Background: The xanthine permease XanQ is a prototype of nucleobase:proton symporters of the ubiquitous family NCS2.
Results: Site-directed replacements of Asn-93 and Phe-94 in the third transmembrane segment of XanQ distort the specificity for substrate.
Conclusion: Asn-93 interacts with xanthine-coordinating residues in the binding pocket.
Significance: Dissecting specificity determinants in family NCS2 is crucial for understanding evolution of purine uptake.
Keywords: Escherichia coli, Homology Modeling, Membrane Transport, Purine, Site-directed Mutagenesis, Cysteine-scanning Analysis
Abstract
The xanthine permease XanQ of Escherichia coli is used as a study prototype for function-structure analysis of the ubiquitous nucleobase-ascorbate transporter (NAT/NCS2) family. Our previous mutagenesis study of polar residues of XanQ has shown that Asn-93 at the middle of putative TM3 is a determinant of substrate affinity and specificity. To study the role of TM3 in detail we employed Cys-scanning mutagenesis. Using a functional mutant devoid of Cys residues (C-less), each amino acid residue in sequence 79–107 (YGIVGSGLLSIQSVNFSFVTVMIALGSSM) including TM3 (underlined) and flanking sequences was replaced individually with Cys. Of 29 single-Cys mutants, 20 accumulate xanthine to 40–110% of the steady state observed with C-less, six (S88C, F94C, A102C, G104C, S106C) accumulate to low levels (10–30%) and three (G83C, G85C, N93C) are inactive. Extensive mutagenesis reveals that Gly-83 and, to a lesser extent, Gly-85, are crucial for expression in the membrane. Replacements of Asn-93 disrupt affinity (Thr) or permit recognition of 8-methylxanthine which is not a wild-type ligand (Ala, Ser, Asp) and utilization of uric acid which is not a wild-type substrate (Ala, Ser). Replacements of Phe-94 impair affinity for 2-thio and 6-thioxanthine (Tyr) or 3-methylxanthine (Ile). Single-Cys mutants S84C, L86C, L87C, and S95C are highly sensitive to inactivation by N-ethylmaleimide. Our data reveal that key residues of TM3 cluster in two conserved sequence motifs, 83GSGLL87 and 93NFS95, and highlight the importance of Asn-93 and Phe-94 in substrate recognition and specificity; these findings are supported by structural modeling on the recently described x-ray structure of the uracil-transporting homolog UraA.
Introduction
The Nucleobase-Ascorbate Transporter (NAT)2 or Nucleobase-Cation Symporter-2 (NCS2) family is evolutionarily ubiquitous and encompasses more than 2,000 putative members in all major taxa of organisms. Despite their relevance to the recognition and uptake of several frontline purine-related drugs, only 16 members have been characterized experimentally; these are specific for the cellular uptake of uracil, xanthine, or uric acid (microbial, plant and non-primate mammalian genomes) or vitamin C (mammalian genomes) (1–3). Structure-function relationships have been studied extensively in two members, the prokaryotic XanQ, a specific, high-affinity xanthine:H+ symporter from Escherichia coli (4–9), and the eukaryotic UapA, a high-affinity uric acid/xanthine:H+ symporter from Aspergillus nidulans (10–14). Mutagenesis data have revealed striking similarities of key NAT determinants between the two transporters, implying that few residues conserved throughout the family or displaying distinct conservation patterns may be invariably critical for function and underlie specificity differences.
Recently, Lu et al. solved the first x-ray structure for a NAT homolog (the uracil permease UraA), which represents a novel structural fold (1). Homology modeling shows that most of the residues identified as important in both XanQ (7–9) and UapA (11–14) fall in TM1, TM3, TM8, and the NAT-signature motif (TM10) and are either in the putative binding site or at the periphery forming a shelter around substrate in the core domain of the protein (supplemental Fig. S1). These residues are probably crucial in defining the molecular basis of NAT substrate recognition and selectivity. In addition, more dynamic structural elements located in the gate domain (1) may contribute to affinity and specificity by controlling access of substrates to the binding site and implementing the appropriate conformational changes; the current genetic and biochemical evidence on UapA (12–14) and XanQ (6) suggests that residues involved in such gating mechanisms are found in the last transmembrane segment (TM14) (6, 12, 13), in cytoplasmic loop TM12-TM13 (13) and in sequence regions flanking TM1 (13, 14).
The xanthine-specific permease XanQ has been subjected to a systematic Cys-scanning and site-directed mutagenesis study to address the role of each amino acid residue (5–9). Of more than 180 residues analyzed thus far (Fig. 1), a small set emerges as crucial for the mechanism at positions where a native residue is functionally irreplaceable (Glu-272, Asp-304, Gln-324, Asn-325), replaceable with a limited number of side chains (His-31, Asn-93, Asp-276, Ala-279, Thr-280, Ile-432) or sensitive to alkylation of a substituted Cys with N-ethylmaleimide leading to inactivation (Gly-275, Ala-323, Asn-326, Gly-327, Val-328, Ile-329, Thr-332, Gly-333, Ser-336, Asn-430). Structural modeling shows that, with two exceptions, these functionally important residues fall in TMs of the core domain and correspond to UraA residues which are implicated with substrate binding (Glu-272, Gln-324, Asp-276, Ala-323) or involved in crucial hydrogen bonding (Asn-325, His-31) or disposed to the cytoplasmic halves of TM10 and TM8 which contain the key binding residues (see supplemental Fig. S1). Site-directed alkylation analysis of XanQ suggests that Gln-324 and Asn-325 may participate directly in the XanQ binding site (8), while His-31 and Asn-93 are essential for the proper binding affinity and selectivity, as evidenced from ligand inhibition assays (7). In the light of the homologous UraA structure (1), it appears that the effect of His-31 (TM1) might be indirect, through its interaction with Asn-325 (TM10), while Asn-93 (TM3) is at the binding pocket and may be involved in more direct interactions with substrate or substrate-binding residues (supplemental Fig. S1).
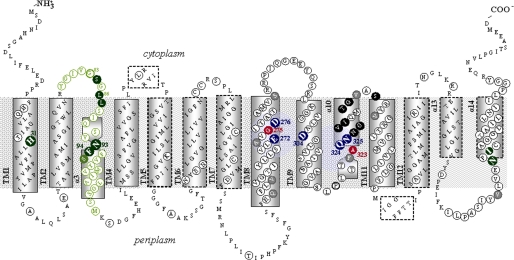
FIGURE 1.
