Abstract
Perturbation of cell cycle regulation is a characteristic feature of infection by many DNA and RNA viruses, including Coronavirus infectious bronchitis virus (IBV). IBV infection was shown to induce cell cycle arrest at both S and G2/M phases for the enhancement of viral replication and progeny production. However, the underlying mechanisms are not well explored. In this study we show that activation of cellular DNA damage response is one of the mechanisms exploited by Coronavirus to induce cell cycle arrest. An ATR-dependent cellular DNA damage response was shown to be activated by IBV infection. Suppression of the ATR kinase activity by chemical inhibitors and siRNA-mediated knockdown of ATR reduced the IBV-induced ATR signaling and inhibited the replication of IBV. Furthermore, yeast two-hybrid screens and subsequent biochemical and functional studies demonstrated that interaction between Coronavirus nsp13 and DNA polymerase δ induced DNA replication stress in IBV-infected cells. These findings indicate that the ATR signaling activated by IBV replication contributes to the IBV-induced S-phase arrest and is required for efficient IBV replication and progeny production.
Keywords: DNA Damage, DNA Polymerase, Viral Protein, Viral Replication, Virus, Coronavirus, DNA Polymerase δ, DNA Replication Stress, Interaction, nsp13
Introduction
DNA damage response is a signal transduction pathway that coordinates cell cycle transition, DNA replication, and repair in response to DNA damage or replication stress (1, 2). It is essential for maintenance of genome integrity and cell survival. DNA damage response is primarily mediated by two related protein kinases, the ataxia-telangiectasia mutated (ATM)2 and ATM/Rad3-related (ATR). ATM is activated as a result of double-stranded breaks (DSBs) and is recruited to DSBs by Mre11-Rad50-NBS1 complex (3, 4). ATR, on the other hand, is activated by a wide range of DNA damage, including stalled DNA replication forks and the subsequent single-stranded lesion (ssDNA), base adducts, ultraviolet (UV)-induced nucleotide damage, and double-stranded breaks during S phase (1, 5). ATR is recruited to replication factor A (RPA)-coated ssDNA by ATR-interacting protein (ATRIP) (6, 7). When ATM or ATR is recruited to sites of damage, they phosphorylate and activate different substrates, including checkpoint kinase-2 (CHK2) and CHK1, respectively (1, 8, 9). A large number of overlapping substrates of ATM and ATR that might be involved in DNA damage response has been presented (e.g. H2AX, RPA32, p53, BRCA1) (10, 11). Those signaling modules finally lead to cell cycle arrest to allow for DNA repair or apoptosis in cases of severe DNA damage. In contrast to ATM, ATR prevents replication fork collapse at stalled replication forks and is essential for cell viability (7, 12–18).
Many DNA viruses and retroviruses, including Epstein-Barr virus, herpes simplex virus 1, human Papillomavirus 16, human immunodeficiency virus (HIV), adenovirus, simian virus 40 (SV40), and Polyomavirus, are known to eliminate, circumvent, or exploit various aspects of cellular DNA damage response machinery to maximize their own replication. In RNA virus families, however, the only example so far is hepatitis C virus (19–22). Hepatitis C virus NS3/4A interacts with ATM, induces cytoplasmic translocation of ATM, and increases the sensitivity to irradiation (22). Activation of ATM and Chk2 was also shown to promote hepatitis C virus RNA replication (20). In this study we report that Coronavirus infection induces DNA replication stress through interaction of its nonstructural protein 13 (nsp13) with DNA polymerase δ (Pol δ).
The Pol δ activity is crucial for chromosome replication and DNA repair and plays an essential role in genome stability. Current evidence specifies Pol ϵ as the leading strand DNA polymerase and Pol δ as the lagging strand polymerase during undisturbed DNA replication (23). Human Pol δ consists of four subunits, p125, p50, p68, and p12. Subunit p125 harbors the polymerase and exonuclease active sites, p50 is tightly associated with the p125 subunit, p68 is associated with p50, and p12 binds to both p125 and p50 (23–26).
Coronaviruses are a diverse group of large, enveloped, single-stranded, and positive-sense RNA viruses that cause a variety of economically important diseases affecting humans and animals (27). In 2003, severe acute respiratory syndrome Coronavirus (SARS-CoV) emerged as a dangerous pandemic agent that caused a highly contagious health threat with a fatality rate of 10% (28). Infectious bronchitis virus (IBV), a prototype Coronavirus, is the etiological agent of infectious bronchitis that impairs the respiratory and urogenital tracts of chickens (29). IBV infection perturbs cell cycle progression and arrests cell at the S and G2/M phases (30, 31). Data present in this report demonstrate that induction of DNA damage response is one of the mechanisms used by IBV to induce cell cycle arrest. Furthermore, the ATR/Chk1 pathway is shown to be activated in IBV-infected cells, and interaction between Coronavirus nsp13 and Pol δ may induce the DNA replication stress in IBV-infected cells.
EXPERIMENTAL PROCEDURES
Viruses and Cells
Fibroblasts IBRhTERT (wild type control) and F02-98hTERT (ATR-Seckel), obtained as a kind gift from Penny Jeggo (Genome Damage and Stability Centre, University of Sussex, UK) (32, 33), were cultured in complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. African green monkey kidney cell line Vero, human cervical cancer cell line HeLa, and human lung carcinoma cell line H1299 were obtained from the American Type Culture Collection (ATCC) and cultured in complete DMEM (Invitrogen) or RPMI 1640 (Hyclone) supplemented with 10% newborn calf serum (Sterile), penicillin (100 units/ml), and streptomycin (100 μg/ml) and maintained at 37 °C in humidified 5% CO2.
Vero-adapted IBV Beaudette strain (p65, DQ001339), IBV-HA-RdRp, and IBV-Luc were propagated and titrated on Vero cells (34–36). Virus stock was prepared by two repeated freeze-thaw cycles and kept at −80 °C until use. Titers of the virus stocks were determined by the 50% tissue culture infective doses (TCID50) as previously described (37). A recombinant IBV with HA-tagged nsp13 (IBV-HA-hel) was obtained by using an established infectious cDNA system (38). An HA tag (YPYDVPDYA) was inserted between Asp5462 and Ser5463 at the N terminus of nsp13. UV-inactivated IBV was made as previously described using a CL-1000 cross-linker (UVP) (30, 39).
Chemicals, Antibodies, and Reagents
Caffeine (Sigma) was dissolved in DMEM (100 mm stock solution). The ATM inhibitor KU-55933 (Merck), DNA-PK inhibitor NU7026 (Merck), Schisandrin B (SchB, Shanghai TauTo Biotech Co LTD, China), and CGK733 (Sigma) were dissolved in DMSO and stored at −80 °C. 5-Bromo-2′-deoxyuridine 5′-triphosphate (BrdU) (Sigma) was dissolved in sterilized water (2.5 mm) and stored at −20 °C.
Antibodies against Chk1Ser317, H2AX, γ-H2AX, ATR, ATMSer1981, and Chk2Thr68 were purchased from Cell Signaling Technology (Beverly, MA). Phosphor RPA32Ser4/8 antibody was from Bethyl Laboratories Inc. Antibodies against ATM, BrdU, and RPA32 were from Abcam (Cambridge, UK). Antibodies against Myc and FLAG were from Sigma, and antibodies against actin were from Santa Cruz (Santa Cruz, CA). Polyclonal antibodies against IBV N and S proteins were raised in rabbits (40, 41). Mouse antibodies against HA for indirect immunofluorescence assays was from ETC (Singapore). Antibodies against p125 for indirect immunofluorescence and Western blot assays were from BD Biosciences. Horseradish peroxidase (HRP)-linked goat anti-rabbit secondary antibodies and HRP-linked goat anti-mouse secondary antibodies were purchased from Dako (Glostrup, Denmark). Alexa Fluor 488-linked anti-rabbit IgG and Alexa Fluor 594-linked anti-mouse IgG were from Molecular Probes.
Plasmid Construction
Plasmid pGBKT7-Snsp13 encoding the full-length SARS-CoV nsp13 (1–601 aa) was constructed by cloning an 1802-bp PCR fragment from SARS-CoV strain sin2774 (AY283798) (16,151–17,953 nucleotides) into vector pGBKT7 using BamHI and XhoI sites. The same fragment was cloned into pXJ-FLAG, giving rise to pXJFLAG-Snsp13. The Myc-tagged SARS-CoV nsp13 was PCR-amplified from pGBKT7-Snsp13 and subcloned into pXJ41 with a neomycin selection marker. Plasmid pGBKT7-nsp13 encoding the full-length IBV nsp13 (1–600 aa) were constructed by cloning a 1.8-kb PCR fragment from IBV Beaudette strain (p65, DQ001339) (15,132–16,931 nucleotide) into vector pGBKT7. The same fragment was cloned into vector pXJ-FLAG and pXJ-Myc, forming pXJFLAG-nsp13 and pXJmyc-nsp13, respectively. Plasmids pGBKT7-Snsp13 and pGBKT7-nsp13 were used as bait expression vector to screen a cDNA library from HeLa cells as previously described (36).
To express the full-length p125 subunit of Pol δ in mammalian cells, pXJmyc-p125 was constructed. An RT-PCR fragment covering the p125 ORF (NM_002691) was amplified from H1299 cells using the primer pair 5′-CGTAGGCTGTGGCGGGAAACGCTGTT-3′ and 5′-GCAAGGTCACCAGGCCTCAGGTCCAG-3′. This RT-PCR product was cloned into pCR®2.1, giving rise to Topo-p125. A PCR fragment with Topo-p125 as the template was amplified using primers 5′-GCGGGATCCGCTGTTAGAAGCGGGATGGATGGCAA-3′ and 5′-CCGGCTCGAGCAAGGTCACCAGGCCTCAGGTCCAG-3′ and cloned into pXJ-myc, forming pXJmyc-p125. Plasmid pGEX-p125C was constructed by cloning a PCR fragment encoding the C-terminal 172 amino acids of p125 (935–1107 aa) into pGEX-5X-3. All constructs were confirmed by sequencing.
Drug Treatment and Luciferase Reporter Assay
Inhibitors KU-55399, NU7026, CGK733, caffeine, and SchB were tested for their effect on IBV replication by measuring the luciferase activities in cells infected with IBV-Luc. The inhibitors were added to cells 1 h before infection and kept in the media during the infection. Cells of 90–95% confluence grown on 12-well plates were infected with IBV-Luc at an m.o.i. of 1 and were harvested for luciferase reporter assay at the indicated time points (0–16 h) post-infection. Luciferase reporter assay was performed according to the manufacturer's manual (Promega).
