interacting domain known as the death domain superfamily. NALP3 inflammasome is assembled via a pyrin domain (PYD)/PYD interaction between ASC and NALP3 and a caspase recruitment domain/caspase recruitment domain interaction between ASC and Caspase-1. As a first step toward elucidating the molecular mechanisms of inflammatory caspase activation by formation of inflammasome, we report the crystal structure of the PYD from NALP3 at 1.7-Å resolution. Although NALP3 PYD has the canonical six-helical bundle structural fold similar to other PYDs, the high resolution structure reveals the possible biologically important homodimeric interface and the dynamic properties of the fold. Comparison with other PYD structures shows both similarities and differences that may be functionally relevant. Structural and sequence analyses further implicate conserved surface residues in NALP3 PYD for ASC interaction and inflammasome assembly. The most interesting aspect of the structure was the unexpected disulfide bond between Cys-8 and Cys-108, which might be important for regulation of the activity of NALP3 by redox potential.
Keywords: Autoimmunity, Death Domain, Inflammation, Innate Immunity, Protein Structure
Introduction
The inflammasome is a macromolecular complex responsible for the innate immune response against infection with bacterial and viral pathogens (1, 2). It is assembled in response to upstream intracellular sensors of pathogens. The inflammasome functions as a platform to recruit Caspase-1, providing proximity for self-activation (3). Activated Caspase-1 by formation of inflammasome processes the inactive inflammatory cytokines pro-interleukin1β and pro-interleukin18 to active, leading to NF-κB activation and elicitation of innate immunity (4, 5).
Currently, four distinct inflammasomes have been identified as follows: the NALP1 inflammasome, the NALP2 inflammasome, the NALP3 inflammasome, and AIM2 inflammasome (6). The most studied among these is the NALP3 inflammasome, which is activated by several bacterial ligands, nucleic acids, synthetic antiviral compounds, cellular toxins, and some Toll-like receptor agonists (7–9). The critical function of the NALP3 inflammasome in the cell has been well indicated by studying a relationship between mutations within the NALP3 gene with autoinflammatory diseases. Muckle-Wells syndrome, familial cold autoinflammatory syndrome (10), and chronic infantile neurological cutaneous and articular syndrome are linked to the NALP3 mutations (11).
NALP3 (NACHT, leucine-rich region, and PYD2 domains containing protein 3), ASC (apoptosis-associated speck-like protein containing a caspase-recruitment domain), and Caspase-1 are three protein components that form the NALP3 inflammasome. NALP3 contains N-terminal PYD (pyrin domain), a central NACHT (NAIP, CIITA, HET-E, and TP1), and a C-terminal leucine-rich region and performs an important role in sensing signal and oligomerization (12). ASC is a bipartite adaptor protein containing an N-terminal PYD domain and a C-terminal CARD domain and plays a role in connecting NALP3 and Caspase-1 (13, 14). Caspase-1 is a prototypical inflammatory caspase that is responsible for the maturation of the inflammatory cytokines interleukin1β and interleukin18 (15, 16). It possesses the CARD domain at its N terminus for protein/protein interaction (16).
The assembly of the NALP3 inflammasome depends on the protein interacting domain known as the death domain superfamily, which is composed of the following four subfamilies: death domain (DD), death effector domain (DED), caspase-recruitment domain (CARD), and PYD (17, 18). Death domain superfamily is the one of the biggest families of protein domains and highly prevalent in apoptotic and inflammatory signaling proteins (19). NALP3 inflammasome is assembled via a PYD/PYD interaction between ASC and NALP3 and a CARD/CARD interaction between ASC and Caspase-1 (3, 20). Despite the fundamental importance of the inflammasome, including NALP3 inflammasome in innate immunity and many immune disorders, limited structural information is available. Moreover, several reported PYD structures were derived from studies by nuclear magnetic resonance (NMR). These include the PYD of NALP1 (21), NALP7 (22), ASC (23, 24), and POP1 (25). Structural studies of the PYD domain and the PYD/PYD interaction have been difficult because PYD domains are subject to self-aggregation under physiological conditions (26). Although several DD superfamily complex structures have been reported, no structural information is available for PYD complexes. The structural studies of many PYDs are essential for understanding the molecular basis of inflammation signaling. Here, we report the first crystal structure of NALP3 PYD at 1.7 Å resolution. Although NALP3 PYD has the canonical six-helical bundle structural fold similar to other PYDs, the high resolution structure reveals possible biologically important homodimeric interface and the dynamic properties of the fold. Comparison with other PYD structures showed both similarities and differences that may be functionally relevant. Structural and sequence analyses further implicated conserved surface residues in NALP3 PYD, many of which are hydrophobic and charged, for ASC interaction and inflammasome assembly. The most interesting aspect of the structure was the unexpected disulfide bond between Cys-8 and Cys-108, which might be important for regulation of the activity of NALP3 by redox potential.