Revised topology model of XanQ permease. The model is derived from homology threading using the UraA structure (3QE7) as a template (see supplemental Fig. S1) and initial topology predictions for XanQ based on program TMHMM and the accessibility of loops to hydrophilic reagents (8). The α-helical segments are indicated in rectangles with continuous (core domain) or broken-line perimeters (gate domain). Irreplaceable residues of XanQ (as well as Asp-276 where the carboxyl group is essential) are numbered and bolded and shown in blue. NEM-sensitive positions are shown in dark background. Ala-323 and Gly-275 where the NEM sensitivity is enhanced by substrate binding are shown in red. Residues analyzed with site-directed mutagenesis are circled. Residues of TM3 analyzed in this study are shown in green.
The core, substrate-coordinating domain of NAT transporters (1) consists of two inverted repeats which are discontinuous in sequence (TM1–4 and TM8–11) (Fig. 1). In the context of our previous mutagenesis studies (5–9), we have analyzed thoroughly the C-terminal repeat of the core domain of XanQ permease (TM8–11) but no systematic mutagenesis has been exerted at the N-terminal repeat (TM1–4) (Fig. 1). In this report, we address for the first time the role of a continuous sequence region in the N-terminal repeat, namely the entire TM3 and flanking sequence regions, by employing systematic site-directed mutagenesis in the XanQ sequence 79–107 (Fig. 1) and analysis of a set of 51 engineered mutants (supplemental Table S1). We show that residues important for function or expression in the membrane cluster in two sequence motifs, one at the cytoplasmic end of TM3 (GxGLx) and one at the middle (NFx). The latter motif includes Asn-93 and Phe-94, both of which are important in substrate recognition and specificity. The results are discussed with respect to models of XanQ and other homologs built on the recently described x-ray structure of UraA (1).
EXPERIMENTAL PROCEDURES
Materials
[8-3H]Xanthine (27.6 Ci mmol−1) and [8-14C]uric acid (51.5 mCi mmol−1) were purchased from Moravek Biochemicals. Non-radioactive nucleobases and analogues were from Sigma. Oligodeoxynucleotides were synthesized from BioSpring GmbH. High fidelity Taq Polymerase (Phusion High Fidelity PCR System) was from Finnzymes. Restriction endonucleases used were from Takara. N-ethylmaleimide (NEM) was from Sigma. Horseradish peroxidase (HRP)-conjugated avidin was from Amersham Biosciences. All other materials were reagent grade and obtained from commercial sources.
Bacterial Strains and Plasmids
E. coli K-12 was transformed according to Inoue (15). TOP10F′ (Invitrogen) was used for initial propagation of recombinant plasmids. T184 (16) harboring pT7–5/xanQ (4) with given replacements was used for IPTG-inducible expression from the lacZ promoter/operator.
DNA Manipulations
Construction of expression plasmids and BAD (biotin-acceptor domain)-tagged versions of XanQ has been described (4). For construction of Cys-less XanQ, the five native-Cys codons were replaced simultaneously with Ser codons, using two-stage (multiple overlap/extension) PCR on the template of wild-type XanQ tagged at C terminus with the BAD tag (5). For construction of mutants, two-stage PCR was performed on the template of Cys-less or wild-type XanQ, as indicated. The entire coding sequence of all engineered constructs was verified by double-strand DNA sequencing in an automated DNA sequencer (MWG-Biotech) (supplemental Table S1).
Growth of Bacteria
E. coli T184 harboring given plasmids was grown aerobically at 37 °C in Luria-Bertani medium containing streptomycin (0.01 mg/ml) and ampicillin (0.1 mg/ml). Fully grown cultures were diluted 10-fold, allowed to grow to mid-logarithmic phase, induced with IPTG (0.5 mm) for an additional 2 h at 37 °C, harvested, and washed with appropriate buffers.
Transport Assays and Kinetic Analysis
E. coli T184 were assayed for active transport of [3H]xanthine (1 μm), by rapid filtration, at 25 °C, pH 7.5, as described (4). For kinetic uptake measurements, initial rates were assayed in T184 cells, at 5–20 s, in the concentration range of 0.1–100 μm [3H]xanthine. Selected mutants were also assayed for transport of [14C]uric acid (0.04- 2 mm), using the paralog YgfU as a positive control.3 For assaying the effect of NEM on xanthine uptake activity, T184 cells were preincubated with NEM at the indicated conditions, excess reagents and ligands were removed by centrifugation, and transport assays were performed in the presence of phenazine methosulfate (PMS) (0.2 mm) and potassium ascorbate (20 mm) (5). For ligand competition experiments, uptake of [3H]xanthine (1 μm) was assayed in the absence or presence of unlabeled analogues (1 mm) (4). For kinetic analysis, putative inhibitors were used in the concentration range of 0.001–2 mm, and data were fitted to the equation y = B + (T - B)/(1 + 10((log IC50-log x) h)) for sigmoidal dose-response (variable slope), where x is the concentration variable, y (activity) values range from T (top) to B (bottom) and h is the Hill coefficient, using Prism4, to obtain IC50 values; in all cases, the Hill coefficient was close to −1, consistent with presence of one binding site. Ki values for each analog were calculated from the Cheng and Prusoff equation Ki = IC50/[1+(L/Km)] (where L is the permeant concentration and Km is the value obtained for this permeant), assuming a simple model of competitive inhibition with the binding site of the transporter (5); competitive inhibition has been shown with certain analogues (1-methyl, 2-thio, and 8-methylxanthine) by assaying their effect on Km and Vmax for wild type and selected mutants, and showing that Vmax remains unaltered (5–8).
Immunoblot Analysis
Membrane fractions were prepared from 10-ml cultures of E. coli T184 harboring given plasmids and subjected to SDS-PAGE (12%), as described (4). Proteins were electroblotted to poly(vinylidene difluoride) membranes (Immobilon-PVDF; Pall Corp.). XanQ-BAD was probed with avidin-HRP. Signals were developed with enhanced chemiluminescence (ECL).
In Silico Analysis
Comparative sequence analysis of NAT/NCS2 homologs was based on BLAST-p search and ClustalW alignment; the most recent genome annotations were used for retrieving sequence data. Initial analysis of transmembrane topology was performed using program TMHMM (17). Homology threading was performed using the known x-ray structure of UraA as a template (PDB code 3QE7) on the web-based SWISSPROT modeling server (18); results were displayed and analyzed with PyMOL v1.4 (Schrödinger, LLC) or VMD (19), as indicated.
