Background: Cytoplasmic dynein performs a great variety of cellular functions using a diversity of regulators.
Results: NudE and dynactin compete for a common site within the dynein complex.
Conclusion: This mechanism prevents dual regulation by dynactin and LIS1 and suggests a major new mode of regulatory control.
Significance: This is the first insight into coordination of cytoplasmic dynein regulators.
Keywords: Dynein, Microtubules, Molecular Cell Biology, Molecular Motors, Protein-Protein Interactions, Dynactin, Lis1
Abstract
Cytoplasmic dynein is responsible for a wide range of cellular roles. How this single motor protein performs so many functions has remained a major outstanding question for many years. Part of the answer is thought to lie in the diversity of dynein regulators, but how the effects of these factors are coordinated in vivo remains unexplored. We previously found NudE to bind dynein through its light chain 8 (LC8) and intermediate chain (IC) subunits (1), the latter of which also mediates the dynein-dynactin interaction (2). We report here that NudE and dynactin bind to a common region within the IC, and compete for this site. We find LC8 to bind to a novel sequence within NudE, without detectably affecting the dynein-NudE interaction. We further find that commonly used dynein inhibitory reagents have broad effects on the interaction of dynein with its regulatory factors. Together these results reveal an unanticipated mechanism for preventing dual regulation of individual dynein molecules, and identify the IC as a nexus for regulatory interactions within the dynein complex.
Introduction
Cytoplasmic dynein is a 1.2 MDa protein complex that functions as the predominant microtubule minus end-directed molecular motor in most cell types. It is involved in a very wide range of cellular roles, but the underlying basis for its great functional diversity is poorly understood. A variety of regulatory factors appear to tailor the motor protein for specific cellular roles. These factors are responsible for recruitment of dynein to appropriate sites within the cell, proper temporal activation of motor activity, and modulation of mechanochemical behavior to accommodate different cellular tasks.
The most well-studied dynein regulatory factors are dynactin (3–7) and the LIS1-NudE/L complex (8–15). Each of these systems is involved both in dynein cargo recruitment and mechanochemical regulation, though via different mechanisms. Dynactin is itself a ∼1 MDa multi-subunit complex. It was initially found to be required for dynein vesicular transport in vitro (4, 16), and to recruit the motor to mitotic kinetochores, and vesicular organelles (5, 17). Dynactin has also been found to increase dynein processivity by up to 2-fold in single molecule in vitro assays (7, 18, 19). The mechanism responsible for this effect is incompletely understood. Processivity of mammalian dynein is stimulated in both the plus- and minus-end directions along microtubules (20, 21), though yeast dynein with or without dynactin is primarily unidirectional (19). Although the microtubule binding CAP-Gly domain of the dynactin p150Glued subunit had been assumed to contribute to the enhancement of dynein processivity, recent studies showed no effect after its removal. Nonetheless, it was still required for complete dynactin function in vivo (6, 19, 22, 23).
LIS1 and its binding partners NudE and NudEL form a tripartite complex with dynein (15). LIS1 and NudE/L play critical roles in a subset of dynein functions, many of which appear to involve high-load dynein mediated transport. LIS1 is required for nuclear migration in neural progenitors and post mitotic neurons in vertebrates, and for nucleokinesis in several organisms (24–27). LIS1 and its interactors have also been implicated in translocation or reorientation of the entire microtubule cytoskeleton during mitosis and cell migration, as well as in centrosome and kinetochore dynamics, (1, 25, 28–32). The range of cellular functions involving LIS1 and NudE/L and their extent of overlap with dynactin-requiring functions remains incompletely resolved. Aspects of vesicular transport that involve dynactin were found not to require LIS1 (32, 33), though general (34–38) or conditional (39) roles for LIS1, NudE, and NudEL have been reported in other studies.
NudE and NudEL have been implicated in recruiting cytoplasmic dynein to cargo (1, 30, 40–42) as well as in recruiting LIS1 to dynein (15). We recently identified effects of LIS1 and NudE/L on dynein motor activity, and found them to be complex and distinct from those reported for dynactin (15). LIS1 stabilized the dynein-MT interaction during the transition state of the cross-bridge cycle, resulting in persistent force production under load. NudE alone inhibited the dynein-MT interaction. Strikingly, the tripartite complex of LIS1, NudE, and dynein transformed the motor to a persistent force-producing state and enhanced multiple motor transport under load (15). This behavior is likely to be important in cellular scenarios requiring dynein to produce force against large opposing loads, such as nuclear migration (25).
Dynactin, NudE, and NudEL each interact with the tail region of the dynein complex. Dynactin binds via the central region of its p150Glued subunit to the N terminus of the dynein intermediate chain (IC)3 (2, 43, 44). NudE and NudEL have been found to bind to both the dynein IC and LC8 subunits (1, 15). NudE and NudEL were initially reported to contain a C-terminal dynein-interaction site (12), but a separate N-terminal site has also recently been reported as well (45, 46).
The current study was initiated to define the nature of the NudE-dynein interaction in greater detail. We find the primary binding site for NudE to lie within the dynein IC N terminus, the same region implicated in dynactin binding (2, 43). We observe clear competition between NudE and dynactin for dynein, identifying a novel mechanism for coordinating dynein regulators. The common interaction site is also a target for frequently used inhibitory probes, and our results, therefore, have important implications for phenotypic analysis of dynein function in vivo.