MATERIALS AND METHODS
Protein Expression and Purification
The expression and purification methods used in this study have been described elsewhere in detail. In summary, human NALP3 PYD (amino acids 3–110) was expressed in Escherichia coli BL21 (DE 3) by overnight induction at 20 °C. The protein contained a C-terminal His tag and was purified by nickel affinity and gel filtration chromatography. Superose 6 gel filtration column 10/30 (GE healthcare) that had been pre-equilibrated with a solution of 20 mm sodium citrate at pH 5.0 and 150 mm NaCl was used. The protein was concentrated to 6–8 mg/ml.
MALS
The molar mass of NALP3 PYD was determined by MALS. NALP3 PYD was injected onto a Superdex 200 HR 10/30 gel filtration column (GE Healthcare) equilibrated in a buffer containing 20 mm sodium citrate, pH 5.0, and 150 mm NaCl. The chromatography system was coupled to a MALS detector (mini-DAWN EOS) and a refractive index detector (Optilab DSP) (Wyatt Technology).
Crystallization and Data Collection
The crystallization conditions were initially screened at 20 °C by the hanging drop vapor-diffusion method using various screening kits. Initial crystals were grown on the plates by equilibrating a mixture containing 1 μl of protein solution (6–8 mg ml−1 protein in 20 mm sodium citrate, pH 5.0, 150 mm NaCl) and 1 μl of a reservoir solution containing 0.2 m ammonium sulfate, 0.1 m sodium acetate trihydrate, pH 4.6, 30% polyethylene glycol monomethyl ether 2.000 against 0.4 ml of reservoir solution. Crystallization was further optimized by searching over a range of concentrations of protein, PEG MME 2.000, ammonium sulfate. Selenomethionine-substituted NALP3 PYD was produced using previously established method (27) and crystallized similarly. A single wavelength anomalous diffraction data set was collected at the selenium peak wavelength at the BL-4A beamline at Pohang Accelerator Laboratory, Republic of Korea. Data processing and scaling were carried out in the HKL2000 package (28). A 2.2-Å native data set was also collected.
Structure Determination and Analysis
Selenium positions were found with HKL2MAP (29) using the data set collected at the peak wavelength. Phase calculation and phase improvement were performed at a resolution of 1.7 Å using the programs SOLVE and RESOLVE (30). Approximately 80% of the structure was auto-traced. Model building and refinement were performed in COOT (31) and Refmac5 (32), respectively. Water molecules were added automatically with the ARP/wARP function in Refmac5 and then were examined manually for reasonable hydrogen bonding possibilities. The final atomic model contains residues 5–94 for A chain and 5–110 for B chain. The quality of the model was checked using PROCHECK and was found to be good. A total of 96.4% of the residues are located in the most favorable region, and 3.6% are in the allowed regions of the Ramachandran plot. The data collection and refinement statistics are summarized in Table 1. Ribbon diagrams and molecular surface representations were generated using the program PyMOL (45).
TABLE 1.
Crystallographic statistics
| Selenomethionine | Native | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, and c | 42.0, 60.0, 51.5 Å | 42.0, 60.0, 51.5 Å |
| β | 107.7 | 107.6 |
| Resolution | 50 to 1.7 Å | 50 to 2.2 Å |
| Rsyma | 9.2% (30.0%) | 3.5% (6.6%) |
| I/σIa | 34.5 (6.8) | 44.0 (27.4) |
| Completenessa | 98.3% (97.1%) | 98.9% (98.0%) |
| Redundancya | 3.8 (3.8) | 8.9 (8.9) |
| Refinement | ||
| Resolution | 50–1.7 Å | |
| No. of reflections used (completeness) | 26,472 (100%) | |
| Rwork/Rfree | 18.5/23.5% | |
| No. of atoms | ||
| Protein | 1608 | |
| Water and other small molecule | 254 | |
| Average B-factors | ||
| Protein | 13.3 Å2 | |
| Water and other small molecule | 28.3Å2 | |
| Root mean square deviations | ||
| Bond lengths | 0.026Å | |
| Bond angles | 2.142° | |
| Ramachandran plot | ||
| Most favored regions | 96.4% | |
| Additional allowed regions | 3.6% | |
a The highest resolution shell is shown in parentheses.
Sequence Alignment
The amino acid sequence of PYDs was analyzed using ClustalW.
Protein Data Bank Accession Codes
The coordinates and structural factors have been deposited in Protein Data Bank with accession code 3QF2.