RESULTS
Revisiting the Topology of TM3 Based on the Structure of UraA
A structural model of XanQ built on the recently released x-ray structure of the uracil-transporting homolog UraA (1) shows that the transporter contains 14 TMs (Fig. 1) and its topology differs from the one proposed previously based on prediction algorithms and cysteine-accessibility analysis (8, 9). The main difference is at the C-terminal half (TMs 8–14). In particular, the sequence region downstream TM8 which was predicted to contain a single long TM9a/b with two amphipathic helices and a short inter-helical loop corresponding to the NAT-signature motif (9) is now modeled as three sequential TMs (TM9–11) with the NAT-signature motif falling in TM10 (Fig. 1). This difference shifts accordingly the numbering of the three C-terminal TMs which are now TM12, TM13, and TM14. In addition, the modeled structure dictates that the α-helices of TM13 and TM14 are only halfway into the membrane (Fig. 1) while TM10, as well as TM3, have the unusual feature of including a short β-strand preceded by an extended, unwound region and followed by a short α-helix (α3 or α10) which also constitutes half of the TM segment. The two β-strands of TM3 and TM10 are antiparallel and located at the center of the structure (supplemental Fig. S1) and, along with the connecting loops and neighboring helices, they provide a shelter for substrate binding in the structure of the UraA homolog (1). The new topology and structural models of XanQ imply that residues at the junction between the β-strand and the helical region of TM3 are in proximity with key binding residues of TM8 and TM10 (supplemental Fig. S1). On the other hand, it is intriguing that, in contrast with TM10 which contains the conserved NAT-signature motif (8), the sequence of TM3 is relatively poorly conserved among NAT members (Fig. 2 and supplemental Table S3).
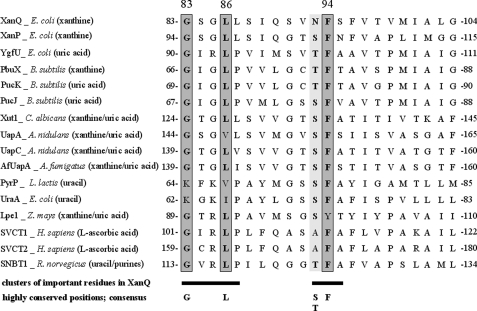
FIGURE 2.
Alignment of TM3 sequences in functionally known NAT homologs. Shown is a consensus of 16 characterized NAT/NCS2 transporters, including Escherichia coli XanQ (P67444), XanP (P0AGM9), YgfU (Q46821) (see footnote 2), and UraA (P0AGM7), Bacillus subtilis PbuX (P42086), PucK (O32140), and PucJ (O32139), Lactococcus lactis PyrP (AAK05701), Aspergillus nidulans UapA (Q07307) and UapC (P487777), A. fumigatus AfUapA (XP748919), Candida albicans Xut1 (AAX2221), Zea mays Lpe1 (AAB17501), Homo sapiens SVCT1 (SLC23A1) (AAH50261), and SVCT2 (SLC23A2) (Q9UGH3) and Rattus norvegicus SNBT1 (AB511909). The full-length sequences were aligned using ClustalW and part of this alignment including the residues of interest is presented. Major substrates of each transporter are shown in parentheses.
Active Xanthine Transport
Using a functional XanQ permease devoid of Cys residues (C-less), each amino acid residue in sequence 79YGIVGSGLLSIQSVNFSFVTVMIALGSSM107 including TM3 (underlined) and flanking sequences was replaced individually with Cys. After verification of the sequence, each single-Cys mutant was transformed into E. coli T184 and assayed for its ability to catalyze active xanthine transport. As shown in Fig. 3A, of the 29 single-Cys mutants, 19 transport xanthine at rates between 40 and 105% of C-less permease, three (L86C, L87C, A102C) transport at rates of about 30%, four (S88C, F94C, G104C, S106C) display very low but detectable rates (5–20%) and three (G83C, G85C, N93C) display rates that are indistinguishable from cells containing no xanQ insert. Steady state levels of xanthine accumulation also show the same general picture (Fig. 3B); of the 29 single-Cys mutants, 20 accumulate xanthine to 40–110% of the steady state observed with C-less, six (L86C, S88C, F94C, A102C, G104C, S106C) accumulate to low levels (10–30%) and three (G83C, G85C, N93C) are inactive.
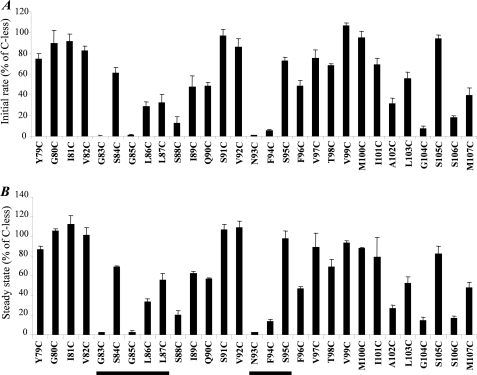
FIGURE 3.
Active xanthine transport activities of single Cys mutants. E. coli T184 harboring pT7–5/xanQ(C-less-BAD) with given mutations were grown aerobically at 37 °C in complete medium to mid-logarithmic phase, induced with IPTG (0.5 mm) for 2 h, normalized to 0.7 mg of protein per ml of cell suspension, and assayed for transport of [3H]xanthine (1 μm) at 25 °C. A, Initial rates of uptake measured at 5–20 s. Control values obtained from T184 harboring pT7–5 alone (0.018 nmol mg−1 min−1 on average) were subtracted from the sample measurements in all cases. Results are expressed as a percentage of the rate of Cys-less XanQ (1.9 nmol mg−1 min−1 on average) with standard deviations from three independent determinations. B, steady state levels of xanthine accumulation (reached at 1–10 min for most mutants). Control values obtained from T184 harboring pT7–5 alone (0.012 nmol mg−1 on average) were subtracted from the sample measurements in all cases. Results are expressed as a percentage of the level of Cys-less XanQ (0.8 nmol mg−1 on average) with standard deviations from three independent determinations.
Expression in the Membrane
Immunoblot analysis of BAD-tagged single-Cys permeases shows that negligible activity of mutants G83C and G85C is associated with negligible or marginal expression in the membrane. All other mutants are expressed to high or moderate levels comparable with those of C-less XanQ (Fig. 4).
FIGURE 4.