EXPERIMENTAL PROCEDURES
DNA Cloning and Protein Purification
Full-length mouse NudE was cloned into pGEX6P-1 (GE Biosciences) with N-terminal HA- and C-terminal His tags. NudE fragments were also cloned into this vector. p150Glued fragments were cloned from a full-length rat construct into pGEX6P-1 with an N-terminal FLAG-tag and human LC8 (accession number NM_003746) was also cloned into this vector. Dynein IC fragments from rat were also cloned into pGEX6P-1 with a Myc tag at the C terminus, or into pCDNA 3.1 (IC2C 1–260 and 123–280) or pEGFP (IC2C 1–100) for mammalian cell expression. For expression in bacteria, constructs were transformed into BL21-CodonPlus RIPL competent cells (Agilent Technologies) and expressed in Luria broth or Terrific broth. Protein production was induced by addition of 0.1–0.5 mm IPTG, and the culture was moved to 18 °C overnight. Bacteria were broken by sonication, and proteins were purified by batch incubation of a high speed supernatant with glutathione resin (GE Biosciences) in lysis buffer (PBS, 1 mm DTT, protease inhibitor mixture (Sigma), 1% Triton X-100) for 1–2 h at 4 °C. The beads were collected and washed extensively with lysis buffer in a column. The beads were then washed into PMEG buffer (100 mm PIPES, 5 mm EGTA, 4 mm MgCl, 0.1 mm EDTA, 0.9 m glycerol, 1 mm DTT, pH 7.0) for freezing, or washed into PreScission protease cleavage buffer (50 mm Tris-HCl pH 7.0, 150 mm NaCl, 1 mm EDTA), and incubated with PreScission protease (GE Biosciences) overnight to cleave off the GST moiety according to the manufacturer's instructions. The cleaved proteins were collected, concentrated using Amicon concentrators (Amicon) and flash frozen in liquid nitrogen for storage. For full-length NudE, the protein was then incubated with Talon resin for 1 h at 4 °C, the beads were washed extensively and eluted in PMEG containing 350 mm imidazole. Protein containing fractions were pooled, concentrated, and flash frozen in liquid nitrogen. Cytoplasmic dynein was purified from rat brain tissue as described (47) and frozen in liquid nitrogen. This dynein preparation is essentially free of dynactin (15). Baculovirus expressed LIS1 was purified as described (15).
Protein Biochemistry
Protein interaction experiments were performed in buffer A: 50 mm HEPES (pH 7.4), 150 mm NaCl, 2 mm DTT, 0.2 mg/ml BSA, 0.1% Nonidet P-40. For GST pull-downs, proteins were bound to glutathione-agarose and incubated with interactors in 350 μl total volume for 1 h at 4 °C. For HA or FLAG pull-downs, tagged proteins were first incubated with an equal molar amount of monoclonal anti-HA or anti-FLAG antibodies for 1 h on ice. The protein-antibody complexes were then bound to protein A beads (Invitrogen) for 1 h at 4 °C, washed twice to remove unbound protein, and then used for pull-downs as above. Beads were washed four times with 350 μl of buffer A before being processed for SDS-PAGE analysis. For pull-downs from brain lysate, a high speed supernatant (47) of rat brain was used. In competition experiments, beads coated with dynein interactors were incubated with ∼4 nm purified brain dynein for 1 h at 4 °C, followed by three buffer washes of 350 μl each. The beads were then resuspended to volume, and the indicated amount of competitor protein was added for an additional hour, followed by four 350 μl washes. Beads were resuspended in 50 μl and processed for gel analysis. Western blots were processed on an Odyssey IR scanner (LI-COR Biosciences). Densitometry was performed using ImageJ software (NIH).
Antibodies
Antibodies used in this study were: monoclonal anti-dynein intermediate chain clone 74.1 (a gift from Dr. Kevin Pfister), or clone 70.1 (Sigma). Monoclonal and polyclonal anti-FLAG, anti-HA, anti-Myc, anti-GFP, and anti-LC8 (Abcam) and monoclonal anti-LIS1 (Sigma). Polyclonal anti-p150Glued (D'art) (48) and polyclonal anti-NudE/NudEL (1).
Pepscan
A membrane array spotted with overlapping dodecapeptides was generated based on mouse NudE (GenBankTM accession number Q9CZA6) C-terminal residues 192–344 (JPT Peptide Techonologies, Berlin). Each spot contains ∼5 nmol of a 12-amino acid long peptide that is covalently linked to a cellulose-β-alanine membrane. The sequence of peptides in adjacent spots are shifted C-terminally by two residues such that two neighboring spots overlap by ten residues (Fig. 2B). Before use, the membrane was reconstituted at room temperature in methanol for 5 min, followed by three 10-min washes with TBS. Blocking was performed for 1 h with 5% milk in TBS-T followed by a 1-h incubation simultaneously with primary monoclonal LC8 antibody at a dilution of 1:2500 and secondary anti-rabbit Alexa Fluor 680 (Invitrogen, A10043) at a dilution of 1:10,000 in 5% milk in TBS-T at room temperature. The membrane was then scanned using Odyssey Imaging System (LI-COR) to assess nonspecific interactions of the antibodies. 200 nm of recombinant purified human LC8 in 5% milk in TBS-T was incubated with the membrane overnight at 4 °C and for 1 h at room temperature the following day, followed by three 10-min washes with TBS-T, sequential probing with primary and secondary antibodies, and scanning as before. Scans of the membrane before and after incubation with LC8 were compared to identify the residues in the C terminus of NudE that are involved in binding LC8.
