Abstract
Type 1 diabetes is characterized by local inflammation (insulitis) in the pancreatic islets causing β-cell loss. The mitochondrial pathway of apoptosis is regulated by the balance and interaction between Bcl-2 members. Here we clarify the molecular mechanism of β-cell death triggered by the pro-inflammatory cytokines tumor necrosis factor (TNF)-α and interferon (IFN)-γ. The combination of TNF-α + IFN-γ induced DP5, p53 up-regulated modulator of apoptosis (PUMA), and Bim expression in human islets and rodent β-cells. DP5 and PUMA inactivation by RNA interference partially protected against TNF-α + IFN-γ-induced β-cell apoptosis. DP5 knock-out mice had increased β-cell area, and isolated islets from these mice were resistant to cytokine exposure. Bim expression was transcriptionally regulated by STAT1, and its activation triggered cleavage of caspases. Silencing of Bim protected rodent and human β-cells to a large extent against TNF-α + IFN-γ, indicating a major role of this BH3-only activator protein in the mechanism of apoptosis. Our data support a highly regulated and context-dependent modulation of specific Bcl-2 members controlling the mitochondrial pathway of β-cell apoptosis during insulitis.
Keywords: Cell Death, Cytokine, Diabetes, Mitochondrial Apoptosis, STAT Transcription Factor, Bcl-2 Proteins, Beta Cells, Type 1 Diabetes
Introduction
Immune-mediated pancreatic β-cell loss, mainly due to apoptosis, is a hallmark of type 1 diabetes (T1D)5 (1, 2). In the early stages of the disease, infiltrating macrophages and T-cells release pro-inflammatory cytokines such as interleukin (IL)-1β, tumor necrosis factor (TNF)-α, and interferon (IFN)-γ, which, together with cell-to-cell death effectors (granzyme B, FasL, etc.), contribute to the induction of β-cell apoptosis and the buildup of insulitis (1–4). In vitro studies demonstrated that the combination of different pro-inflammatory cytokines, but not each of them alone, activates β-cell apoptosis (1). It is conceivable that the cytokine combination and distribution in the vicinity of the β-cells vary during T1D development (3). The individual genetic background, immune assault timing, and degree of islet infiltration may also affect cytokine composition during insulitis. Therefore, a clear understanding of the apoptotic β-cell pathways activated downstream of different cytokine combinations is needed to individualize therapies aiming to prevent β-cell destruction in T1D.
Apoptosis was originally described as a physiological mechanism of cell death that allows cell turnover and tissue reorganization, but later evidence indicated that it is also an important mechanism of cell demise during viral infection and autoimmune diseases (5, 6). Different protein modulators, effectors, and pathways regulate the decision to undergo apoptotic cell death (6). Apoptosis can be activated by two major mechanisms: the extrinsic and intrinsic pathways (6). The extrinsic pathway is characterized by engagement of death receptors and caspase-8 cleavage/activation. In the second mechanism (intrinsic), the mitochondria play a key role in the triggering of cell death. Transcriptional and post-transcriptional modulation and protein-protein interaction of Bcl-2 members determine the cell outcome in this pathway (7–9). After an apoptotic stimulus, the sensitizer Bcl-2 proteins (DP5, Bik, Bad, and/or Noxa) are transcriptionally or post-transcriptionally activated and interact through their Bcl-2 homology 3 (BH3) domain with the anti-apoptotic Bcl-2 members (Bcl-2, Bcl-XL, Bcl-W, and A1). This interaction releases BH3-only activator proteins (Bid, Bim, and/or PUMA) that directly bind and induce conformational changes in the multichannel pro-death proteins Bax and Bak (9–11). Activated Bax translocates from the cytosol to the mitochondria and together with Bak forms pores in the mitochondrial membrane, releasing pro-apoptotic proteins such as cytochrome c to the cytoplasm (10, 12) and/or apoptosis-inducing factor (AIF) to the nucleus (13). Once in the cytoplasm, cytochrome c interacts with apoptotic protease activating factor to form the apoptosome, leading to pro-caspase cleavage and activation and subsequent cell death (14).
We have recently shown that the pro-inflammatory cytokines IL-1β + IFN-γ induce the BH3-only sensitizer DP5 and the BH3-only activator PUMA, leading to β-cell death (9, 15, 16). Less is known about the pathway of apoptosis and Bcl-2 proteins modulated by TNF-α + IFN-γ. It was previously described that mouse islets lacking the transcription factor STAT1 are resistant to TNF-α + IFN-γ-induced apoptosis (17), but the downstream molecular effector(s) remain(s) unknown. Against this background, we presently performed an extensive study using human islets, DP5 knock-out mice, rat fluorescence-activated cell sorting (FACS)-purified primary β-cells, and INS-1E cells to clarify the mechanisms underlying TNF-α + IFN-γ-induced β-cell demise. The findings obtained indicate that TNF-α + IFN-γ utilize the BH3-only activator Bim as a key pro-apoptotic effector downstream of STAT1 induction, suggesting a characteristic modulation of Bcl-2 pathways by different inflammatory mediators.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatment
Human islets were isolated in Pisa (Italy) from non-diabetic organ donors, with the approval of the local ethical committee. The islets were isolated by enzymatic digestion and density gradient purification (18) and cultured in M199 medium containing 5.5 mm glucose. Donor age was 57 ± 14 years, and the preparations contained 50 ± 16% β-cells (n = 10), as evaluated by immune staining for insulin (methods as in Ref. 19). Primary islets were also isolated from adult Wistar rats (Charles River Laboratories Belgium, Brussels, Belgium), DP5 knock-out mice previously generated on 129SV/C57BL/6 mixed background (20), or wild type littermate mice. Animals were bred and used in accordance with the guidelines of the Belgian Regulations for Animal Welfare or with the Canadian Council of Animal Care guidelines. All experimental protocols used were approved by the Ethical Committee for Animal Experiments of the ULB or by the Animal Care Committee of Mount Sinai Hospital. Primary rat β-cell isolation and culture were carried out as published previously (21, 22). Islet isolation was achieved by collagenase digestion, and primary rat β-cells were obtained after hand picking of islets using a stereomicroscope, islet dispersion, and FACS sorting (FACSAria, BD Biosciences). Sorted cells were plated on polylysine-coated dishes and precultured in Ham's F-10 (Invitrogen, Paisley, UK) medium for 48 h before any further experimental procedures. Mouse islet isolation was performed as described previously (23). The insulin-producing INS-1E cell line (a kind gift from Professor C. Wollheim, Centre Médical Universitaire, Geneva, Switzerland) was cultured in RPMI 1640 medium (Invitrogen) supplemented with 5% fetal calf serum (FCS), 0.1 mm sodium pyruvate, 1 mm Hepes, and 0.5 μm 2-mercaptoethanol (Invitrogen) (24). Recombinant rat (100 units/ml for INS-1E cells and 500 units/ml for primary rat β-cell), mouse (500 units/ml), or human (500 units/ml) IFN-γ (R&D Systems, Abingdon, UK), recombinant murine TNF-α (1000 units/ml, Innogenetics, Gent, Belgium), human recombinant IL-1β (50 units/ml for mouse islets and 10 units/ml for INS-1E cells, a kind gift from Dr. C. W. Reynolds, NCI, National Institutes of Health, Bethesda, MD), and IL-1 receptor antagonist (IL-1ra; 1000 units/ml, R&D Systems) were used. Cytokine concentrations were selected based on our previous time course and dose-response studies (1, 24).