Immunoblot analysis of single Cys mutants. Membranes were prepared from IPTG-induced cultures of E. coli T184 harboring pT7–5/xanQ(C-less-BAD) with given mutations. Samples containing ∼100 μg of membrane protein were subjected to SDS-PAGE (12%) and immunoblotting using HRP-conjugated avidin. A, representative blots of single-Cys mutants and Cys-less XanQ. Membranes prepared from cells harboring pT7–5 alone exhibited no immunoreactive material. Prestained molecular mass standards (in kDa; Bio-Rad, low range) are shown on the left. B, quantitative estimation of the expression level of each mutant as a percentage of Cys-less expression derived from the relative density of the corresponding band, as measured with program Quantity One (Bio-Rad). Results shown are the means of four determinations from two independent experiments, with standard deviations that were <15%.
Expression and Transport Analysis of Mutants in Wild-type Background
From the Cys-scanning transport analysis described above, 9 positions of inactive or low activity mutants were delineated. We analyzed these positions further by engineering the most conservative site-directed replacement mutants to introduce an amino acid other than Cys in the wild-type XanQ background. For positions of aliphatic or aromatic native residues (Leu-86, Phe-94, Ala-102), we constructed and assayed mutants L86M(wt), L86V(wt), F94Y(wt), F94I(wt), A102G(wt), and A102S(wt) which exhibited wild-type expression in the membrane and xanthine uptake activities of about 40% (L86V), 70% (F94I), 100% (A102G), 150% (L86M, A102S), and 170% (F94Y) relative to wild type (Fig. 5A). For positions of a native Gly (Gly-83, Gly-85, Gly-104), we constructed and analyzed mutants G83A(wt), G83P(wt), G85A(wt), G85P(wt), G104A(wt), and G104P(wt); of them, G83A, G83P, and G85P display negligible expression in the membrane and are inactive, G85A displays compromised expression and approximately wild-type levels of activity, G104A and G104P are expressed to high levels but yield low activity (xanthine uptake rates and levels of 20–40% relative to wild type) (Fig. 5B). For positions of a native Ser (Ser-88, Ser-106), we constructed and assayed mutants S88A(wt), S88T(wt), S88V(wt), S106A(wt), S106T(wt) and S106V(wt), all of which display wild-type protein levels in the membrane; S88T, S106A, S106T, and S106V are highly active (xanthine uptake rates of 140–160% relative to wild type and accumulation levels of about 100%), while S88A is 50% active and S88V yields marginal activity (2% of wild type) (Fig. 5C). Finally, replacements of Asn-93 with amino acids that are found in other NAT members (Ala, Ser, Thr, Asp) yield wild-type protein levels and activities that are higher than wild type for N93S (xanthine uptake rate 180%; accumulation level 120%), intermediate for N93A (xanthine uptake rate 60%; accumulation level 70%) and low for N93T and N93D (10–20% of wild type) (Fig. 5D; see also Ref. 7).
FIGURE 5.
Expression and xanthine uptake activities of site-directed mutants at positions 83, 85, 86, 88, 93, 94, 102, 104, and 106. E. coli T184 harboring pT7–5/xanQ(wild-type-BAD) with given mutations were grown, induced and subjected to immunoblot analysis of membrane fractions (upper panels) or assayed for transport of [3H]xanthine (1 μm, 25 °C) (lower panels), exactly as described in the legends to Figs. 3 and 4. Open and closed histogram bars represent initial rate and steady-state values, respectively.
Effect of N-ethylmaleimide on Transport Activity
The effect of N-ethylmaleimide (NEM), a membrane-permeable sulfhydryl reagent, on the initial rate of xanthine transport for each active single-Cys mutant is presented in Fig. 6. When incubated with 2 mm NEM, it is found that four mutants are inhibited to near completion, S84C by 90%, L87C by 98%, S95C by 99%, and L86C by 100%, an additional five mutants (I89C, S91C, F96C, A102C, G104C) are inhibited by 40–70%, and activity of the remaining 16 single-Cys mutants is not altered significantly (<1.5-fold enhancement or inhibition) (Fig. 6). S84C, L86C, L87C, and S96C were then assayed in the concentration range of 1 μm to 2 mm, and the concentration resulting in half-maximal inhibition (IC50) was determined, in the absence or presence of substrate (xanthine) (Fig. 7). The results show that L86C is highly sensitive to NEM (IC50,25 μm), while L87C, S84C and S96C display 4.5- to 11.5-fold lower sensitivity (IC50 values,112 μm, 132 μm and 286 μm, respectively); the sensitivity of L86C is not affected significantly in the presence of substrate (Fig. 7).
FIGURE 6.
Effect of NEM on the xanthine transport activity of single Cys mutants. E. coli T184 harboring pT7–5/xanQ(C-less-BAD) with given mutations were grown, induced and assayed for transport of [3H]xanthine (1 μm, 25 °C). Cells had been pre-incubated in the absence or presence of 2 mm NEM for 10 min at 25 °C. Transport assays were performed in the presence of 20 mm potassium ascorbate and 0.2 mm PMS. Rates are presented as percentages of the rate measured in the absence of NEM with standard deviations from three independent determinations shown. Average and standard deviation values of C-less control are also shown as interrupted line and gray horizontal bars, respectively. Values were not determined (N.D.) for mutants G83C, G85C, N93C, or F94C, which display very low or negligible initial rates (6% of C-less).
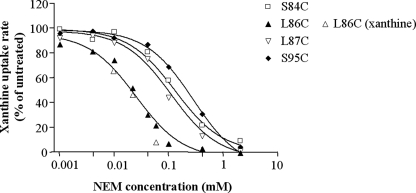
FIGURE 7.
Sensitivity of L86C, L87C, S84C, and S95C to NEM. E. coli T184 harboring pT7–5/xanQ(C-less-BAD) with given mutations were grown, induced and assayed for transport of [3H]xanthine (1 μm, 25 °C), after preincubation with NEM (0.001–2 mm, 10 min) in the absence or presence of 1 mm xanthine, as indicated. The data shown were fitted to the equation y = B + (T − B)/(1 + 10 ((log IC50-log x) h)) for sigmoidal dose-response (variable slope), where x is the concentration variable, y (activity) values range from T (top) to B (bottom) and h is the Hill coefficient, using Prism4, and the IC50 values obtained for NEM inhibition were 25 μm (L86C), 112 μm (L87C), 132 μm (S84C), and 286 μm (S95C). The IC50 obtained for NEM inhibition of L86C in the presence of xanthine was identical with the corresponding value obtained in the absence of xanthine.
