Background: The functional relevance of ST6Gal-I down-regulation during monocyte activation/macrophage differentiation is not well understood.
Results: Cell and transgenic mouse models suggest that ST6Gal-I-mediated sialylation of TNFR1 blocks TNFα-induced apoptosis.
Conclusion: ST6Gal-I down-regulation may limit monocyte/macrophage lifespan by sensitizing cells to apoptosis via TNFR1 hyposialylation.
Significance: This is the first determination that TNFR1 function is regulated by its glycan structure.
Keywords: Apoptosis, Macrophages, Sialic Acid, Sialyltransferase, Tumor Necrosis Factor (TNF), Death Receptor
Abstract
Macrophages play a central role in innate immunity, however mechanisms regulating macrophage survival are not fully understood. Herein we describe a novel apoptotic pathway involving α2-6 sialylation of the TNFR1 death receptor by the ST6Gal-I sialyltransferase. Variant glycosylation of TNFR1 has not previously been implicated in TNFR1 function, and little is known regarding the TNFR1 glycan composition. To study sialylation in macrophages, we treated U937 monocytic cells with PMA, which stimulates both macrophage differentiation and apoptosis. Interestingly, macrophage differentiation induces ST6Gal-I down-regulation, leading to reduced α2-6 sialylation of selected receptors. To prevent loss of α2-6 sialylation, we forced constitutive expression of ST6Gal-I, and found that this strongly inhibited PMA-induced apoptosis. Given that PMA-mediated apoptosis is thought to result from up-regulation of TNFα, which then activates TNFR1, we next evaluated the α2-6 sialylation of TNFR1. U937 cells with forced ST6Gal-I displayed TNFR1 with elevated α2-6 sialylation, and this was associated with diminished TNFα-stimulated apoptosis. Correspondingly, removal of α2-6 sialylation from TNFR1 through either neuraminidase treatment or expression of ST6Gal-I shRNA markedly enhanced TNFα-mediated apoptosis. To confirm the physiologic importance of TNFR1 sialylation, we generated overexpressing ST6Gal-I transgenic mice. Peritoneal macrophages from transgenic lines displayed TNFR1 with elevated α2-6 sialylation, and these cells were significantly protected against TNFα-stimulated apoptosis. Moreover, greater numbers of thioglycollate-induced peritoneal cells were observed in transgenic mice. These collective results highlight a new mechanism of TNFR1 regulation, and further intimate that loss of α2-6 sialylation during macrophage differentiation may limit macrophage lifespan by sensitizing cells to TNFα-stimulated apoptosis.
Introduction
The sialyltransferase family of glycosyltransferases catalyzes the addition of the negatively charged sugar, sialic acid, to various glycoconjugates on glycoproteins and glycolipids (1, 2). The trans-Golgi ST6Gal-I2 sialyltransferase (β-galactoside α2-6-sialyltransferase-1) adds an α2-6-linked sialic acid to the termini of N-linked glycans on glycoproteins bound for the plasma membrane or secretion (3). The importance of ST6Gal-I in the immune system was established by the finding that ST6Gal-I-deficient mice have impaired thymopoiesis and granulopoiesis (4, 5), as well as defective B cell maturation and antibody production (6). Intriguingly, ST6Gal-I expression is dynamically regulated in some immune cell populations. For example, our group has reported that ST6Gal-I is down-regulated during the differentiation of monocytes along the macrophage lineage, which in turn leads to the loss of α2-6 sialylation from selected cell surface receptors including the β1 integrin (7–9).
To study the role of ST6Gal-I in monocyte behavior we previously employed a standard in vitro model system. Specifically, U937 promonocytic cell lines were treated with the protein kinase C (PKC) activator, phorbol-12-myristate-13-acetate (PMA), which is known to stimulate monocyte activation as well as macrophage differentiation. These experiments revealed that PMA, acting through a PKC/ras/ERK signaling pathway, induced up-regulation of the BACE1 β-secretase, which then directed cleavage and shedding of ST6Gal-I from cells (9). ST6Gal-I was also reported by Taniguchi et al. (10) to be down-regulated through transcriptional mechanisms in PMA-treated HL60 myeloid cells. We hypothesize that shedding and transcriptional repression cooperate to maintain low levels of ST6Gal-I (and consequently α2-6 sialylation) in differentiated cells. Importantly, primary human CD14+ monocytes differentiated to macrophages using human serum rather than PMA also exhibited ST6Gal-I down-regulation (9), indicating that loss of ST6Gal-I-dependent α2-6 sialylation is associated with cell differentiation, and not an isolated effect of PMA. Taken together these results suggest that ST6Gal-I down-regulation may be critical for some aspect of the monocyte/macrophage phenotype.