RESULTS AND DISCUSSION
NALP3 PYD Structure
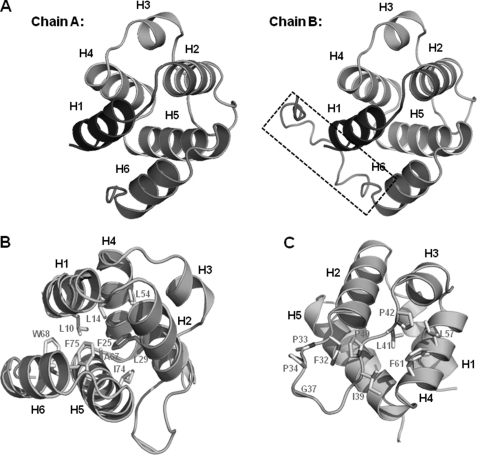
The 1.7 Å crystal structure of NALP3 PYD was solved using a single wavelength anomalous diffraction method and refined to an Rwork = 18.5% and Rfree = 23.5%. The high resolution structure of NALP3 PYD showed that it includes six helices, H1 to H6, which form the canonical anti-parallel six-helical bundle fold characteristic of the DD superfamily (Fig. 1A). There were two monomers in the asymmetric unit, called chain A and chain B (Fig. 1A). Model of chain B was built from residues 3 to 110, and that of chain A was built from residues 3 to 96. Extra residues built in chain B at the C terminus are indicated by a dashed box (Fig. 1A, right panel). They formed a symmetric dimer. Helix 3 (H3) that was not well defined in the structure of NALP1 PYD is obviously detected in this structure. The length of H3 is shorter than that of other helices. The N and C termini of NALP3 PYD reside at the same side of the molecule. The six helices consisting of residues 6–16, 21–31, 43–48, 51–62, 64–78, and 81–90 are numbered H1, H2, H3, H4, H5, and H6, respectively (Fig. 1A). The helix bundle tightly packed by a central hydrophobic core included Leu-10, Ala-11, Tyr-13, and Leu-14 from H1; Phe-25 and Leu-29 from H2; Leu-54, Ala-55, and Met-58 from H4; Ala-67, Ile-74, and Phe-75 from H5; and Ala-87 from H6 (Fig. 1B). The second hydrophobic cluster that was essential for the stabilizing H3 to the core of the NALP7 PYD was also detected at the NALP3 PYD structure (Fig. 1C). Residues Phe-32, Ile-39, Pro-40, Leu-41, Pro-42, Leu-57, and Phe-61 are involved in the formation of the second hydrophobic patch for NALP3 PYD (Fig. 1C). Residues buried within the core of NALP3 PYD and residues for second hydrophobic cluster are conserved among different PYDs that are supposed to interact with ASC (Fig. 2A). Five loops, including residues 17–20 (H1-H2 loop), 32–42 (H2-H3 loop), 49–50 (H3-H4 loop), 63–64 (H4-H5 loop), and 79–80 (H5-H6 loop), connect the six helices.
FIGURE 1.
Crystal structure of NALP3 PYD. A, ribbon diagram of NALP3 PYD. Chain A and chain B are shown separately. Helices are labeled. Extra structure of C terminus on chain B is shown by a black dashed box. B, conserved central hydrophobic core. Hydrophobic residues essential for the formation of this core that stabilizes H1 to H6 (except H3) are shown as a gray stick. C, second hydrophobic patch. Helices are labeled, and residues involved in the formation of exposed second hydrophobic patch are shown as a gray stick.
FIGURE 2.
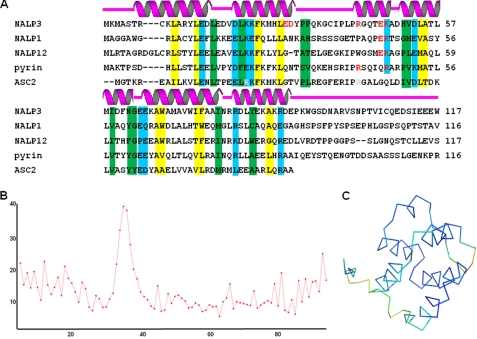
Structural based sequence alignment and B-factor distribution. A, structure-based sequence alignment of NALP3 PYD with other ASC-binding proteins. Secondary structures (helices H1 to H6) are shown above the sequences. Residues at the hydrophobic core are shaded in yellow; conserved exposed hydrophobic and charged residues are shaded in green and cyan, respectively. Residues involved in the interaction between chain A and chain B are colored in red. B, B-factor distribution. Whole residue B-factors are plotted. The x and y axes indicate residue number and B-factor, respectively. C, B-factor distribution on the structure. Warm and cold colors indicate high and low B-factors, respectively.
