Background: ENU mutagenesis was used to generate new animal models of diabetes.
Results: We identified two novel mutations in glucokinase, with glucose >400 mg/dl in homozygotes, and differential responsiveness to glucokinase activators.
Conclusion: Increased GCK thermolability is a major cause of hyperglycemia in Gck mutant mice.
Significance: Chemical genetics creates new models to study glucose homeostasis and diabetes drugs.
Keywords: Animal Models, Diabetes, Glucokinase, Liver, Mutagenesis
Abstract
We performed genome-wide mutagenesis in C57BL/6J mice using N-ethyl-N-nitrosourea to identify mutations causing high blood glucose early in life and to produce new animal models of diabetes. Of a total of 13 new lines confirmed by heritability testing, we identified two semi-dominant pedigrees with novel missense mutations (GckK140E and GckP417R) in the gene encoding glucokinase (Gck), the mammalian glucose sensor that is mutated in human maturity onset diabetes of the young type 2 and the target of emerging anti-hyperglycemic agents that function as glucokinase activators (GKAs). Diabetes phenotype corresponded with genotype (mild-to-severe: Gck+/+ < GckP417R/+, GckK140E/+ < GckP417R/P417R, GckP417R/K140E, and GckK140E/K140E) and with the level of expression of GCK in liver. Each mutant was produced as the recombinant enzyme in Escherichia coli, and analysis of kcat and tryptophan fluorescence (I320/360) during thermal shift unfolding revealed a correlation between thermostability and the severity of hyperglycemia in the whole animal. Disruption of the glucokinase regulatory protein-binding site (GCKK140E), but not the ATP binding cassette (GCKP417R), prevented inhibition of enzyme activity by glucokinase regulatory protein and corresponded with reduced responsiveness to the GKA drug. Surprisingly, extracts from liver of diabetic GCK mutants inhibited activity of the recombinant enzyme, a property that was also observed in liver extracts from mice with streptozotocin-induced diabetes. These results indicate a relationship between genotype, phenotype, and GKA efficacy. The integration of forward genetic screening and biochemical profiling opens a pathway for preclinical development of mechanism-based diabetes therapies.
Introduction
Type 2 diabetes mellitus (T2DM)3 is an escalating cause of metabolic disease involving interactions between genetic and environmental factors. At the cellular level, both insulin resistance and pancreatic β-cell failure contribute to disease onset and progression, validating strategies to augment insulin secretion and/or enhance insulin action as the cornerstones of mechanism-based therapeutics. Clues to elucidating the genetic basis of diabetes have emerged with genetic studies of Maturity Onset Diabetes of the Young (MODY) (1–6), which are syndromic variants characterized by a dominant mode of inheritance that account for 2–5% of T2DM. Recent genome-wide association studies (7–13) have implicated additional factors in the pathogenesis of T2DM in humans; however, obstacles in such studies have been the polygenic nature of the disease, the small effect of most gene variants, and a lack of experimental models that recapitulate human disease mechanisms for the preclinical development of anti-hyperglycemic therapeutics.
We have used the alkylating agent, N-ethyl-N-nitrosourea (ENU), which produces single nucleotide mutations, to perform an unbiased forward genetic screen to identify and characterize genes involved in glucose homeostasis and to model aspects of human diabetes (14). An advantage of chemical mutagenesis in comparison with generation of knock-out models is the opportunity to produce an allele series that yields a spectrum of effects on glucose homeostasis because mutations may generate gain-of-function, dominant-negative, and partial loss-of-function alleles (15, 16). The allele series produced by ENU mutagenesis can in turn provide preclinical models to test the efficacy and mechanism of small molecule anti-hyperglycemic drugs, whereas gene knock-out models may be refractory to many of these agents.
Here, we report on the identification and functional analysis of two novel alleles of glucokinase (Gck) encoding the rate-limiting enzyme in hepatic glucose catabolism and glycogen synthesis in mice and humans. GCK is also expressed in the α-, β-, and δ-cells of the pancreas in addition to the L and K cells of the intestine, the pituitary, and the hypothalamus. In human subjects, loss-of-function alleles of glucokinase cause hyperglycemia by increasing the glucose threshold for insulin release, resulting in a reduced insulin/glucose ratio, whereas gain-of-function alleles lower this threshold and cause hyperinsulinism (17).
GCK (or hexokinase IV) is most abundant in hepatic parenchymal cells where its transcription is stimulated by insulin (18); it functions to modulate both glycolysis and glycogen synthesis, and it also plays an essential role in the pancreas as the β-cell glucose sensor. Thus, stimulation of GCK activity may impact both β-cell insulin secretion and hepatic glucose production. GCK exhibits unique biophysical features as follows: it is half the size of other hexokinases; it has a lower affinity for glucose; it works in the presence of the product; and it displays sigmoidal kinetics of glucose phosphorylation. Although the ATP affinity of hexokinases is uniformly high, new GCK activator (GKA) drugs increase the Vmax and glucose affinity of GCK through binding to an allosteric site, different from the glucose- or ATP-binding sites (19). Binding of GKAs is thought to stabilize the active conformation of GCK, which appears to be more compact in the crystal structure (Fig. 1, E and F). The existence of an allosteric site suggests the presence of an endogenous ligand, possibly an activator. Here we present the following: 1) results of a genome-wide mutagenesis screen for diabetes in C57BL/6J mice; 2) analysis of kinetic, thermostability, and physiological profiles of two new GCK mutant enzymes identified in this screen using recombinant forms of the enzyme; 3) inhibitory effects of liver lysates from animals expressing the mutations on GCK activity; and 4) genotype-phenotype response to GKA drugs in vitro and in vivo.
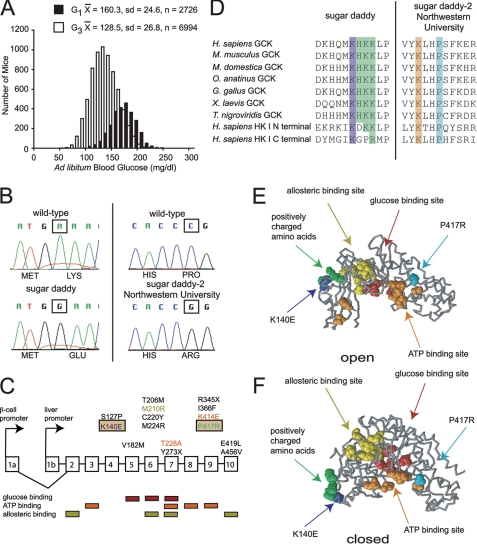
FIGURE 1.
Generation and location of glucokinase mutations. A, histogram of ad libitum fed blood glucose for the mice screened. Black bars, G1 mice, n = 2726; white bars, G3 mice, n = 6994. B, chromatograms for the sugar daddy and the sugar daddy-2 Northwestern University mutations. The sugar daddy mutation, located in exon 4, is an A to G transition, which causes the missense mutation K140E. The sugar daddy-2 Northwestern University mutation, located in exon 9, is a C to G transversion, which causes the missense mutation P417R. C, gene structure for glucokinase. Shown above the panel are missense and nonsense glucokinase mutations that have been isolated from, or introduced into, mice. Indicated below the panel are locations important for substrate binding (41). D, multiple sequence alignments for glucokinase and hexokinase I. Human (Homo sapiens, gi:4503951 and gi:116242516), mouse (Mus musculus, gi:1008870), opossum (Monodelphis domestica, gi:126303415), platypus (Ornithorhynchus anatinus, gi:149557345), chicken (Gallus gallus, gi:148743500), frog (X. laevis, gi:148236406), pufferfish (Tetraodon nigroviridis, gi:47226566). E, protein structure for the open conformation of GCK, Protein Data Bank code 1V4T (32). F, protein structure for the closed conformation of GCK, Protein Data Bank code 1V4S (32). Protein structures created with Rasmol (52). Protein Data Bank codes 1V4T and 1V4S obtained from the RCSB (53). Red, glucose-binding site; orange, ATP-binding site; yellow, allosteric binding site; green, positively charged amino acids; dark blue, Lys-140; turquoise, Pro-417.
EXPERIMENTAL PROCEDURES
Animal Protocols
Male C57BL/6J (B6) mice were mutagenized with ENU to produce G0 mice. The G0 mice were then bred with wild-type female B6 mice to produce mutant generation 1 (G1). G1 mice were either screened for dominant mutations or used for further breeding. G1 mice were bred with wild-type B6 mice to produce G2 females. G2 females were backcrossed to their G1 fathers to produce G3 mice. G3 mice were screened for dominant and homozygous recessive mutations. The screen consisted of measuring glucose in whole blood drawn from tails of nonfasted 8-week-old male mice. We used male mice because they are more diabetes-prone than females; they tend to have higher glucose levels than females, and they usually have higher postprandial increases in glycemia. Mice of interest expressed glucose values above 200 mg/dl. To determine whether the hyperglycemia was heritable, these animals were mated with two wild-type B6 females, producing backcross progeny for further screening. If progeny (of either sex) from these matings also showed a hyperglycemic phenotype, this suggested that the phenotype was heritable and the trait was considered dominant or semi-dominant. All animal care and use procedures were in accordance with guidelines of the Northwestern University Institutional Animal Care and Use Committee.
Sequencing
Primers to amplify the exonic sequence of Gck (supplemental Table 2) were designed with Primer 3 (20). The same primers were used for PCR amplification and sequencing. PCR was performed on genomic DNA with the proofreading Pyrococcus furiosus DNA polymerase. PCR products were purified with the QIAquick® gel extraction kit (Qiagen) or the QIAquick® PCR purification kit (Qiagen). The ABI Big Dye Terminator, version 3.1, cycle sequencing kit and the ABI 3730 high throughput DNA sequencer were used for the sequencing reaction and gel sequencing at the Northwestern Genomics Core Facility.