Transport Kinetics and Ligand Specificity Profiles of Mutants at Asn-93, Phe-94, and Ser-95
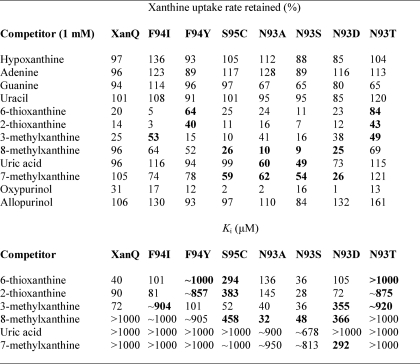
The sum of our scanning data (Figs. 3–6) indicates that structurally or functionally important residues, including positions replaceable with few or no other side chains and/or sensitive to inactivation by NEM, are contained in two short sequences, one at the cytoplasmic end of TM3 (GSGLLS) and one at the middle (NFS), which are relatively well conserved (GxGLx and NFx) in NAT family (Fig. 2). Interestingly, the second sequence motif (NFx) lies at the junction between the β-strand and the α-helix of TM3 (Fig. 1), close to the putative binding site (1) and contains Asn-93, which is found only as Ser, Thr, Asp or Ala in other members, and Phe-94, which is almost invariably conserved (Fig. 2 and supplemental Table S3). To understand the contribution of this motif to the substrate and ligand recognition profile of XanQ, we assayed mutants at positions Asn-93, Phe-94, and Ser-95 for transport kinetics (Table 1) and inhibition of [3H]xanthine uptake in the presence or absence of a series of purine analogues (Table 2). Kinetic analysis reveals that replacement of Phe-94 with Ile or Tyr yields 3- or 8-fold higher Vmax for xanthine uptake but lower affinity (2-fold increased Km) while replacement of Ser-95 with Cys yields 3.5-fold higher Vmax without affecting Km (Table 1). In comparison, replacement of Asn-93 with Ala or Ser yields higher affinity than wild type (2-fold decreased Km) with equivalent (N93A) or 4.5-fold higher Vmax (N93S); the low activity of mutants N93T and N93D is associated with low affinity (2-fold increased Km) and low Vmax, respectively (Table 1; see also Ref. 7). On the ligand competition analysis, the replacements of Phe-94 were found to impair affinity for 2- and 6-thioxanthine (F94Y) or 3-methylxanthine (F94I), while a lower, but not impaired affinity for 2- and 6-thioxanthine was also observed with S95C. On the other hand, replacements of Asn-93 were found to either impair affinity for all analogues (N93T) or allow the recognition of analogues that are not recognized by wild type and differ from xanthine only at the imidazole moiety (8- or 7-methylxanthine, uric acid). Most strikingly, N93A and N93S allow high-affinity recognition of 8-methylxanthine (Ki 32 and 48 μm, respectively); N93D recognizes both 8- and 7-methylxanthine, but with low affinity (Ki 366 and 292 μm, respectively) (Table 2). In addition, N93S and, to a lesser extent, N93A display the ability to recognize uric acid (8-oxy-xanthine), albeit with very low affinity (Ki of about 678 and 900 μm, respectively) (Table 2). This latter property is highly unusual for a XanQ mutant; of more than 35 mutants tested in this respect (5–9), marginal recognition of uric acid in competition assays has also been observed only with G333R (5), H31Q (7), Q258N (7), and V334C (8).
TABLE 1.
Km and Vmax values of XanQ mutants at positions Asn-93, Phe-94, and Ser-95
E. coli T184 expressing the corresponding constructs were assayed for initial rates of xanthine uptake at 5–20 s, in the concentration range of 0.1–100 μm; negative control values obtained from T184 harboring vector pT7–5 alone were subtracted from the sample measurements in all cases. Kinetic parameters were determined from non-linear regression fitting to the Michaelis-Menten equation using Prism4; values represent the means of three independent determinations with standard deviations shown. ND, assays were performed, but kinetic values were not determined due to very low uptake rates (≤0.1 nmol mg−1 min−1). All mutants and the wild-type XanQ version used contained a C-terminal BAD. Data on the Asn-93 mutants are from Ref. 7.
| Permease | Km | Vmax | Vmax/Km |
|---|---|---|---|
| μm | nmol min−1mg−1protein | μl min−1mg−1 | |
| XanQ(wt) | 4.5 ± 0.5 | 7.9 ± 1.0 | 1756 |
| XanQ(C-less) | 5.5 ± 0.6 | 10.2 ± 0.9 | 1855 |
| F94I(wt) | 9.4 ± 1.5 | 50.2 ± 3.0 | 5340 |
| F94Y(wt) | 9.5 ± 1.7 | 147.8 ± 9.8 | 15557 |
| S95C | 5.1 ± 1.6 | 33.9 ± 2.7 | 6647 |
| N93A(wt) | 2.4 ± 0.7 | 5.0 ± 0.4 | 2083 |
| N93S(wt) | 2.1 ± 0.9 | 16.7 ± 1.8 | 7952 |
| N93T(wt) | 11.6 ± 2.9 | 5.0 ± 0.4 | 431 |
| N93D(wt) | 2.3 ± 0.3 | 0.5 ± 0.1 | 217 |
TABLE 2.
Specificity profile of mutants at positions Asn-93, Phe-94, and Ser-95
E. coli T184 expressing the corresponding constructs were assayed for initial rates of [3H]xanthine (1 μm) uptake at 5–20 sec, in the absence or presence of the indicated unlabeled nucleobases and analogues. For initial assessment of the inhibitory effect, xanthine (1 μm) uptake assays were performed in the presence of a 1000-fold excess (1 mm) of unlabeled competitor and the percentage of the uptake rate retained was determined. The uptake value obtained in the absence of competitor was taken as 100%. Values represent the means of three determinations, with standard deviations <20%. For kinetic inhibition analysis, xanthine (1 μm) transport assays were performed in the presence of 0.001–2 mm of competitor and IC50 and Ki values were determined as described under “Experimental Procedures.” Most significant differences from the wild-type profile are highlighted in bold. All mutants and the wild-type XanQ version used contained a C-terminal BAD. The mutants used are in the wild-type background except S95C, which is in the C-less permease background. The low affinity observed with S95C for 8-methylxanthine (Ki 458 μm) is attributable to a comparably low affinity observed with C-less which distinguishes C-less from wild-type XanQ (not shown; see also Ref. 6).