Confocal Fluorescence Microscopy
Cells that were transfected with plasmids or infected with IBV-HA-RdRp and IBV-HA-hel viruses were fixed with 4% formaldehyde, respectively. Cells were permeabilized with 100% methanol and blocked with PBST (1× PBS with 0.3% Triton X-100) containing 5% normal goat serum. Cells were then incubated with primary antibodies diluted in 1× PBST containing 5% normal goat serum. The target proteins were detected with 1:300 diluted primary antibodies. After washing three times, cells were incubated with 1:200 diluted secondary antibodies (Alexa Fluor 488 anti-rabbit IgG and Alexa Fluor 594 anti-mouse IgG). Images were taken using an Olympus confocal microscope (FLUOVIEW FV1000).
GST Pulldown Assays
Plasmids pGEX-5X1 and pGEX-p125C were transformed into bacteria BL21 competent cells. GST and GST-p125C fusion proteins were expressed in BL21 by induction with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 3 h and purified using the GST purification module (Amersham Biosciences) according to manufacturer's instructions. The 35S-labeled nsp13 and Snsp13 were expressed by in vitro translation in wheat germ extracts in the presence of [35S]methionine based on the protocol recommended by the manufacturer (Promega). To precipitate in vitro synthesized polypeptides, 30 μl of GST-Sepharose 4B or GST-p125C-Sepharose 4B beads were added into and incubated with a mixture solution of 10 μl of 35S-labeled translation products in wheat germ diluted with 90 μl of lysis buffer (140 mm NaCl, 10 mm Tris-HCl (pH 8.0), and 0.5% Nonidet P-40)) for 1 h at room temperature. The Sepharose 4B beads were washed seven times with lysis buffer and boiled with 2× SDS loading buffer for 4 min. The eluted precipitates were then subjected to SDS-PAGE and detected by autoradiography.
Co-immunoprecipitation and Western Blot
Cells were transfected with the appropriate plasmids by Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. At 28 h post-transfection, cells were lysed in buffer containing 10 mm Tris-HCl (pH 8.0), 150 mm NaCl, 4 mm EDTA, 0.5% Triton X-100 plus 1 tablet/40 ml of protease inhibitors (Roche Applied Science). Lysates were clarified by adding 20 μl of protein A-Sepharose beads, and the precleared supernatants were incubated with 2 μl of rabbit anti-Myc antibodies (Sigma) at 4 °C for 1 h. After absorption of the precipitates on 30 μl of protein A-Sepharose beads for 1 h, the resin was washed 7 times with lysis buffer and boiled with 2× SDS loading buffer to elute the precipitates. The proteins were then subjected to SDS-PAGE followed by immunoblotting analysis using anti-FLAG-HRP (Sigma) or anti-FLAG-HRP (Sigma) antibodies.
To confirm the interaction between p125 and nsp13 in IBV-infected cells, co-immunoprecipitation was performed as previously described with minor modifications (42). In brief, Vero cells were infected with either wild type IBV or IBV-HA-hel at an m.o.i. of 1 and harvested at 16 h post-infection. Cells from one 175-cm2 flask were harvested with 2 ml of lysis buffer (50 mm Tris-HCl (pH 7.4), 500 mm NaCl, 0.5% Nonidet P-40, 0.5% Triton X-100) plus 10 μg/ml RNase A and 1 tablet/40 ml of protease inhibitors (Roche Applied Science). Immunoprecipitation was carried out with EZviewTM Red Anti-HA Affinity Gel (Sigma).
The precipitates from co-immunoprecipitation or total cell lysates dissolved in 2× SDS loading buffer were subjected to SDS-PAGE and transferred to PVDF membranes (Bio-Rad). Membrane was blocked in blocking buffer (5% fat-free milk powder in PBST buffer) for 1 h and incubated with 1:2000 diluted primary antibodies in blocking buffer for 2 h at room temperature. After washing three times with PBST, the membrane was incubated with 1:2000 diluted anti-mouse or anti-rabbit IgG antibodies conjugated with horseradish peroxidase (DAKO) in blocking buffer for 1 h at room temperature. After washing the membrane three times with PBST, polypeptides were detected with a chemiluminescence detection kit (ECL, Amersham Biosciences) according to the manufacturer's instructions.
Analysis of S-phase Cells and Host Cell DNA Replication by Indirect Immunofluorescence
Dual-indirect immunofluorescence was used to determine the S-phase cell populations in both mock- and virus-infected cells. The intake of BrdU into the actively replicating DNA was used to estimate the S-phase cells and the expression of IBV S protein as a marker for IBV-infected cells. In brief, Vero cells grown on 4-well chamber slides were infected with IBV at an m.o.i. of 1, and 10 μm BrdU was added 1 h before harvest. Cells were harvested at 4, 8, 12, 16, 20, and 24 h post-infection and fixed with 70% ice-cold ethanol for 1 h followed by incubation in 3 n HCl for 30 min at room temperature to denature DNA. Cells were then washed with PBS and blocked with 10% goat serum in PBST and incubated with primary antibody solution (anti-mouse BrdU and anti-rabbit IBV S protein for 1 h at room temperature. After thorough washing with PBST, cells were incubated with 1:200 diluted secondary antibodies (Alexa Fluor 488 anti-rabbit IgG and Alexa Fluor 594 anti-mouse IgG). The cells were also counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Images were visualized using an Olympus confocal microscope (FLUOVIEW FV1000). The percentage of S-phase nucleus in IBV-infected cells at each time point was recorded by counting BrdU-stained nuclei among DAPI-stained nuclei in IBV S protein-stained cells. About 100–150 events were recorded for each sample, and each experiment was repeated three times.
Transfection of siRNA
Short interfering RNA targeting ATR (siATR, 5′-AACGAGACUUCUGCGAUUG CTT-3′) and a negative control siRNA targeting EGFP (siEGFP 5′-GCAACGUGACCCUGAAGUUCAT-3′) were purchased from Sigma. H1299 cells were plated 24 h before transfection to reach 30% confluence next day, and DharmaFECT® 2 transfection reagent (Dharmacom) was used to deliver siRNA duplexes into cells according to the manufacturer's instructions. Transfection was repeated 24 h later, and cells were infected with IBV at an m.o.i. of 0.5 at 34 h post the second transfection.
Establishment of Stable Snsp13-overexpressing Cells
To establish cell lines with stable expression of Myc-tagged Snsp13, pXJ41-Snsp13 was transfected into Vero cells using Effectene transfection reagent (Invitrogen) according to the manufacturer's protocol. At 48 h post-transfection, the transfected cells were selected in DMEM supplemented with 1 mg/ml G418 antibiotic (Sigma) until formation of G418-resistant colonies. After ∼30 days, the G418-resistant clones were picked and amplified. The amplified cell clones were harvested and analyzed by Western blot analysis, and cell clones overexpressing Snsp13 were selected for subsequent studies. In parallel, Vero cells transfected with pXJ41 were selected under G418 antibiotic to serve as control cells.
Flow Cytometry
To determine cell cycle status, nuclear DNA content was measured by using propidium iodide (PI) staining and fluorescence-activated cell sorting (FACS) analysis. Briefly, cells were detached with trypsin and washed with PBS. The cell pellets were fixed with 70% ethanol. After resuspending in PBS containing 20 μg/ml RNase A and 50 μg/ml PI, cells were incubated at room temperature for 1 h and subjected to FACS analysis. A total of 10,000 events were analyzed for each sample, and each experiment was repeated three times.
For intracellular staining using conjugated secondary antibody, H1299 cells grown in a 10-cm dish at about 90% confluence were transfected with pXJ-Myc and pXJ-Myc-nsp13. At 40 h post-transfection, cells were harvested by trypsin treatment and fixed with 80% ethanol. After blocking with PBST containing 5% BSA for 30 min, cells were incubated with primary anti-Myc mouse monoclonal antibody (Sigma) in a 1:200 dilution in blocking buffer for 1 h. Cells were washed twice with blocking buffer and incubated with a 1:200 dilution of fluorescent-tagged goat anti-mouse secondary antibody. After washed with PBS, cells were stained for 1 h with PI staining solution (PBS with 50 μg/ml PI and 25 μg/ml RNase A). Flow cytometric analysis was performed using a FACScan flow cytometer, and data acquisition was performed with WinMDI Version 2.8.
Densitometry
The intensities of RNA and protein bands were quantified using ImageJ program according to the manufacturer's instruction.
RESULTS
IBV Infection Induces a DNA Damage Response in Cultured Cells
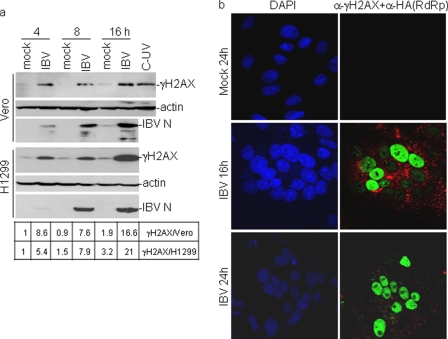
In previous studies IBV infection was shown to induce cell cycle arrest at the S and G2/M phases (30, 31). To test if a DNA damage response was induced in IBV-infected cells, the phosphorylation status of H2AX was first examined. Phosphorylation of H2AX on Ser139 (γH2AX) was a well known marker for DNA damage response. It occurs in response to double-strand breaks as well as DNA replication stress and may play an important role in either recruitment or stabilization of DNA repair proteins to damage site (43, 44). Levels of γH2AX in mock- and IBV-infected H1299 were measured by Western blotting at various time points post-infection and showed the presence of γH2AX in IBV-infected, but not mock-infected, H1299 cells as early as at 4 h post-infection (Fig. 1a). At 8 h post-infection, a similar level of γH2AX continued to be observed in the infected cells, and a higher level of γH2AX was observed at 16 h post-infection (Fig. 1a). Quantification of the corresponding bands by densitometry showed an approximate 8-fold induction of γH2AX in virus-infected cells at 4 and 8 h post-infection and 20-fold induction at 16 h post-infection (Fig. 1a). In IBV-infected Vero cells, a similar pattern of induction of γH2AX was also observed from 4 to 16 h post-infection (Fig. 1a), demonstrating that induction of H2AX phosphorylation by IBV infection was not cell type-specific.
FIGURE 1.