Western Blot
After cell culture and treatment, cells were lysed using the Laemmli buffer, and total protein extracts were extracted and resolved by SDS-PAGE, transferred unto a nitrocellulose membrane, and immunoblotted with the antibodies indicated in supplemental Table S1. The proteins were revealed with a secondary anti-rabbit or anti-mouse horseradish peroxidase-labeled anti-IgG (1/5000, Lucron Bioproducts, De Pinte, Belgium). The protein bands were visualized using an enhanced chemiluminescence (ECL) kit (SuperSignal West Femto, Thermo Scientific, Pierce) and quantified using the Aida1D analysis software (Fujifilm, London, UK). The intensity values for the proteins were corrected by the values of the housekeeping protein α-tubulin and are shown as -fold induction versus the control sample (considered as 1).
Small Interfering RNA (siRNA) Treatment
Cells were transfected overnight with 30 nm of siRNAs (Dharmacon, Chicago, IL; Santa Cruz Biotechnology, Santa Cruz, CA; or Invitrogen) or with an inactive control siRNA (Qiagen, Venlo, Netherlands) using DharmaFECTTM lipid reagent (Dharmacon), LipofectamineTM 2000 reagent (Invitrogen), and Opti-MEM® (Invitrogen). The inactive control siRNA affects neither insulin release (25) nor gene expression (26). A target inhibition of 50–90% was obtained, similar to our previous studies (15, 19, 22, 26). The list of siRNAs for PUMA, DP5, Bim, Bcl-XL, STAT1, and PTPN2 used is shown in supplemental Table S2.
Cell Viability Assay
The percentage of β-cell death was determined after a 15-min incubation with the DNA binding dyes propidium iodide (PI, 5 μg/ml; Sigma) and Hoechst 33342 (HO, 10 μg/ml; Sigma) (19, 24, 27, 28). A minimum of 500 cells was counted in each experimental condition. Viability was evaluated by two independent observers (one being blind to sample identity). The agreement between findings obtained by the two observers was 90%. Results are expressed as the percentage of dead cells, calculated as the number of apoptotic plus necrotic cells/total number of cells counted × 100. Apoptosis was also measured by an enzyme-linked immunosorbent assay (ELISA), using a cytoplasmic histone-associated DNA fragment detection kit (Roche Diagnostics, Vilvoorde, Belgium). Results are expressed as arbitrary units of optical densities. Apoptosis was confirmed by additional methods including mitochondrial Bax translocation, cytochrome c release, and caspase-3 and -9 cleavage as suggested (29).
Real Time RT-PCR
After exposure to cytokines, INS-1E cells, rat primary β-cells, and human islets were lysed and harvested; poly(A) mRNA was isolated using the Dynabeads mRNA DIRECT kit (Invitrogen) and reverse-transcribed as described previously (22, 24). Quantitative RT-PCR was done by using SYBR Green fluorescence on a LightCycler machine (Roche Diagnostics, Manheim, Germany), and values were compared with a standard curve. Expression values of the genes of interest were corrected by the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and were normalized by the control value in the experimental series considered as 1. GAPDH expression is not modified by the present cytokine combinations in INS-1E cells (27) or when compared with the stable housekeeping genes OAZ1 and SRP14 (30) in primary β-cells or human islets (supplemental Fig. S1, A and B). Primer sequences for rat and human PUMA, Bim, DP5, OAZ1, SRP14, and GAPDH and rat Bcl-XL, Bcl-2, STAT1, and PTPN2 are described in supplemental Table S3.
Immunofluorescence
β-Cells were plated on polylysine-coated coverslips and treated with cytokines for at least 48 h. The cells were then fixed with 4% paraformaldehyde and permeabilized with 0.3% Triton X-100. The fixed cells were incubated overnight at 4 °C with the following primary antibodies: rabbit anti-Bax (1/200, Santa Cruz Biotechnology), anti-AIF, (1/200, Cell Signaling, Danvers, MA), anti-Cox (1/200, Cell Signaling), anti-ATP synthase β (1/1000, Sigma), and mouse anti-cytochrome c (1/200, BD Biosciences). The secondary antibody used for visualization was either FITC-conjugated or rhodamine-conjugated secondary antibodies raised against rabbit or mouse (Jackson ImmunoResearch, West Grove, PA), diluted at 1/200. Nuclei were counterstained with HO (10 μg/ml; Sigma) before mounting of coverslips. Pictures were made using an inverted fluorescent microscope (Zeiss Axiovert 200, Oberkochen, Germany).
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed as described (15). Briefly, extracts were precleared by a 1-h incubation with protein A/herring sperm DNA at 4 °C. Samples were incubated overnight at 4 °C with anti-STAT1 antibody (Santa Cruz Biotechnology, 1:200) or preimmune goat serum as negative control. Immunocomplexes were conjugated with protein A/herring sperm DNA and washed. Samples were then eluted, proteinase K-treated, and incubated overnight at 65 °C in high salt solution to reverse the cross-link reaction. DNA fragments were analyzed by standard PCR, and input DNA was analyzed simultaneously for normalization. PCR was performed with the following primer pair for the STAT1 binding site −686 to −385 of the Bim promoter: 5′-TCCCACCACCAGCTCTGCAC-3′ and 5′-GCTCCCTTGGTTTGCGGAGC-3′.
Statistical Analysis
Data are expressed as means ± S.E. of the indicated number of independent experiments. A significant difference between experimental conditions was assessed by a two-tailed paired t test or ANOVA followed by paired t test with the Bonferroni correction. p values < 0.05 were considered statistically significant.
RESULTS
The Cytokine Combination TNF-α + IFN-γ Triggers β-Cell Apoptosis through the Mitochondrial Pathway of Cell Death
β-Cell exposure to TNF-α + IFN-γ induced mitochondrial Bax translocation, cytochrome c release, and DNA fragmentation (Fig. 1, A and B; supplemental Fig. S2, A–D), but not nuclear AIF translocation (Fig. 1A; supplemental Fig. S2A). In primary rat β-cells and INS-1E cells, AIF is anchored to the inner mitochondrial membrane and needs to be actively processed to translocate to the nucleus (13); this process occurs independently of mitochondrial Bax translocation and cytochrome c release. Consistently with cytochrome c release from the mitochondria, TNF-α + IFN-γ triggered the cleavage and activation of caspases -9, -7, and -3 (Fig. 1C). Total pro-caspase-7 protein expression was up-regulated after cytokine combination (Fig. 1C), but the ratio of cleaved/pro-caspase 7 increased by 5–7-fold after 16–24 h (p < 0.05; data not shown). Taken together, these results suggest that the intrinsic pathway of apoptosis is activated in β-cells following exposure to TNF-α + IFN-γ. Importantly, we confirmed that TNF-α combined with IFN-γ induces cleavage of caspase-3 and triggers apoptosis in dispersed human islets (Fig. 2, A and B; supplemental Fig. S2E). This apoptotic effect of TNF-α + IFN-γ was not prevented by IL-1ra in INS-1E cells. Conversely, IL-1ra protected against IL-1β + IFN-γ-induced cell death (Fig. 2C). Thus, TNF-α + IFN-γ directly induce apoptosis in our experimental model, without the need of IL-1β production by the target cells (31).
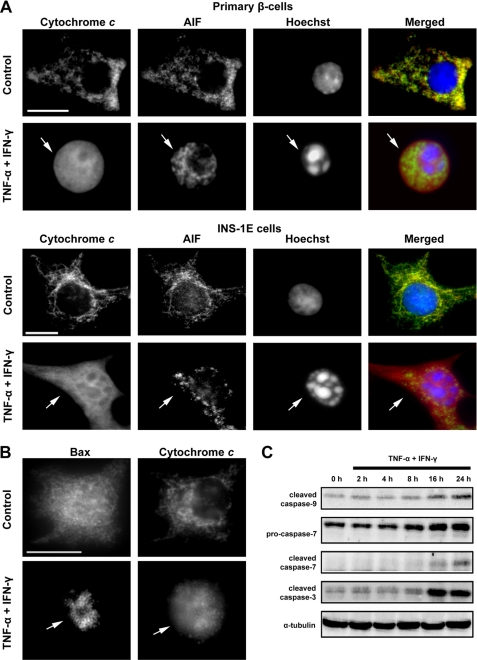
FIGURE 1.