Mutants N93S and N93A Transport Both Xanthine and Uric Acid
Mutants N93S and N93A, as well as other XanQ mutants that are either inactive with respect to xanthine uptake or able to recognize uric acid and/or 8-methylxanthine in transport competition assays (5–9) were assayed directly for transport of [14C]uric acid (0.04–2 mm), using the uric acid-transporting paralog YgfU from E. coli as a positive control (supplemental Fig. S2). It was found that only N93A and N93S are able to transport [14C]uric acid at clearly detectable rates and levels. However, this uric acid uptake activity is only detectable at 2 mm and is still very low relative to the YgfU control (<1%) (supplemental Fig. S2). These data preclude further kinetic analysis or determination of Km and Vmax values for uric acid with mutants N93A and N93S.
DISCUSSION
The results in this study show that important residues of TM3 of the XanQ permease cluster in two short sequence motifs, one at the cytoplasmic entrance of the segment (GxGLx) and one at the middle (NFx), and highlight the importance of the second motif (NFx) in substrate recognition and selectivity. The NFx motif consists of Asn-93, which had been identified as crucial for affinity and specificity in a previous study (7) and is now shown to be unique in preventing aberrant recognition of substrates that differ from xanthine in the imidazole moiety (uric acid, 8-methylxanthine), Phe-94, which is crucial for the recognition of analogues that differ in the pyrimidine moiety (2-thio and 6-thioxanthine, 3-methylxanthine), and Ser-95, which affects specificity less pronouncedly but is highly sensitive to inactivation of the corresponding Cys-replacement mutant by site-directed alkylation. The GxGLx motif consists of Gly-83, which is irreplaceable with respect to expression of XanQ in the membrane (Figs. 4 and 5), Gly-85, which is also associated with optimal expression of the protein in the membrane, and Ser-84, Leu-86, and Leu-87, which are highly sensitive to inactivation by site-directed alkylation (Fig. 7). It is interesting that, although the overall TM3 sequence is poorly conserved in NAT family, the sequence of the two motifs highlighted as structurally and/or functionally important follows a prominent conservation pattern, with Gly-83 and Phe-94 being essentially invariable, Leu-86 alternating only with hydrophobic amino acids of comparable volume (Ile, Met, Val), Gly-85 alternating only with Arg or Lys, and the positions of Asn-93 and Ser-95 occupied with either a polar (Ser, Thr, Asn) or a small residue (Ala, Gly) (Fig. 2 and supplemental Table S3).
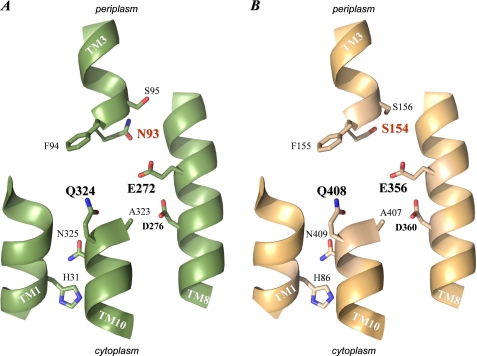
The probable structural arrangement of TM3 relative to other TMs of the core domain of XanQ which contain important residues (TM1, TM8, TM10) has been modeled based on the x-ray structure of the uracil-transporting homolog UraA (1) and is presented in Fig. 8A. It is evident that the NFx motif, which constitutes the beginning and initial turn of helix α3 immediately after the short β-strand of TM3 (Fig. 1), is at the center of the structure, in close proximity with the side chains of Glu-272 (TM8) and Gln-324 (TM10) which correspond to the two carboxyl groups (Glu-241 and Glu-290) forming direct hydrogen bonds with substrate (uracil) in the structural prototype UraA (1). In addition, Phe-94, the second residue of the NFx motif, corresponds to Phe-73 which is also pivotal in coordinating the UraA substrate, through both hydrogen bonding (involving the amide nitrogen of the main chain at Phe-73) and van der Waals interactions (1). Asn-93, which corresponds to a non-bonding Ser residue in UraA, is at a strategic position in the XanQ model due to its closer proximity with the essential Glu-272 (TM8). Ser-95, the third residue of the NFx motif, is distant from the presumed binding pocket but oriented on the same face as Asn-93 (Fig. 8A). In the light of the above arrangement, our data on the importance of Phe-94 and Asn-93 for the proper recognition of the pyrimidine and the imidazole moiety of substrate, respectively (Table 2), might reflect direct interactions of these residues with xanthine and/or xanthine-coordinating residues at the binding site. Molecular dynamics simulations of the interaction of XanQ and relevant mutants with xanthine or analogues will be needed to draw more specific conclusions; such studies have been initiated in our laboratory.4
FIGURE 8.
Arrangement of TM1, TM3, TM8, and TM10 in the modeled structures of XanQ and UapA permeases. The sequences of XanQ and UapA were threaded on the known x-ray structure of UraA (1) using the SWISSPROT modeling server. The structural models were displayed with PyMOL v1.4. For clarity, only parts of TM1, TM3, TM8, and TM10 are shown and the β-strand regions of TM3 and TM10 have been excluded from the figure (see supplemental Fig. S1). The side chains of functionally important residues of XanQ (A) and corresponding residues of UapA (B) are shown as thick sticks. Note that only one such side chain differs between XanQ (N93) and UapA (S154) (shown with red label).