The structure of NALP3 PYD is highly compact and ordered with an average B factor of 13.3 Å2 (Table 1). Plotting individual B factors for each residue showed that H2, H3, H4, and H5 have the lowest B factors, whereas the H2-H3 loop has the highest B factor (Fig. 2, B and C). The C-terminal sequences are also more flexible with higher B factors. This trend in B factor distribution may be consistent with the lack of further polar interactions between the H2 and H3 loop and the remaining part of the structure. As expected, residues that are buried in the hydrophobic core tend to have lower B factors, both in their main chains and their side chains. In contrast, the more exposed residues may have low main chain B factors but often have higher side chain B factors. The positional spread of the NMR structures of NALP1 PYD and NALP7 PYD shows a similar distribution of flexibility, suggesting that this dynamic behavior may be universal for all PYDs (21, 22).
The structure of NALP3 PYD provides the first step toward elucidating the inflammasome-mediated Caspase-1 activation. Conserved hydrophobic residues are essential for the formation of the overall fold. The second hydrophobic patch that allows for the formation of H3 and anchors it to H2, which detected the structure of NALP7 PYD but not that of NALP1 PYD, was also detected at NALP3 PYD. The hydrophobicity of this patch is not as strong as that of NALP7 PYD. This second hydrophobic patch is important for the formation of H3.
Dimer Interface within NALP3 PYD
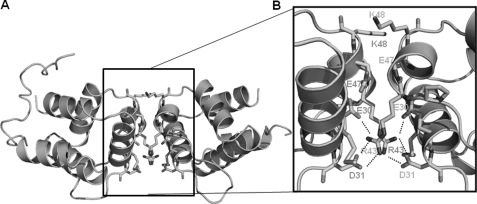
The structure of NALP3 PYD also reveals interesting insights on the novel homodimeric interfaces that may be formed by crystallographic packing. The two NALP3 PYDs in the asymmetric unit are packed as a symmetric dimer whose interface is predominantly electrostatic interaction (Fig. 3A). A total dimer surface buries 641 Å2 (a monomer surface area of 320 Å2), which represents 5.5% of dimer surface area. The principal contacts are made by Glu-30 (on H2), Asp-31 (on H2), and Arg-43 (H3) from one NALP3 PYD molecule (chain A) and by same residues from the second molecule (chain B) (Fig. 3B). Arg-43 of H3 on chain A forms a salt bridge with Glu-30 and Asp-31 of H3 on chain B (Fig. 3B) and vice versa. Because the isolated NALP3 PYD behaves as a monomer or dimer in solution depending on the salt concentration and pH, as judged by size-exclusion chromatography and MALS, stoichiometry of NALP3 PYD is hard to determine (supplemental Fig. 1). Thus, this dimeric structure might be coincidentally formed by crystal packing. However, it is still possible that this dimer is a biologically meaningful dimer. However, it is still possible that this dimer is a biologically meaningful dimer because the DD superfamily openly forms a stable dimer for the function (33). In addition, several reports showed that monomer/dimer transition can be detected at the DD superfamily (34). Although the oligomerized form of NALP3 PYD was detected, the majority of NALP3 PYD exists as a monomer in a physiological condition based on MALS experiment. The calculated monomeric molecular mass of NALP3 PYD, including the C-terminal His tag, was 12.685 Da, and the calculated molecular mass from MALS was 12.019 Da (0.7% fitting error), with a polydispersity of 1.000 (supplemental Fig. 1).
FIGURE 3.
Dimeric interface of the structure of NALP3 PYD. A, dimeric structure of NALP3 PYD. B, close-up view of the interacting residues in the interface between two dimers. Helices are labeled, and residues involved in the contact are shown as sticks. Salt bridges formed between Arg-43 from one chain and Glu-30 and Asp-31 from counterparts are shown as dashed lines.
Comparison with Other PYD Structures
A structural homology search with DALI (35) showed that NALP3 PYD is highly similar to other PYDs (Table 2). The top eight matches, with Z-scores from 13.8 to 9.0, are six PYDs, ASC (23), ASC2 (25), NALP7 (22), NALP10, MNDA, and NALP1 (21), and two DEDs, vFLIP (36, 37) and FADD DED (38). Detecting two DED structures as a structurally similar domain with NALP3 PYD indicates that the structure of PYD is relatively close to that of DED among the death domain superfamily. A structure-based sequence alignment showed that most of the residues buried in the hydrophobic core of NALP3 PYD are also conserved among these highly divergent PYD structures, suggesting a conserved hydrophobic core across all PYDs (Fig. 2).
TABLE 2.