TNF-α + IFN-γ induce the mitochondrial pathway of apoptosis in β-cells. A, representative immunofluorescence images of cytochrome c (red) and AIF (green) localization in purified primary rat β-cells and INS-1E cells treated for 48 or 14 h, respectively, with TNF-α + IFN-γ. Nuclear morphology is shown by Hoechst staining (blue). Arrows indicate compaction of the nuclear chromatin and cytochrome c release from the mitochondria. Bar, 10 μm. B, subcellular localization of Bax after exposure of purified primary rat β-cells for 48 h to TNF-α + IFN-γ. Arrows indicate mitochondrial Bax translocation. Bar, 10 μm. C, time course of caspase activation in INS-1E cells exposed to cytokines for the indicated time points. Data shown are representative of 2–3 independent experiments.
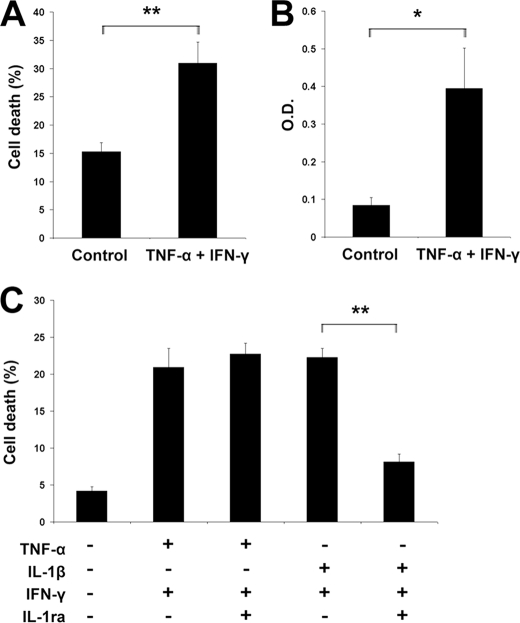
FIGURE 2.
Human β-cells trigger apoptosis after TNF-α + IFN-γ exposure. A and B, dispersed human islets were treated with TNF-α + IFN-γ for 48 h, and apoptosis was evaluated by HO/PI staining (A) or by ELISA using a cytoplasmic histone-associated DNA fragment detection kit (B). Two-tailed paired t test: *, p < 0.05; **, p < 0.01. O.D., optical density. C, INS-1E cells were cultured for 16 h in the presence of TNF-α + IFN-γ, IL-1β + IFN-γ, alone, or in combination with IL-1ra. IL-1ra prevents IL-1β + IFN-γ- but not TNF-α + IFN-γ-induced β-cell death. Two-tailed paired t test: **, p < 0.01. Data shown are representative of 3–4 independent experiments.
The BH3-only Protein DP5 Is Induced by TNF-α + IFN-γ and Activates β-Cell Apoptosis
DP5, also known as Harakiri, belongs to the BH3-only sensitizer subgroup, and its expression was observed in human neurons and pancreas (11, 32). We previously demonstrated that IL-1β + IFN-γ induce activation of DP5 via JNK/c-Jun and nitric oxide (NO) production in β-cells, which triggers the classic intrinsic apoptotic pathway (15). We presently observed that islets from four human preparations exposed to TNF-α + IFN-γ increased DP5 mRNA expression (Fig. 3A). DP5 mRNA up-regulation was also observed in primary rat β-cells and in insulin-producing INS-1E cells (Fig. 3, B and C). DP5 induction by TNF-α + IFN-γ (2–5-fold) was less marked than the extent of DP5 overexpression observed with the combination of IL-1β + IFN-γ (3–20-fold) (15). Knockdown of DP5 by two different siRNAs (supplemental Fig. S3) decreased TNF-α + IFN-γ-induced β-cell death (Fig. 3D). Because the present and previous (15) in vitro studies indicated that DP5 participates in β-cell apoptosis, we next studied the effects of DP5 inactivation in an in vivo model, namely DP5 knock-out mice (20). β-Cell area was increased in pancreases from DP5-deficient mice when compared with wild type littermates (Fig. 3E), but glucose tolerance and islet insulin secretion were similar (supplemental Fig. S4, A–D). Importantly, islets isolated from mice lacking DP5 were partially protected against TNF-α + IFN-γ (Fig. 3F) and against TNF-α + IFN-γ + IL-1β (data not shown). All together, these data indicate that DP5 inactivation partially prevents the triggering of apoptosis by pro-inflammatory cytokines.
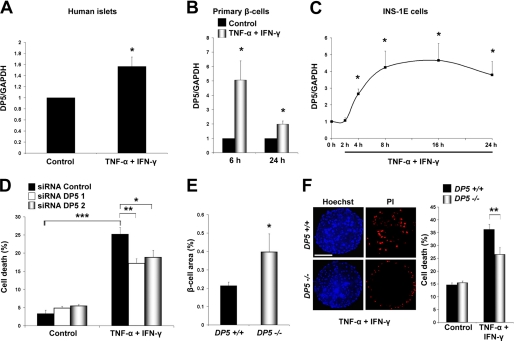
FIGURE 3.
Role of DP5 activation in β-cell apoptosis induced by TNF-α + IFN-γ. A, expression of DP5 mRNA in human islets isolated from four organ donors, under control condition or following a 48-h treatment with TNF-α + IFN-γ as indicated. DP5 expression was determined by real time RT-PCR and is represented as -fold induction when compared with control (non-treated cells). Two-tailed paired t test: *, p < 0.05. B, DP5 mRNA expression in purified rat β-cells treated with cytokines. Real time RT-PCR for DP5 was performed at the indicated time points after cytokine addition. Two-tailed paired t test: *, p < 0.05. C, time course of DP5 mRNA expression after cytokine exposure was evaluated by real time RT-PCR in INS-1E cells. Data are means ± S.E. Two-tailed paired t test: *, p < 0.05 versus control (time 0 h). D, INS-1E cells were transfected with inactive siRNA (Control) or two different DP5 siRNAs (siRNA DP5 1 and siRNA DP5 2), and cells were allowed to recover for 36 h; cytokines were then added, and cell death was measured by HO/PI after 16 h of cytokine addition. Data shown are means ± S.E. of 5 independent experiments. ANOVA followed by paired t test with the Bonferroni correction: *, p < 0.05; **, p < 0.01; ***, p < 0.001. E, immunostaining results for insulin in pancreas sections from wild type and DP5 knock-out mice were summed using an optimized positive pixel count algorithm and normalized per total pancreas area (square millimeters) analyzed per mouse (n = 4). Two-tailed paired t test: *, p < 0.05. F, islets from DP5 knock-out or wild type mice were exposed to the cytokine combination TNF-α + IFN-γ for 48 h. The viability of islet cell preparations was assessed by HO/PI. Cell death was counted in at least 24 islets derived from four mice per condition. Bar, 60 μm. Two-tailed paired t test: **, p < 0.01. Data shown are representative of 3–5 independent experiments.