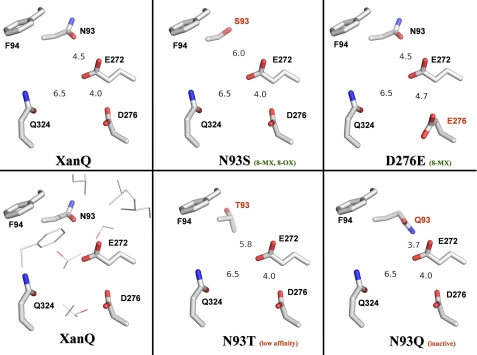
The apparently unique role of Asn-93 in defining the specificity of XanQ for xanthine (Table 2) and its counter-selectivity against uric acid (supplemental Fig. S2) is corroborated by both the structural evidence (Fig. 8) and the distinct conservation pattern of this residue in NAT transporters (Fig. 2). In general, Asn-93 is poorly conserved as an amidic side chain even in close XanQ relatives (supplemental Table S3) but its polar character is conserved invariably in the known nucleobase-transporting members (Asn, Thr, or Ser) while the ascorbate-transporting SVCTs have an Ala at this position (Fig. 2). Furthermore, all the dual-selectivity uric-acid/xanthine transporters (Xut1, UapA, UapC, AfUapA, Lpe1) have a Ser at the position corresponding to Asn-93 in XanQ (Fig. 2). To understand the structural relevance of this difference, we have built structural models for the above homologs and compared them with the one of XanQ and selected mutants (Fig. 8 and data not shown). In all dual-selectivity NATs, the Ser replacing Asn-93 is distal from the conserved, substrate-relevant glutamate of TM8 (minimal distance between oxygen atoms, 6.0 Å) while Asn-93 in XanQ is significantly closer (distance between oxygen atoms, 4.5 Å). This difference is most prominent in the model of UapA which perfectly conserves all the other side chains of functionally important residues of TM1, TM3, TM8, or TM10, except Asn-93 (Fig. 8B). In this homolog, Ser-154 (the UapA residue corresponding to Asn-93) is oriented away from the carboxyl group of Glu-356 (corresponding to Glu-272 of TM8) and leaves more space between TM3 and TM8 in the substrate binding pocket (Fig. 8, A and B). Thus, occupation of the Asn-93 position by Ser-154 (TM3) may relax a constraint for the recognition of analogues modified at position 8 of the imidazole moiety of xanthine and allow binding and transport of uric acid (8-oxy-xanthine), which modifies the NAT selectivity toward a less stringent, dual-substrate profile. Accordingly, the XanQ mutants replacing Asn-93 with Ser (or Ala) yield efficient recognition of 8-methylxanthine and low but significant uptake of [14C]uric acid, mimicking in part the fungal, dual-selectivity NATs (20). On the other hand, replacement of Asn-93 with Thr in mutant N93T leads to low affinity for all xanthine analogues (Table 2), possibly due to interference of the methyl group of Thr-93 in the vicinity of the essential Glu-272 (Fig. 9). An even more dramatic effect has been observed with mutant N93Q which is completely inactive (7); in this case, the amide of Gln-93 is at bonding distance from Glu-272 (Fig. 9) and may disrupt essential hydrogen-bonding interactions of this residue in the binding pocket.
FIGURE 9.
Spatial relationship between Asn-93, Glu-272, and Asp-276 in XanQ permease. The figure is based on models of XanQ and selected mutants built on the structure of the UraA homolog (see Fig. 8). The modeled side chains of Asn/Ser/Thr/Gln-93 (TM3), Phe-94 (TM3), Glu-272 (TM8), Asp/Glu-276 (TM8), and Gln-324 (TM10) are displayed with PyMOL v1.4. Numbers represent minimal distances in Å between N93-E272 (oxygen-oxygen, or carbon-oxygen in N93T, or nitrogen-oxygen in N93Q), D276-E272 (oxygen-oxygen), and Q324-E272 (oxygen-oxygen). Other side chains vicinal to E272 (at distances 4.0–6.3 Å from the carboxyl oxygen atoms) are shown without labels in the bottom left panel; these are I158, L162 (TM5), L268, S269 (TM8), T320, T321, A323 (TM10), and F376 (TM12). Characteristic properties of each mutant are indicated in parenthesis (see discussion). The abbreviations are: 8-MX, high-affinity recognition of 8-methylxanthine; 8–0X, transport of uric acid (8-oxy-xanthine).
Mutants N93A and N93S are unique not only because they can bind and transport uric acid (supplemental Fig. S2), but also because they recognize 8-methylxanthine with high affinity (Ki values, 32 μm and 48 μm, respectively). Wild-type XanQ does not recognize 8-methylxanthine as a ligand (4) and our extensive site-directed mutagenesis study (Fig. 1) has revealed only one additional mutant, namely D276E (TM8), which enables binding of 8-methylxanthine with high affinity (Ki, 79 μm) (9). Interestingly, Asp-276, where presence of a carboxyl group is needed for decent activity (9), is also close to the essential Glu-272 which lies one α-helical turn upstream (Fig. 8). Structural modeling shows that, similarly to what happens with N93S, the D276E replacement shifts the polar group of this position away from the carboxyl group of Glu-272 (Fig. 9). Thus, in both cases, the high affinity for 8-methylxanthine may derive from relaxation of a constraint for the accommodation of the 8-methyl group in the binding pocket which involves interactions of the essential Glu-272. However, the specificity profile of D276E differs from the one of N93S (or N93A) in that the D276E replacement leads, additionally, to abrogation of the recognition of analogues at the pyrimidine moiety of substrate (9), implying that Asp-276 has a more stringent role. In the solved structure of the uracil-transporting homolog UraA, Glu-272 (invariant in NAT family) corresponds to Glu-241 which directly hydrogen-bonds with substrate and Asp-276 (conserved partially in NAT family) corresponds to His-245 which forms a water-mediated hydrogen bond with uracil and has been suggested to be essential for the proton symport (1).
The role of TM3 had not been addressed with mutagenesis in any other NAT transporter until recently, when polar and conserved residues of TM3 were subjected to Ala-scanning mutagenesis in the fungal uric-acid/xanthine transporter UapA (14). Of eight residues mutagenized in the latter study (Ser-145, Ser-149, Ser-154, Phe-155, Ser-156, Ser-159, Ser-162; see Fig. 2), only Ser-154 (corresponding to Asn-93 in XanQ) was found to be critical for activity, since its replacement with Ala, or even Asn, resulted in low-activity mutants (14). The specificity profiles of S154A and S154N were found to be different from wild-type UapA with most prominent differences being a 5-fold higher affinity for xanthine observed with S154N, a 7-fold lower affinity for uric acid observed with S154A, and a 3-fold lower affinity for 9-methylxanthine observed with both mutants (14). These changes are rather minor compared with the clear-cut specificity effects seen with XanQ mutants N93S and N93A (Table 2 and supplemental Fig. S2). However, the UapA results are similar in the sense that all specificity-related effects of S154A and S154N refer to analogues deviating at the imidazole moiety of substrate and to the distinction between xanthine and uric acid. For example, the mirror image replacement S154N which introduces the side chain found in XanQ renders UapA more efficient to bind xanthine and shifts the dual-selectivity profile to the xanthine-selective direction (Km or Ki values, 8 μm for uric acid versus 1.5 μm for xanthine) (14).