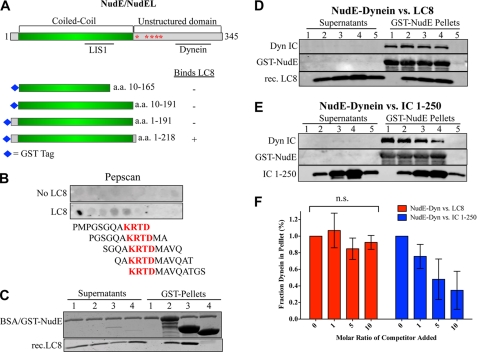
FIGURE 2.
Characterization of LC8-NudE interaction. A, diagram of NudE/NudEL showing structural and functional domains, and fragments used for pull-downs. B, Pepscan array of overlapping dodecapeptides covering NudE amino acids 192–344 incubated with purified LC8, followed by primary anti-LC8 and secondary antibodies. Sequence common to positive peptides is highlighted in red. C, CBB-stained gel of LC8 pull-downs using GST-NudE and its fragments. Lane 1, beads alone; lane 2, GST-NudE; lane 3, GST-NudE1–218; lane 4, GST-NudE1–191. D, immunoblot of purified dynein pulled down with GST-NudE followed by incubation with excess recombinant LC8. Lanes 1–4, 0×, 1×, 5×, or 10× molar ratio (over NudE) of LC8; lane 5, beads alone. E, same as in D using excess recombinant IC 1–250-myc instead of LC8. F, quantification of D and E. The dynein IC signal in lanes 2–4 was normalized to the IC signal in lane 1. Error bars represent S.D. (n = 3 experiments), n.s., not statistically significant.
Inhibition of CC1/NudE-Dynein Interaction with Antibodies
Monoclonal anti-dynein intermediate chain antibodies 74.1 and 70.1 were incubated with rat brain purified cytoplasmic dynein at ∼10-fold molar excess for 60 min at 4 °C with gentle rotating. Antibody-dynein complexes or dynein alone were subsequently incubated with bacterially expressed GST-CC1 or GST-NudE on glutathione beads or beads alone for 90 min at 4 °C with gentle rotating in buffer A. In a similar experiment, GST-CC1 or GST-NudE either alone or after preincubation with a polyclonal anti-NudE/L antibody for 60 min at 4 °C were incubated with rat brain purified cytoplasmic dynein for 90 min at 4 °C with gentle rotating in buffer A. After the incubation periods, unbound dynein was separated from the beads by centrifugation. The beads were washed three times with buffer (15-fold bead volume) and resuspended in protein sample buffer. Coomassie-stained gels were scanned using the Odyssey IR system.
RESULTS
NudE Binds to the Dynein Intermediate Chain N Terminus
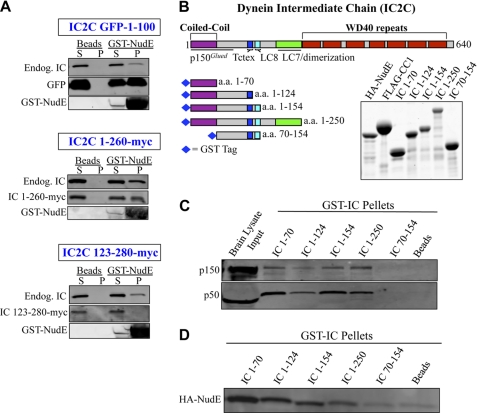
In a previous study we screened an array of dynein and dynactin subunits for NudE binding, and identified interactions with the dynein IC and LC8 subunits (1, 15). To gain further insight into the nature of these interactions we first determined where NudE bound within the dynein IC. This subunit consists of a short N-terminal α-helical coiled-coil, followed by binding sites for dynein's three LC classes, a dimerization domain, and finally by a WD40 domain responsible for dynein heavy chain (HC) binding (49, 50) (Fig. 1B). Expression of the WD40 domain was toxic in HeLa cells, as previously reported (43), but the N-terminal fragments expressed well. Pull-downs with GST-NudE localized IC binding activity to the first 100 a.a. of rat IC2C (Fig. 1A). GST-NudE also interacted with the endogenous dynein complex as evidenced by the presence of the full-length dynein IC in the pulldowns (1). To test for a direct NudE-IC interaction, we expressed a series of GST-tagged N-terminal IC constructs in Escherichia coli (Fig. 1B). All constructs that contained the N-terminal coiled-coil domain were capable of interacting with native dynactin complex in rat brain lysates (Fig. 1C) indicating the proteins were correctly folded. The IC constructs were screened for interactions with purified HA-NudE, which mapped NudE binding to IC amino acids 1–70 (Fig. 1, B–D). NudE bound to each of six alternatively spliced variants of rat IC1 and IC2 (supplemental Fig. S1B), which all contain a common N-terminal predicted coiled-coil sequence, but diverge immediately downstream (2, 49) (supplemental Fig. S1A), arguing that IC isoform composition is unlikely to affect the IC-NudE interaction.
FIGURE 1.
Binding site for NudE on the dynein IC. A, GST NudE pull-downs of rat dynein IC2C fragments expressed in HeLa cells, immunoblotted with antibodies to GFP and Myc tags, and anti-IC antibody (74.1) to indicate endogenous dynein, labeled endog-IC. S, supernatant; P, pellet. B, diagram of the dynein IC2C polypeptide and GST-IC fragments used in this study, and a Coomassie Brilliant Blue (CBB)-stained electrophoretic gel of the purified bacterially expressed protein fragments. C, GST-IC pull-downs of dynactin from rat brain cytosol. Retention of native dynactin complex was assayed using antibodies against both p150Glued and p50/dynamitin subunits. D, GST-IC pull-downs of purified bacterially expressed HA-NudE.