PUMA, a BH3-only Activator, Participates in TNF-α + IFN-γ-induced β-Cell Death
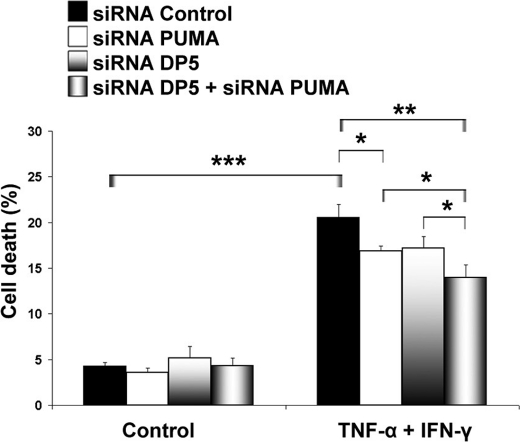
PUMA plays a major role for pancreatic β-cell apoptosis in the context of IL-1β + IFN-γ exposure (16). PUMA transcript expression was induced by ∼6-fold in human islet preparations, primary rat β-cells, and INS-1E cells following treatment with TNF-α + IFN-γ (Fig. 4, A–C). Up-regulation of PUMA protein was demonstrated in INS-1E cells (Fig. 4D). SMARTpool (a combination of four different siRNAs) and a single sequence siRNA were used to silence PUMA, achieving a 70% reduction in mRNA expression as measured by real time RT PCR (Fig. 4E) and a 50% decrease in protein level as observed by Western blot analysis (Fig. 4F). PUMA knockdown protected INS-1E cells against TNF-α + IFN-γ-induced apoptosis (Fig. 4G). Simultaneous knockdown of both DP5 and PUMA failed to fully protect INS-1E cells against cytokines (Fig. 5). Together, these data demonstrate that DP5 and PUMA contribute to the control of β-cell death induced by TNF-α + IFN-γ. The partial protective effect obtained by knocking down these two proteins, however, suggests that alternative Bcl-2 proteins participate in β-cell apoptosis following exposure to TNF-α and IFN-γ.
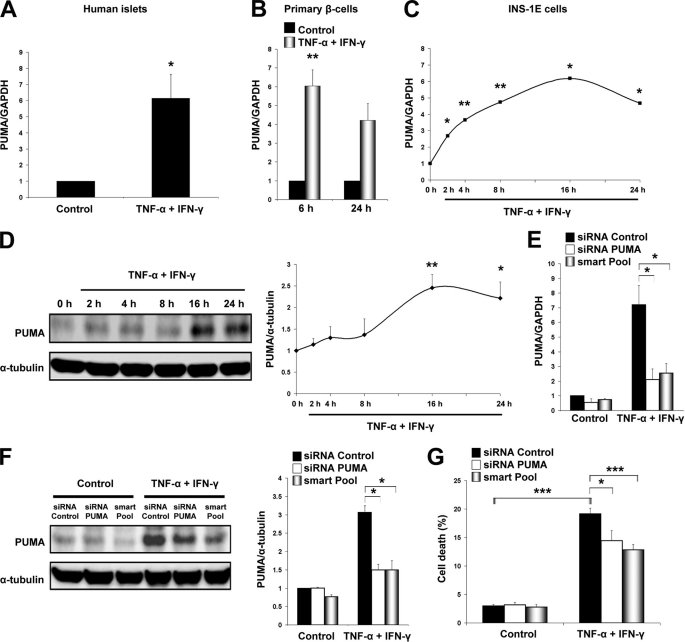
FIGURE 4.
PUMA knockdown partially protects β-cells from TNF-α + IFN-γ-induced apoptosis. A, expression of PUMA as determined by real time RT-PCR in human islets after a 48-h TNF-α + IFN-γ treatment as indicated. Values are shown as -fold induction when compared with control (non-cytokine-treated islets). Two-tailed paired t test: *, p < 0.05. B, PUMA mRNA expression in purified rat β-cells treated with cytokines. Real time RT-PCR for PUMA was performed at the indicated time points after cytokine addition. Two-tailed paired t test: **, p < 0.01. C, time course of PUMA mRNA expression in INS-1E cells after TNF-α + IFN-γ exposure, evaluated by real time RT-PCR. Two-tailed paired t test: *, p < 0.05; **, p < 0.01. D, Western blot demonstrating increased expression of PUMA protein in TNF-α + IFN-γ-treated INS-1E cells. Quantification of PUMA band intensities is indicated at the right side as a ratio to α-tubulin loading. Two-tailed paired t test: *, p < 0.05; **, p < 0.01. E, INS-1E cells were transfected with inactive siRNA (Control), a single sequence siRNA against PUMA (siRNA PUMA), or PUMA SMARTpool siRNAs (smart Pool). Cells were allowed to recover for 36 h before exposure to cytokines. Real time RT-PCR for PUMA and GAPDH mRNA expression was performed 16 h after cytokine exposure. Two-tailed paired t test: *, p < 0.05. F, Western blot analysis of PUMA expression in INS-1E cells after siRNA transfection, 36 h of recovery, and subsequent 16 h of cytokine treatment. Two-tailed paired t test: *, p < 0.05. G, PUMA knockdown partially prevents INS-1E death after 16 h of cytokine treatment as measured by HO/PI staining. ANOVA followed by paired t test with the Bonferroni correction: *, p < 0.05; ***, p < 0.001. Data shown are means ± S.E. of 3–5 independent experiments.
FIGURE 5.
Knockdown of DP5, PUMA, or combination partially prevents TNF-α + IFN-γ-induced cell death in INS-1E cells. Cytokines were added for 16 h to INS-1E cells previously transfected with siRNAs as indicated, and cell death was measured by HO/PI staining. ANOVA followed by paired t test with the Bonferroni correction: *, p < 0.05; **, p < 0.01; ***, p < 0.001. Data shown are representative of 5 independent experiments.
The BH3-only Activator Bim Is Transcriptionally Activated by STAT1 in TNF-α + IFN-γ-exposed β-Cells
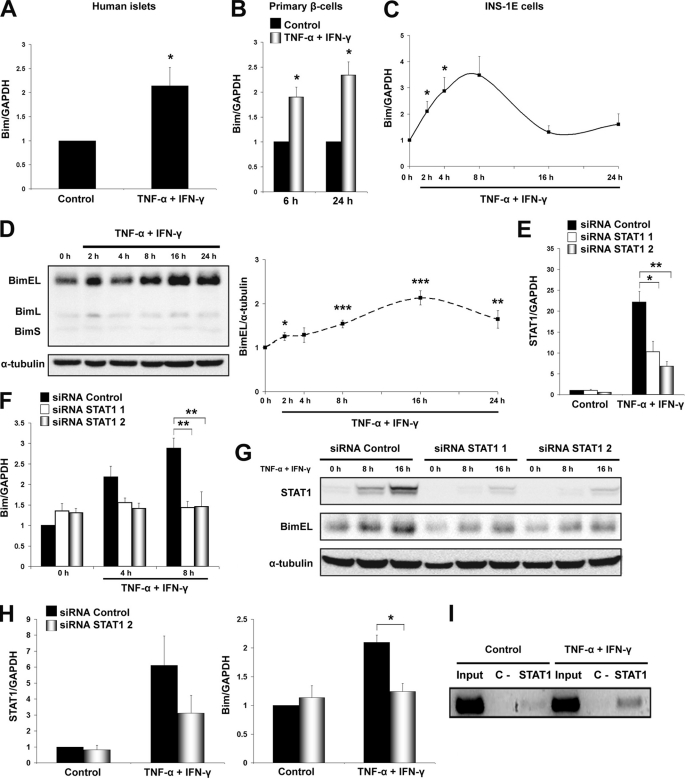
Bim, together with PUMA, has been previously reported to play a key role in β-cell apoptosis induced by high glucose (33). On the other hand, Bim translation is not modulated by IL-1β + IFN-γ in β-cells (16). We next evaluated whether Bim is activated in the context of TNF-α + IFN-γ. Bim is mainly localized at the mitochondria of the β-cells (supplemental Fig. S5A), and protein localization is not affected in INS-1E cells after TNF-α + IFN-γ exposure (data not shown). Treatment of islets derived from human donors, primary rat β-cells, and INS-1E cells with TNF-α + IFN-γ resulted in up-regulation of total Bim transcript level (Fig. 6, A–C). Thus, we analyzed by Western blot the different protein isoforms generated by alternative mRNA splicing of Bim (BimEL, BimL, and BimS). Among the three isoforms, BimEL and BimL were significantly induced from 8 to 24 h in INS-1E cells after cytokine treatment (Fig. 6D; supplemental Fig. S5B). IFN-γ alone up-regulated Bim mRNA expression to the same extent as TNF-α + IFN-γ (supplemental Fig. S5C). The cytokine IFN-γ activates signal transduction pathways that involve the tyrosine Janus kinases JAK1 and JAK2, which phosphorylate and induce the dimerization of the transcription factor STAT1 (34). To evaluate the role of STAT1 for Bim induction, we knocked down STAT1 by using two different siRNAs, achieving more than 60% silencing in INS-1E cells as demonstrated by mRNA and protein analysis (Fig. 6, E and G; supplemental Fig. S6A). Silencing of STAT1 decreased TNF-α + IFN-γ-induced Bim mRNA and protein expression in INS-1E and primary β-cells (Fig. 6, F–H; supplemental Fig. S6A) and protected both INS-1E cells and primary β-cells against TNF-α + IFN-γ-induced apoptosis (supplemental Fig. S6B). ChIP analysis confirmed STAT1 direct binding to the Bim promoter in positions −686 to −385; this region harbors two STAT1 binding sites, TTCtacGAA and TTCttgGAA (Fig. 6I). The phosphatase PTPN2, a candidate gene for T1D (35), was previously shown to down-regulate IFN-γ signal transduction in β-cells (22). PTPN2 knockdown increased phosphorylation and activation of STAT1 in β-cells (22) and, in