Induction of γH2AX by IBV infection of Vero and H1299 cells. a, Western blot analysis of γH2AX in IBV-infected Vero and H1299 cells is shown. Vero and H1299 cells were mock-infected or -infected with IBV at an m.o.i. of 2 and harvested at 4, 8, and 16 h post infection. Cells were lysed and analyzed by Western blot with specific anti-γH2AX antibodies. The same membranes were also probed with anti-actin as a loading control. Viral replication was confirmed by Western analysis of N protein with anti-IBV N polyclonal antibodies. The intensity of each γH2AX band was determined by densitometry and is shown as -fold induction after normalization to actin. The signal for each γH2AX band from mock-infected cells at 4 h post-infection is treated as 1. b, immunofluorescent staining of γH2AX in IBV-infected Vero cells is shown. Vero cells were mock-infected (mock) or infected with IBV-HA-RdRp (IBV) at an m.o.i. of 1. At 16 and 24 h post-infection, cells were fixed and double-immunostained with specific rabbit anti-γH2AX and mouse anti-HA antibodies. Green represents γH2AX, and red represents HA-tagged RdRp expressed in IBV-infected cells.
Indirect immunofluorescent staining of IBV-infected Vero cells with antibodies specific for γH2AX was then conducted to study the induction kinetics of γH2AX at late stages of the infection cycle. To specifically and sensitively detect and characterize events occurring during viral replication cycle, a recombinant IBV (IBV-HA-RdRp) with an HA tag at the N terminus of nsp12 protein (RdRp) was used to infect Vero cells (36). The mock-infected cells were fixed at 24 h post-infection, and IBV-infected cells were fixed at 16 and 24 h post-infection. Cells were dual-stained with antibodies against γH2AX and HA, and the nuclei were stained with DAPI. At 16 h post-infection, γH2AX was observed in the infected cells expressing the HA-tagged RdRp with a bright, even pan-nuclear staining pattern instead of γH2AX foci (Fig. 1b). This staining pattern resembles the class I or II γH2AX staining pattern induced by UV irradiation in the S-phase cells (45). Recently, pan-nuclear phosphorylation of H2AX was detected in cells in the presence of replication stress or upon treatment with Chk1 inhibitors (46–49). More γH2AX-positive cells were observed at 24 h post-infection (Fig. 1b), indicating gradually increased accumulation of γH2AX in the infected cells over time at the late stages of the infection cycle. Collectively, these results suggest that IBV infection may activate a DNA damage response, probably due to replication stress, in the infected cells.
IBV Infection Activates the ATR-dependent DNA Damage Response Pathway
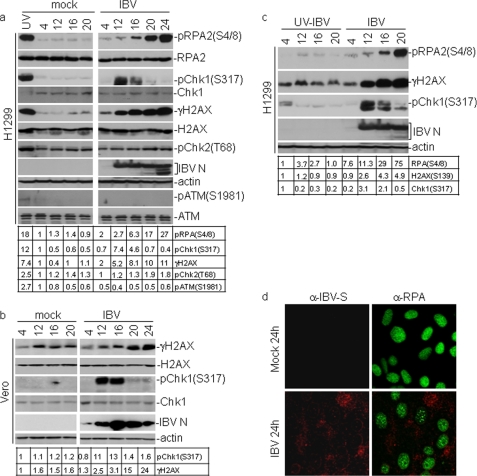
To identify which signaling pathway of the DNA damage response is activated by IBV infection, the phosphorylation status of substrates specifically modified by either ATR or ATM was analyzed. Chk1 is the best studied ATR substrate, and its phosphorylation by ATR on Ser317 and Ser345 is a reliable indicator of ATR/Chk1 activation (50, 51). Phosphorylation of RPA2, a component of the heterotrimeric RPA complex, on Ser4/8 is also specifically catalyzed by ATR but not by ATM and DNA-PKc (52). The specific residues for the ATM kinase activity include Chk2 on Thr68 and ATM on Ser1981 (53, 54). Phosphorylation of Chk1 on Ser317, RPA2 on Ser4/8 and Chk2 on Thr68 was first examined in IBV-infected H1299 cells by Western blotting with specific antibodies. The replication of IBV was monitored by checking the expression of nucleocapsid (N) protein of IBV. Upon IBV infection of H1299 cells, a 7.4-fold increased detection of Chk1Ser317 was obtained at 12 h post-infection, but the signal was gradually decreased from 4.6-fold induction at 16 h post-infection to almost undetectable in the following two time points (Fig. 2a). Phosphorylation of RPA on Ser4/8 became detectable (2–3-fold induction) at 12 h post-infection and accumulated to a 27-fold induction at 24 h post-infection (Fig. 2a). In contrast to the ATR targets, the phosphorylated form of Chk2 on Thr68 and ATM on Ser1981 was not visibly increased upon infection at all time points (Fig. 2a). Treatment of cells with UV irradiation clearly induced Chk1Ser317, RPASer4/8, and γH2AX, showing that the ATR/Chk1 pathway is intact in H1299 cells. These results indicate that the ATR, but not ATM, signaling branch is activated in IBV-infected H1299 cells. Similar studies were then conducted in Vero cells, showing that ATR was activated as indicated by the strong phosphorylation of Chk1 on Ser317 (Fig. 2b). However, in cells incubated with UV-inactivated IBV, the levels of Chk1 phosphorylation on Ser317 only slightly increased at 4 h post-infection and disappeared at the later time points (Fig. 2c). Similarly, no obvious phosphorylation of RPA2 at Ser4/8 was detected in cells incubated with UV-inactivated IBV (Fig. 2c), demonstrating that activation of the ATR signaling pathway by IBV infection is dependent on active virus replication. Taken together, these results confirm that IBV replication activates the ATR signaling pathway rather than the ATM checkpoint.
FIGURE 2.
Activation of an ATR-dependent cellular DNA damage response by IBV replication. a, activation of the ATR signaling by IBV infection is shown. H1299 cells were mock- or IBV-infected at an m.o.i. of 1. At 12, 16, 20, and 24 h post-infection, cells were harvested, and total lysates were subjected to immunoblotting assay. The levels of phosphor-Chk1 (pChk1(Ser317)), total Chk1, γH2AX, total H2AX, phosphor-RPA2 (pRPA2(Ser4/8)), total RPA, phosphor-Chk2 (pChk2(Thr68)), total ATM, phosphor-ATM (pATM(Ser1981), actin, and IBV N were determined with appropriate antibodies. Actin served as a loading control, and IBV N protein served as a marker of IBV infection. H1299 cells treated by UV light with a wavelength of 254 nm (100 J/m2) and allowed to recover for 0.5 and 2 h were used as positive controls for phosphorylation of ATM (0.5-h recovery) and ATR (2-h recovery) substrates, respectively. The intensity of each γH2AX, pChk1(Ser317), pChk2(Thr68), or pRPA2(Ser4/8) band was determined by densitometry and is shown as -fold induction after normalization to actin. The signal for each phosphor protein band from mock-infected cells at 4 h post-infection is treated as 1. b, activation of the ATR signaling by IBV infection of Vero cells. c, activation of ATR signaling is dependent on active IBV replication. H1299 cells were infected with live or UV-inactivated IBV (UV-IBV) at an m.o.i. of 1, and the phosphorylation status of ATM and ATR substrates was analyzed by Western blotting. The intensity of each γH2AX, pChk1(Ser317), or pRPA2(Ser4/8) band was determined by densitometry and is shown as -fold induction after normalization to actin. The signal for each phosphor protein band from UV-IBV-infected cells at 4 h post-infection is treated as 1. d, immunofluorescent staining of total RPA in IBV-infected Vero cells is shown. Vero cells were mock-infected or infected with IBV-HA-RdRp (IBV) at an m.o.i. of 1. At 24 h post-infection, cells were fixed and double-immunostained with specific mouse anti-RPA and rabbit anti-IBV S antibodies. Green represents RPA, and red represents IBV S.
Whether IBV infection would lead to the formation of RPA foci was then examined. RPA is a single-strand DNA-binding protein and may play critical roles in DNA synthesis, damage repair, and recombination (55, 56). RPA2, a component of the heterotrimeric RPA complex, formed nuclear foci in response to various DNA damage signals including UV irradiation and hydroxyurea treatment (57, 58). Formation of nuclear foci by RPA2 would be an additional indicator of DNA damage response in IBV-infected cells. For this purpose, Vero cells were either mock- or IBV-infected for 24 h, fixed, and stained using mouse anti-RPA and rabbit anti-IBV S antibodies, respectively. The staining patterns were then examined by confocal microscopy, showing obvious RPA foci in IBV-infected, but not in mock-infected, cells at 24 h post-infection (Fig. 2d).
ATR Inhibitors Suppress the IBV-induced DNA Damage Response
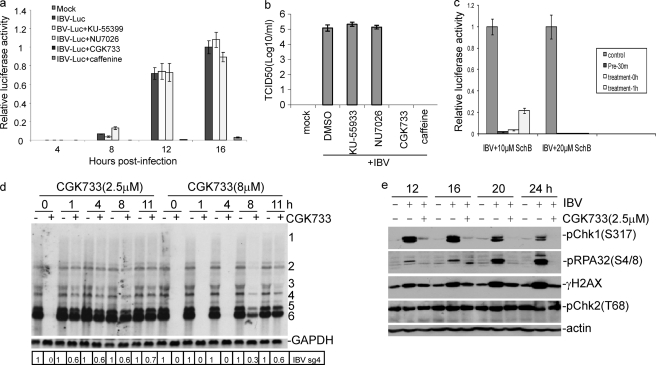
The induction of the ATR signaling pathway by IBV replication was then investigated by inhibition of the ATR kinase activity in IBV-infected cells with CGK733, a specific inhibitor of ATM/ATR kinases (59). Other chemical inhibitors of ATM, ATR, and DNA-PKc, including caffeine, KU-55933 (a specific ATM kinase inhibitor), and NU7026 (a specific DNA-PKc inhibitor), were also tested (60, 61). Caffeine inhibits the ATM activity in the concentration below 5 mm and ATR in the concentration above 10 mm in cultured cells (62). To fully inhibit the kinase activities, CGK733, KU-55933, and NU7026 were used in the concentration of 10 μm and caffeine in the concentration of 10 mm. The effects of these inhibitors on IBV replication were first tested by infection of cells with IBV-Luc, a recombinant IBV expressing the firefly luciferase. The luciferase activity in whole cell lysates of IBV-infected cells was measured and used as a marker for the replication efficiency of IBV (34), and virus titers at peak time point as a marker for viral prorogation were also determined by TCID50. H1299 cells were infected with IBV-Luc in the presence of either an inhibitor or DMSO and harvested at 4, 8, 12, and 16 h post-infection, respectively, to measure the relative luciferase activity. As shown in Fig. 3a, the luciferase activity was almost completely lost in all time points in the presence of either 10 μm CGK733 or 10 mm caffeine, indicating that CGK733 and caffeine strongly suppress the replication of IBV. Concurrently, production of progeny viruses in CGK733- and caffeine-treated cells was also inhibited at 16 h post-infection (Fig. 3b). However, specific ATM inhibitor KU-55933 and specific DNA-PKc inhibitor NU7026 in the concentration of 10 μm rendered no inhibitory effect on IBV replication and production as indicated by 100% of the relative luciferase activities and viral titers (Fig. 3, a and b). During the course of this study, SchB, a dibenzocyclooctadiene derivative isolated from Fructus Schisandrae, was reported to be a specific ATR inhibitor (63). Similar to CGK733 and caffeine, the addition of 10–20 μm SchB to IBV-infected H1299 cells either pre- or post-infection showed potent inhibition of IBV replication (Fig. 3c). These results suggest that the ATR pathway may play a role in IBV replication and production.