Mapping
To map the sugar daddy line, heterozygous B6 male mice were crossed to wild-type female A/J or DBA/2J mice to produce F1 progeny. F1 mice were tested for blood glucose at 8 weeks of age, and female mice with glucose values over 200 mg/dl were chosen to backcross to a homozygous mutant B6 male, to produce heterozygous or homozygous N2 mice. For mapping purposes, mice with blood glucose values less than 300 mg/dl were phenotyped as heterozygous, and mice with blood glucose values greater than 400 mg/dl were phenotyped as homozygous. Two mice with blood glucose values between 300 and 400 mg/dl were not included in the mapping analysis. A panel of 37 simple sequence length polymorphism markers (supplemental Table 3) was used to genotype these N2 mice.
Genotyping
Once the causative SNP was identified through sequencing, primers for real time PCR genotyping were designed using primer 3 (20). Two forward primers were used with a single reverse primer for each genotype. For GckK140E, the primers were 5′-GGACAAGCATCAGAAGG-3′ (forward mutant), 5′-GGACAAGCATCAGAAGA-3′ (forward wild-type), and 5′-AGGGAAGGAGAAGGTGAAGC-3′ (reverse). For GckP417R, the primers used were 5′-CGTGTACAAGCTGCACCG-3′ (forward mutant), 5′-CGTGTACAAGCTGCACCC-3′ (forward wild-type), and 5′-AGGATCGGCTCACAAAAGC-3′ (reverse).
Glucose Tolerance Tests
Glucose tolerance tests were performed in 5–7-month-old mice following a 16-h fast. d-Glucose was given intraperitoneally at a dose of 2 g/kg body weight. Blood was obtained from a snip at the tip of the tail at 0, 15, 30, 60, and 120 min. Serum was obtained by centrifugation of whole blood. Serum glucose was measured using an Analox GM 7. Serum insulin was measured using insulin ELISA kits from Crystal Chem.
Kinetic Analysis of Recombinant Human and Hepatic Mouse GCK
Recombinant wild-type and mutant human β-cell GCKs were generated and expressed as GST fusion proteins in Escherichia coli as described previously (21). GST-GCK fusion proteins were cleaved with factor Xa. Mouse livers were dissected from ad libitum fed mice, snap-frozen in liquid nitrogen, and then stored frozen at −80 °C until used. Frozen livers were thawed and homogenized in 25 mm Hepes buffer (pH 7.4) containing 150 mm KCl, 2 mm DTT, and 1 mm EDTA. A pyridine nucleotide-coupled assay was used to measure the kinetics of glucose phosphorylation. The composition of the assay reagent was as follows: 100 mm Hepes buffer (pH 7.4), 5 mm ATP (sodium salt), 6 mm MgCl2, 0.1% BSA, 150 mm KCl, 1 mm DTT, 45 mm 5-thio-d-glucose-6-phosphate (to inhibit other interfering hexokinases, most importantly hexokinase I), and different glucose concentrations (0, 0.5, 1.5, 3.0, 6.0, 13, 20, 40, and 60 mm), both in the presence and absence of 30 mm of a GKA. Near-saturating GKA was employed to assess whether the assay was specific for GCK. A 4–5-fold lowering of the glucose S0.5 (where S0.5 indicates the concentration necessary for half-maximal enzyme activity) and a 1.5-fold increase in Vmax in the presence of the GKA was indicative of the GCK being measured. The medium contained 2.5 IU/ml glucose-6-phosphate dehydrogenase. Mixing experiments with recombinant human wild-type GCK were routinely carried out to assess recovery or the possibility of interference. Calculations were done as described previously and resulted in information on Vmax, glucose S0.5, and the Hill coefficient (nH), both in the presence and absence of GKA.
Thermolability of Recombinant Human GCK
Thermolability of the GCK mutants, GCKK140E and GCKP417R, and GCKWT protein was assessed by measuring enzyme activity after a temperature step or by following tryptophan fluorescence as the temperature of the cuvette was stepped up. For the activity assay, GST-GCK fusions were used and enzymes were incubated in a water bath at 30, 32.5, 35, 37.5, 40, 42.5, 45, 47.5, 50, and 52.5 °C for 30 min. GCK activity was then determined spectroscopically as described above. The effect of temperature on tryptophan fluorescence was studied with pure GST-free recombinant GCK using a Fluorolog-3–21 Jobin-Yvon Spex Instrument SA (Edison, NJ) equipped with a 450-watt xenon lamp for excitation and a cooled R2658P Hamamatsu photomultiplier tube for detection. Thermolability studies were performed by recording fluorescence intensity during the course of a stepwise increase of the temperature (usually from 10 to 65 °C) using about 1 μm enzyme in 5 mm phosphate buffer at pH 7.3 with 100 mm KCl and 1 mm DTT. Tryptophan fluorescence decreases with a rise of temperature and is red-shifted. The kinetics of the melting curve as indicated by the intensity ratios at 320 to 360 nm and the Tm, the temperature at the inflection point of the melting curve, were recorded.
GKRP Inhibition of Recombinant Human GCK
GKRP is a hepatic GCK inhibitor of glucose. This inhibition is increased in the presence of sorbitol 6-phosphate. The analysis was carried out with glucose and ATP concentrations adjusted to account for differences in the kinetic constants. GCK activity was then determined spectroscopically both with and without sorbitol 6-phosphate, as described previously (22).
Western Blot Analysis
Total liver protein was extracted using radioimmunoprecipitation assay buffer and protease inhibitor mixture (Sigma). Tissues were homogenized in radioimmunoprecipitation assay buffer, and after a 10-min centrifugation at 4 °C, the resulting supernatants were frozen until further analysis. The protein concentrations were determined spectroscopically by the DC assay (Bio-Rad). For Western blotting analysis, the protein samples were denatured at 90 °C and separated on a 9% (GCK) or 10% (GKRP and actin) SDS-polyacrylamide gel. The proteins were then transferred to a nitrocellulose membrane (Immobilon, Millipore), and immunoblotting was performed with antibodies against GCK (1:2000, Santa Cruz Biotechnology), GKRP (1:2000, Santa Cruz Biotechnology), and actin (1:400, Santa Cruz Biotechnology).
Quantification of mRNA by Real Time PCR
mRNA was extracted from frozen liver tissue using Tri Reagent (Molecular Research Center, Inc.) according to manufacturer's protocol. cDNAs were synthesized from mRNA using the high capacity cDNA reverse transcription kit (Applied Biosystems). Real time PCR analysis with SYBR® Green Master Mix (Applied Biosystems) was performed and analyzed using an Applied Biosystems 7900 fast real time PCR system. Relative expression levels were determined using ΔΔCT method to normalize target gene mRNA to Gapdh.
In Vivo Studies with GKA
Two- to 5-month-old male mice were fasted for 2 h; the vehicle for the GKA RO0281675 was administered by oral gavage, and serum was collected at −120, 0, 15, 30, 60, 120, and 180 min. The following week, mice were fasted for 2 h; RO0281675 was administered by oral gavage at a dose of 50 mg/kg, and serum was collected at −120, 0, 15, 30, 60, 120, and 180 min. Serum glucose was measured using an Analox GM 7. Serum insulin was measured using insulin ELISA kits from Crystal Chem. The zero time point was used as base line, and the area under the curve was calculated from 0 to 180 min.
Data Analysis
Data are expressed as means ± S.E. One- and two-way ANOVA and repeated measures ANOVA were performed using NCSS 97. Tukey-Kramer post hoc tests were performed for variables that were significant in the ANOVA at p 0.05. Paired t tests were performed using Excel with corrections to the p value made for the number of tests performed. Unpaired t tests were performed with corrections to the p value made for the number of tests performed.
RESULTS
Generation of ENU-mutagenized Mice and Diabetes Screening
We used the chemical mutagen ENU to identify genes involved in glucose regulation and to generate new mouse models of diabetes. The dose of ENU used has been reported to produce a mutation rate between 2.6 × 10−7 and 1.04 × 10−6 mutations/base pair in B6 mice (23–27). As has been argued previously (28), such a mutation rate would produce multiple mutations in a single mouse, but the mutations would be spaced widely enough that they would sort independently during meiosis. Over 30 hyperglycemic mice were identified in a screen of 2726 8-week-old G1 male and 6994 G3 B6 mice (Fig. 1A). To determine inheritance of hyperglycemia in these animals, we backcrossed the hyperglycemic founders to wild-type B6 mice and identified those progeny with glucose remaining over 200 mg/dl (>2 standard deviations above the population mean) for further analysis. Altogether, we established 13 lines with transmissible hyperglycemia, all of which were either dominant or semi-dominant. In two of the lines, we observed that hyperglycemia was more severe in progeny of intercrossed littermates than were observed in the screening population, consistent with a semi-dominant mode of inheritance.
Identification of Missense Mutations in Gck
After establishing lines with both dominant and semi-dominant inheritance, we obtained genomic DNA from the tails of hyperglycemic mice and sequenced this to search for mutations in genes known to cause monogenic diabetes in humans, including Gck (MODY2), the most common MODY in populations of European descent, in addition to other known MODY genes (Hnf4α, Hnf1α, Pdx1, Hnf1β, and Neurod1), as well as Ins1 and Ins2 because Ins2 has previously been associated with diabetes (29, 30), and both subunits of the ATP-sensitive K+ channel (Abcc8 and Kcnj11). Candidate gene sequencing resulted in the identification of two lines with missense mutations located within exons 4 and 9 in the Gck gene. The first mutation, termed sugar daddy (GckSgrd),4 resulted in an A to G transition within the coding region producing a K140E substitution localized to the glucokinase regulatory protein (GKRP)-binding site within GCK (Fig. 1B). The mutation in a second line, sugar daddy-2 Northwestern University (GckSgrd-2Nwu), resulted in a C to G transversion and produced a P417R substitution within the ATP binding domain of GCK (Fig. 1B). A subsequent low resolution genome scan of the sugar daddy line using 22 N2 mice also mapped the mutation to the proximal region of chromosome 11, where the glucokinase gene is located (supplemental Fig. 1 and supplemental Table 3).