line with the above observations, increased cytokine-induced Bim expression (supplemental Fig. S6C). Interestingly, Bim protein expression does not change after IL-1β + IFN-γ exposure (supplemental Fig. S7A) (16), although Bim transcript is induced by this cytokine combination at a near similar level as with TNF-α + IFN-γ (supplemental Fig. S7B). IL-1β + IFN-γ trigger severe ER stress in β-cells (15), whereas TNF-α + IFN-γ did not increase expression of the ER stress marker XBP-1s (supplemental Fig. S7C) and only slightly up-regulated Chop transcript level (supplemental Fig. S7D). Because ER stress inhibits protein translation (36), we next used two different chemical ER stressors, namely cyclopiazonic acid and tunicamycin, to test whether ER stress affects Bim protein up-regulation. The two chemical ER stressors prevented TNF-α + IFN-γ-induced Bim protein expression in INS-1E cells (supplemental Fig. S7E), suggesting that severe ER stress may explain why IL-1β + IFN-γ fail to induce Bim translation.
FIGURE 6.
Bim induction is STAT1-dependent in TNF-α + IFN-γ-treated β-cells. A, expression of Bim mRNA in human islets isolated from six organ donors, under control condition or following a 48-h treatment with TNF-α + IFN-γ as indicated. Bim expression was determined by real time RT-PCR and is represented as -fold induction when compared with control (non-treated cells). Two-tailed paired t test: *, p < 0.05. B, Bim mRNA expression in purified rat β-cells treated with cytokines. Real time RT-PCR for Bim was performed at the indicated time points after TNF-α + IFN-γ addition. Two-tailed paired t test: *, p < 0.05. C, time course of Bim mRNA expression in INS-1E cells after TNF-α + IFN-γ exposure, as evaluated by real time RT-PCR. Two-tailed paired t test: *, p < 0.05. D, time course of Bim protein expression after TNF-α + IFN-γ treatment of INS-1E cells. Cell lysates were subjected to Western blotting with antibodies detecting BimEL, BimL, BimS, or α-tubulin as loading control (left side of the figure). Quantification of BimEL bands is shown on the right side as -fold induction of control after correction for α-tubulin. Quantification of BimL and BimS is shown in supplemental Fig. S5B. Two-tailed paired t test: *, p < 0.05; ***, p < 0.001. E, INS-1E cells were transfected with inactive siRNA (siRNA Control) or two different STAT1 siRNAs (siRNA STAT1 1 and siRNA STAT1 2); cells were allowed to recover for 48 h and were then treated with cytokines. Real time RT-PCR for STAT1 and GAPDH expression was performed after 8 h of cytokine exposure. Two-tailed paired t test: *, p < 0.05; **, p < 0.01. F, siRNA-mediated STAT1 knockdown decreased TNF-α + IFN-γ-induced Bim mRNA expression in INS-1E cells as determined by real time RT-PCR. Two-tailed paired t test: **, p < 0.01. G, INS-1E cells were transfected with inactive (siRNA Control) or two different active STAT1 siRNAs (siRNA STAT1 1 and siRNA STAT1 2) and allowed to recover for 36 h; STAT1 and BimEL expression was determined by Western blot at 16 h after cytokine exposure. Densitometric quantification of the bands is shown in supplemental Fig. S6A. H, primary rat β-cells were transfected with inactive siRNA (siRNA Control) or siRNA STAT1 2; cells were allowed to recover for 48 h and were then treated with cytokines. Real time RT-PCR for STAT1, Bim, and GAPDH expression was performed after 48 h of cytokine exposure. Two-tailed paired t test: *, p < 0.05. I, ChIP analysis revealed binding of STAT1 to the Bim promoter region −686 to −385, where two STAT1 binding sites are located, after 6 h of cytokine treatment as assessed by PCR. Preimmune goat serum was used as negative control (C−). Data shown are means ± S.E. or representative blots of 3–7 independent experiments.
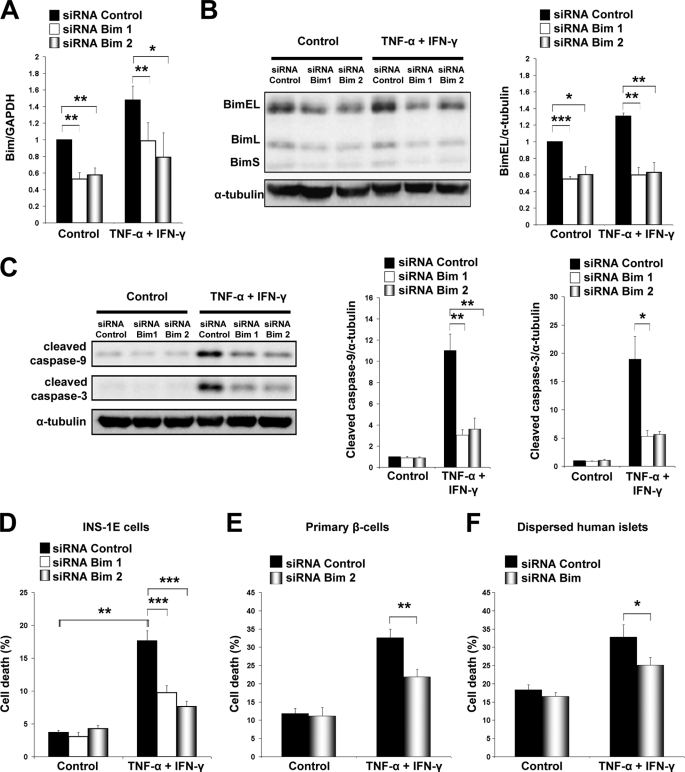
Bim Activation Is a Major Event for Apoptosis Induced by TNF-α + IFN-γ in β-Cells
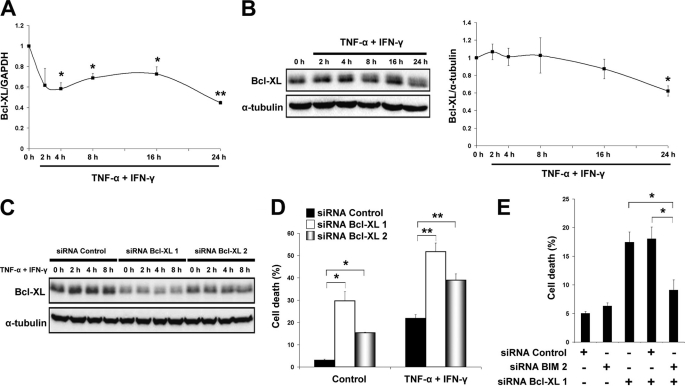
Bim was successfully silenced at both mRNA and protein levels by using two different siRNAs in INS-1E cells (Fig. 7, A and B). Bim knockdown resulted in 70–80% down-regulation of cleaved caspase-9 and -3 after cytokine treatment (Fig. 7C). Inactivation of Bim consistently prevented TNF-α + IFN-γ-induced cell death in INS-1E cells (Fig. 7D; supplemental Fig. S8A). The protective effect of Bim inactivation was significantly higher for the cytokine combination TNF-α + IFN-γ than for IL-1β + IFN-γ (supplemental Fig. S7F). Importantly, primary rat β-cells and dispersed human islets were also protected against TNF-α + IFN-γ-induced apoptosis by Bim knockdown (Fig. 7, E and F; supplemental Fig. S8, B–D). Parallel knockdown of DP5 and Bim did not increase the anti-apoptotic effects of Bim alone (supplemental Fig. S9). In other cell types, Bim binds to and is inhibited by the anti-apoptotic proteins Bcl-2 and Bcl-XL (11). We thus characterized the role of these pro-survival Bcl-2 members in the context of TNF-α + IFN-γ-induced β-cell apoptosis. Bcl-2 mRNA and protein expression was not modulated by TNF-α + IFN-γ (supplemental Fig. S10, A and B). Bcl-XL, on the other hand, was decreased at both mRNA and protein levels (Fig. 8, A and B). This effect was TNF-α-dependent because this cytokine alone down-regulated Bcl-XL (supplemental Fig. S11). To address the role of Bcl-XL inactivation, we silenced this protein by using two different siRNAs (Fig. 8C). Bcl-XL knockdown increased β-cell death in both basal and cytokine-treated conditions (Fig. 8D). Interestingly, simultaneous knockdown of Bcl-XL and Bim reversed the lethal effect of Bcl-XL silencing (Fig. 8E). These results suggest that both Bim up-regulation (by IFN-γ) and inactivation of Bcl-XL (by TNF-α) contribute to Bim release and activation, a critical step for TNF-α + IFN-γ-mediated β-cell death.