Dynein LC8 Binds Directly to NudE
Although LC8 interacts with both NudE and dynein, the specific role of the LC in the NudE-dynein interaction is unknown. LC8 interacts with full-length NudE ((1); this study), but not with the truncation mutants NudE10–165, NudE10–191, and NudE1–191 (Fig. 2A and C; data not shown), consistent with binding of LC8 only to full-length, but not an N-terminal NudEL fragment (45). To map the site of LC8 binding more precisely we probed an array of overlapping dodecapeptides covering the C-terminal region of NudE (Fig. 2A, amino acids 192–344) using purified bacterially expressed LC8 (51, 52). We observed binding to 5 sequential peptides in the region amino acids 192–211, yielding a minimal binding sequence of amino acids 200–203 (KRTD) within NudE (Fig. 2B). In support of this result we found LC8 to bind NudE1–218, but not NudE1–191 (Fig. 2C). The LC8 binding sequence does not contain the canonical LC8 binding motif identified in many other LC8 interactors (53), but whether this indicates a unique mode of LC8 binding is uncertain (Refs. 54, 55). We also found the NudE-dynein interaction to be unaffected by exposure of NudE to LC8 before, during, or after dynein-NudE binding (Figs. 2, D and F, supplemental Figs. S2, A and B). These results contrasted with the ability of excess IC 1–250 to disrupt the dynein-NudE interaction (Fig. 2, E and F). These data reveal that LC8 can bind directly and apparently independently to the IC and NudE polypeptides. Furthermore, the data indicate that dynein interacts with NudE predominantly via the ICs, rather than LC8.
Overlap between Dynactin and NudE Binding Sites
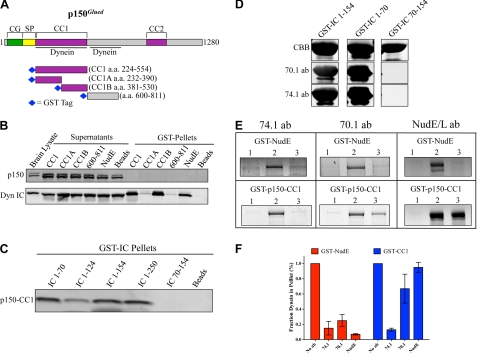
Dynactin binds to dynein through a direct p150Glued-IC interaction (2, 43, 44). The CC1 coiled-coil region (3, 43, 56) (Fig. 3A) of p150Glued and a downstream region (amino acids 600–811) (57) have each been implicated in dynein binding. To determine which region of p150Glued interacts with the native dynein complex, we performed pull-downs from rat brain lysate using a series of bacterially expressed GST-tagged p150Glued constructs (Fig. 3A). The CC1 region as well as a C-terminal subfragment, CC1B, each clearly pulled down brain cytoplasmic dynein, whereas an N-terminal CC1 subfragment, CC1A, as well as the p150Glued 600–811 fragment did not (Fig. 3B). None of the fragments pulled down dynactin, indicating that CC1 cannot exchange with full-length p150Glued in the dynactin complex. These results are consistent with previous work (19, 43, 56) and demonstrate that the CC1 region can bind to the native dynein complex in cytosol.
FIGURE 3.
NudE and p150Glued bind to a common site on the dynein IC. A, diagram of the p150Glued subunit of dynactin showing domain architecture and previously mapped dynein interaction sites. CG, CAP-Gly; SP, serine-proline rich region; CC1-CC2, coiled-coil domains. Right, CCB stained gel of p150Glued fragments used in this study. B, GST-p150Glued fragments were used in pull-downs from rat brain cytosol and dynein and dynactin dynactin binding was assayed by immunoblotting with anti-IC and p150Glued antibodies, respectively. Comparable levels of dynein were pulled down with GST-NudE. Note the endogenous dynactin complex was absent from both p150Glued and NudE pull-downs. C, GST-IC fragments (Fig. 1B) were assayed for their ability to interact directly with purified FLAG-CC1. D, GST-IC fragments were probed with the 74.1 or 70.1 monoclonal antibodies by Western blot. All three GST-IC fragments are visualized by CBB staining of the gel. E, purified dynein alone (lanes 2) or after preincubation with the indicated antibodies (lanes 3) was bound to beads containing either GST-NudE or GST-CC1. Additionally, GST-NudE or GST-CC1 were preincubated with a polyclonal NudE antibody before dynein was added (right). The amount of dynein remaining bound to the beads was assayed by CBB staining. Dynein heavy chain (∼530 kDa) was not retained on beads alone (lanes 1), but was retained by GST-NudE and GST-CC1 in the absence of antibodies (lanes 2). F, quantification of E. The amount of dynein remaining bound to the beads in the presence of the antibodies is plotted relative to control beads with no antibody added. Error bars represent S.D.
We then used CC1 to map its interaction site within the dynein ICs. CC1 binding specifically required the presence of the IC N-terminal 70 amino acids (Fig. 3C), as we observed for NudE (Fig. 1D). Furthermore, CC1 bound directly to each of the six IC splice variants (supplemental Fig. S1B). These results identify a common IC region for NudE and dynactin binding, and a common range of interacting IC isoforms. As a further test of this conclusion we utilized two well characterized and widely used function-blocking anti-IC monoclonal antibodies, 74.1 and 70.1 (58–60). Each antibody recognized IC2C amino acids 1–70 as judged by Western blotting (Fig. 3D), localizing the epitopes within this region. Purified rat brain cytoplasmic dynein was incubated with an excess of each antibody and then exposed to beads coated with GST-NudE or GST-CC1. Alternatively, the GST-NudE and GST-CC1 beads were pre-exposed to a polyclonal anti-NudE antibody (1), followed by addition of dynein. Both the NudE-dynein and CC1-dynein interactions were strongly inhibited by the 74.1 antibody, whereas 70.1 preferentially inhibited NudE-dynein binding (Fig. 3, E and F). As expected the NudE antibody specifically disrupted the binding of dynein to NudE, (1), but had no effect on the dynein-CC1 interaction. Similar results were obtained if the antibodies were incubated with a preformed dynein-CC1 or dynein-NudE complex (data not shown). These results reveal that dynactin and NudE bind to overlapping, but not identical sites within the conserved N-terminal coiled-coil domain of the IC.