Mutations in any part of the glucokinase molecule have been found to affect glycemia (Fig. 1C). Sequence alignment using ClustalW (31) revealed that both the Lys-140 and Pro-417 residues are highly conserved among human, mouse, and Xenopus. Remarkably, Lys-140 is located adjacent to a trio of basic amino acids (His-141, Lys-142, and Lys-143) that are also phylogenetically conserved and include a positively charged surface distinct from the substrate site (32, 33). This basic patch within GCK has been proposed to be a critical site for GKRP binding (Fig. 1, D–F) (34).
Confirmation of the Mutations, Synthetic Effect and Noncomplementation
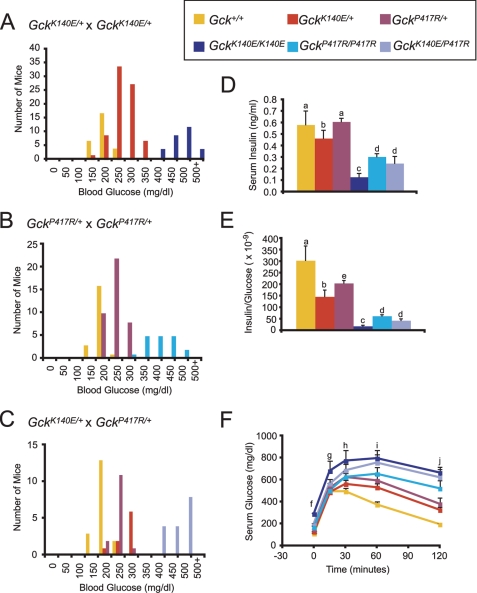
To further analyze the relationship between Gck genotype and glucose levels, we established intercrosses between affected heterozygous GckK140E and GckP417R littermate mice following one generation of backcrossing of the G1 (GckP417R) and G3 (GckK140E) animals on the B6 strain. Mice were born at the expected Mendelian ratio for each of the five genotypes (Gck+/+, GckK140E/+, GckK140E/K140E, GckP417R/+, and Gck P417R/P417R). The observed births included the following: 1) GckK140E/+ × GckK140E/+ (1:2:1, Gck+/+/GckK140E/+/GckK140E/K140E); 2) GckP417R/+ × GckP417R/+ (1:2:1, Gck+/+/GckP417R/+/GckP417R/P417R) (Fig. 2, A and B). When blood glucose was analyzed in progeny of both sexes, we observed a gradient of blood glucose levels that corresponded with gene dosage (GckK140E/K140E > 458 mg/dl, GckP417R/P417R 386 ± 14 mg/dl > GckK140E/+ 245 ± 5 mg/dl, GckP417R/+ 228 ± 5 mg/dl > Gck+/+ 172 ± 5 mg/dl from the GckK140E/+ × GckK140E/+ cross, and 170 ± 5 mg/dl from the GckP417R/+ × GckP417R/+ cross, p < 0.05 for genotype using a one-way ANOVA). The higher level of blood glucose in homozygous mutant mice compared with heterozygous Gck mutants is consistent with a semi-dominant mode of inheritance for both GckK140E and GckP417R alleles, mirroring the haploinsufficiency found in human forms of the disease.
FIGURE 2.
Genotype-phenotype relationship of glucokinase mutations. A, histogram of ad libitum fed blood glucose levels for mice from GckK140E/+ × GckK140E/+ matings. Gck+/+, n = 25 (expected = 28.75); GckK140E/+, n = 65 (expected = 57.5); GckK140E/K140E, n = 25 (expected = 28.75); χ2 = 1.96, p = 0.38. B, histogram of ad libitum fed blood glucose levels for mice from GckP417R/+ × GckP417R/+ matings. Gck+/+, n = 20 (expected = 19.5); GckP417R/+, n = 40 (expected = 39); GckP417R/P417R, n = 18 (expected = 19.5); χ2 = 0.153846, p = 0.926. C, histogram of ad libitum fed blood glucose levels for mice from GckK140E/+ × GckP417R/+ matings. Gck+/+, n = 18 (expected = 14); GckK140E/+, n = 9 (expected = 14); GckP417R/+, n = 14 (expected = 14); GckK140E/P417R, n = 16 (expected = 14); χ2 = 3.285714, p = 0.3496. D, ad libitum fed serum insulin levels. n = 6–8. E, ad libitum fed serum insulin/serum glucose of mice shown in D. F, glucose tolerance test for all genotypes. n = 5–7. Yellow, Gck+/+; red, GckK140E/+; maroon, GckP417R/+; dark blue, GckK140E/K140E; turquoise, GckP417R/P417R; light blue, GckK140E/P417R. Error bars, S.E., a, different from GckK140E/K140E, GckP417R/P417R, GckK140E/P417R; b, different from GckK140E/K140E; c, different from Gck+/+, GckK140E/+, GckP417R/+; d, different from Gck+/+, GckP417R/+; e, different from Gck+/+, GckK140E/K140E; f, Gck+/+, GckK140E/+, different from GckK140E/K140E; g, Gck+/+, GckK140E/+, GckP417R/+, GckP417R/P417R different from GckK140E/K140E; h, Gck+/+ different from GckK140E/K140E, GckK140E/P417R, GckK140E/+ different from GckK140E/K140E; i, Gck+/+ different from all mutants. GckK140E/+, GckP417R/+ different from GckK140E/K140E, GckP417R/P417R; j, Gck+/+ different from all mutants, GckK140E/+, GckP417R/+ different from GckK140E/K140E, GckP417R/P417R, GckK140E/P417R. p < 0.05.
We also found that compound heterozygous mice generated by intercrosses of GckK140E/+ and GckP417R/+ mutants exhibited elevated ad libitum fed glucose levels (446 ± 12 mg/dl) comparable with those of the homozygous mice (Fig. 2C). Failure of the two mutant variants to complement each other (i.e. normalize blood glucose levels) provides evidence that diabetes in these two lines of mice is caused by different alleles of the same gene.
Gck Genotype Corresponds with Dynamics of Glucose and Insulin Homeostasis in Vivo
Because GCK acts as a glucose sensor and serves as a determinant of the glucose-stimulated insulin release (GSIR) threshold within the pancreas, we next examined serum insulin and glucose levels both ad libitum and following glucose challenge. Ad libitum fed 8-week-old male homozygous and compound heterozygous mutant mice displayed significant hypoinsulinemia (Fig. 2D). Furthermore, even the heterozygous mutant mice exhibited relative hypoinsulinemia, reflected in a reduced ratio of insulin to glucose (Fig. 2E). Finally, homozygous mutant mice displayed significantly impaired glucose tolerance compared with wild-type controls following intraperitoneal glucose, and glucose tolerance in heterozygous mutants was intermediate between the homozygous and wild-type mice (Fig. 2F). Compound heterozygous mice displayed a magnitude of glucose intolerance similar to that of homozygous mutants (Fig. 2F).
Structure-Function Analysis and Thermal Unfolding Properties of GCK
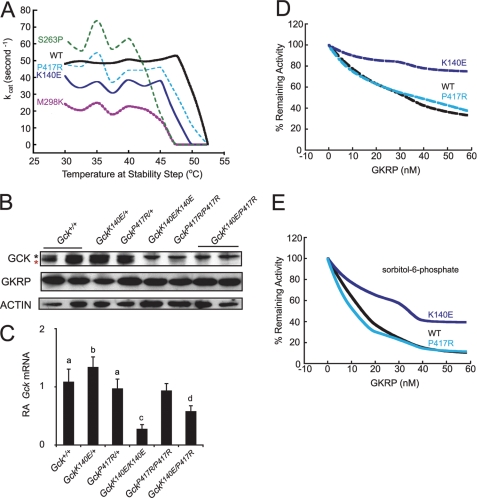
To determine the effect of each ENU mutation on GCK activity and structure, we produced both GCKK140E and GCKP417R as recombinant proteins in E. coli and determined catalytic rates by phosphorylation studies (Fig. 4A and Table 1) (35) and thermal stability using fluorescence monitoring (Fig. 3 and Table 2). The K140E mutation caused a 36% decrease in kcat (40.0 ± 1.36 versus 62.8 ± 4.11). In addition, the K140E variant had a nonsignificant trend toward decreased affinity for glucose, increasing the glucose S0.5 by about 45% from 7.45 ± 0.26 to 10.8 ± 1.13, although there was no change of glucose affinity for P417R. In contrast, P417R caused a 4-fold decrease in affinity for Km(ATP) (2.08 ± 0.15 versus 0.46 ± 0.02). Although both mutations trended toward slightly reduced cooperativity (Hill coefficient, nH), responsiveness to GKA was preserved. The activity index, a composite measure of GCK activity, was 1.37 ± 0.16 for GCKWT, 0.78 ± 0.08 for GCKK140E, and 1.60 ± 0.14 for GCKP417R. Surprisingly, the apparent thresholds for GSIR predicted from these activity indices would be close to 5 mm glucose for wild type and the P417R line and ∼6 mm for the K140E line of mutant mice (36), within physiological range of mammalian glucose concentration.
FIGURE 4.
GCK thermostability and responsiveness to GKRP. A, effect of temperature on enzyme activity. Enzymes were diluted in buffer containing glucose at the S0.5 of the particular enzyme and incubated for 30 min at different temperatures before kinetic analysis. Black, GCKWT; dark blue, GCKK140E; turquoise, GCKP417R; green, GCKS263P; purple, GCKM298K. B, representative Western blot of GCK and GKRP protein levels. Red star, major isoform of GCK; black star, minor isoform of GCK. C, quantification of liver Gck mRNA levels by real time PCR. RA, relative abundance. Gck+/+, n = 6; GckK140E/+, n = 6; GckP417R/+, n = 5; GckK140E/K140E, n = 5; GckP417R/P417R, n = 5; and GckK140E/P417R, n = 6. Error bars, S.E. a, different from GckK140E/K140E; b, different from GckK140E/K140E and GckK140E/P417R; c, different from Gck+/+, GckK140E/+, and GckP417R/+; d, different from GckK140E/+. p < 0.05. D, effect of human GKRP on wild-type and mutant GCK in the absence of sorbitol 6-phosphate. Black, GCKWT; dark blue, GCK K140E; turquoise, GCKP417R. E, effect of human GKRP on wild-type and mutant GCK in the presence of 10 mm sorbitol 6-phosphate. Black, GCKWT; dark blue, GCKK140E; Turquoise, GCKP417R.