Structural similarity search using DALI (35)
r.m.s.d. means root mean square deviation.
| Proteins and accession numbers | Z-score | r.m.s.d. | Identity | Refs. |
|---|---|---|---|---|
| Å | % | |||
| ASC PYD (1ucp) | 13.8 | 2.0 | 22 | 23 |
| ASC2 (2hm2) | 13.3 | 1.9 | 28 | 25 |
| NALP7 PYD (2km6) | 12.2 | 2.2 | 26 | 22 |
| vFLIP DED (2f1s) | 10.1 | 2.0 | 13 | 36 |
| NALP10 PYD (2do9) | 10.0 | 2.0 | 26 | Not published |
| MNDA PYD (2dbg) | 9.8 | 2.3 | 14 | Not published |
| NALP1 PYD (1pn5) | 9.3 | 2.1 | 26 | 21 |
| FADD DED (1fad) | 9.0 | 2.0 | 18 | 38 |
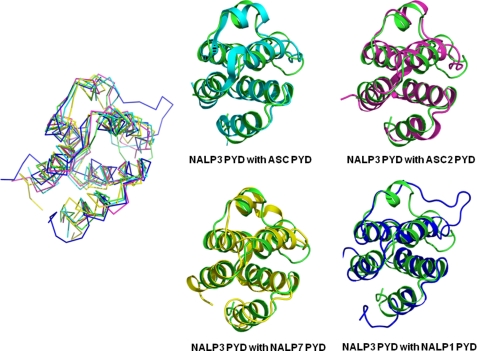
Pairwise structural alignments between NALP3 PYD and these other PYDs showed that the helices in NALP3 PYD slightly differ from them in their lengths and orientations (Fig. 4). For example, in comparison with NALP7 and NALP1, helix H2 of NALP3 PYD shows differences in orientations. The conspicuous differences are detected at the loops connecting the helices, especially those at H2-H3 (Fig. 4). The location of the H2-H3 loop, especially compared with NALP1, is completely opposite, indicating the dynamic property of this loop among various PYDs. A relatively small helix of H3 is also an interesting feature that is only detected on the structure of PYDs.
FIGURE 4.
Superposition of NALP3 PYD with their structural homologues. NALP3 PYD and four structural homologues are superimposed (green, NALP3 PYD; cyan, ASC PYD; magenta, ASC2 PYD; yellow, NALP7 PYD; blue, NALP1 PYD). Pairwise structural comparisons are also performed. NALP3 PYD is colored in green, and each counterpart is colored in cyan for ASC PYD, magenta for ASC2 PYD, yellow for NALP7 PYD, and blue for NALP1 PYD.
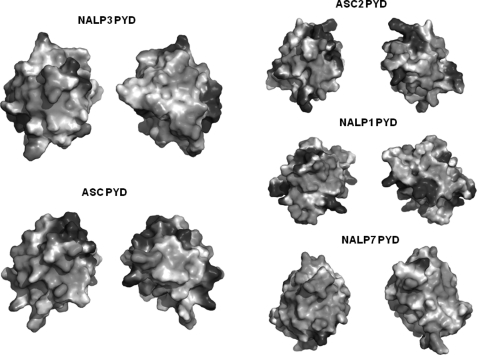
NALP3 PYD has similar gross features in its electrostatic surface, in comparison with other ASC-binding PYDs such as NALP1 PYD and ASC2 PYD (Fig. 5). Similar to the charged nature of most of the other ASC-binding PYDs, NALP3 PYD surface is also the mixture of positively and negatively charged features. This is the case for both sides of the molecule (Fig. 5). Because PYDs are protein interaction modules, their surface features dictate their mode of interactions with partners. In the case of ASC2 PYD, although no direct structural information is available, it has been implicated that its charged surface is important for interaction with ASC PYD (39). Intensive mutagenesis study showed that the positively charged residues, Lys-21 and Arg-41, on the H2 and H3 helices of ASC2 PYD might be counterparts in the interaction with the negatively charged patch containing Asp-6, Glu-13, Asp-48, and Asp-54 on the H1 and H4 helices of ASC PYD (39). For the complex formation of the other death domain superfamily, the charged interactions are also known to be important. In the structure of a DD complex, between the Drosophila proteins Pelle and Tube (40), the interaction is mixed, with both hydrophobic and hydrophilic components. In the Apaf-1·Caspase-9 CARD complex, the interaction is mediated largely by charge complementarity (41). A hydrophobic patch formed by Ile-39, Pro-40, Leu-41, and Pro-42 at the H2-H3 loop and Leu-57 and Phe-61 at H4 and detected at the structures of NALP7, ASC, and ASC2 was also conserved on the NALP3 PYD. This hydrophobic patch exposed to the surface may also have functional significance in the assembling of the inflammasome for Caspase-1 activation. Based on the analysis of the electrostatic surface of NALP3, it might be possible to use both charged and hydrophobic interactions that have been found for the interaction of the other death domain superfamily.
FIGURE 5.