Dynactin and NudE Compete for Binding to Dynein
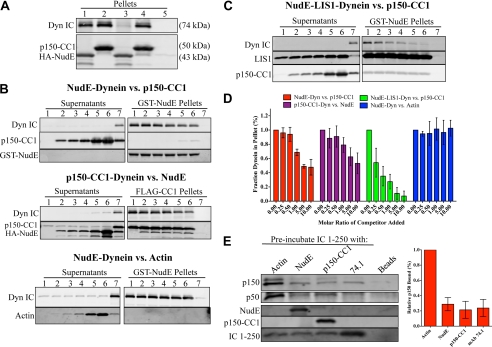
In view of these results we tested relative abilities of NudE and dynactin to bind dynein. When simultaneously mixed with equal concentrations of NudE and CC1, purified brain dynein was pulled down predominantly by the latter (Fig. 4A), indicating CC1 has a higher affinity for dynein than NudE in this assay. We also prebound dynein to GST-NudE or FLAG-CC1, and then tested for displacement of dynein by the reciprocal binding partner (Fig. 4B). FLAG-CC1 caused a clear concentration-dependent displacement of up to 53 ± 22% (n = 3 experiments) of the dynein bound to GST-NudE (Fig. 4, B and D), and a similar amount of dynein (47 ± 11%, n = 3 experiments) was displaced by NudE from FLAG-CC1 (Fig. 4, B and D). The added NudE and CC1 showed no evidence of specific binding to the beads, arguing against simultaneous binding of the two factors to dynein or an interaction between NudE and CC1. As further evidence for the specificity of this assay, purified actin had no effect on dynein binding to GST-NudE (Fig. 4, B and D). We also tested the ability of NudE to interfere with the interaction between dynein and the complete dynactin complex pulled down from rat brain lysate using beads coated with GST-IC 1–250 (Fig. 4E). Preincubation of the IC beads with HA-NudE largely abrogated their ability to pull-down dynactin (Fig. 4E), confirming that NudE competes with endogenous dynactin for binding to IC. Preincubation of the beads with FLAG-CC1 or the 74.1 anti-IC antibody also blocked the interaction with dynactin as expected, while preincubation with actin had no effect (Fig. 4E). Together, these data demonstrate that NudE and dynactin compete for a common binding site at the N terminus of the dynein IC.
FIGURE 4.
Dynactin and NudE compete for binding to the dynein IC. A, purified dynein was incubated with a 10-fold molar excess of either HA-NudE alone (lane 1), FLAG-CC1 alone (lane 2), or both proteins simultaneously (lanes 3 and 4) and then immunoprecipitated with either HA (lanes 1 and 3) or FLAG (lanes 2 and 4) antibodies. HA-NudE or FLAG-CC1 alone pull down similar amounts of dynein (lanes 1 and 2). However, when all three proteins are incubated simultaneously, only FLAG-CC1 is able to pull down dynein, indicating that FLAG-CC1 outcompetes HA-NudE for dynein. Dynein is not retained on beads alone (lane 5). B, dynein was prebound to GST-NudE or FLAG-CC1 on beads. The complexes were then exposed to increasing amounts of the competitor protein (0×, 0.25×, 0.5×, 1×, 5×, 10× molar ratio to protein on the beads in lanes 1–6, respectively). The amount of dynein remaining bound to the beads after a 1-h incubation is assayed by immunoblotting for the IC. The nonspecific protein actin was unable to compete dynein away from GST-NudE. No proteins were retained on beads alone (lanes 7). For quantification, see D. C, purified LIS1 protein was included in the competition assay between GST-NudE and FLAG-CC1. The LIS1 protein contains a FLAG tag, is approximately the same molecular weight as FLAG-CC1, and thus appears in the FLAG-CC1 blots. D, quantification of the competition effects shown in B and C (n 3 experiments, error bars represent S.D.). Values are set relative to the amount of dynein bound in the absence of competitor (lane 1). E, purified IC 1–250-myc was preincubated with the indicated reagents, followed by rat brain cytosol. The ability to interact with dynactin in the lysate was assayed by immunoblotting for the p150Glued and p50/dynamitin subunits. The p150Glued signals in the pellets, relative to the condition preblocked with actin, are quantified to the right (n = 3 experiments, error bars represent S.D.).
We previously proposed that NudE acts as a scaffold to mediate the interaction of LIS1 with dynein (1, 15). We therefore tested whether LIS1 affects the competition between NudE and dynactin for dynein. Purified dynein and LIS1 were bound to GST-NudE coated beads, followed by incubation with increasing amounts of FLAG-CC1. Unexpectedly, inclusion of LIS1 in the competition assay facilitated the release of dynein from the GST-NudE beads by FLAG-CC1 (Fig. 4, C and D).