FIGURE 3.
Effects of PIKK, ATM/ATR, ATM, and DNA-PKc inhibitors on IBV replication and infectivity. a, analysis of the Inhibitory effects of KU-55399, NU7026, CGK733, and caffeine on IBV replication by luciferase assay is shown. H1299 cells were pretreated with an inhibitor or DMSO for 1 h before and during infection with IBV-Luc at an m.o.i. of 1. At indicated time points, cells were collected, and luciferase activities were measured. The relative luciferase activities of all samples were normalized to the luciferase reading from cells infected with IBV for 16 h with DMSO treatment. b, analysis of the inhibitory effects of KU-55399, NU7026, CGK733, and caffeine on IBV replication by TCID assay is shown. The virus titers of samples with peak luciferase activities were determined by TCID50 assay. The data are the average of three independent experiments, and error bars denote S.D. c, analysis of the Inhibitory effects of SchB on IBV replication by luciferase assay is shown. H1299 cells were pre- or post-treated with 10 and 20 μm SchB and infected with IBV-Luc at an m.o.i. of 1. At 16 h post-infection, cells were collected, and luciferase activities were measured. The relative luciferase activities of all samples were normalized to the luciferase reading from cells infected with IBV with DMSO treatment. d, Northern blotting analysis of the effect of CGK733 on the early steps of IBV replication is shown. DMSO (−) or 2.5 or 8 μm CGK733 (+) was added to IBV-infected H1299 cells at the indicated time points post-infection until cells were harvested for total RNA extraction at 16 h post-infection. Total RNA (10 μg) was separated on 1% agarose and transferred to a Hybond N+ membrane. Hybridization was performed with a Digoxigenin-labeled DNA probe specific for IBV 3′-UTR. Numbers on the right indicate individual subgenomic RNA. The intensity of each IBV sgRNA4 band was determined by densitometry, and the ratio of each band from the inhibitor-treated cells to that from the untreated cells after normalization to GAPDH is shown. The signal for each RNA band from the untreated infected cells at each time point is shown as 1. e, shown is the effect of CGK733 on the phosphorylation levels of ATR substrates induced by IBV replication. H1299 cells were mock-infected or infected with IBV at an m.o.i. of 1 and treated with CGK733 at 1 h post IBV infection. Cells were harvested at 12, 16, 20, 24 h post-infection, and total lysates were subjected to Western blotting analysis with specific antibodies against γH2AX, Chk1Ser317, RPASer4/8. Actin and IBV N protein were detected as loading and IBV infection controls, respectively.
The effect of CGK733 on IBV-induced phosphorylation of Chk1, RPA, and H2AX was then examined. To determine the optimal concentration of CGK733 and the time for adding the inhibitor to the infected cells post-infection that efficiently inhibit the ATR activity but render minimal inhibitory effect on virus replication, 2.5 and 8 μm CGK733 were added to the infected cells at 0, 1, 4, 8, and 11 h post-infection. Cells were harvested at 16 h post-infection, total RNA was prepared and subjected to Northern blot analysis. As shown in Fig. 3d, the addition of 2.5 μm CGK733 at 0 h post-infection and 8 μm at 0, 1, and 4 h post-infection totally inhibited the replication of IBV. A variable degree of inhibition on viral replication was observed in other times points when 2.5 and 8 μm CGK733 were added to the cultured media (Fig. 3d).
As only minor inhibition of viral replication was observed when 2.5 μm CGK733 was added at 1 h post-infection, H1299 cells were infected with IBV, and 2.5 μm CGK733 was added at 1 h post-infection and kept in the cultured medium until cells were harvested at 12, 16, 20, and 24 h post-infection. The levels of phosphorylated Chk1Ser317, Chk2Thr68, H2AXSer139, and RPASer4/8 in IBV-infected cells in the presence or absence of CGK733 were determined by Western blot analysis, showing that CGK733 was able to significantly, but not completely, reverse the accumulation of Chk1Ser317, γH2AX, and RPASer4/8 induced by IBV infection from 12 to 24 h post-infection (Fig. 3e).
Suppression of the ATR Signaling Pathway Inhibits the Replication of IBV
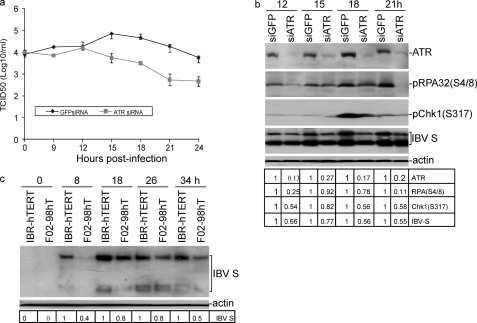
To provide more direct evidence that ATR kinase was involved in the regulation of IBV replication, H1299 cells were transfected with siATR or siEGFP as a negative control. At 58 h post-transfection, cells were infected with IBV at an m.o.i. of 0.5 and harvested at 0, 9, 12, 15, 18, 21, and 24 h post-infection. The effect of ATR knockdown on IBV replication was studied and showed an ∼10-fold lower TCID50 value of IBV in ATR knockdown cells compared with those in the control cells (Fig. 4a). The expression levels of ATR, Chk1Ser317, RPA Ser4/8, and IBV S proteins in the infected cells were then determined by Western blotting analysis and showed 73–87% knockdown of ATR at protein levels compared with cells transfected with siEGFP (Fig. 4b). ATR knockdown resulted in reduced accumulation of RPA2Ser4/8 and Chk1Ser317, further supporting that induction of DNA damage signaling in response to IBV infection is through the activation of ATR kinase. ATR knockdown resulted in reduced accumulation of IBV S protein expression in total cell lysates (Fig. 4b). The reduced levels of IBV replication in ATR knockdown cells, as indicated by less expression of IBV S protein (Fig. 4b) and virus titers from 15 to 24 h post-infection (Fig. 4a), supported the idea that ATR may play an important role in IBV replication.
FIGURE 4.
Functional requirement of ATR for efficient IBV replication. a, effect of siRNA-mediated knockdown of ATR on progeny IBV production is shown. H1299 cells were transfected with siATR or siGFP as control. At 58 h post-transfection, cells were infected with IBV at m.o.i. of 0.5 and harvested at the indicated time points post-infection. Culture media containing viral particles were collected for virus titer by TCID50 assay. b, the effect of siRNA-mediated ATR knockdown on IBV-induced DNA damage response and IBV replication is shown. H1299 cells were transfected with siATR or siGFP as control. At 58 h post-transfection, cells were infected with IBV at an m.o.i. of 0.5 and harvested at the indicated time points post-infection. Cells lysates were prepared and analyzed by Western blot assay. The intensity of each of the ATR, pRPA(Ser4/8), pChk1(Ser317), and IBV S bands was determined by densitometry, and the ratio of the bands from the knockdown and control cells is shown after normalization to actin. The signal for each band in the control cells at each time point is shown as 1. c, shown is the effect of ATR knock-out on IBV replication. IBRhTERT cells, a fibroblasts cell line with wild type ATR (ATR+/+) and F02-98hTERT, an ATR-deficient cell line (ATR−/−), were infected with IBV at an m.o.i. of 1 and harvested at the indicated time points post-infection. Total cell lysates were immunoblotted with IBV S antibodies. The intensity of each IBV S band was determined by densitometry, and the ratio of each band from the knock-out cells to that from wild type cells is shown after normalization to actin. The signal for each band from wild type cells at each time point is treated as 1.
The implication of the ATR signaling pathway in IBV replication was further analyzed in ATR-deficient fibroblast F02-98hTERT (ATR−/−) and control IBRhTERT (ATR+/+) cells (32, 33, 64). These cells were infected with IBV and harvested at 0, 8, 18, 26, and 34 h post-infection. IBV S protein from total cell lysates was measured by Western blotting assay and showed a 20–60% reduction of IBV S protein in IBV-infected F02-98hTERT (ATR−/−) cells compared with that in IBV-infected wild type IBRhTERT (ATR+/+) cells (Fig. 4c). The reduced accumulation of IBV S protein in F02-98hTERT (ATR−/−) cells clearly showed the decreased replication of IBV in ATR-deficient fibroblast. This result further supports the conclusion that efficient IBV replication may be dependent on the presence of an active ATR signaling pathway.
IBV Infection Extends Cell Cycle Arrest at S Phase and Is Coupled with Host Cell DNA Replication
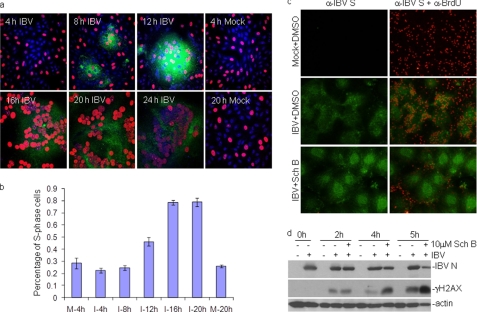
To more precisely determine the proportion of cells arrested at S phase and the relationship between viral RNA replication at the cytoplasm and host DNA replication at the nucleus, dual-indirect immunofluorescence was used to determine the effect of IBV replication on S-phase progression by comparing the percentage of nuclei with BrdU incorporation into the actively replicating DNA in mock- and IBV-infected cells. Results shown in Fig. 5a demonstrated that IBV infection of Vero cells induced the formation of syncytial cells from 8 h post-infection, and more extensive syncytium formation was observed over the infection time. At the same time, BrdU-positive nuclei were observed in most IBV-infected cells. The percentages of DNA-replicating cells (S-phase nuclei) in all IBV S-positive syncytial cells at 4, 8, 12, 16, and 20 h post-infection were determined by counting the BrdU-positive cells among total cells as indicated by nuclear staining with DAPI. Meanwhile, the percentages of S-phase cells in mock-infected cells were also determined by the same way. A gradual increase of S-phase cell populations was observed in the infected cells over time (Fig. 5b). At 16 and 20 h post-infection, ∼80% of IBV-infected cells were BrdU-positive, significantly higher than the 20% of S-phase cells in the infected cells at 4 h post-infection and the mock infection control, respectively (Fig. 5b). At 24 h post-infection, BrdU staining was significantly reduced (Fig. 5b), consistent with the previous report that IBV infection leads to the reduction in BrdU uptake in cells undergoing S phase by BrdU/PI dual-stained FACS analysis (30).