Electrostatic surface representation of NALP3 PYD and their structural homologues. The left side of the NALP3 PYD surface and the remaining PYDs are shown in the same orientations as in Fig. 3. The opposite side of NALP3 PYD and the other PYDs are rotated by 180° along the vertical axis (y).
Conserved Surface of NALP3 PYD and Potential ASC Interaction Site for Assembly of Inflammasome
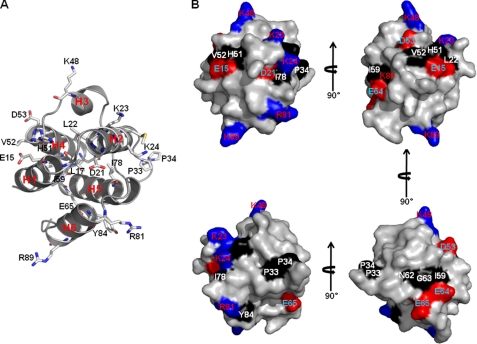
A number of residues on the surface are conspicuously conserved among NALP3 PYD from other PYD domains that are supposed to interact with ASC PYD (Fig. 2). These include Leu-17, Leu-22, Pro-33, Pro-34, His-51, Val-52, Ile-59, Gly-63, Ile-78, and Tyr-84, which are conserved as large hydrophobic residues on the surface of NALP3 PYD. In addition to hydrophobic residues, 10 surface-charged residues, Glu-15, Asp-21, Lys-23, Lys-24, Lys-48, Asp-53, Glu-64, Glu-65, Arg-81, and Lys-89, are conserved among different PYDs. Mapping of these residues onto the NALP3 PYD surface shows that two hydrophobic patches are located in two opposite directions, and more charged regions are dispersed all over the surface of NALP3 PYD (Fig. 6). The mapping analysis suggests that this extensive surface of NALP3 PYD, including both a hydrophobic central region and a more charged peripheral region, may be involved in interaction with ASC PYD. Lys-48, which is a critical residue for self-dimerization, may also be involved in the heterotypic interaction.
FIGURE 6.
Mapping of conserved exposed residues onto the NALP3 PYD. A, ribbon diagram of NALP3 PYD with conserved surface residues. B, molecular surface representation of NALP3 PYD with conserved exposed residues colored and labeled. Positively charged residues and negatively charged residues are colored in blue and red, respectively. Conserved exposed hydrophobic residues are colored in black. Four orientations of NALP3 PYD surface are shown.
Because NALP3 PYD is involved in the assembly of the oligomeric inflammasome, it is possible that this mapped region of the NALP3 PYD surface interacts with multiple molecules of ASC, which is detected for the assembly of other caspase-activating complexes such as PIDDosome, DISC. In the case of PIDDosome assembly, most of the surface on the RIP-associated Ich-1 (RAIDD)/CED homologous protein with death domain was involved in the interaction with p53-induced protein with death domain (PIDD) (42). In the case of FADD, the adaptor protein involved in the assembly of the DISC for extrinsic cell death, initially residues in H2 and H3 helices of FADD DD were shown to be important for Fas interaction. Subsequently, an extensive surface of FADD DD, with residues spreading all six helices of its structure, also was shown to participate in assembly of the DISC (43). In NALP3 PYD, the conserved surface residues distribute to many regions of the protein sequence indicating that NALP3 interaction with ASC for inflammasome formation may be mediated by using most surfaces of NALP3 PYD. The involvement of most surfaces of the death domain in the assembly of PIDDosome and DISC supports this idea. Multiple complex formations of death domains are a critical step for assembly of those complexes. Therefore, these conserved residues may be involved in the self-association and the interaction with NALP3 binding partner, ASC. However, it is possible that the assembly of the inflammasome and the other caspase-activating complexes such as DISC, apoptosome, and PIDDosome may be quite different in detail, including stoichiometry, nature of the interface, and region of the molecular surface. More structural studies are needed to unravel the molecular architectures of these complexes. In summary, the structure of NALP3 PYD provides a first step toward elucidating the molecular basis of formation of the NALP3 inflammasome.