TABLE 1.
Kinetic constants of pure, GST-free, recombinant GCK expressed in E. coli
GKA is RO0274375. Means ± S.E. of results of the analysis from three to four protein expression preparations are presented.
| Parameter | GCKWT | GCKK140E | GCKP417R |
|---|---|---|---|
| kcat (s−1) | 62.8 ± 4.11 | 40.0 ± 1.36a | 48.3 ± 7.40 |
| kcat fold increase with GKA | 1.64 ± 0.04 | 1.73 ± 0.07 | 1.46 ± 0.02 |
| Glucose S0.5 (mm) | 7.45 ± 0.26 | 10.8 ± 1.13 | 6.59 ± 0.32 |
| Glucose S0.5 fold decrease with GKA | 3.61 ± 0.45 | 4.52 ± 0.48 | 4.66 ± 0.25 |
| nH | 1.77 ± 1.10 | 1.57 ± 0.13 | 1.52 ± 0.04 |
| nH/nHGKA | 1.11 ± 0.06 | 0.99 ± 0.10 | 1.01 ± 0.02 |
| Km(ATP) (mm) | 0.46 ± 0.02 | 0.35 ± 0.08 | 2.08 ± 0.15a |
| Km(ATP)/Km(ATP)GKA | 1.02 ± 0.19 | 1.14 ± 0.17 | 1.12 ± 0.03 |
| Relative activity index | 1.37 ± 0.16 | 0.78 ± 0.08a | 1.60 ± 0.14 |
| Relative activity index fold increase with GKA | 23.0 ± 2.14 | 22.7 ± 1.69 | 15.0 ± 0.56a |
a p < 0.025 by unpaired t test in comparison with GCKWT.
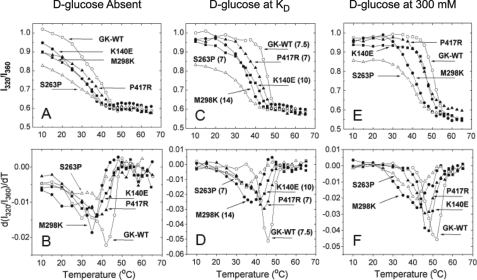
FIGURE 3.
Temperature denaturation of control and mutant GCKs measured by tryptophan fluorescence. The results of five different enzymes are as follows: 1) white circles, stable control; 2) moderately unstable mutants: black squares GCKK140E and black triangles GCKP417R; and 3) severely unstable mutants: white triangles GCKS263P and black circles GCKM298K, which have relative activity indices of 0.65 and 0.58, respectively, extrapolating to thresholds of GSIR of about 6 mm. The ratios of 320 nm to 360 nm portions of the fluorescence spectra are shown in the upper panels, and the differential forms in the lower panels. A. and B, in d-glucose-free buffer; C and D with d-glucose at the respective KD in mm; E and F, at 300 mm d-glucose. The excitation wavelength was 295 nm. Samples contained 5 mm phosphate (pH 7.3), 100 mm KCl, 1 mm DTT, and 3 mm enzyme.
TABLE 2.
Melting temperature of pure, GST-free, GCKWT and GCK mutants measured by temperature-dependent tryptophan fluorescence change
The Tm values represent the inflection points of the d(I320/I360)/dT versus temperature plots of B, D, and F of Fig. 3, n = 1.
| d-Glucose condition | GCKWT | GCKP417R | GCKK140E | GCKM298K | GCKS263P |
|---|---|---|---|---|---|
| Absent | 42.9 | 38.4 | 37.2 | 34.9 | 32.0 |
| KD | 46.0 | 44.0 | 41.9 | 37.2 | 35.0 |
| 300 mm | 50.9 | 46.7 | 47.2 | 41.7 | 41.7 |
Because the observed hyperglycemia could not be explained by such subtle changes in kinetics, we next tested the hypothesis that altered structure of GCK might contribute to the mutant phenotype, by monitoring fluorescence and catalytic activity during thermal shift experiments. Increased thermolability also has been observed in analyses of some Gck mutations associated with human MODY2 (37, 38), possibly altering expression and function of the enzyme in pancreatic islets and the liver. Three tryptophan residues in GCK produce a spectral shift from blue to red during unfolding. Tryptophan fluorescence was therefore monitored when both WT and mutant GCK variants were exposed to rising temperatures. When compared with GCKWT, the Tm values for GCKK140E and GCKP417R decreased by 4.1 and 2 °C, respectively (Fig. 3 and Table 2), indicating more rapid unfolding of the two mutant proteins. The temperature dependencies of the kinetic activity tests also showed that the kcat began decreasing at a temperature ∼3 °C lower for GCKK140E and GCKP417R than for GCKWT (Fig. 4A). The thermolability of the ENU mutants K140E and P417R was intermediate between that of wild-type GCK and two severely affected mutants, M298K and S263P, discovered in diabetic patients with MODY2. This is clearly shown by the relative positions of thermolability profiles in Figs. 3 and 4A, which were obtained using very different assay principles. The results illustrate that variations of functional and structural protein instability must be considered in the biochemical genetic evaluations of GCK-linked impairments of glucose homeostasis.
That structural instability contributed to impaired GCK function in the whole animal was corroborated by direct kinetic analysis of GCK activity of liver lysates (Table 3). Lysates from GckK140E/+ and GckP417R/+ mice showed a reduced Vmax compared with wild-type mice (0.49 ± 0.35 and 0.73 ± 0.16 versus 1.19 ± 0.27 units/g tissue). The glucose S0.5 values were in the normal range in all heterozygous genotypes, and the Hill coefficient was unchanged, whereas there was no measurable GCK activity in the lysates from homozygous or compound heterozygous mice.
TABLE 3.
Kinetic constants of GCK from liver lysates
The means ± S.E., n = 2–6; ND, not determinable; GKA, RO0274375. One-way ANOVA and two-way repeated measures ANOVA were used, p < 0.05. GckK140E/K140E, GckP417R/P417R, and GckK140E/P417R were qualitatively different from Gck+/+, GckK140E/+, and GckP417R/+ for Vmax, S0.5, and nH and so could not be included in the statistical analysis. Although there was a statistically significant main effect of GKA treatment for Vmax (p < 0.05), Tukey-Kramer post hoc testing did not identify differences within genotypes.
| Parameter | Gck+/+ | GckK140E/+ | GckP417R/+ | GckK140E/K140E | GckP417R/P417R | GckK140E/P417R |
|---|---|---|---|---|---|---|
| Serum glucose (mg/dl) | 204 ± 10.5a | 315 ± 52.4a | 272 ± 13.9a | 708 ± 44.9b | 580 (563, 597)b | 628 ± 85.0b |
| Vmax (units/g) | 1.19 ± 0.27 | 0.49 ± 0.35 | 0.73 ± 0.16 | < 0.05 | <0.05 | <0.05 |
| Vmax with GKA (units/g) | 2.15 ± 0.47 | 0.91 ± 0.35 | 1.06 ± 0.25 | ND | ND | ND |
| Glucose S0.5 (mm) | 8.64 ± 0.62c | 7.81 ± 0.86c | 9.16 ± 0.87c | ND | ND | ND |
| Glucose S0.5 with GKA (mm) | 2.80 ± 0.42d | 3.25 ± 0.57d | 3.10 ± 0.70d | ND | ND | ND |
| nH | 1.77 ± 0.11 | 1.82 ± 0.19 | 1.57 ± 0.15 | ND | ND | ND |
| nH with GKA | 1.83 ± 0.27 | 2.02 ± 0.46 | 1.71 ± 0.28 | ND | ND | ND |
| % Recovery | 62.7 ± 6.52a,c | 44.3 ± 17.6e | 64.0 ± 10.8a,c | 5.75 ± 0.73c,e | 5.00c,e (5.00, 5.00) | 6.67 ± 1.72e |
| % Recovery with GKA | 107.0 ± 12.4d,f | 99.0 ± 16.7d | 110.0 ± 4.9d,f | 49.0 ± 13.5e,d | 70.0d (84.0, 55.0) | 52.0 ± 9.8e |
a This is different from GckK140E/K140E, GckP417R/P417R, and GckK140E/P417R in the same row.
b This is different from Gck+/+, GckK140E/+, and GckP417R/+ in the same row.
c This is different with GKA.
d This is different without GKA.
e This is different from Gck+/+ and GckP417R/+ in the same row.
f This is different from GckK140E/K140E and GckP417R/P417R in the same row.
In the process of enzymatic analysis, we performed mixing experiments in which liver lysates from the mutants were combined with the wild-type recombinant enzyme. Surprisingly, recombinant wild-type protein showed drastically reduced activity when mixed with the liver extracts from the homozygous or compound heterozygous mutants (62.7 ± 6.52 for Gck+/+ versus 5.75 ± 0.73 for GckK140E/K140E, 5.00 for GckP417R/P417R, and 6.67 ± 1.72 for GckK140E/P417R) (Table 3). This decrease in recovery of wild-type enzyme in the presence of liver lysates suggests either that the mutant GCK interferes with the activity of the wild-type protein or that there is a GCK inhibitor in liver of diabetic mice. The latter explanation is consistent with the observation that recombinant mutant GCKs did not inhibit the recombinant wild-type enzyme. Furthermore, similar inhibition of recombinant GCK was observed with liver lysates prepared from animals with severe streptozotocin-induced diabetes.5 When the assay was performed with saturating levels of the GKA RO0274375, the low recovery of exogenous GCK standard in the presence of liver extract was significantly increased (to 107.0 ± 12.4 for GCKWT, 49.0 ± 13.5 for GCKK140E/K140E, 70.0 for GCKP417R/P417R, and 52.0 ± 9.8 for GCKK140E/P417R) (Table 3). However, no GCK activity was detectable in liver extracts from homozygous and compound heterozygous mutant mice when tested in the presence of the drug, although the drug was effective in assays of extracts from heterozygous mutant mice (Vmax with GKA 2.15 ± 0.47 for Gck+/+, 0.91 ± 0.35 for GckK140E/+, and 1.06 ± 0.25 for GckP417R/+) (Table 3).