DISCUSSION
How cytoplasmic dynein can contribute to an extremely diverse array of intracellular functions has remained a long-standing question. The discovery of dynein regulatory complexes has shed light on how the motor might be targeted to and otherwise adapted for various duties, but it is currently unclear how the regulatory complexes might work together to coordinate dynein function. Our finding that dynactin and NudE compete for binding to dynein provides the first insight into how the activities of distinct dynein regulators may be coordinated.
Our results also identify the IC as an important nexus for dynein regulation. In contrast we found the dynein LC8 subunit not to contribute to the NudE-dynein interaction, but, rather, that NudE interacts with LC8 independently of dynein. This is consistent with a growing body of literature supporting a dynein-independent role for LC8 (61).
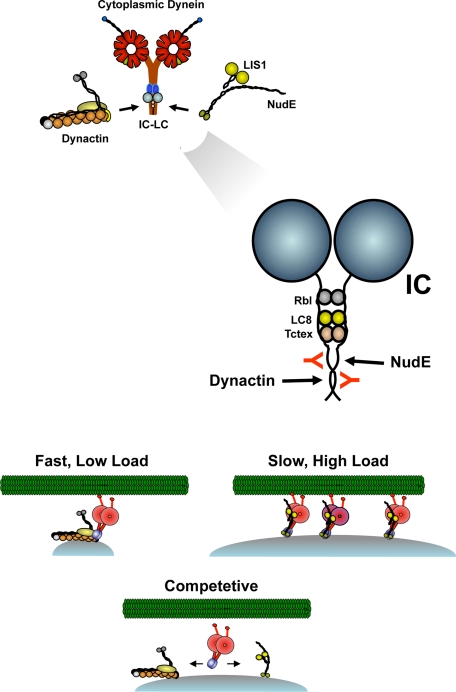
Our results indicate most dramatically that dynein molecules cannot be simultaneously occupied by dynactin and NudE. This arrangement is novel to the motor protein field, and further implies separate functional pools of dynein motors within the cell. Dynactin or NudE-LIS1 regulated dyneins appear to have distinct mechanochemical outputs (15, 18–20), and the mutually exclusive interactions of the regulators with the dynein ICs revealed in the current study strongly suggest that individual dynein molecules can be tailored to fit specific transport roles (Fig. 5). In view of its stimulatory effect on dynein processivity, dynactin might be specifically required for fast, long-range transport of smaller vesicular and macromolecular cargoes. Conversely, NudE-LIS1 prolongs the dynein force producing state, resulting in increased forces under multi-motor conditions (15). This mode of regulation seems certainly to be required for high load forms of dynein transport, such as nuclear migration (Fig. 5) (25, 26).
FIGURE 5.
Model for mutually exclusive binding of dynactin and NudE to dynein. Dynactin and NudE are shown at top to interact with the base of the dynein complex within and the coiled-coil N-terminal region of the ICs (expanded view). The 74.1 and 70.1 monoclonal antibodies (red) bind within the same region. Dynactin has been proposed to regulate dynein processivity for fast, low-load transport, whearas NudE-LIS1 promote multi-motor transport of high-loads. The competition for dynein is proposed here to prevent dual regulation of individual dynein complexes and, potentially, to provide a mechanism to allow dyneins to shift between high-load and long travel distance modes of transport.
Whether dynactin and NudE-LIS1 are segregated to different forms of cargo for different modes of transport remains to be seen. Equally interesting is the possibility that NudE-LIS1-dynein and dynactin-dynein coexist on common cellular cargoes, adapting transport to distinct subcellular environments (Fig. 5). Indeed, recent evidence suggests that teams of multiple dyneins move membranous cargoes in vivo (62–64). Finally, it is also possible that the balance of a dynein regulatory factors may be subject to regulation in vivo, an issue of considerable further interest. Indeed, phosphorylation of the dynein IC has been reported to affect the affinity of this subunit for dynactin (65), and might conceivably contribute to switching between dynein regulatory factors. Further experiments are needed to test this hypothesis.
Such a shift between regulatory modes may require new tools to assay properly. Dynactin and NudE/NudEL-LIS1 each control aspects of dynein recruitment to subcellular cargo, as well as dynein mechanochemical activity. This dual role will make it necessary to quantify relative effects on the number of dynein molecules associated with cargo versus the nature of dynein regulation. Changes in the affinity of dynein for dynactin relative to NudE and NudEL could alter the number of cargo-associated dyneins or shuttle dyneins between high-force and long travel distance regulators, or both.
Surprisingly, the addition of LIS1 caused dynein to be released from NudE more easily in the presence of FLAG-CC1 (Fig. 4, C and D). This result is unexpected given NudE established role in recruiting LIS1 to dynein (1, 15, 45, 46). These are the first results suggesting that LIS1 may affect the NudE-dynein interaction and suggest further complexity in the interaction between dynein and its regulators. Additional information on the structural nature of the various dynein complexes will be needed to clarify this issue.
Our study also reveals broader effects for commonly used dynein inhibitory probes than has been assumed. The dynactin CC1 fragment, as well as the 74.1 and 70.1 monoclonal antibodies, have been favored reagents for cytoplasmic dynein inhibition in vivo. Our data indicate that the first two of these should interfere with both dynactin and NudE-LIS1 binding, while the third interferes preferentially with NudE. However, some of the more readily assayed dynein functions require both types of regulatory factor. For this reason, physiological assays for the specificity of the dynein and dynactin inhibitory agents may require more quantitative in vivo assays for dynein behavior than are currently available (39). Although the effects of each reagent provide insight into dynein function, their implications for understanding dynein regulation now appear less clear. Further development of probes specific for cytoplasmic dynein and for its individual regulatory factors will be needed to address these issues.