Disulfide Bond between Cys-8 and Cys-108 and Possible Role for Regulation of the Activity of NALP3 by Redox Potential
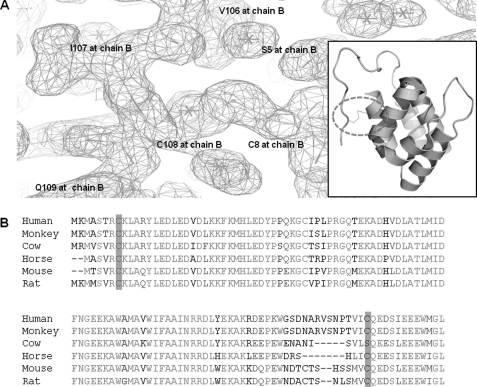
Disulfide bond between Cys-8 and Cys-108 was unexpectedly detected in our structure. The disulfide bond observed in the structure was formed between H1 and long loops connecting the PYD domain and the nucleotide-binding site domain (Fig. 7A). Cys-8 and Cys-108 that participate in the formation of the disulfide bond were conserved crossed species (Fig. 7B) indicating that this disulfide bond might be important for the function of NALP3. Recently, inflammation activation by reactive oxygen species has been identified. ROS generated by an NADPH oxidase participated in NALP3 inflammasome activation (44). Although ROS, generated by NAPDH oxidase after microbe phagocytosis, is known to constitute one of the most well known danger signals and is a candidate of activator for NALP3 inflammasome, the activation mechanism by ROS is not clear. Based on our structure, it is possible that the regulation of the formation of a disulfide bond by ROS is a critical point for NALP3 inflammasome activation. Particularly, it might be speculated that NALP3 is autoinhibited by not being formed by a disulfide bond. Upon activation by ROS, NALP3 became activated by forming a disulfide bond and changing the conformation of the structure of NALP3. The relationship between an unexpectedly identified disulfide bond and a redox potential that is known to be an activator of NALP3 inflammasome has to be identified by further research.
FIGURE 7.
Disulfide bond between Cys-8 and Cys-108 and conserved cysteine residue cross-species. A, electron density map is shown and disulfide bond is shown as sticks. Disulfide bond on the ribbon diagram is also shown in the boxed figure. The dotted circle indicates the position of the disulfide bond. B, conserved cysteine residues that are involved in the disulfide bond. The conserved cysteine residue cross-species are highlighted.
Supplementary Material
Acknowledgments
We thank Drs. Hao Wu and Chao Zheng at Cornell University (Weill Medical School) for generating MALS data and Ji-Joon Song at Korea Advanced Institute of Science and Technology for helpful discussions.
This work was supported by Grants 2011-0003406 and 2011-0025697 from the Basic Science Research Program through the National Research Foundation of Korea, Ministry of Education, Science, and Technology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The atomic coordinates and structure factors (code 3QF2) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PYD
- pyrin domain
- CARD
- caspase-recruitment domain
- DD
- death domain
- DED
- death effector domain
- MALS
- multiangle light scattering
- FADD
- Fas-associated protein with death domain
- DISC
- death-inducing signaling complex
- ROS
- reactive oxygen species.
REFERENCES
- 1. Schroder K., Tschopp J. (2010) Cell 140, 821–832 [DOI] [PubMed] [Google Scholar]
- 2. Eisenbarth S. C., Flavell R. A. (2009) EMBO Mol. Med. 1, 92–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martinon F., Burns K., Tschopp J. (2002) Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 4. Faustin B., Lartigue L., Bruey J. M., Luciano F., Sergienko E., Bailly-Maitre B., Volkmann N., Hanein D., Rouiller I., Reed J. C. (2007) Mol. Cell 25, 713–724 [DOI] [PubMed] [Google Scholar]
- 5. Agostini L., Martinon F., Burns K., McDermott M. F., Hawkins P. N., Tschopp J. (2004) Immunity 20, 319–325 [DOI] [PubMed] [Google Scholar]
- 6. Fernandes-Alnemri T., Yu J. W., Datta P., Wu J., Alnemri E. S. (2009) Nature 458, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kolly L., Busso N., Palmer G., Talabot-Ayer D., Chobaz V., So A. (2010) Immunology 129, 178–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu-Bryan R. (2010) Immunol. Cell Biol. 88, 20–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martinon F., Pétrilli V., Mayor A., Tardivel A., Tschopp J. (2006) Nature 440, 237–241 [DOI] [PubMed] [Google Scholar]
- 10. Hoffman H. M., Mueller J. L., Broide D. H., Wanderer A. A., Kolodner R. D. (2001) Nat. Genet. 29, 301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feldmann J., Prieur A. M., Quartier P., Berquin P., Certain S., Cortis E., Teillac-Hamel D., Fischer A., de Saint Basile G. (2002) Am. J. Hum. Genet. 71, 198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martinon F. (2008) J. Leukocyte Biol. 83, 507–511 [DOI] [PubMed] [Google Scholar]