FIGURE 5.
Inhibition of host cell DNA replication in IBV-infected cells by an ATR inhibitor. a, a representative three-color overlay images for mock- and IBV-infected Vero cells at indicated time points post-IBV infection is shown. Host cell DNA replication (S-phase nuclei) in IBV-infected cells was determined by immunostaining with anti-BrdU (red) and anti-IBV S (green) antibodies. DAPI staining (blue) was used to visualize the nucleus. b, induction and extension of cell cycle arrest at S phase during IBV infection is shown. Histogram bars represent the mean percentage of S-phase nuclei in mock- and IBV-infected cells at the indicated time points (±S.D., n = 3) as shown in a. The percentage of S-phase nuclei in IBV-infected cells was determined by counting the BrdU-positive nuclei (red, indicating S phase nuclei) among DAPI-staining nuclei (blue, indicating total nuclei) in IBV-infected cells (green, indicating IBV S-positive cells). About 100–150 nuclei within BrdU/DAPI/IBV S overlay images from three independent experiments for each sample were counted. c, inhibition of host cell DNA replication by SchB in IBV-infected cells is shown. Vero cells were infected with IBV at an m.o.i. of 1. At 14 h post-infection, 10 μm SchB or DMSO was added to IBV-infected cells and incubated for further 4 h until cells were harvested for immunostaining. At 1 before harvesting, cells were labeled with 10 μm BrdU, fixed, and stained with BrdU (red) and IBV-S (green) antibodies. Images were visualized using an Olympus fluorescence microscope. Shown are representative dual-stained BrdU/IBV S images from DMSO-treated mock-infected cells (Mock+DMSO), DMSO-treated IBV-infected cells (IBV+DMSO), and SchB-treated IBV-infected cells (IBV+SchB). d, induction of H2AX phosphorylation by SchB in IBV-infected cells is shown. Cells were treated as described in C, and total cell lysates were immunoblotted with the indicated antibodies.
Next, 10 μm SchB were added to IBV-infected cells at 14 h post-infection, and its effect on host cell DNA replication at 4 h post-treatment was analyzed by dual-indirect immunofluorescent staining. As shown in Fig. 5c, active DNA replication was observed in IBV-infected cells in the absence of the ATR-specific inhibitor. In the presence of 10 μm SchB, however, much reduced intake of BrdU was observed in the infected cells, although incorporation of BrdU into host cell DNA in neighboring uninfected cells was still observed (Fig. 5c). Considering the potent inhibitory effects of ATR inhibitors on IBV replication, these data indicate that activation of the ATR pathway by IBV infection and the consequent cell cycle arrest at S phase are beneficial to host cell DNA replication in the nucleus and viral RNA replication in the cytoplasm.
ATR and H2AX cooperate in maintaining genome stability under replication stress. Inhibition of the ATR activity may lead to the accumulation of γH2AX by ATM and DNA breaks upon replication fork stalling (15, 17, 47). The possibility that ATR inhibition during IBV infection at the S-phase arrested stage may result in even higher replication stress, and increased accumulation of γH2AX was tested by treating IBV-infected cells with 10 μm SchB at 14 h post-infection for 2, 4, and 5 h. The induction of γH2AX was detected by Western blot and showed moderate increases of γH2AX in IBV-infected cells in the absence of the inhibitor (Fig. 5d). However, a robust increase of γH2AX was detected in IBV-infected cells after 4–5 h of SchB treatment (Fig. 5d). This result suggests that IBV-induced replication stress resembles the hydroxyurea and oncogene-induced replication stress upon ATR inhibition (65).
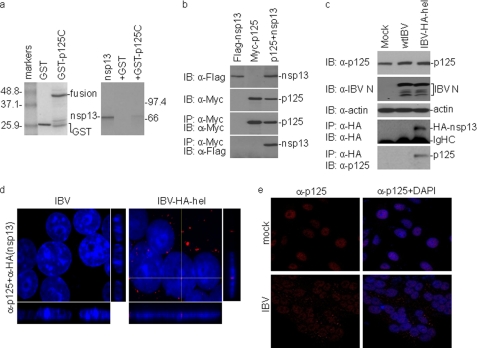
IBV nsp13 Interacts with the p125 Subunit of DNA Pol δ in Vitro and in IBV-infected Cells
By yeast two-hybrid screening, the C-terminal portion (p125C, aa 935–1107) of the p125 catalytic subunit of DNA Pol δ was identified as a potential partner of IBV and SARS-CoV nsp13. A GST pulldown assay was performed to see if IBV nsp13 could interact with GST-p125C directly in vitro. For this purpose, p125C was expressed in bacteria as a GST fusion protein (GST-p125C) (Fig. 6a) and then bound to glutathione-Sepharose 4B beads. Co-precipitation of 35S-labeled IBV nsp13 with GST-p125C or GST alone showed direct binding of IBV nsp13 to GST-p125C in vitro (Fig. 6a), whereas GST alone did not interact with the protein (Fig. 6a).
FIGURE 6.
Interaction of IBV nsp13 with the p125 subunit of DNA Pol δ. a, shown is the physical interaction between IBV nsp13 and p125C in GST pulldown assays. Input GST and GST fusion protein (GST-p125C) were separated on SDS-12% polyacrylamide gel and stained with Coomassie Blue (left panel). GST and GST-p125C were used to pull down the 35S-labeled, in vitro synthesized IBV nsp13. The precipitates (right panel, second and third lanes) and the in vitro translation products (right panel, first lane) were detected by autoradiography. GST was used as a negative control. b, shown is co-immunoprecipitation (IP) of IBV nsp13 with full-length p125 in cells overexpressing the two proteins. HeLa cells were transiently transfected with DNA constructs coding for FLAG-nsp13 (first lane), Myc-p125 (second lane), or both (third lane). Cells were lysed with lysis buffer at 28 h post-transfection, and co-immunoprecipitation was carried out using specific antibodies against Myc. The precipitates (lower two panels) and total cell lysates (upper two panels) were immunoblotted (IB) with the indicated antibodies. c, shown is co-immunoprecipitation of the endogenous p125 with IBV nsp13 in IBV-infected cells. Vero cells were either mock-infected (first lane) or infected with wild type IBV (second lane) or IBV-HA-hel (third lane) at an m.o.i. of 1 and harvested with lysis buffer at 16 h post-infection. Total cell lysates were used for immunoprecipitation assay with anti-HA antibody, and the immunoprecipitates were analyzed by immunoblotting with indicated antibodies (bottom two panels). Western blot analysis of the input lysates is also shown (upper three panels). d, shown is partial localization of IBV nsp13 to the nucleus in IBV-infected cells. Vero cells were infected with either wild type IBV (left panel) or IBV-HA-hel (right panel) at an m.o.i. of 1. At 16 h post-infection, cells were fixed and immunostained with mouse anti-HA antibody, and nuclei were stained with DAPI. Red represents HA-tagged nsp13 expressed in IBV-infected cells. Shown are representative images of Z-section with immunostaining for HA-nsp13 and nuclear staining by DAPI. e, partial relocalization of p125 from the nucleus to the cytoplasm in IBV-infected cells. Vero cells were infected with IBV and fixed at 24 h post-infection. Cells were immunostained with mouse anti-p125 antibody. Red and blue represent p125 and the nuclear staining by DAPI, respectively, in IBV-infected cells.
Next, interaction of the full-length p125 with IBV nsp13 was studied by co-immunoprecipitation assay in mammalian cells. To facilitate detection of both proteins, p125 was tagged with an Myc and nsp13 FLAG tag at their N termini. Co-immunoprecipitation was performed in cells transfected with plasmids either expressing the two proteins individually or together. The expression of these constructs was first examined by Western blot with a monoclonal antibody against either Myc or FLAG and showed similar levels of expression of both proteins either alone or together (Fig. 6b). Cell lysates were then subjected to immunoprecipitation with anti-Myc monoclonal antibody, and the bound proteins were examined by Western blot with either a FLAG or Myc monoclonal antibody. As shown in Fig. 6b, FLAG-nsp13 could be efficiently co-immunoprecipitated with Myc-p125.
Interaction of IBV nsp13 with the p125 subunit in IBV-infected cells was then studied by co-immunoprecipitation assay. For efficient and specific detection of IBV nsp13 protein, an HA tag was fused to the N terminus of the protein, generating a recombinant IBV, IBV-HA-hel. Similar to the recombinant IBV with an HA tag at the N terminus of nsp12 (36, 66), IBV-HA-hel displayed very similar growth characteristic as wild type IBV (data not shown). Indeed, a similar amount of IBV N protein was detected in Vero cells infected with wild type (wtIBV) and IBV-HA-hel (Fig. 6c). A co-immunoprecipitation study showed that the endogenous p125 subunit was precipitated only in cells infected with IBV-HA-hel but not wtIBV (Fig. 6c), confirming that interaction between IBV nsp13 and p125 did occur in virus-infected cells.
The subcellular localization of IBV nsp13 was then studied by infection of Vero cells with IBV-HA-hel and immunofluorescent staining with anti-HA monoclonal antibody. Examination of the stained cells by Z-section confocal microscopy showed that, consistent with previous observations of viral RNA replication-transcription complex (67–69), the majority of the HA-tagged nsp13 protein was localized to the perinuclear region of the infected cells (Fig. 6d). However, a small proportion of the protein was localized to the nuclei of the infected cells (Fig. 6d). The effect of IBV infection on the subcellular localization of Pol δ was then examined by immunofluorescent staining of IBV-infected cells at 20 h post-infection with anti-p125 polyclonal antibodies. As shown in Fig. 6e, the protein was exclusively localized to the nuclei in mock-infected cells. In IBV-infected cells, however, a minor but significant portion of the protein was relocated to the cytoplasm (Fig. 6e).