GckK140E and GckP417R Mutations Decrease GCK Expression
To test the hypothesis that the decrease in phosphorylating activity in the mutant liver lysates was due to a decrease in GCK expression, we analyzed GCK protein levels by immunoblotting liver lysates from mutant and wild-type mice. Western blots of mouse liver extracts showed that the homozygous mutant and compound heterozygous mutant mice have markedly reduced amounts of GCK protein (Fig. 4B). Because expression of hepatic GCK is tightly regulated by insulin, we tested whether the decrease in GCK protein is due to a decrease in transcription of Gck mRNA using quantification of real time PCR. As shown in Fig. 2, D and E, GckK140E/K140E, GckP417R/P417R, and GckK140E/P417R mice have low relative and absolute concentrations of serum insulin, with GckK140E/K140E mice having the lowest. We found that hepatic Gck mRNA abundance was significantly decreased in GckK140E/K140E and in GckK140E/P417R mice (Fig. 4C). A trend toward decreased mRNA abundance was also observed in GckK140E/K140E livers for the insulin-transcribed genes Fasn and Srebf1, although the differences did not reach statistical significance (supplemental Fig. 3). In contrast, Pck1, a gene for which insulin inhibits transcription, was increased in mRNA abundance in liver lysate from GckK140E/K140E mice (supplemental Fig. 3).
The marked reduction of Gck mRNA in GckK140E/K140E mice, and to a lesser degree, GckK140E/P417R mice, is consistent with reduced insulin-induced transcription of the Gck gene in the livers of these mice (Fig. 4B). However, Gck mRNA levels in GckP417R/P417R were not significantly reduced, suggesting that post-translational rather than transcriptional deficits caused by hypoinsulinemia account for the diabetes in these animals.
K140E Mutation Alters GCK Binding to GKRP
In the liver, GCK is regulated by its sequestration in the nucleus through binding to GKRP, a protein that has also been implicated in human T2DM (8). We therefore tested whether the two mutant GCKs displayed altered binding to GKRP, as measured by decreased activity in the presence of GKRP, alone, and also with the addition of its activator, sorbitol 6-phosphate. Wild-type and P417R mutant GCK showed the expected reduction of phosphorylation activity with increasing amounts of GKRP, both in the presence and in the absence of sorbitol 6-phosphate (Fig. 4, D and E). In contrast, GKRP showed greatly reduced capacity to inhibit the K140E mutant, whether the potentiating sorbitol 6-phosphate was present or not (Fig. 4, D and E). Importantly, GKRP knock-out (39, 40) or reduced GKRP responsiveness of GCK (41) results in much lower GCK activity in the liver because the storage of inactivated but functional enzyme in the nucleus is prevented.
GKA RO0281675 Treatment Partially Rescues the Mutant Phenotype in Vivo
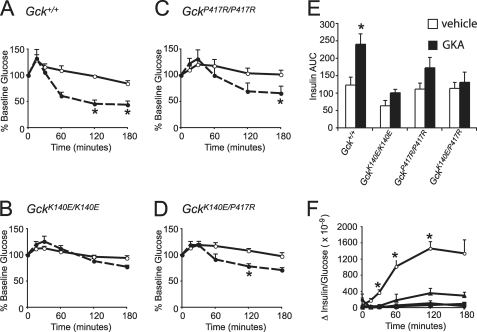
To determine whether the GCK mutations could be rescued using a small molecule allosteric activator, we treated 2–5-month-old male mice with the GKA RO0281675. Acute treatment with this drug has been previously reported to make wild-type animals hypoglycemic and to normalize the blood glucose levels in a variety of diabetic animal models (19). Our wild-type mice became hypoglycemic with a single dose of RO0281675, with a similar time course to that previously reported (Fig. 5A). Treatment with RO0281675 also significantly lowered the blood glucose levels of GckP417R/P417R and GckK140E/P417R mice (Fig. 5, C and D). However, there was no significant difference between vehicle treatment and GKA treatment for GckK140E/K140E mice (Fig. 5B). GKA treatment significantly increased the insulin area under the curve compared with vehicle treatment for wild-type mice but not for any of the mutant mice (Fig. 5E). Similarly, treatment with GKA significantly increased the insulin to glucose ratio compared with vehicle treatment for the wild-type mice but not for any of the mutant mice (Fig. 5F).
FIGURE 5.
Response to the GKA RO0281675 (50 mg/kg). A, glucose as a % of base line for Gck+/+, n = 5. Error bars, S.E. White circles, vehicle treatment; black circles, GKA. *, p < 0.01. B, glucose as a % of base line for GckK140E/K140E, n = 6. Error bars, S.E. White circles, vehicle treatment; black circles, GKA. C, glucose as a % of base line for GckP417R/P417R, n = 5. Error bars, S.E. White circles, vehicle treatment; black circles, GKA. *, p < 0.01. D, glucose as a % of base line for GckK140E/P417R, n = 5. Error bars, S.E. White circles, vehicle treatment; black circles, GKA. *, p < 0.01. E, insulin area under the curve (AUC). Gck+/+, n = 5; GckK140E/K140E, n = 6; GckP417R/P417R, n = 5; GckK140E/P417R, n = 4. Error bars, S.E. White bars, vehicle treatment; black bars, GKA. *, p < 0.05. F, difference in serum insulin/serum glucose between vehicle and GKA treatment. White circles, Gck+/+, n = 5; black squares, GckK140E/K140E, n = 6; black triangles, GckP417R/P417R, n = 5; black circles, GckK140E/P417R, n = 4. Error bars, S.E. *, p < 0.007.
DISCUSSION
We performed a deliberate chemical mutagenesis screen to study gene pathways involved in glucose homeostasis and to create preclinical models to test anti-hyperglycemic therapeutics. The GckK140E and GckP417R mutant alleles that were identified are informative because they localize to the same domain as mutations causing human MODY2, and they are viable as both the heterozygous and homozygous genotypes causing a gradient from moderate to severe diabetes. Because GckK140E and GckP417R animals harboring point mutations survive into adulthood, they can be used in longitudinal studies of disease progression, overcoming the lethality in homozygous mice caused by deletion of Gck in global and conditional knock-outs (42–46) and knock-in of a human Gck point mutation (41). Thus, the Gck allele series generated by the present ENU screen highlights advantages of utilizing a forward genetic strategy to complement reverse genetic approaches to model diabetes mellitus.
GckK140E and GckP417R mice also provide insight into the biochemical basis of glucokinase diabetes. In vitro analyses of human Gck mutants have been used to assess the effect of changes in GCK catalytic activity on insulin release. Such analyses involve use of kinetic constants to derive a relative activity index ((GCK kcat/glucose S0.5nH)·(2.5/(2.5 + Km(ATP))). Lower or higher relative activity indices are closely correlated with higher or lower thresholds for GSIR, respectively. Based upon in vitro glucose phosphorylation studies, we observed that both GCKK140E and GCKP417R exhibited only modest changes in relative activity, indicating that altered kinetics of glucose phosphorylation do not lead to hyperglycemia in the present Gck mutant lines. An alternative explanation for the development of hyperglycemia in GckK140E and GckP417R mice is that the mutations alter the thermostability and/or interactions with the heterologous GKRP (41), which stabilizes the enzyme through sequestration in the nucleus.
Consistent with an effect of the point mutations on protein conformation, we found that both GCKK140E and GCKP417R protein exhibited increased sensitivity to temperature shift with respect to kinetic (kcat) and fluorescence (I320/360) measurements. In addition, the reduced GCK catalytic activity in mutant liver lysates suggested decreased expression of GCK protein. Indeed, Western blots revealed reduced abundance of homozygous and compound heterozygous GCK protein, consistent with its decreased stability. Collectively, our studies indicate that the predominant cause of hyperglycemia in the GckK140E and GckP417R variants involved changes in protein expression caused by reduced stability. Because the total absence of GCK in the liver in conditional knock-outs (45, 47) exhibited a milder hyperglycemic phenotype than that of GckK140E/K140E, GckP417R/P417R, and GckK140E/P417R mice, and because total loss of pancreatic GCK is lethal (44, 45), it is likely that partial loss of GCK function within both the liver and pancreatic islets contributes to hyperglycemia in GckK140E and GckP417R mutant mice.
GckK140E mutant mice also elucidate the role of the heterologous liver regulatory factor, GKRP, in glucokinase diabetes. In low glucose conditions, GKRP sequesters GCK within the nucleus, preventing its proteolytic degradation; with the influx of glucose, GKRP is displaced resulting in relocalization of GCK to the cytoplasm where it is again active. Importantly, recombinant GCKK140E mutant showed markedly decreased GKRP inhibition in both the absence and presence of the potentiator sorbitol 6-phosphate. Reduced binding to GKRP results in degradation of hepatocyte GCK because of abrogation of its nuclear sequestration (41). In the three-dimensional structure of GCK, the lysine at position 140 is located in proximity to a patch of the following basic amino acids (32): His-141, Lys-142, and Lys-143. Mutagenesis of these conserved amino acids decreased GKRP binding (34) and GCK nuclear entry (48); similarly, in vitro binding of GCKK140E to GKRP was reduced (Fig. 4, D and E). Our mixing experiments with the whole liver lysate also suggested the existence of a previously unrecognized GCK inhibitor. This GCK inhibitor decreased the catalytic activity of wild-type recombinant enzyme (Table 3) and was also observed in liver lysates from streptozotocin-treated mice. We speculate that insulin deficiency may result in overexpression of a GCK inhibitor that may in turn contribute to the progression of diabetes mellitus.