- 13. Shiohara M., Taniguchi S., Masumoto J., Yasui K., Koike K., Komiyama A., Sagara J. (2002) Biochem. Biophys. Res. Commun. 293, 1314–1318 [DOI] [PubMed] [Google Scholar]
- 14. Masumoto J., Taniguchi S., Ayukawa K., Sarvotham H., Kishino T., Niikawa N., Hidaka E., Katsuyama T., Higuchi T., Sagara J. (1999) J. Biol. Chem. 274, 33835–33838 [DOI] [PubMed] [Google Scholar]
- 15. Thornberry N. A., Bull H. G., Calaycay J. R., Chapman K. T., Howard A. D., Kostura M. J., Miller D. K., Molineaux S. M., Weidner J. R., Aunins J., et al. (1992) Nature 356, 768–774 [DOI] [PubMed] [Google Scholar]
- 16. Ghayur T., Banerjee S., Hugunin M., Butler D., Herzog L., Carter A., Quintal L., Sekut L., Talanian R., Paskind M., Wong W., Kamen R., Tracey D., Allen H. (1997) Nature 386, 619–623 [DOI] [PubMed] [Google Scholar]
- 17. Park H. H., Lo Y. C., Lin S. C., Wang L., Yang J. K., Wu H. (2007) Annu. Rev. Immunol. 25, 561–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park H. H. (2011) Apoptosis 16, 209–220 [DOI] [PubMed] [Google Scholar]
- 19. Damiano J. S., Reed J. C. (2004) Curr. Drug Targets 5, 367–374 [DOI] [PubMed] [Google Scholar]
- 20. Franchi L., Eigenbrod T., Muñoz-Planillo R., Nuñez G. (2009) Nat. Immunol. 10, 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hiller S., Kohl A., Fiorito F., Herrmann T., Wider G., Tschopp J., Grütter M. G., Wüthrich K. (2003) Structure 11, 1199–1205 [DOI] [PubMed] [Google Scholar]
- 22. Pinheiro A. S., Proell M., Eibl C., Page R., Schwarzenbacher R., Peti W. (2010) J. Biol. Chem. 285, 27402–27410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liepinsh E., Barbals R., Dahl E., Sharipo A., Staub E., Otting G. (2003) J. Mol. Biol. 332, 1155–1163 [DOI] [PubMed] [Google Scholar]
- 24. de Alba E. (2009) J. Biol. Chem. 284, 32932–32941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Natarajan A., Ghose R., Hill J. M. (2006) J. Biol. Chem. 281, 31863–31875 [DOI] [PubMed] [Google Scholar]
- 26. Jang T. H., Park H. H. (2011) J. Biotechnol. 151, 335–342 [DOI] [PubMed] [Google Scholar]
- 27. Hendrickson W. A., Horton J. R., LeMaster D. M. (1990) EMBO J. 9, 1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Otwinowski Z. (1990) DENZO Data Processing Package, Yale University, New Haven, CT [Google Scholar]
- 29. Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 30. Terwilliger T. (2004) J. Synchrotron Radiat. 11, 49–52 [DOI] [PubMed] [Google Scholar]
- 31. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 33. Coussens N. P., Mowers J. C., McDonald C., Nuñez G., Ramaswamy S. (2007) Biochem. Biophys. Res. Commun. 353, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Srimathi T., Robbins S. L., Dubas R. L., Hasegawa M., Inohara N., Park Y. C. (2008) Biochemistry 47, 1319–1325 [DOI] [PubMed] [Google Scholar]
- 35. Holm L., Sander C. (1995) Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 36. Li F. Y., Jeffrey P. D., Yu J. W., Shi Y. (2006) J. Biol. Chem. 281, 2960–2968 [DOI] [PubMed] [Google Scholar]
- 37. Yang J. K., Wang L., Zheng L., Wan F., Ahmed M., Lenardo M. J., Wu H. (2005) Mol. Cell 20, 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eberstadt M., Huang B., Chen Z., Meadows R. P., Ng S. C., Zheng L., Lenardo M. J., Fesik S. W. (1998) Nature 392, 941–945 [DOI] [PubMed] [Google Scholar]
- 39. Srimathi T., Robbins S. L., Dubas R. L., Chang H., Cheng H., Roder H., Park Y. C. (2008) J. Biol. Chem. 283, 15390–15398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiao T., Towb P., Wasserman S. A., Sprang S. R. (1999) Cell 99, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qin H., Srinivasula S. M., Wu G., Fernandes-Alnemri T., Alnemri E. S., Shi Y. (1999) Nature 399, 549–557 [DOI] [PubMed] [Google Scholar]
- 42. Park H. H., Logette E., Raunser S., Cuenin S., Walz T., Tschopp J., Wu H. (2007) Cell 128, 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang L., Yang J. K., Kabaleeswaran V., Rice A. J., Cruz A. C., Park A. Y., Yin Q., Damko E., Jang S. B., Raunser S., Robinson C. V., Siegel R. M., Walz T., Wu H. (2010) Nat. Struct. Mol. Biol. 17, 1324–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dostert C., Pétrilli V., Van Bruggen R., Steele C., Mossman B. T., Tschopp J. (2008) Science 320, 674–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.