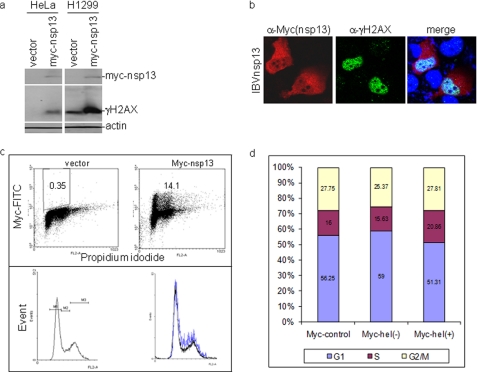
Interaction of IBV nsp13 with p125 Induces DNA Damage Response and Cell Cycle Arrest at S Phase
The possibility that interaction of nsp13 with p125 may interfere with the host cell DNA duplication was tested by examining the effect of nsp13 overexpression on H2AX phosphorylation and host cell cycle progression. A significant induction of γH2AX by overexpression of Myc-nsp13 was observed in both HeLa and H1299 cells (Fig. 7a). Immunofluorescent staining of H1299 cells overexpressing Myc-nsp13 showed that the protein was predominantly localized to the cytoplasm (Fig. 7b). Staining of the same cells with anti-γH2AX antibody showed much brighter nuclear staining of the positively transfected cells (Fig. 7b). Overlapping of the two images showed that a certain proportion of the Myc-nsp13 protein was expressed in the nuclei of the transfected cells (Fig. 7b).
FIGURE 7.
Induction of DNA damage response and cell cycle arrest at S phase by IBV nsp13. a, induction of γH2AX by overexpression of IBV nsp13 is shown. H1299 and HeLa cells were transfected with pXJ-Myc and pXJ-Myc-nsp13, respectively, using Lipofectamine 2000 Transfection kit. Cells were harvested at 48 h post transfection, lysed, and analyzed by Western blot with specific anti-γH2AX antibodies. The same membranes were also probed with anti-actin and anti-Myc antibodies. b, immunofluorescent staining of γH2AX in cells overexpressing Myc-nsp13 is shown. H1299 cells were transfected with pXJ-Myc and pXJ-Myc-nsp13 using the CalPhos Mammalian Transfection kit. At 48 h post-transfection, cells were fixed and double immuno-stained with mouse anti-Myc and rabbit anti-γH2AX antibodies. c, induction of cell cycle arrest at S phase by overexpression of IBV nsp13 is shown. H1299 cells were transfected with pXJ-Myc and pXJ-Myc-nsp13 and incubated for 40 h. Cells were harvested by trypsin treatment, fixed with 80% ethanol, and stained with mouse anti-Myc antibody and PI for FACS. The DNA contents of cells were measured by PI, and the Myc-nsp13 (+) cells were viewed with anti-Myc antibody. A total of 100.000 cells were counted in each experiment. Results of representative experiments performed in triplicate are shown. FITC, fluorescein isothiocyanate, d, cell cycle profiles in cells overexpressing Myc-nsp13 are shown. Statistical analysis was carried out by t test (p < 0.001, n = 3).
Cell cycle profiles of two intercultural populations were analyzed by flow cytometry in H1299 cells transfected with either Myc-nsp13 or an empty Myc-tag vector by using the Myc tag as an indicator of positively transfected cells. The transfection efficiency of plasmids in H1299 cells was assessed by flow cytometry, and the biparametric analysis of Myc-nsp13 immunofluorescence versus DNA content was carried out to show cell cycle profiles of cells overexpressing nsp13. As shown in Fig. 7, c and d, overexpression of nsp13 induced cell cycle arrest at S phase. The percentage of S-phase cells was increased to 20.86% in Myc-nsp13-positive cells compared with 15.63% in Myc-nsp13 negative cells and 16% in Myc control cells (Fig. 7d). Statistical analysis confirmed that the S-phase cell population in nsp13-expressing cells is significantly higher than that in the control cells (p < 0.001, n = 3).
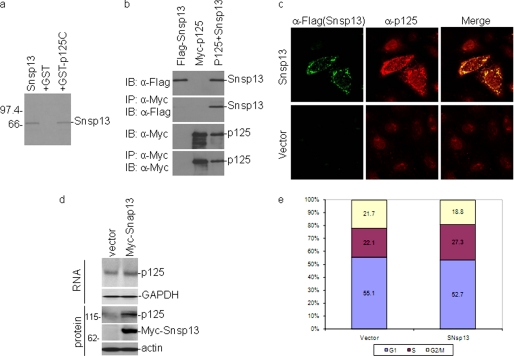
SARS-CoV nsp13 Interacts with p125 and Induces S-phase Arrest and DNA Damage Response
As SARS-CoV nsp13 (Snap13) was also shown to interact with the C-terminal portion of p125 by a yeast two-hybrid screen, GST pulldown and co-immunoprecipitation assays were performed to confirm the interaction. As shown in Fig. 8a, a GST pulldown assay showed that SARS-CoV nsp13 did interact with GST-p125C directly in vitro (Fig. 8a), whereas GST alone did not interact with the protein (Fig. 8a). Similarly, immunoprecipitation assays showed that FLAG-Snsp13 could be co-immunoprecipitated with Myc-p125 in mammalian cell (Fig. 8b).
FIGURE 8.
Interaction of SARS-CoV nsp13 with the p125 subunit of Pol δ and induction of DNA damage response and cell cycle arrest. a, physical association of SARS-CoV nsp13 (Snsp13) with p125C in GST pulldown assay is shown. GST and GST-p125C were used to pull down the 35S-labeled, in vitro translated Snsp13. The precipitates and the in vitro translation products were detected by autoradiography. The GST protein was used as a negative control. b, shown is co-immunoprecipitation (IP) of Snsp13 with full-length p125 in mammalian cells overexpressing the two proteins. HeLa cells were transiently transfected with DNA constructs coding for FLAG-Snsp13 (first lane 1), myc-p125 (second lane), or both (third lane). Cells were lysed with lysis buffer at 28 h post-transfection, and co-immunoprecipitation was carried out using specific antibodies against Myc. The precipitates and total cell lysates were immunoblotted (IB) with indicated antibodies. c, partial relocalization of p125 from the nucleus to the cytoplasm in cells overexpressing Snsp13 is shown. HeLa cells were transfected with plasmid pXJFLAG-Snsp13 or empty vector pXJFLAG and fixed at 24 h post-transfection. Cells were co-immunostained with mouse anti-p125 and rabbit anti-FLAG antibodies. Red and green represent endogenous p125 and the FLAG-tagged Snsp13, respectively. Cells transfected with an empty vector expressing the FLAG tag alone were used as a negative control. d, shown is a Western blot analysis of cells stably expressing Snsp13. The expression of the endogenous p125 and Myc-Snsp13 in Snsp13-expressing stable cells was determined by Western blot analysis with anti-p125 and anti-Myc antibodies. The expression of the endogenous p125 and GAPDH at the mRNA level in Snsp13-expressing cells was determined by Northern blot analysis. e, induction of S-phase arrest in cells stably expressing SARS-CoV nsp13 is shown. Cell cycle profiles of stable Snsp13-expressing cell clones (SNsp13) and control cells (vector) were determined by flow cytometry analysis with PI staining. Unsynchronized cells grown for 24 h were fixed and stained for FACS analysis. A total of 10,000 cells were counted in each experiment, and the results represent the means of seven repeated experiments. Statistical analysis was carried by t test (p < 0.001, n = 7).
The effect of Snsp13 overexpression on the subcellular localization of endogenous p125 in HeLa cells was then examined by fluorescence microscopy. As shown in Fig. 8c, the FLAG-tagged Snsp13 protein was mainly localized to the cytoplasm with some nuclear staining. In cells transfected with the empty vector, p125 was mainly in the nuclei as previously described (Fig. 8c). In cells expressing the FLAG-tagged Snsp13, strong cytoplasmic staining of p125 was observed (Fig. 8c). The two images partially overlapped in both the cytoplasm and the nucleus (Fig. 8c).
A stable cell line with Myc-tagged-Snsp13 overexpression in Vero was made successfully. Overexpression of Myc-Snsp13 at the protein level was confirmed by Western blotting, and its overexpression induced a slightly increased expression of endogenous p125 at the RNA level as confirmed by Northern blotting (Fig. 8d). Quantification by Image J analysis and real-time RT-PCR showed a 1.35-fold increase of the endogenous p125 in the stable cell clone expressing Myc-Snsp13. This result coincides with a previous report that UV irradiation induced p125 expression at the mRNA level (70). As shown in Fig. 8e, analysis of the cell cycle profiles by flow cytometry demonstrated that overexpression of Snsp13 also significantly induced cell cycle arrest at S phase (p < 0.001, n = 7), indicating that Snsp13 may inhibit DNA replication on the replication fork.
DISCUSSION
Manipulation of cell cycle progression and induction of apoptosis are two common strategies used by viruses to regulate their infection cycles. In cells infected with Coronavirus, cell cycle perturbation and apoptosis were observed in several reports (30, 31, 71). In a previous study, IBV infection was shown to impose a growth inhibitory effect on cultured cells by inducing cell cycle arrest at S and G2/M phases in both p53-null and wild type cells (31). This cell cycle arrest may be catalyzed by modulation of various cell cycle regulatory genes and accumulation of hypophosphorylated RB but was independent of p53. In this study we further show that activation of cellular DNA damage response is one of the mechanisms exploited by Coronavirus to induce cell cycle arrest. An ATR-dependent DNA damage response was shown to be activated by IBV infection, as implicated by induction of phosphorylation of multiple downstream targets of ATR, including Chk1, H2AX, and RPA. Suppression of the ATR kinase activity by chemical inhibitors and by siRNA-mediated knockdown of ATR reduced the IBV-induced ATR signaling and inhibited the replication of IBV.