The ENU glucokinase mutants also elucidate mechanisms of allosteric regulation of GCK. Indeed, the discovery of small molecule GKAs that augment the activity of GCK both in liver and pancreas has led to speculation that GCK is subjected to allosteric regulation under physiological circumstances. However, the effect of GKA on multitissue mutants with severe diabetes has not been previously established because these agents have been tested only in the setting of gene deletion models with Gck haploinsufficiency (i.e. hemizygous knock-out mice) (46, 49). If homozygous Gck knock-outs were viable, they presumably would not respond to GKA, because such animals would lack the drug target, endogenous GCK, on which the activator could act. Our in vitro results indicate that catalytic activities of both GCKK140E and GCKP417R are similarly augmented, indicating that at the biochemical level GKA action does not involve residues within the GKRP-binding site (K140E) nor the ATP-binding site (P417R) (Table 1). Because previous studies in Gck−/+ knock-outs indicated that GKA increased insulin secretion in islets (46, 49) and decreased in vivo glucose levels (49), we reasoned that GKA treatment of multitissue GckK140E and GckP417R mutant mice might improve both pancreatic insulin secretion and hepatic glucose disposal. However, when a related compound, RO0281675, was administered in vivo to the mice, we observed significant allelic differences in the response to GKA in the mutant mice. Single dose RO0281675 treatment significantly decreased serum glucose of control animals and both GckP417R/P417R and GckK140E/P417R mice but not of GckK140E/K140E mice, and although the glucose levels of GckP417R/P417R and GckK140E/P417R mice were lowered, they did not reach normoglycemia. Thermolability of the mutant GCK would cause reduced GCK levels and therefore less available targets for GKA.
GKA treatment did not increase insulin area under the curve for any of the mutant genotypes. One possibility is that a subtle change in insulin secretion, albeit below the threshold of statistical significance, may be sufficient to decrease glucose concentrations. Such a scenario would suggest that GKA is working in both the pancreas and the liver to decrease glucose levels. An alternative possibility is that the allosteric activator exerts its anti-hyperglycemic actions in the mutant mice through effects on hepatic GCK, rather than pancreatic GCK. It is possible that the anti-hyperglycemic actions that were observed in the mutant mice were due to competition of the GKA with the endogenous GCK inhibitor within the liver, the existence of which was suggested by our mixing studies (Table 3). If so, then the lack of effect of the GKA observed in the GckK140E/K140E mice may be due to some combination of decreased GCK expression and increased levels of an inhibitor compared with GckP417R/P417R and GckK140E/P417R mice. A further implication would be that augmenting insulin levels, either through administration of insulin or an insulin secretagogue, may potentiate GKA activity significantly by reducing expression of the GCK inhibitor, and also by increasing expression of the Gck gene within liver.
Because the present monogenic diabetes models maintain a wide range of blood sugar levels for long periods, it will be possible to use these to study the effect of persistent high glucose levels on pancreatic islet cell function and proliferation independent of increased glucose metabolism of these cells (50, 51). The Gck ENU models thus enable testing from gene to phenotype to drug.
Supplementary Material
Acknowledgments
We thank Karen Wu, Eun Joo Lee, Jenny Yin, Tony Yuan, and Reema Ghatnekar for their help with genotyping. We also thank Weimin Song, Chiaki Omura, Emily Nicholl, Alayna Corden, Joseph Doering, Catherine Zhang, and Steven Sukert for their help with data collection, Alayna Corden for help with figure production, and Kathryn Moynihan Ramsey for helpful comments regarding this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants U01 MH61915 from NIMH (to J. S. T.) and R01 DK022122 from NIDDK (to F. M. M.). This work was also supported by the University of Chicago Diabetes Research and Training Center Grant DK-20595 (to J. B.) and the Chicago Biomedical Consortium with support from The Searle Funds at the Chicago Community Trust Grant C-007 (to J. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Tables 1–3.
Nomenclature of the mutations was assigned by Mouse Genome Informatics at The Jackson Laboratory. The two lines were designated sugar daddy (Sgrd) and sugar daddy-2 Northwestern University (Sgrd-2Nwu). We refer to these throughout this text as based upon the corresponding amino acid substitutions (Sgrd, GckK140E; Sgrd-2Nwu, GckP417R).
F. Matschinsky, unpublished data.
- T2DM
- type 2 diabetes mellitus
- ENU
- N-ethyl-N-nitrosourea
- Gck
- glucokinase
- MODY
- maturity onset diabetes of the young
- GKA
- GCK activator
- GKRP
- glucokinase regulatory protein
- GSIR
- glucose-stimulated insulin release
- ANOVA
- analysis of variance.
REFERENCES
- 1. Vionnet N., Stoffel M., Takeda J., Yasuda K., Bell G. I., Zouali H., Lesage S., Velho G., Iris F., Passa P., et al. (1992) Nature 356, 721–722 [DOI] [PubMed] [Google Scholar]
- 2. Froguel P., Vaxillaire M., Sun F., Velho G., Zouali H., Butel M. O., Lesage S., Vionnet N., Clément K., Fougerousse F., et al. (1992) Nature 356, 162–164 [DOI] [PubMed] [Google Scholar]
- 3. Hattersley A. T., Turner R. C., Permutt M. A., Patel P., Tanizawa Y., Chiu K. C., O'Rahilly S., Watkins P. J., Wainscoat J. S. (1992) Lancet 339, 1307–1310 [DOI] [PubMed] [Google Scholar]
- 4. Nishi S., Stoffel M., Xiang K., Shows T. B., Bell G. I., Takeda J. (1992) Diabetologia 35, 743–747 [DOI] [PubMed] [Google Scholar]
- 5. Yamagata K., Furuta H., Oda N., Kaisaki P. J., Menzel S., Cox N. J., Fajans S. S., Signorini S., Stoffel M., Bell G. I. (1996) Nature 384, 458–460 [DOI] [PubMed] [Google Scholar]
- 6. Yamagata K., Oda N., Kaisaki P. J., Menzel S., Furuta H., Vaxillaire M., Southam L., Cox R. D., Lathrop G. M., Boriraj V. V., Chen X., Cox N. J., Oda Y., Yano H., Le Beau M. M., Yamada S., Nishigori H., Takeda J., Fajans S. S., Hattersley A. T., Iwasaki N., Hansen T., Pedersen O., Polonsky K. S., Bell G. I., et al. (1996) Nature 384, 455–458 [DOI] [PubMed] [Google Scholar]
- 7. Sladek R., Rocheleau G., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., Balkau B., Heude B., Charpentier G., Hudson T. J., Montpetit A., Pshezhetsky A. V., Prentki M., Posner B. I., Balding D. J., Meyre D., Polychronakos C., Froguel P. (2007) Nature 445, 881–885 [DOI] [PubMed] [Google Scholar]
- 8. Saxena R., Voight B. F., Lyssenko V., Burtt N. P., de Bakker P. I., Chen H., Roix J. J., Kathiresan S., Hirschhorn J. N., Daly M. J., Hughes T. E., Groop L., Altshuler D., Almgren P., Florez J. C., Meyer J., Ardlie K., Bengtsson Boström K., Isomaa B., Lettre G., Lindblad U., Lyon H. N., Melander O., Newton-Cheh C., Nilsson P., Orho-Melander M., Råstam L., Speliotes E. K., Taskinen M. R., Tuomi T., Guiducci C., Berglund A., Carlson J., Gianniny L., Hackett R., Hall L., Holmkvist J., Laurila E., Sjögren M., Sterner M., Surti A., Svensson M., Svensson M., Tewhey R., Blumenstiel B., Parkin M., Defelice M., Barry R., Brodeur W., Camarata J., Chia N., Fava M., Gibbons J., Handsaker B., Healy C., Nguyen K., Gates C., Sougnez C., Gage D., Nizzari M., Gabriel S. B., Chirn G. W., Ma Q., Parikh H., Richardson D., Ricke D., Purcell S. (2007) Science 316, 1331–133617463246 [Google Scholar]