FIGURE 7.
Bim knockdown protects β-cells from TNF-α + IFN-γ-induced apoptosis. A, INS-1E cells were transfected with inactive siRNA (siRNA Control) or two different siRNAs against Bim (siRNA Bim 1 and siRNA Bim 2). Cells were allowed to recover for 48 h before treatment with cytokines. Real time RT-PCR for Bim and GAPDH mRNA expression was performed 16 h after TNF-α + IFN-γ treatment. Two-tailed paired t test: *, p < 0.05; **, p < 0.01. B, Western blot analysis of Bim expression in INS-1E cells after siRNA transfection, 36 h of recovery, and subsequent 16 h of cytokine treatment. Densitometric quantification of BimEL bands (with correction for the housekeeping protein α-tubulin) is shown in the right panel. Two-tailed paired t test: *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, INS-1E cells were transfected with control or Bim siRNAs, cells were allowed to recover for 36 h, and Western blot for cleaved caspase-9 and -3 was performed 16 h after cytokine addition. Densitometric quantification is shown in the right panels. Two-tailed paired t test: *, p < 0.05; **, p < 0.01. D, INS-1E cells were transfected with siRNAs against Bim or control. Cells were allowed to recover for 36 h, and after that, they were treated with TNF-α + IFN-γ for 16 h as indicated; cell death was measured by HO/PI. ANOVA followed by paired t test with the Bonferroni correction: **, p < 0.01; ***, p < 0.001. E and F, Bim knockdown decreased TNF-α + IFN-γ-induced apoptosis in rat primary β-cells (E) and dispersed human islets (F). Cell death was evaluated by HO/PI 48 h after cytokine treatment. Two-tailed paired t test: *, p < 0.05; **, p < 0.01. Data shown are means ± S.E. of 3–5 independent experiments.
FIGURE 8.
Bcl-XL antagonizes Bim but is inhibited by TNF-α + IFN-γ in β-cells. A, time course analysis of Bcl-XL mRNA expression after cytokine exposure in INS-1E cells, as evaluated by real time RT-PCR. Two-tailed paired t test: *, p < 0.05; **, p < 0.01. B, Western blot for Bcl-XL was performed at the indicated time points of cytokine exposure. Densitometric quantification of bands is shown on the right as -fold induction of control after correction for α-tubulin. Two-tailed paired t test: *, p < 0.05. C, two different siRNAs against Bcl-XL (siRNA Bcl-XL 1 and siRNA Bcl-XL 2) or control siRNAs were introduced into INS-1E cells, cells were allowed to recover for 48 h, and Bcl-XL protein expression was measured by Western blot at the indicated time points after cytokine exposure. D, Bcl-XL knockdown triggered apoptosis and increased TNF-α + IFN-γ-induced INS-1E cell death (36 h of recovery after siRNA transfection and then 16 h of cytokine exposure). Two-tailed paired t test: *, p < 0.05; **, p < 0.01. E, INS-1E cells were transfected with siRNA control, siRNA Bim 2, and/or siRNA Bcl-XL 1, and cell death was measured by HO/PI after 24 h. Two-tailed paired t test: *, p < 0.05. Data shown are means ± S.E. of 3–5 independent experiments.
DISCUSSION
The composition of and interaction between pro-inflammatory cytokines probably vary during the evolution of insulitis in early T1D (3). This might explain the different level of protection achieved by the blockage of TNF-α or IL-1β in rodent models of autoimmune diabetes (1, 37). Therefore, the understanding of apoptotic networks induced downstream of each cytokine combination might contribute to future individualized therapies. We presently demonstrate that activation of the BH3-only protein Bim is critical for TNF-α + IFN-γ-induced β-cell death.
TNF-α combined with IFN-γ induces DP5 expression in human islets and in rat primary and clonal β-cells. DP5 participates in β-cell apoptosis, as demonstrated by the observation that its knockdown partially prevents cell death induced by TNF-α + IFN-γ. Islets derived from DP5 knock-out mice are also protected against this cytokine combination. Importantly, DP5 inactivation does not affect insulin secretion, as was previously reported with the BH3-only sensitizer Bad (38). The group of BH3-only sensitizer proteins is composed of DP5, Bad, Noxa, Bik, and other Bcl-2 members (9, 11). Bad was described to be activated by calcineurin in TNF-α + IFN-γ-treated MIN6N8 cells (39). We also evaluated the putative response of Noxa and Bik in β-cells, but these proteins were not induced by TNF-α + IFN-γ (data not shown). On the other hand, TNF-α + IFN-γ induce transcriptional down-regulation of the anti-apoptotic Bcl-XL (present data), which may contribute (together with DP5 overexpression and Bad dephosphorylation) to “sensitize” β-cells to apoptosis. Inactivation of Bcl-XL is dependent on TNF-α alone, and this phenomenon was also observed in other cell types (40). The anti-apoptotic protein Bcl-2 is expressed at low levels in human islets (41) and was not modulated in INS-1E cells by TNF-α + IFN-γ (present data).