Distinct strategies, including activation of DNA damage response and recruitment of DNA repair proteins to viral replication centers were found to be used by different viruses to manipulate host DNA damage response and viral replication (19, 21). As activation of DNA damage response can promote apoptosis, viruses have also evolved ways to tailor the damage signaling by targeting a specific sensor or a repair protein for degradation or mislocalization to suppress the downstream signaling. For example, Epstein-Barr virus lytic replication induces ATM-dependent DNA damage checkpoint signaling, leading to the clustering of phosphorylated ATM and Mre11-Rad50-Nbs1 (MRN) complexes as well as homologous recombinational repair (HRR) factors such as RPA, Rad51, and Rad52 to sites of viral genome synthesis in the nucleus (72, 73). The nuclear antigen 3C (EBNA3C) of Epstein-Barr virus could release G2/M cell cycle checkpoint by direct interaction with Chk2 (74). Human Papillomavirus 16 E7 oncoprotein attenuates DNA damage checkpoint control by increasing the proteolytic turnover of claspin (75). SV40 large T antigen is a target of ATM kinase, and this ATM-mediated phosphorylation of T-antigen is required for optimal viral DNA synthesis (77). Inhibition of ATM activity decreases SV40 DNA replication and delays recruitment of T-antigen and DNA repair proteins to viral replication foci (76–78). SV40 infection was also shown to activate the ATR-Δp53-p21-mediated intra-S checkpoint to maintain the host cells in S phase, an optimal environment for SV40 replication (79). In our previous study arrest of cell cycle at both S and G2/M phase by IBV was shown to enhance viral replication (31). It appears that this may be one of the strategies exploited by Coronavirus to create a more conducible environment for its own replication. The observation that inhibition of the ATR pathway dramatically reduces the replication of IBV in this study lends further support to this conclusion. IBV infection induces cell cycle arrest at S phase as indicated by BrdU incorporation in the infected cells. Abrogation of the ATR kinase activity by chemical inhibitors reverses the accumulation of S-phase cells with BrdU incorporation induced by IBV replication. These findings indicate that the ATR signaling activated by IBV replication contributes to the IBV-induced S phase arrest and is required for efficient IBV replication and production. On the other hand, the observation that inhibition of ATR activity reduces DNA replication in IBV-arrested S phase is consistent with previous reports that deletion of Chk1 and inhibition of ATR activity by caffeine result in reduced BrdU incorporation in cells after release from aphidicolin treatment (16, 80).
How could cellular DNA replication, an exclusive nuclear event, be coupled with the replication of an RNA virus in the cytoplasm? Many DNA viruses have evolved mechanisms to override normal checkpoint control and force cells into S phase to ensure an abundant supply of nucleotides and other essential replication factors during their replication cycles (21, 81). Evidence provided in this study demonstrated that the same strategy was also exploited by Coronavirus, an RNA virus that exclusively replicates its RNA in the cytoplasm. The extended S phase may provide a more conducible cellular environment for both DNA replication in the nucleus and viral RNA replication in the cytoplasm. It is likely that some host factors in the DNA replication machinery may be directly involved in the replication of viral RNA in the cytoplasm. As the cellular conditions in the S phase favor the replication of cellular DNA, the factors that participate in DNA replication would be relatively more abundant in the nucleus. These factors would also facilitate viral RNA replication in the cytoplasm by virus-induced translocation from the nucleus to the cytoplasm. In this study partial relocation of Pol δ from the nucleus to the cytoplasm was observed in IBV-infected cells. In a previous study, DDX1, an essential host cell factor involved in RNA metabolism, was also shown to be relocated from the nucleus to the cytoplasm and is involved in Coronavirus RNA replication in IBV-infected cells (36).
In this study the p125 catalytic subunit of Pol δ was identified as an interacting partner of Coronavirus nsp13 replicase protein. Pol δ is primarily a lagging strand polymerase during DNA replication across the entire nuclear genome (23, 82). Lagging strand DNA replication is believed to proceed in several discrete stages; they are initiation by DNA primase, limited elongation of the RNA primer by Pol α, switching of the primer terminus from Pol α to Pol δ, elongation by Pol δ, and maturation of the elongated Okazaki fragment by Pol δ, FEN1, and DNA ligase. Each transition may be mediated by a specific protein or protein complex and has to occur with very high efficiency. In a mammalian cell, this process occurs 20–50 million times during every cell cycle (23). Pol δ also participates in DNA repair as a gap-filling polymerase, including nucleotide excision repair, mismatch repair, and base excision repair (70, 83–88). ATR and ATR orthologues in yeast may stabilize replication forks that have stalled as the result of DNA polymerase inhibition (13–15, 17). Polymerase inhibition activates ATR through the generation of ssDNA (5, 6, 89). Interaction between Coronavirus nsp13 and Pol δ may lead to an increased reliance on ATR for replication fork stability. However, the underlying mechanisms for generation of extensive ssDNA are yet to be elucidated. One possibility is that partial relocation of Pol δ from the nucleus to the cytoplasm, as observed in this study, in IBV-infected cells may reduce the local concentration of Pol δ in the nucleus and slow down the elongation of lagging chain, resulting in the formation of ssDNA. Alternatively, physical interaction between nsp13 and Pol δ may render direct inhibition on host DNA replication.
It would be interesting to know if other RNA viruses would also exploit this pathway during their replication cycles. Preliminary studies showed that ATR inhibitors exhibited potent inhibitory effects on dengue virus replication in cultured cells (data not shown), suggesting that dengue virus, a member of the Flavivirus family, may also utilize the ATR pathway to complete efficiently its replication cycle. Systematic analysis of other RNA viruses and the underlying mechanisms would reveal if this is a general phenomenon during the replication of RNA viruses. Nevertheless, as potent inhibitory effects on IBV replication by ATR inhibitors were observed in virus-infected cells, it would be plausible to explore the possibility of designing anti-viral therapeutic approaches based on these inhibitors. As long term admission of ATR inhibitors may render side effects on host DNA metabolism, such an approach would be particularly useful for an acute viral infection, such as infections caused by dengue virus and Coronavirus.
Footnotes
- ATM
- ataxia-telangiectasia mutated
- ATR
- ATM/Rad3-related
- RPA
- replication factor A
- CHK1 and -2
- checkpoint kinase-1 and -2, respectively
- SV40
- simian virus 40
- nsp13
- nonstructural protein 13
- SARS-CoV
- severe acute respiratory syndrome Coronavirus
- IBV
- infectious bronchitis virus
- TCID50
- 50% tissue culture infective dose
- PI
- propidium iodide
- Pol δ
- polymerase δ
- SchB
- Schisandrin B
- aa
- amino acids
- m.o.i.
- multiplicity of infection.
REFERENCES
- 1. Cimprich K. A., Cortez D. (2008) Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nyberg K. A., Michelson R. J., Putnam C. W., Weinert T. A. (2002) Annu. Rev. Genet. 36, 617–656 [DOI] [PubMed] [Google Scholar]
- 3. Andegeko Y., Moyal L., Mittelman L., Tsarfaty I., Shiloh Y., Rotman G. (2001) J. Biol. Chem. 276, 38224–38230 [DOI] [PubMed] [Google Scholar]
- 4. Lee J. H., Paull T. T. (2005) Science 308, 551–554 [DOI] [PubMed] [Google Scholar]
- 5. Costanzo V., Gautier J. (2003) Cell Cycle 2, 17. [DOI] [PubMed] [Google Scholar]
- 6. Zou L., Elledge S. J. (2003) Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 7. Paulsen R. D., Cimprich K. A. (2007) DNA Repair 6, 953–966 [DOI] [PubMed] [Google Scholar]
- 8. Bartek J., Lukas J. (2003) Cancer Cell 3, 421–429 [DOI] [PubMed] [Google Scholar]
- 9. Reinhardt H. C., Yaffe M. B. (2009) Curr. Opin. Cell Biol. 21, 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 11. Stokes M. P., Rush J., Macneill J., Ren J. M., Sprott K., Nardone J., Yang V., Beausoleil S. A., Gygi S. P., Livingstone M., Zhang H., Polakiewicz R. D., Comb M. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19855–19860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C. S., Foiani M. (2001) Nature 412, 557–561 [DOI] [PubMed] [Google Scholar]
- 13. Tercero J. A., Diffley J. F. (2001) Nature 412, 553–557 [DOI] [PubMed] [Google Scholar]
- 14. Casper A. M., Nghiem P., Arlt M. F., Glover T. W. (2002) Cell 111, 779–789 [DOI] [PubMed] [Google Scholar]
- 15. Brown E. J., Baltimore D. (2003) Genes Dev. 17, 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zachos G., Rainey M. D., Gillespie D. A. (2003) EMBO J. 22, 713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chanoux R. A., Yin B., Urtishak K. A., Asare A., Bassing C. H., Brown E. J. (2009) J. Biol. Chem. 284, 5994–6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Segurado M., Tercero J. A. (2009) Biol. Cell 101, 617–627 [DOI] [PubMed] [Google Scholar]
- 19. Lilley C. E., Schwartz R. A., Weitzman M. D. (2007) Trends Microbiol. 15, 119–126 [DOI] [PubMed] [Google Scholar]
- 20. Ariumi Y., Kuroki M., Dansako H., Abe K., Ikeda M., Wakita T., Kato N. (2008) J. Virol. 82, 9639–9646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chaurushiya M. S., Weitzman M. D. (2009) DNA Repair 8, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lai C. K., Jeng K. S., Machida K., Cheng Y. S., Lai M. M. (2008) Virology 370, 295–309 [DOI] [PubMed] [Google Scholar]