The GxGLx motif at the cytoplasmic end of TM3 (Figs. 1 and 2) is far from the presumed binding pocket (supplemental Fig. S1). This motif contains residues that are critical for the structural integrity and expression of the transporter in the membrane (Gly-83, Gly-85). The conformational flexibility of Gly residues (especially of the irreplaceable Gly-83; Fig. 4) may be needed to control insertion and/or stability of the protein product through dictating architecture of the crucial TM3 segment. Similarly, Pro-318 at the beginning of TM10 (Fig. 1) has been delineated as essential for the proper expression of XanQ in the membrane (5). It appears that the unique folds of TM3 and TM10, which contain unwound intramembrane regions stabilized by a multitude of buried hydrogen bonds within the core NAT domain (1) are highly sensitive to distortion by abrogating flexibility or rigidity at key positions (Gly-83 or Pro-318, respectively) which, in turn, would disrupt the overall structure. In addition, the GxGLx motif of XanQ permease contains three residue positions where a Cys replacement is highly sensitive to inactivation by NEM (S84C, L86C, L87C). Sensitivity of L86C, in particular (IC50,25 μm), is one of the highest observed for a single-Cys XanQ, comparable only with the ones of G275C (TM8), A323C, G327C, T332C, G333C (TM10) and N430C (TM14), which are in the IC50 range of 10–35 μm (6–9). The alkylation sensitivity of the GxGLx-motif mutants may reflect important conformational interactions of the cytoplasmic part of TM3 with other regions of XanQ that are yet to be defined. Such interactions, involving alkylation-sensitive positions at the periphery of the binding pocket, can be crucial for implementing the appropriate conformational changes in the alternating access mechanism, as seen with other secondary active transporters (21–23).
Supplementary Material
Acknowledgment
We thank Dr. George Papamokos for help in preparing supplemental Fig. S1.
This report is part of a research project co-funded by the European Union, European Social Fund (ESF) and National Sources, in the framework of program “Hraklitos II” through the Operational Program “Education and Lifelong Learning” of the Hellenic Ministry of Education, Lifelong Learning and Religious Affairs, in the context of the National Strategic Reference Framework (NSRF).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1–S3.
K. Papakostas and S. Frillingos, manuscript in preparation.
G. Papamokos and S. Frillingos, unpublished information.
- NAT
- nucleobase-ascorbate transporter
- TM
- transmembrane segment
- NEM
- N-ethylmaleimide
- HRP
- horseradish peroxidase
- IPTG
- isopropyl 1-thio-β,d-galactopyranoside
- BAD
- biotin-acceptor domain
- Cys-less (C-less)
- permease devoid of native Cys residues
- PMS
- phenazine methosulfate.
REFERENCES
- 1. Lu F., Li S., Jiang Y., Jiang J., Fan H., Lu G., Deng D., Dang S., Zhang X., Wang J., Yan N. (2011) Nature 472, 243–246 [DOI] [PubMed] [Google Scholar]
- 2. Yamamoto S., Inoue K., Murata T., Kamigaso S., Yasujima T., Maeda J. Y., Yoshida Y., Ohta K. Y., Yuasa H. (2010) J. Biol. Chem. 285, 6522–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsukaguchi H., Tokui T., Mackenzie B., Berger U. V., Chen X. Z., Wang Y., Brubaker R. F., Hediger M. A. (1999) Nature 399, 70–75 [DOI] [PubMed] [Google Scholar]
- 4. Karatza P., Frillingos S. (2005) Mol. Membr. Biol. 22, 251–261 [DOI] [PubMed] [Google Scholar]
- 5. Karatza P., Panos P., Georgopoulou E., Frillingos S. (2006) J. Biol. Chem. 281, 39881–39890 [DOI] [PubMed] [Google Scholar]
- 6. Papakostas K., Georgopoulou E., Frillingos S. (2008) J. Biol. Chem. 283, 13666–13678 [DOI] [PubMed] [Google Scholar]
- 7. Karena E., Frillingos S. (2009) J. Biol. Chem. 284, 24257–24268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Georgopoulou E., Mermelekas G., Karena E., Frillingos S. (2010) J. Biol. Chem. 285, 19422–19433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mermelekas G., Georgopoulou E., Kallis A., Botou M., Vlantos V., Frillingos S. (2010) J. Biol. Chem. 285, 35011–35020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diallinas G., Valdez J., Sophianopoulou V., Rosa A., Scazzocchio C. (1998) EMBO J. 17, 3827–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koukaki M., Vlanti A., Goudela S., Pantazopoulou A., Gioule H., Tournaviti S., Diallinas G. (2005) J. Mol. Biol. 350, 499–513 [DOI] [PubMed] [Google Scholar]
- 12. Papageorgiou I., Gournas C., Vlanti A., Amillis S., Pantazopoulou A., Diallinas G. (2008) J. Mol. Biol. 382, 1121–1135 [DOI] [PubMed] [Google Scholar]
- 13. Kosti V., Papageorgiou I., Diallinas G. (2010) J. Mol. Biol. 397, 1132–1143 [DOI] [PubMed] [Google Scholar]
- 14. Amillis S., Kosti V., Pantazopoulou A., Mikros E., Diallinas G. (2011) J. Mol. Biol. 411, 567–580 [DOI] [PubMed] [Google Scholar]
- 15. Inoue H., Nojima H., Okayama H. (1990) Gene 96, 23–28 [DOI] [PubMed] [Google Scholar]
- 16. Teather R. M., Bramhill J., Riede I., Wright J. K., Fürst M., Aichele G., Wilhelm V., Overath P. (1980) Eur. J. Biochem. 108, 223–231 [DOI] [PubMed] [Google Scholar]
- 17. Granseth E., Daley D. O., Rapp M., Melén K., von Heijne G. (2005) J. Mol. Biol. 352, 489–494 [DOI] [PubMed] [Google Scholar]
- 18. Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graphics 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 20. Goudela S., Karatza P., Koukaki M., Frillingos S., Diallinas G. (2005) Mol. Membr. Biol. 22, 263–275 [DOI] [PubMed] [Google Scholar]
- 21. Kaback H. R., Dunten R., Frillingos S., Venkatesan P., Kwaw I., Zhang W., Ermolova N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 491–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tavoulari S., Frillingos S. (2008) J. Mol. Biol. 376, 681–693 [DOI] [PubMed] [Google Scholar]
- 23. Jiang X., Nie Y., Kaback H. R. (2011) Biochemistry 50, 1634-1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.