PUMA and Bim belong to the BH3-only activator subgroup and must be released from anti-apoptotic Bcl-2 proteins to induce Bax and Bak conformational changes, mitochondrial pore formation, and apoptosis (9, 10). We have previously shown that IL-1β combined with IFN-γ activates an early induction of NF-κB, which is responsible for PUMA up-regulation (16). TNF-α + IFN-γ induce PUMA mRNA and protein expression, and PUMA inactivation partially decreases cytokine-induced β-cell apoptosis. Nevertheless, double knockdown of DP5 and PUMA fails to fully protect β-cells against TNF-α + IFN-γ, suggesting that an additional BH3-only activator protein is involved in the pathway of apoptosis. Herein we identified Bim as a key regulator of β-cell death induced by TNF-α + IFN-γ. Thus, inactivation of Bim by siRNAs prevents to a large extent TNF-α + IFN-γ-mediated β-cell apoptosis, as shown by caspase-9 and -3 cleavage, cytoplasmic histone-associated DNA fragment detection, and HO/PI staining. We observed that inactivation of Bcl-XL induced β-cell death, but this was reversed by concomitant knockdown of the downstream pro-apoptotic protein Bim. Furthermore, knockdown of both DP5 and Bim did not augment the protection provided by Bim knockdown alone. Together, these observations are compatible with the concept of a direct or hierarchical pathway of apoptosis (9–11), where DP5 acts as an upstream sensitizer inhibiting Bcl-XL and thus releasing the BH3-only activator Bim.
Bim has a less marked protective effect in β-cell death induced by IL-1β + IFN-γ and is not translationally induced by this cytokine combination, suggesting that different pro-inflammatory cytokines induce a context-dependent modulation of BH3-only proteins in β-cells. IL-1β combined with IFN-γ triggers ER stress via NO formation, leading to inhibition of protein translation (42), whereas chemical ER stressors prevent Bim protein induction by TNF-α + IFN-γ (present data). It is conceivable that activation of severe ER stress by IL-1β + IFN-γ, but not by TNF-α + IFN-γ, explains why IL-1β + IFN-γ fail to induce Bim protein expression despite the fact that both cytokine combinations up-regulate Bim mRNA levels. In other words, TNF-α + IFN-γ induce a mild ER stress, which enables Bim translation and favors this particular pathway of apoptosis, whereas IL-1β + IFN-γ induce a more pronounced ER stress that inhibits the protein synthesis of Bim, allowing alternative pathways of cell death to prevail. It has been previously reported that ER stress induces apoptosis via up-regulation of Bim in thymocytes, macrophages, and kidney and breast epithelial cells (43). We did not observe, however, Bim protein induction following β-cell exposure to chemical ER stressors, indicating that the pathways by which ER stress leads to apoptosis might be cell-specific.
As described above, Bim is induced at the transcriptional level following cytokine exposure. The transcription factor STAT1 has a major role in the apoptotic mechanism induced by cytokines in β-cells (17, 26, 44, 45). This transcription factor is activated by IFN-γ downstream of JAK. Knockdown of STAT1 by two different siRNAs prevents Bim mRNA and protein induction, whereas silencing of PTPN2, a phosphatase that provides a negative feedback on STAT1 activation (22), enhances Bim expression. In line with these results, STAT1-dependent Bim activation was also suggested in chronic lymphocytic leukemia cells after IL-21 exposure (46). We propose, therefore, that STAT1-mediated Bim overexpression is a key mechanism by which IFN-γ contributes to β-cell apoptosis. These intriguing observations provide a putative link between a candidate gene for T1D (PTPN2), a cytokine-induced transcription factor (STAT1), and an effector BH3-only activator protein (Bim).
In conclusion, we demonstrate that TNF-α combined with IFN-γ induces Bim protein expression. This event, together with DP5 and PUMA up-regulation and Bcl-XL inactivation, is critical for pancreatic β-cell death following exposure to the pro-inflammatory cytokines TNF-α and IFN-γ. Our findings suggest the novel concept that induction of β-cell apoptosis by different cytokine combinations (i.e. IL-1β + IFN-γ or TNF-α + IFN-γ) have a characteristic “signature” of β-cell death. This knowledge may contribute toward the future development of individualized therapies to protect β-cells in the early stages of T1D.
Supplementary Material
Acknowledgments
We thank M. A. Neef, G. Vandenbroeck, M. Urbain, R. Makhnas, A. E. Musuaya, and S. Mertens of the Laboratory of Experimental Medicine (Université Libre de Bruxelles, Brussels, Belgium) for excellent technical support.
This work was supported by grants from the Communauté Française de Belgique – Actions de Recherche Concertées (ARC), the Fonds National de la Recherche Scientifique (FNRS) (Belgium), the Belgium Program on Interuniversity Poles of Attraction initiated by the Belgium State (IUAP P6/40), the Juvenile Diabetes Research Foundation International (JDRFI Grant 17-2009-106), and the European Union (Project Naimit, in the Framework Programme 7 of the European Community).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S11.
- T1D
- type 1 diabetes
- BH3
- Bcl-2 homology 3
- PUMA
- p53 up-regulated modulator of apoptosis
- AIF
- apoptosis-inducing factor
- ER
- endoplasmic reticulum
- IL-1ra
- IL-1 receptor antagonist
- HO
- Hoechst
- PI
- propidium iodide
- ANOVA
- analysis of variance.
REFERENCES
- 1. Eizirik D. L., Mandrup-Poulsen T. (2001) Diabetologia 44, 2115–2133 [DOI] [PubMed] [Google Scholar]
- 2. Thomas H. E., McKenzie M. D., Angstetra E., Campbell P. D., Kay T. W. (2009) Apoptosis 14, 1389–1404 [DOI] [PubMed] [Google Scholar]
- 3. Eizirik D. L., Colli M. L., Ortis F. (2009) Nat. Rev. Endocrinol. 5, 219–226 [DOI] [PubMed] [Google Scholar]
- 4. Thomas H. E., Trapani J. A., Kay T. W. (2010) Cell Death Differ 17, 577–585 [DOI] [PubMed] [Google Scholar]
- 5. Kerr J. F., Wyllie A. H., Currie A. R. (1972) Br. J. Cancer 26, 239–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hotchkiss R. S., Strasser A., McDunn J. E., Swanson P. E. (2009) N. Engl. J. Med. 361, 1570–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Galonek H. L., Hardwick J. M. (2006) Nat. Cell Biol. 8, 1317–1319 [DOI] [PubMed] [Google Scholar]
- 8. Brunelle J. K., Letai A. (2009) J. Cell Sci. 122, 437–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gurzov E. N., Eizirik D. L. (2011) Trends Cell Biol. 21, 424–431 [DOI] [PubMed] [Google Scholar]
- 10. Kim H., Tu H. C., Ren D., Takeuchi O., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2009) Mol. Cell 36, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim H., Rafiuddin-Shah M., Tu H. C., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2006) Nat. Cell Biol. 8, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 12. Gross A., Jockel J., Wei M. C., Korsmeyer S. J. (1998) EMBO J. 17, 3878–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Norberg E., Orrenius S., Zhivotovsky B. (2010) Biochem. Biophys. Res. Commun. 396, 95–100 [DOI] [PubMed] [Google Scholar]