- 23. Burgers P. M. (2009) J. Biol. Chem. 284, 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Podust V. N., Chang L. S., Ott R., Dianov G. L., Fanning E. (2002) J. Biol. Chem. 277, 3894–3901 [DOI] [PubMed] [Google Scholar]
- 25. Li H., Xie B., Zhou Y., Rahmeh A., Trusa S., Zhang S., Gao Y., Lee E. Y., Lee M. Y. (2006) J. Biol. Chem. 281, 14748–14755 [DOI] [PubMed] [Google Scholar]
- 26. Pohler J. R., Otterlei M., Warbrick E. (2005) BMC Mol. Biol. 6, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Woo P. C., Lau S. K., Huang Y., Yuen K. Y. (2009) Exp. Biol. Med. (Maywood) 234, 1117–1127 [DOI] [PubMed] [Google Scholar]
- 28. Ksiazek T. G., Erdman D., Goldsmith C. S., Zaki S. R., Peret T., Emery S., Tong S., Urbani C., Comer J. A., Lim W., Rollin P. E., Dowell S. F., Ling A. E., Humphrey C. D., Shieh W. J., Guarner J., Paddock C. D., Rota P., Fields B., DeRisi J., Yang J. Y., Cox N., Hughes J. M., LeDuc J. W., Bellini W. J., Anderson L. J. (2003) N. Engl. J. Med. 348, 1953–1966 [DOI] [PubMed] [Google Scholar]
- 29. Cavanagh D. (2007) Vet. Res. 38, 281–297 [DOI] [PubMed] [Google Scholar]
- 30. Dove B., Brooks G., Bicknell K., Wurm T., Hiscox J. A. (2006) J. Virol. 80, 4147–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li F. Q., Tam J. P., Liu D. X. (2007) Virology 365, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'Driscoll M., Ruiz-Perez V. L., Woods C. G., Jeggo P. A., Goodship J. A. (2003) Nat. Genet. 33, 497–501 [DOI] [PubMed] [Google Scholar]
- 33. Burdak-Rothkamm S., Rothkamm K., Prise K. M. (2008) Cancer Res. 68, 7059–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shen H., Fang S. G., Chen B., Chen G., Tay F. P., Liu D. X. (2009) J. Virol. Methods 160, 48–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fang S. G., Shen S., Tay F. P., Liu D. X. (2005) Biochem. Biophys. Res. Commun. 336, 417–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu L., Khadijah S., Fang S., Wang L., Tay F. P., Liu D. X. (2010) J. Virol. 84, 8571–8583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamada Y., Liu D. X. (2009) J. Virol. 83, 8744–8758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fang S., Chen B., Tay F. P., Ng B. S., Liu D. X. (2007) Virology 358, 136–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang X., Liao Y., Yap P. L., Png K. J., Tam J. P., Liu D. X. (2009) J. Virol. 83, 12462–12472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiao H., Xu L. H., Yamada Y., Liu D. X. (2008) PLoS One 3, e1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li F. Q., Xiao H., Tam J. P., Liu D. X. (2005) FEBS Lett. 579, 2387–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang K., Ye Y., Xu Z., Zhang X., Hou Z., Cui Y., Song Y. (2010) Cancer Genet. Cytogenet. 200, 40–46 [DOI] [PubMed] [Google Scholar]
- 43. Kinner A., Wu W., Staudt C., Iliakis G. (2008) Nucleic Acids Res. 36, 5678–5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ward I. M., Chen J. (2001) J. Biol. Chem. 276, 47759–47762 [DOI] [PubMed] [Google Scholar]
- 45. Marti T. M., Hefner E., Feeney L., Natale V., Cleaver J. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9891–9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Syljuåsen R. G., Sørensen C. S., Hansen L. T., Fugger K., Lundin C., Johansson F., Helleday T., Sehested M., Lukas J., Bartek J. (2005) Mol. Cell. Biol. 25, 3553–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Toledo L. I., Murga M., Zur R., Soria R., Rodriguez A., Martinez S., Oyarzabal J., Pastor J., Bischoff J. R., Fernandez-Capetillo O. (2011) Nat. Struct. Mol. Biol. 18, 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murga M., Bunting S., Montaña M. F., Soria R., Mulero F., Cañamero M., Lee Y., McKinnon P. J., Nussenzweig A., Fernandez-Capetillo O. (2009) Nat. Genet. 41, 891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Toledo L. I., Murga M., Gutierrez-Martinez P., Soria R., Fernandez-Capetillo O. (2008) Genes Dev. 22, 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Heffernan T. P., Simpson D. A., Frank A. R., Heinloth A. N., Paules R. S., Cordeiro-Stone M., Kaufmann W. K. (2002) Mol. Cell. Biol. 22, 8552–8561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao H., Piwnica-Worms H. (2001) Mol. Cell. Biol. 21, 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Olson E., Nievera C. J., Klimovich V., Fanning E., Wu X. (2006) J. Biol. Chem. 281, 39517–39533 [DOI] [PubMed] [Google Scholar]
- 53. Matsuoka S., Rotman G., Ogawa A., Shiloh Y., Tamai K., Elledge S. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10389–10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kozlov S. V., Graham M. E., Peng C., Chen P., Robinson P. J., Lavin M. F. (2006) EMBO J. 25, 3504–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wold M. S. (1997) Annu. Rev. Biochem. 66, 61–92 [DOI] [PubMed] [Google Scholar]
- 56. Iftode C., Daniely Y., Borowiec J. A. (1999) Crit. Rev. Biochem. Mol. Biol. 34, 141–180 [DOI] [PubMed] [Google Scholar]
- 57. Liu J. S., Kuo S. R., Melendy T. (2006) J. Cell. Biochem. 99, 1452–1462 [DOI] [PubMed] [Google Scholar]
- 58. Manthey K. C., Opiyo S., Glanzer J. G., Dimitrova D., Elliott J., Oakley G. G. (2007) J. Cell Sci. 120, 4221–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Won J., Kim M., Kim N., Ahn J. H., Lee W. G., Kim S. S., Chang K. Y., Yi Y. W., Kim T. K. (2006) Nat. Chem. Biol. 2, 369–374 [DOI] [PubMed] [Google Scholar]
- 60. Hickson I., Zhao Y., Richardson C. J., Green S. J., Martin N. M., Orr A. I., Reaper P. M., Jackson S. P., Curtin N. J., Smith G. C. (2004) Cancer Res. 64, 9152–9159 [DOI] [PubMed] [Google Scholar]
- 61. Willmore E., de Caux S., Sunter N. J., Tilby M. J., Jackson G. H., Austin C. A., Durkacz B. W. (2004) Blood 103, 4659–4665 [DOI] [PubMed] [Google Scholar]
- 62. Sarkaria J. N., Busby E. C., Tibbetts R. S., Roos P., Taya Y., Karnitz L. M., Abraham R. T. (1999) Cancer Res. 59, 4375–4382 [PubMed] [Google Scholar]
- 63. Nishida H., Tatewaki N., Nakajima Y., Magara T., Ko K. M., Hamamori Y., Konishi T. (2009) Nucleic Acids Res. 37, 5678–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Alderton G. K., Joenje H., Varon R., Børglum A. D., Jeggo P. A., O'Driscoll M. (2004) Hum. Mol. Genet. 13, 3127–3138 [DOI] [PubMed] [Google Scholar]
- 65. Toledo L. I., Murga M., Fernandez-Capetillo O. (2011) Mol. Oncol. 5, 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fang S. G., Shen H., Wang J., Tay F. P., Liu D. X. (2008) Virology 379, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ivanov K. A., Thiel V., Dobbe J. C., van der Meer Y., Snijder E. J., Ziebuhr J. (2004) J. Virol. 78, 5619–5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Knoops K., Kikkert M., Worm S. H., Zevenhoven-Dobbe J. C., van der Meer Y., Koster A. J., Mommaas A. M., Snijder E. J. (2008) PLoS Biol. 6, e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hagemeijer M. C., Verheije M. H., Ulasli M., Shaltiël I. A., de Vries L. A., Reggiori F., Rottier P. J., de Haan C. A. (2010) J. Virol. 84, 2134–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zeng X. R., Jiang Y., Zhang S. J., Hao H., Lee M. Y. (1994) J. Biol. Chem. 269, 13748–13751 [PubMed] [Google Scholar]
- 71. Liu C., Xu H. Y., Liu D. X. (2001) J. Virol. 75, 6402–6409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kudoh A., Iwahori S., Sato Y., Nakayama S., Isomura H., Murata T., Tsurumi T. (2009) J. Virol. 83, 6641–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kudoh A., Fujita M., Zhang L., Shirata N., Daikoku T., Sugaya Y., Isomura H., Nishiyama Y., Tsurumi T. (2005) J. Biol. Chem. 280, 8156–8163 [DOI] [PubMed] [Google Scholar]
- 74. Choudhuri T., Verma S. C., Lan K., Murakami M., Robertson E. S. (2007) J. Virol. 81, 6718–6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Spardy N., Covella K., Cha E., Hoskins E. E., Wells S. I., Duensing A., Duensing S. (2009) Cancer Res. 69, 7022–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Digweed M., Demuth I., Rothe S., Scholz R., Jordan A., Grötzinger C., Schindler D., Grompe M., Sperling K. (2002) Oncogene 21, 4873–4878 [DOI] [PubMed] [Google Scholar]
- 77. Shi Y., Dodson G. E., Shaikh S., Rundell K., Tibbetts R. S. (2005) J. Biol. Chem. 280, 40195–40200 [DOI] [PubMed] [Google Scholar]
- 78. Zhao X., Madden-Fuentes R. J., Lou B. X., Pipas J. M., Gerhardt J., Rigell C. J., Fanning E. (2008) J. Virol. 82, 5316–5328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rohaly G., Korf K., Dehde S., Dornreiter I. (2010) J. Virol. 84, 10727–10747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dimitrova D. S., Gilbert D. M. (2000) Nat. Cell Biol. 2, 686–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schang L. M. (2003) Prog. Cell Cycle Res. 5, 103–124 [PubMed] [Google Scholar]
- 82. Larrea A. A., Lujan S. A., Nick McElhinny S. A., Mieczkowski P. A., Resnick M. A., Gordenin D. A., Kunkel T. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 17674–17679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Aboussekhra A., Biggerstaff M., Shivji M. K., Vilpo J. A., Moncollin V., Podust V. N., Proti M., Hübscher U., Egly J. M., Wood R. D. (1995) Cell 80, 859–868 [DOI] [PubMed] [Google Scholar]
- 84. Aboussekhra A., Wood R. D. (1995) Exp. Cell Res. 221, 326–332 [DOI] [PubMed] [Google Scholar]
- 85. Halas A., Baranowska H., Policińska Z., Jachymczyk W. J. (1997) Curr. Genet. 31, 292–301 [DOI] [PubMed] [Google Scholar]
- 86. Longley M. J., Pierce A. J., Modrich P. (1997) J. Biol. Chem. 272, 10917–10921 [DOI] [PubMed] [Google Scholar]
- 87. Overmeer R. M., Gourdin A. M., Giglia-Mari A., Kool H., Houtsmuller A. B., Siegal G., Fousteri M. I., Mullenders L. H., Vermeulen W. (2010) Mol. Cell. Biol. 30, 4828–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ogi T., Limsirichaikul S., Overmeer R. M., Volker M., Takenaka K., Cloney R., Nakazawa Y., Niimi A., Miki Y., Jaspers N. G., Mullenders L. H., Yamashita S., Fousteri M. I., Lehmann A. R. (2010) Mol. Cell 37, 714–727 [DOI] [PubMed] [Google Scholar]
- 89. Byun T. S., Pacek M., Yee M. C., Walter J. C., Cimprich K. A. (2005) Genes Dev. 19, 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]