Supplementary Material
Acknowledgments
We thank Drs. Kevin Vaughan and John Williams for contribution of reagents, sharing of unpublished results, and helpful discussion.
This work was supported by National Institutes of Health Grants GM47434 and HD40182 (to R. B. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- IC
- intermediate chain
- HC
- heavy chain
- LC
- light chain.
REFERENCES
- 1. Stehman S. A., Chen Y., McKenney R. J., Vallee R. B. (2007) J. Cell Biol. 178, 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vaughan K. T., Vallee R. B. (1995) J. Cell Biol. 131, 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schroer T. A. (2004) Annu. Rev. Cell Dev. Biol. 20, 759–779 [DOI] [PubMed] [Google Scholar]
- 4. Gill S. R., Schroer T. A., Szilak I., Steuer E. R., Sheetz M. P., Cleveland D. W. (1991) J. Cell Biol. 115, 1639–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Echeverri C. J., Paschal B. M., Vaughan K. T., Vallee R. B. (1996) J. Cell Biol. 132, 617–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim H., Ling S. C., Rogers G. C., Kural C., Selvin P. R., Rogers S. L., Gelfand V. I. (2007) J. Cell Biol. 176, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Culver-Hanlon T. L., Lex S. A., Stephens A. D., Quintyne N. J., King S. J. (2006) Nat. Cell Biol. 8, 264–270 [DOI] [PubMed] [Google Scholar]
- 8. Reiner O., Carrozzo R., Shen Y., Wehnert M., Faustinella F., Dobyns W. B., Caskey C. T., Ledbetter D. H. (1993) Nature 364, 717–721 [DOI] [PubMed] [Google Scholar]
- 9. Hirotsune S., Fleck M. W., Gambello M. J., Bix G. J., Chen A., Clark G. D., Ledbetter D. H., McBain C. J., Wynshaw-Boris A. (1998) Nat. Genetics 19, 333–339 [DOI] [PubMed] [Google Scholar]
- 10. Feng Y., Olson E. C., Stukenberg P. T., Flanagan L. A., Kirschner M. W., Walsh C. A. (2000) Neuron 28, 665–679 [DOI] [PubMed] [Google Scholar]
- 11. Niethammer M., Smith D. S., Ayala R., Peng J., Ko J., Lee M. S., Morabito M., Tsai L. H. (2000) Neuron 28, 697–711 [DOI] [PubMed] [Google Scholar]
- 12. Sasaki S., Shionoya A., Ishida M., Gambello M. J., Yingling J., Wynshaw-Boris A., Hirotsune S. (2000) Neuron 28, 681–696 [DOI] [PubMed] [Google Scholar]
- 13. Mesngon M. T., Tarricone C., Hebbar S., Guillotte A. M., Schmitt E. W., Lanier L., Musacchio A., King S. J., Smith D. S. (2006) J. Neurosci. 26, 2132–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hebbar S., Mesngon M. T., Guillotte A. M., Desai B., Ayala R., Smith D. S. (2008) J. Cell Biol. 182, 1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McKenney R. J., Vershinin M., Kunwar A., Vallee R. B., Gross S. P. (2010) Cell 141, 304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schroer T. A., Sheetz M. P. (1991) J. Cell Biol. 115, 1309–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roghi C., Allan V. J. (1999) J. Cell Sci. 112, 4673–4685 [DOI] [PubMed] [Google Scholar]
- 18. King S. J., Schroer T. A. (2000) Nat. Cell Biol. 2, 20–24 [DOI] [PubMed] [Google Scholar]
- 19. Kardon J. R., Reck-Peterson S. L., Vale R. D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ross J. L., Wallace K., Shuman H., Goldman Y. E., Holzbaur E. L. (2006) Nat. Cell Biol. 8, 562–570 [DOI] [PubMed] [Google Scholar]
- 21. Dixit R., Ross J. L., Goldman Y. E., Holzbaur E. L. (2008) Science 319, 1086–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vaughan P. S., Miura P., Henderson M., Byrne B., Vaughan K. T. (2002) J. Cell Biol. 158, 305–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moore J. K., Sept D., Cooper J. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5147–5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee W. L., Oberle J. R., Cooper J. A. (2003) J. Cell Biol. 160, 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsai J. W., Bremner K. H., Vallee R. B. (2007) Nat. Neurosci 10, 970–979 [DOI] [PubMed] [Google Scholar]
- 26. Tsai J. W., Chen Y., Kriegstein A. R., Vallee R. B. (2005) J. Cell Biol. 170, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiang X., Osmani A. H., Osmani S. A., Xin M., Morris N. R. (1995) Mol. Biol. Cell 6, 297–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grabham P. W., Seale G. E., Bennecib M., Goldberg D. J., Vallee R. B. (2007) J. Neurosci. 27, 5823–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang Y., Yu W., Li Y., Yu L., Zhang Q., Wang F., Yang Z., Du J., Huang Q., Yao X., Zhu X. (2007) Mol. Biol. Cell 18, 2656–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vergnolle M. A., Taylor S. S. (2007) Curr. Biol. 17, 1173–1179 [DOI] [PubMed] [Google Scholar]
- 31. Siller K. H., Doe C. Q. (2008) Dev. Biol. 319, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Faulkner N. E., Dujardin D. L., Tai C. Y., Vaughan K. T., O'Connell C. B., Wang Y., Vallee R. B. (2000) Nat. Cell Biol. 2, 784–791 [DOI] [PubMed] [Google Scholar]