- 14. Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 15. Gurzov E. N., Ortis F., Cunha D. A., Gosset G., Li M., Cardozo A. K., Eizirik D. L. (2009) Cell Death Differ. 16, 1539–1550 [DOI] [PubMed] [Google Scholar]
- 16. Gurzov E. N., Germano C. M., Cunha D. A., Ortis F., Vanderwinden J. M., Marchetti P., Zhang L., Eizirik D. L. (2010) J. Biol. Chem. 285, 19910–19920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim S., Kim H. S., Chung K. W., Oh S. H., Yun J. W., Im S. H., Lee M. K., Kim K. W., Lee M. S. (2007) Diabetes 56, 2561–2568 [DOI] [PubMed] [Google Scholar]
- 18. Lupi R., Dotta F., Marselli L., Del Guerra S., Masini M., Santangelo C., Patané G., Boggi U., Piro S., Anello M., Bergamini E., Mosca F., Di Mario U., Del Prato S., Marchetti P. (2002) Diabetes 51, 1437–1442 [DOI] [PubMed] [Google Scholar]
- 19. Cunha D. A., Hekerman P., Ladrière L., Bazarra-Castro A., Ortis F., Wakeham M. C., Moore F., Rasschaert J., Cardozo A. K., Bellomo E., Overbergh L., Mathieu C., Lupi R., Hai T., Herchuelz A., Marchetti P., Rutter G. A., Eizirik D. L., Cnop M. (2008) J. Cell Sci. 121, 2308–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Imaizumi K., Benito A., Kiryu-Seo S., Gonzalez V., Inohara N., Lieberman A. P., Kiyama H., Nuñez G. (2004) J. Neurosci. 24, 3721–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pipeleers D. G., in't Veld P. A., Van de Winkel M., Maes E., Schuit F. C., Gepts W. (1985) Endocrinology 117, 806–816 [DOI] [PubMed] [Google Scholar]
- 22. Moore F., Colli M. L., Cnop M., Esteve M. I., Cardozo A. K., Cunha D. A., Bugliani M., Marchetti P., Eizirik D. L. (2009) Diabetes 58, 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maida A., Lovshin J. A., Baggio L. L., Drucker D. J. (2008) Endocrinology 149, 5670–5678 [DOI] [PubMed] [Google Scholar]
- 24. Kutlu B., Cardozo A. K., Darville M. I., Kruhøffer M., Magnusson N., Ørntoft T., Eizirik D. L. (2003) Diabetes 52, 2701–2719 [DOI] [PubMed] [Google Scholar]
- 25. Allagnat F., Cunha D., Moore F., Vanderwinden J. M., Eizirik D. L., Cardozo A. K. (2011) Cell Death Differ. 18, 328–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moore F., Naamane N., Colli M. L., Bouckenooghe T., Ortis F., Gurzov E. N., Igoillo-Esteve M., Mathieu C., Bontempi G., Thykjaer T., Ørntoft T. F., Eizirik D. L. (2011) J. Biol. Chem. 286, 929–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ortis F., Cardozo A. K., Crispim D., Störling J., Mandrup-Poulsen T., Eizirik D. L. (2006) Mol. Endocrinol. 20, 1867–1879 [DOI] [PubMed] [Google Scholar]
- 28. Hoorens A., Van de Casteele M., Klöppel G., Pipeleers D. (1996) J. Clin. Invest. 98, 1568–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galluzzi L., Aaronson S. A., Abrams J., Alnemri E. S., Andrews D. W., Baehrecke E. H., Bazan N. G., Blagosklonny M. V., Blomgren K., Borner C., Bredesen D. E., Brenner C., Castedo M., Cidlowski J. A., Ciechanover A., Cohen G. M., De Laurenzi V., De Maria R., Deshmukh M., Dynlacht B. D., El-Deiry W. S., Flavell R. A., Fulda S., Garrido C., Golstein P., Gougeon M. L., Green D. R., Gronemeyer H., Hajnóczky G., Hardwick J. M., Hengartner M. O., Ichijo H., Jäättelä M., Kepp O., Kimchi A., Klionsky D. J., Knight R. A., Kornbluth S., Kumar S., Levine B., Lipton S. A., Lugli E., Madeo F., Malomi W., Marine J. C., Martin S. J., Medema J. P., Mehlen P., Melino G., Moll U. M., Morselli E., Nagata S., Nicholson D. W., Nicotera P., Nuñez G., Oren M., Penninger J., Pervaiz S., Peter M. E., Piacentini M., Prehn J. H., Puthalakath H., Rabinovich G. A., Rizzuto R., Rodrigues C. M., Rubinsztein D. C., Rudel T., Scorrano L., Simon H. U., Steller H., Tschopp J., Tsujimoto Y., Vandenabeele P., Vitale I., Vousden K. H., Youle R. J., Yuan J., Zhivotovsky B., Kroemer G. (2009) Cell Death Differ. 16, 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Jonge H. J., Fehrmann R. S., de Bont E. S., Hofstra R. M., Gerbens F., Kamps W. A., de Vries E. G., van der Zee A. G., te Meerman G. J., ter Elst A. (2007) PLoS One 2, e898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arnush M., Heitmeier M. R., Scarim A. L., Marino M. H., Manning P. T., Corbett J. A. (1998) J. Clin. Invest. 102, 516–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Inohara N., Ding L., Chen S., Núñez G. (1997) EMBO J. 16, 1686–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McKenzie M. D., Jamieson E., Jansen E. S., Scott C. L., Huang D. C., Bouillet P., Allison J., Kay T. W., Strasser A., Thomas H. E. (2010) Diabetes 59, 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eizirik D. L., Moore F., Flamez D., Ortis F. (2008) Biochem. Soc. Trans. 36, 321–327 [DOI] [PubMed] [Google Scholar]
- 35. Wellcome Trust Case Control Consortium (2007) Nature 447, 661–67817554300 [Google Scholar]
- 36. Eizirik D. L., Cardozo A. K., Cnop M. (2008) Endocr. Rev. 29, 42–61 [DOI] [PubMed] [Google Scholar]
- 37. O'Sullivan B. J., Thomas H. E., Pai S., Santamaria P., Iwakura Y., Steptoe R. J., Kay T. W., Thomas R. (2006) J. Immunol. 176, 7278–7287 [DOI] [PubMed] [Google Scholar]
- 38. Danial N. N., Walensky L. D., Zhang C. Y., Choi C. S., Fisher J. K., Molina A. J., Datta S. R., Pitter K. L., Bird G. H., Wikstrom J. D., Deeney J. T., Robertson K., Morash J., Kulkarni A., Neschen S., Kim S., Greenberg M. E., Corkey B. E., Shirihai O. S., Shulman G. I., Lowell B. B., Korsmeyer S. J. (2008) Nat. Med. 14, 144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chang I., Cho N., Kim S., Kim J. Y., Kim E., Woo J. E., Nam J. H., Kim S. J., Lee M. S. (2004) J. Immunol. 172, 7008–7014 [DOI] [PubMed] [Google Scholar]
- 40. Fox S. A., Kusmiaty, Loh S. S., Dharmarajan A. M., Garlepp M. J. (2005) Biochem. Biophys. Res. Commun. 337, 983–991 [DOI] [PubMed] [Google Scholar]
- 41. Thomas D., Yang H., Boffa D. J., Ding R., Sharma V. K., Lagman M., Li B., Hering B., Mohanakumar T., Lakey J., Kapur S., Hancock W. W., Suthanthiran M. (2002) Transplantation 74, 1489–1496 [DOI] [PubMed] [Google Scholar]
- 42. Cardozo A. K., Ortis F., Storling J., Feng Y. M., Rasschaert J., Tonnesen M., Van Eylen F., Mandrup-Poulsen T., Herchuelz A., Eizirik D. L. (2005) Diabetes 54, 452–461 [DOI] [PubMed] [Google Scholar]
- 43. Puthalakath H., O'Reilly L. A., Gunn P., Lee L., Kelly P. N., Huntington N. D., Hughes P. D., Michalak E. M., McKimm-Breschkin J., Motoyama N., Gotoh T., Akira S., Bouillet P., Strasser A. (2007) Cell 129, 1337–1349 [DOI] [PubMed] [Google Scholar]
- 44. Gysemans C. A., Ladrière L., Callewaert H., Rasschaert J., Flamez D., Levy D. E., Matthys P., Eizirik D. L., Mathieu C. (2005) Diabetes 54, 2396–2403 [DOI] [PubMed] [Google Scholar]
- 45. Callewaert H. I., Gysemans C. A., Ladrière L., D'Hertog W., Hagenbrock J., Overbergh L., Eizirik D. L., Mathieu C. (2007) Diabetes 56, 2169–2173 [DOI] [PubMed] [Google Scholar]
- 46. Gowda A., Roda J., Hussain S. R., Ramanunni A., Joshi T., Schmidt S., Zhang X., Lehman A., Jarjoura D., Carson W. E., Kindsvogel W., Cheney C., Caligiuri M. A., Tridandapani S., Muthusamy N., Byrd J. C. (2008) Blood 111, 4723–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.