- 9. Zeggini E., Weedon M. N., Lindgren C. M., Frayling T. M., Elliott K. S., Lango H., Timpson N. J., Perry J. R., Rayner N. W., Freathy R. M., Barrett J. C., Shields B., Morris A. P., Ellard S., Groves C. J., Harries L. W., Marchini J. L., Owen K. R., Knight B., Cardon L. R., Walker M., Hitman G. A., Morris A. D., Doney A. S., McCarthy M. I., Hattersley A. T. (2007) Science 316, 1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scott L. J., Mohlke K. L., Bonnycastle L. L., Willer C. J., Li Y., Duren W. L., Erdos M. R., Stringham H. M., Chines P. S., Jackson A. U., Prokunina-Olsson L., Ding C. J., Swift A. J., Narisu N., Hu T., Pruim R., Xiao R., Li X. Y., Conneely K. N., Riebow N. L., Sprau A. G., Tong M., White P. P., Hetrick K. N., Barnhart M. W., Bark C. W., Goldstein J. L., Watkins L., Xiang F., Saramies J., Buchanan T. A., Watanabe R. M., Valle T. T., Kinnunen L., Abecasis G. R., Pugh E. W., Doheny K. F., Bergman R. N., Tuomilehto J., Collins F. S., Boehnke M. (2007) Science 316, 1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeggini E., Scott L. J., Saxena R., Voight B. F., Marchini J. L., Hu T., de Bakker P. I., Abecasis G. R., Almgren P., Andersen G., Ardlie K., Boström K. B., Bergman R. N., Bonnycastle L. L., Borch-Johnsen K., Burtt N. P., Chen H., Chines P. S., Daly M. J., Deodhar P., Ding C. J., Doney A. S., Duren W. L., Elliott K. S., Erdos M. R., Frayling T. M., Freathy R. M., Gianniny L., Grallert H., Grarup N., Groves C. J., Guiducci C., Hansen T., Herder C., Hitman G. A., Hughes T. E., Isomaa B., Jackson A. U., Jørgensen T., Kong A., Kubalanza K., Kuruvilla F. G., Kuusisto J., Langenberg C., Lango H., Lauritzen T., Li Y., Lindgren C. M., Lyssenko V., Marvelle A. F., Meisinger C., Midthjell K., Mohlke K. L., Morken M. A., Morris A. D., Narisu N., Nilsson P., Owen K. R., Palmer C. N., Payne F., Perry J. R., Pettersen E., Platou C., Prokopenko I., Qi L., Qin L., Rayner N. W., Rees M., Roix J. J., Sandbaek A., Shields B., Sjögren M., Steinthorsdottir V., Stringham H. M., Swift A. J., Thorleifsson G., Thorsteinsdottir U., Timpson N. J., Tuomi T., Tuomilehto J., Walker M., Watanabe R. M., Weedon M. N., Willer C. J., Illig T., Hveem K., Hu F. B., Laakso M., Stefansson K., Pedersen O., Wareham N. J., Barroso I., Hattersley A. T., Collins F. S., Groop L., McCarthy M. I., Boehnke M., Altshuler D. (2008) Nat. Genet. 40, 638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A. U., Wheeler E., Glazer N. L., Bouatia-Naji N., Gloyn A. L., Lindgren C. M., Mägi R., Morris A. P., Randall J., Johnson T., Elliott P., Rybin D., Thorleifsson G., Steinthorsdottir V., Henneman P., Grallert H., Dehghan A., Hottenga J. J., Franklin C. S., Navarro P., Song K., Goel A., Perry J. R., Egan J. M., Lajunen T., Grarup N., Sparsø T., Doney A., Voight B. F., Stringham H. M., Li M., Kanoni S., Shrader P., Cavalcanti-Proença C., Kumari M., Qi L., Timpson N. J., Gieger C., Zabena C., Rocheleau G., Ingelsson E., An P., O'Connell J., Luan J., Elliott A., McCarroll S. A., Payne F., Roccasecca R. M., Pattou F., Sethupathy P., Ardlie K., Ariyurek Y., Balkau B., Barter P., Beilby J. P., Ben-Shlomo Y., Benediktsson R., Bennett A. J., Bergmann S., Bochud M., Boerwinkle E., Bonnefond A., Bonnycastle L. L., Borch-Johnsen K., Böttcher Y., Brunner E., Bumpstead S. J., Charpentier G., Chen Y. D., Chines P., Clarke R., Coin L. J., Cooper M. N., Cornelis M., Crawford G., Crisponi L., Day I. N., de Geus E. J., Delplanque J., Dina C., Erdos M. R., Fedson A. C., Fischer-Rosinsky A., Forouhi N. G., Fox C. S., Frants R., Franzosi M. G., Galan P., Goodarzi M. O., Graessler J., Groves C. J., Grundy S., Gwilliam R., Gyllensten U., Hadjadj S., Hallmans G., Hammond N., Han X., Hartikainen A. L., Hassanali N., Hayward C., Heath S. C., Hercberg S., Herder C., Hicks A. A., Hillman D. R., Hingorani A. D., Hofman A., Hui J., Hung J., Isomaa B., Johnson P. R., Jørgensen T., Jula A., Kaakinen M., Kaprio J., Kesaniemi Y. A., Kivimaki M., Knight B., Koskinen S., Kovacs P., Kyvik K. O., Lathrop G. M., Lawlor D. A., Le Bacquer O., Lecoeur C., Li Y., Lyssenko V., Mahley R., Mangino M., Manning A. K., Martínez-Larrad M. T., McAteer J. B., McCulloch L. J., McPherson R., Meisinger C., Melzer D., Meyre D., Mitchell B. D., Morken M. A., Mukherjee S., Naitza S., Narisu N., Neville M. J., Oostra B. A., Orrù M., Pakyz R., Palmer C. N., Paolisso G., Pattaro C., Pearson D., Peden J. F., Pedersen N. L., Perola M., Pfeiffer A. F., Pichler I., Polasek O., Posthuma D., Potter S. C., Pouta A., Province M. A., Psaty B. M., Rathmann W., Rayner N. W., Rice K., Ripatti S., Rivadeneira F., Roden M., Rolandsson O., Sandbaek A., Sandhu M., Sanna S., Sayer A. A., Scheet P., Scott L. J., Seedorf U., Sharp S. J., Shields B., Sigurethsson G., Sijbrands E. J., Silveira A., Simpson L., Singleton A., Smith N. L., Sovio U., Swift A., Syddall H., Syvänen A. C., Tanaka T., Thorand B., Tichet J., Tönjes A., Tuomi T., Uitterlinden A. G., van Dijk K. W., van Hoek M., Varma D., Visvikis-Siest S., Vitart V., Vogelzangs N., Waeber G., Wagner P. J., Walley A., Walters G. B., Ward K. L., Watkins H., Weedon M. N., Wild S. H., Willemsen G., Witteman J. C., Yarnell J. W., Zeggini E., Zelenika D., Zethelius B., Zhai G., Zhao J. H., Zillikens M. C., Borecki I. B., Loos R. J., Meneton P., Magnusson P. K., Nathan D. M., Williams G. H., Hattersley A. T., Silander K., Salomaa V., Smith G. D., Bornstein S. R., Schwarz P., Spranger J., Karpe F., Shuldiner A. R., Cooper C., Dedoussis G. V., Serrano-Ríos M., Morris A. D., Lind L., Palmer L. J., Hu F. B., Franks P. W., Ebrahim S., Marmot M., Kao W. H., Pankow J. S., Sampson M. J., Kuusisto J., Laakso M., Hansen T., Pedersen O., Pramstaller P. P., Wichmann H. E., Illig T., Rudan I., Wright A. F., Stumvoll M., Campbell H., Wilson J. F., Bergman R. N., Buchanan T. A., Collins F. S., Mohlke K. L., Tuomilehto J., Valle T. T., Altshuler D., Rotter J. I., Siscovick D. S., Penninx B. W., Boomsma D. I., Deloukas P., Spector T. D., Frayling T. M., Ferrucci L., Kong A., Thorsteinsdottir U., Stefansson K., van Duijn C. M., Aulchenko Y. S., Cao A., Scuteri A., Schlessinger D., Uda M., Ruokonen A., Jarvelin M. R., Waterworth D. M., Vollenweider P., Peltonen L., Mooser V., Abecasis G. R., Wareham N. J., Sladek R., Froguel P., Watanabe R. M., Meigs J. B., Groop L., Boehnke M., McCarthy M. I., Florez J. C., Barroso I. (2010) Nat. Genet. 42, 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Voight B. F., Scott L. J., Steinthorsdottir V., Morris A. P., Dina C., Welch R. P., Zeggini E., Huth C., Aulchenko Y. S., Thorleifsson G., McCulloch L. J., Ferreira T., Grallert H., Amin N., Wu G., Willer C. J., Raychaudhuri S., McCarroll S. A., Langenberg C., Hofmann O. M., Dupuis J., Qi L., Segrè A. V., van Hoek M., Navarro P., Ardlie K., Balkau B., Benediktsson R., Bennett A. J., Blagieva R., Boerwinkle E., Bonnycastle L. L., Bengtsson Boström K., Bravenboer B., Bumpstead S., Burtt N. P., Charpentier G., Chines P. S., Cornelis M., Couper D. J., Crawford G., Doney A. S., Elliott K. S., Elliott A. L., Erdos M. R., Fox C. S., Franklin C. S., Ganser M., Gieger C., Grarup N., Green T., Griffin S., Groves C. J., Guiducci C., Hadjadj S., Hassanali N., Herder C., Isomaa B., Jackson A. U., Johnson P. R., Jørgensen T., Kao W. H., Klopp N., Kong A., Kraft P., Kuusisto J., Lauritzen T., Li M., Lieverse A., Lindgren C. M., Lyssenko V., Marre M., Meitinger T., Midthjell K., Morken M. A., Narisu N., Nilsson P., Owen K. R., Payne F., Perry J. R., Petersen A. K., Platou C., Proença C., Prokopenko I., Rathmann W., Rayner N. W., Robertson N. R., Rocheleau G., Roden M., Sampson M. J., Saxena R., Shields B. M., Shrader P., Sigurdsson G., Sparsø T., Strassburger K., Stringham H. M., Sun Q., Swift A. J., Thorand B., Tichet J., Tuomi T., van Dam R. M., van Haeften T. W., van Herpt T., van Vliet-Ostaptchouk J. V., Walters G. B., Weedon M. N., Wijmenga C., Witteman J., Bergman R. N., Cauchi S., Collins F. S., Gloyn A. L., Gyllensten U., Hansen T., Hide W. A., Hitman G. A., Hofman A., Hunter D. J., Hveem K., Laakso M., Mohlke K. L., Morris A. D., Palmer C. N., Pramstaller P. P., Rudan I., Sijbrands E., Stein L. D., Tuomilehto J., Uitterlinden A., Walker M., Wareham N. J., Watanabe R. M., Abecasis G. R., Boehm B. O., Campbell H., Daly M. J., Hattersley A. T., Hu F. B., Meigs J. B., Pankow J. S., Pedersen O., Wichmann H. E., Barroso I., Florez J. C., Frayling T. M., Groop L., Sladek R., Thorsteinsdottir U., Wilson J. F., Illig T., Froguel P., van Duijn C. M., Stefansson K., Altshuler D., Boehnke M., McCarthy M. I. (2010) Nat. Genet. 42, 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vitaterna M. H., Pinto L. H., Takahashi J. S. (2006) Trends Neurosci. 29, 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vanhaesebroeck B., Rohn J. L., Waterfield M. D. (2004) Cell 118, 274–276 [DOI] [PubMed] [Google Scholar]