- 33. Tai C. Y., Dujardin D. L., Faulkner N. E., Vallee R. B. (2002) J. Cell Biol. 156, 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith D. S., Niethammer M., Ayala R., Zhou Y., Gambello M. J., Wynshaw-Boris A., Tsai L. H. (2000) Nat. Cell Biol. 2, 767–775 [DOI] [PubMed] [Google Scholar]
- 35. Lam C., Vergnolle M. A., Thorpe L., Woodman P. G., Allan V. J. (2010) J. Cell Sci. 123, 202–212 [DOI] [PubMed] [Google Scholar]
- 36. Zhang J., Li S., Fischer R., Xiang X. (2003) Mol. Biol. Cell 14, 1479–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang J., Zhuang L., Lee Y., Abenza J. F., Peñalva M. A., Xiang X. (2010) J. Cell Sci. 123, 3596–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lenz J. H., Schuchardt I., Straube A., Steinberg G. (2006) The EMBO J. 25, 2275–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yi J., Ori-McKenney K. M., McKenney R. J., Vershinin M., Gross S. P., Vallee R. B. (2011) J. Cell Biol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guo J., Yang Z., Song W., Chen Q., Wang F., Zhang Q., Zhu X. (2006) Mol. Biol. Cell 17, 680–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liang Y., Yu W., Li Y., Yang Z., Yan X., Huang Q., Zhu X. (2004) J. Cell Biol. 164, 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bolhy S., Bouhlel I., Dultz E., Nayak T., Zuccolo M., Gatti X., Vallee R., Ellenberg J., Doye V. (2011) J. Cell Biol. 192, 855–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. King S. J., Brown C. L., Maier K. C., Quintyne N. J., Schroer T. A. (2003) Mol. Biol. Cell 14, 5089–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Karki S., Holzbaur E. L. (1995) J. Biol. Chem. 270, 28806–28811 [DOI] [PubMed] [Google Scholar]
- 45. Wang S., Zheng Y. (2011) J. Biol. Chem. 286, 587–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zylkiewicz E., Kijańska M., Choi W. C., Derewenda U., Derewenda Z. S., Stukenberg P. T. (2011) J. Cell Biol. 192, 433–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Paschal B. M., Shpetner H. S., Vallee R. B. (1991) Methods Enzymol. 196, 192–201 [DOI] [PubMed] [Google Scholar]
- 48. Vaughan K. T., Tynan S. H., Faulkner N. E., Echeverri C. J., Vallee R. B. (1999) J. Cell Sci. 112, 1437–1447 [DOI] [PubMed] [Google Scholar]
- 49. Myers K. R., Lo K. W., Lye R. J., Kogoy J. M., Soura V., Hafezparast M., Pfister K. K. (2007) J. Neurosci Res. 85, 2640–2647 [DOI] [PubMed] [Google Scholar]
- 50. Ma S., Trivinos-Lagos L., Collins C. A., Chisholm R. L. (1995) Mol. Biol. Cell 6, 155a [Google Scholar]
- 51. Rodríguez-Crespo I., Yélamos B., Roncal F., Albar J. P., Ortiz de Montellano P. R., Gavilanes F. (2001) FEBS Lett. 503, 135–141 [DOI] [PubMed] [Google Scholar]
- 52. Navarro-Lérida I., Martínez Moreno M., Roncal F., Gavilanes F., Albar J. P., Rodríguez-Crespo I. (2004) Proteomics 4, 339–346 [DOI] [PubMed] [Google Scholar]
- 53. Benison G., Karplus P. A., Barbar E. (2007) J. Mol. Biol. 371, 457–468 [DOI] [PubMed] [Google Scholar]
- 54. Lightcap C. M., Sun S., Lear J. D., Rodeck U., Polenova T., Williams J. C. (2008) J. Biol. Chem. 283, 27314–27324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lightcap C. M., Kari G., Arias-Romero L. E., Chernoff J., Rodeck U., Williams J. C. (2009) PLoS ONE 4, e6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Quintyne N. J., Gill S. R., Eckley D. M., Crego C. L., Compton D. A., Schroer T. A. (1999) J. Cell Biol. 147, 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Deacon S. W., Serpinskaya A. S., Vaughan P. S., Lopez Fanarraga M., Vernos I., Vaughan K. T., Gelfand V. I. (2003) J. Cell Biol. 160, 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Steffen W., Karki S., Vaughan K. T., Vallee R. B., Holzbaur E. L., Weiss D. G., Kuznetsov S. A. (1997) Mol. Biol. Cell 8, 2077–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Steffen W., Hodgkinson J. L., Wiche G. (1996) J. Struct. Biol. 117, 227–235 [DOI] [PubMed] [Google Scholar]
- 60. Burkhardt J. K., Echeverri C. J., Nilsson T., Vallee R. B. (1997) J. Cell Biol. 139, 469–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rapali P., Szenes A., Radnai L., Bakos A., Pal G., Nyitray L. (2011) Febs J. 278, 2980–2996 [DOI] [PubMed] [Google Scholar]
- 62. Hendricks A. G., Perlson E., Ross J. L., Schroeder H. W., 3rd, Tokito M., Holzbaur E. L. (2010) Curr. Biol. : CB 20, 697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Soppina V., Rai A. K., Ramaiya A. J., Barak P., Mallik R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19381–19386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shubeita G. T., Tran S. L., Xu J., Vershinin M., Cermelli S., Cotton S. L., Welte M. A., Gross S. P. (2008) Cell 135, 1098–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vaughan P. S., Leszyk J. D., Vaughan K. T. (2001) J. Biol. Chem. 276, 26171–26179 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.