- 16. Patrucco E., Notte A., Barberis L., Selvetella G., Maffei A., Brancaccio M., Marengo S., Russo G., Azzolino O., Rybalkin S. D., Silengo L., Altruda F., Wetzker R., Wymann M. P., Lembo G., Hirsch E. (2004) Cell 118, 375–387 [DOI] [PubMed] [Google Scholar]
- 17. Gloyn A. L. (2003) Hum. Mutat. 22, 353–362 [DOI] [PubMed] [Google Scholar]
- 18. Bedoya F. J., Matschinsky F. M., Shimizu T., O'Neil J. J., Appel M. C. (1986) J. Biol. Chem. 261, 10760–10764 [PubMed] [Google Scholar]
- 19. Grimsby J., Sarabu R., Corbett W. L., Haynes N. E., Bizzarro F. T., Coffey J. W., Guertin K. R., Hilliard D. W., Kester R. F., Mahaney P. E., Marcus L., Qi L., Spence C. L., Tengi J., Magnuson M. A., Chu C. A., Dvorozniak M. T., Matschinsky F. M., Grippo J. F. (2003) Science 301, 370–373 [DOI] [PubMed] [Google Scholar]
- 20. Rozen S., Skaletsky H. J. (2000) in Bioinformatics Methods and Protocols: Methods in Molecular Biology (Krawetz S., Misener S. eds) pp. 365–386, Humana Press Inc., Totowa, NJ [Google Scholar]
- 21. Liang Y., Kesavan P., Wang L. Q., Niswender K., Tanizawa Y., Permutt M. A., Magnuson M. A., Matschinsky F. M. (1995) Biochem. J. 309, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gloyn A. L., Odili S., Zelent D., Buettger C., Castleden H. A., Steele A. M., Stride A., Shiota C., Magnuson M. A., Lorini R., d'Annunzio G., Stanley C. A., Kwagh J., van Schaftingen E., Veiga-da-Cunha M., Barbetti F., Dunten P., Han Y., Grimsby J., Taub R., Ellard S., Hattersley A. T., Matschinsky F. M. (2005) J. Biol. Chem. 280, 14105–14113 [DOI] [PubMed] [Google Scholar]
- 23. Concepcion D., Seburn K. L., Wen G., Frankel W. N., Hamilton B. A. (2004) Genetics 168, 953–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Michaud E. J., Culiat C. T., Klebig M. L., Barker P. E., Cain K. T., Carpenter D. J., Easter L. L., Foster C. M., Gardner A. W., Guo Z. Y., Houser K. J., Hughes L. A., Kerley M. K., Liu Z., Olszewski R. E., Pinn I., Shaw G. D., Shinpock S. G., Wymore A. M., Rinchik E. M., Johnson D. K. (2005) BMC Genomics 6, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sakuraba Y., Sezutsu H., Takahasi K. R., Tsuchihashi K., Ichikawa R., Fujimoto N., Kaneko S., Nakai Y., Uchiyama M., Goda N., Motoi R., Ikeda A., Karashima Y., Inoue M., Kaneda H., Masuya H., Minowa O., Noguchi H., Toyoda A., Sakaki Y., Wakana S., Noda T., Shiroishi T., Gondo Y. (2005) Biochem. Biophys. Res. Commun. 336, 609–616 [DOI] [PubMed] [Google Scholar]
- 26. Takahasi K. R., Sakuraba Y., Gondo Y. (2007) BMC Mol. Biol. 8, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boles M. K., Wilkinson B. M., Wilming L. G., Liu B., Probst F. J., Harrow J., Grafham D., Hentges K. E., Woodward L. P., Maxwell A., Mitchell K., Risley M. D., Johnson R., Hirschi K., Lupski J. R., Funato Y., Miki H., Marin-Garcia P., Matthews L., Coffey A. J., Parker A., Hubbard T. J., Rogers J., Bradley A., Adams D. J., Justice M. J. (2009) PLoS Genet. 5, e1000759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Augustin M., Sedlmeier R., Peters T., Huffstadt U., Kochmann E., Simon D., Schöniger M., Garke-Mayerthaler S., Laufs J., Mayhaus M., Franke S., Klose M., Graupner A., Kurzmann M., Zinser C., Wolf A., Voelkel M., Kellner M., Kilian M., Seelig S., Koppius A., Teubner A., Korthaus D., Nehls M., Wattler S. (2005) Mamm. Genome 16, 405–413 [DOI] [PubMed] [Google Scholar]
- 29. Herbach N., Rathkolb B., Kemter E., Pichl L., Klaften M., de Angelis M. H., Halban P. A., Wolf E., Aigner B., Wanke R. (2007) Diabetes 56, 1268–1276 [DOI] [PubMed] [Google Scholar]
- 30. Støy J., Edghill E. L., Flanagan S. E., Ye H., Paz V. P., Pluzhnikov A., Below J. E., Hayes M. G., Cox N. J., Lipkind G. M., Lipton R. B., Greeley S. A., Patch A. M., Ellard S., Steiner D. F., Hattersley A. T., Philipson L. H., Bell G. I. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15040–15044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 32. Kamata K., Mitsuya M., Nishimura T., Eiki J., Nagata Y. (2004) Structure 12, 429–438 [DOI] [PubMed] [Google Scholar]
- 33. Marotta D. E., Anand G. R., Anderson T. A., Miller S. P., Okar D. A., Levitt D. G., Lange A. J. (2005) Arch. Biochem. Biophys. 436, 23–31 [DOI] [PubMed] [Google Scholar]
- 34. Veiga-da-Cunha M., Courtois S., Michel A., Gosselain E., Van Schaftingen E. (1996) J. Biol. Chem. 271, 6292–6297 [DOI] [PubMed] [Google Scholar]
- 35. Zelent B., Odili S., Buettger C., Zelent D. K., Chen P., Fenner D., Bass J., Stanley C., Laberge M., Vanderkooi J. M., Sarabu R., Grimsby J., Matschinsky F. M. (2011) Biochem. J., doi: 10.1042/BJ20110440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gloyn A. L., Odili S., Buettger C., Njolstad P. R., Shiota C., Magnuson M. A., Matschinsky F. M. (2004) in Glucokinase and Glycemic Disease (Matschinsky F. M., Magnuson M. A. eds) 1st Ed., pp. 92–109 Karger, New York [Google Scholar]
- 37. Burke C. V., Buettger C. W., Davis E. A., McClane S. J., Matschinsky F. M., Raper S. E. (1999) Biochem. J. 342, 345–352 [PMC free article] [PubMed] [Google Scholar]
- 38. Sagen J. V., Odili S., Bjørkhaug L., Zelent D., Buettger C., Kwagh J., Stanley C., Dahl-Jørgensen K., de Beaufort C., Bell G. I., Han Y., Grimsby J., Taub R., Molven A., Søvik O., Njølstad P. R., Matschinsky F. M. (2006) Diabetes 55, 1713–1722 [DOI] [PubMed] [Google Scholar]
- 39. Farrelly D., Brown K. S., Tieman A., Ren J., Lira S. A., Hagan D., Gregg R., Mookhtiar K. A., Hariharan N. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14511–14516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grimsby J., Coffey J. W., Dvorozniak M. T., Magram J., Li G., Matschinsky F. M., Shiota C., Kaur S., Magnuson M. A., Grippo J. F. (2000) J. Biol. Chem. 275, 7826–7831 [DOI] [PubMed] [Google Scholar]
- 41. Pino M. F., Kim K. A., Shelton K. D., Lindner J., Odili S., Li C., Collins H. W., Shiota M., Matschinsky F. M., Magnuson M. A. (2007) J. Biol. Chem. 282, 13906–13916 [DOI] [PubMed] [Google Scholar]
- 42. Bali D., Svetlanov A., Lee H. W., Fusco-DeMane D., Leiser M., Li B., Barzilai N., Surana M., Hou H., Fleischer N. (1995) J. Biol. Chem. 270, 21464–21467 [DOI] [PubMed] [Google Scholar]
- 43. Terauchi Y., Sakura H., Yasuda K., Iwamoto K., Takahashi N., Ito K., Kasai H., Suzuki H., Ueda O., Kamada N. (1995) J. Biol. Chem. 270, 30253–30256 [DOI] [PubMed] [Google Scholar]
- 44. Grupe A., Hultgren B., Ryan A., Ma Y. H., Bauer M., Stewart T. A. (1995) Cell 83, 69–78 [DOI] [PubMed] [Google Scholar]
- 45. Postic C., Shiota M., Niswender K. D., Jetton T. L., Chen Y., Moates J. M., Shelton K. D., Lindner J., Cherrington A. D., Magnuson M. A. (1999) J. Biol. Chem. 274, 305–315 [DOI] [PubMed] [Google Scholar]
- 46. Gorman T., Hope D. C., Brownlie R., Yu A., Gill D., Löfvenmark J., Wedin M., Mayers R. M., Snaith M. R., Smith D. M. (2008) Diabetes Obes. Metab. 10, 885–897 [DOI] [PubMed] [Google Scholar]
- 47. Zhang Y. L., Tan X. H., Xiao M. F., Li H., Mao Y. Q., Yang X., Tan H. R. (2004) Acta Pharmacol. Sin. 25, 1659–1665 [PubMed] [Google Scholar]
- 48. de la Iglesia N., Veiga-da-Cunha M., Van Schaftingen E., Guinovart J. J., Ferrer J. C. (1999) FEBS Lett. 456, 332–338 [DOI] [PubMed] [Google Scholar]
- 49. Nakamura A., Terauchi Y., Ohyama S., Kubota J., Shimazaki H., Nambu T., Takamoto I., Kubota N., Eiki J., Yoshioka N., Kadowaki T., Koike T. (2009) Endocrinology 150, 1147–1154 [DOI] [PubMed] [Google Scholar]
- 50. Terauchi Y., Takamoto I., Kubota N., Matsui J., Suzuki R., Komeda K., Hara A., Toyoda Y., Miwa I., Aizawa S., Tsutsumi S., Tsubamoto Y., Hashimoto S., Eto K., Nakamura A., Noda M., Tobe K., Aburatani H., Nagai R., Kadowaki T. (2007) J. Clin. Invest. 117, 246–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Salpeter S. J., Klein A. M., Huangfu D., Grimsby J., Dor Y. (2010) Development 137, 3205–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sayle R. A., Milner-White E. J. (1995) Trends Biochem. Sci. 20, 374. [DOI] [PubMed] [Google Scholar]
- 53. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.