In addition to regulating cell differentiation, PMA treatment of monocytes induces apoptosis (11–14), therefore in the current study we evaluated whether ST6Gal-I down-regulation had an effect on death receptor signaling. It has been shown that PMA-directed apoptosis occurs as a consequence of increased PKC-dependent expression and secretion of tumor necrosis factor α (TNFα), which then binds TNF receptors (TNFR) to induce cell death (12–16). There are two principal receptors that bind TNFα, TNFR1, and TNFR2. These receptors are part of a larger TNFR superfamily that also includes Fas (CD95) and TRAIL-R (DR4 and DR5). Only TNFR1 is considered a canonical death receptor because TNFR2 does not contain the death domain (17, 18). Engagement of TNFR1 with TNFα can give rise to two opposing biological outcomes, survival and apoptosis, depending upon the formation of distinct intracellular signaling complexes on the TNFR1 cytosolic domain.
Monocyte/macrophage apoptosis plays a key role in many physiologic and pathophysiologic processes including atherosclerosis and immune disorders. Signaling through TNFR1 represents a predominant apoptotic pathway in macrophages, however the underlying mechanisms that regulate the balance between macrophage survival and apoptosis remain ill-defined. Although intensive research has been directed at understanding aberrant TNFR1 signaling, the contribution of variant glycosylation to TNFR1 function has not previously been addressed. In the current study we report that TNFR1 is a substrate for ST6Gal-I in both human monocytic cells and primary macrophages harvested from overexpressing ST6Gal-I transgenic mice. More importantly, our results show that ST6Gal-I-mediated α2-6 sialylation of TNFR1 inhibits apoptosis, thus suggesting a new paradigm in death receptor signaling.
EXPERIMENTAL PROCEDURES
Cell Culture and Cell Lines
Human monocytic U937 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 1% amphotericin B, and 1% penicillin/streptomycin. Cells constitutively expressing either ST6Gal-I (ST6) or empty vector (EV) were established using a lentiviral vector, and stably transduced cells were obtained and characterized as previously reported (9). Endogenous ST6Gal-I in the parental U937 cells (Par) was knocked down by introducing two different shRNA sequences using lentiviral vectors from Sigma-Aldrich. The cells were selected by addition of 10 μg/ml of puromycin (Sigma-Aldrich), and then maintained in 0.5 μg/ml puromycin. The puromycin was removed from the medium at least 2 days in advance of all experiments. Each of the established cell lines used for these studies represents a pooled population of stably transduced clones.
PMA and TNFα Induced Apoptosis
U937 cells were seeded in 6-well plates (ultra low attachment, Corning Inc.) in complete DMEM medium overnight. Following this incubation, cells were treated for 24 h with either 100 ng/ml of PMA (Sigma-Aldrich) or a combination of 10 ng/ml TNFα (R&D Systems) plus 5 μm cycloheximide (CHX, from Sigma-Aldrich). Cells were then analyzed for apoptosis by using flow cytometry or FLICA staining as described below.
Neuraminidase Treatment
Cell lysates were incubated with 100 units of Clostridium perfringens neuraminidase (New England Biolabs) in 50 mm sodium citrate buffer (pH 6.0) for 1 h at 37 °C. The treated lysates were then immunoblotted for TNFR1. To test whether sialylation status affects cell apoptosis, 5 × 105 cells were treated with or without 100 units of neuraminidase in PBS (pH 7.4) for 1 h at 37 °C. After washing three times with PBS, cells were treated with or without 10 ng/ml TNFα plus 5 μm cycloheximide for 24 h, and then evaluated for apoptosis.
Flow Cytometric Analysis for Apoptosis
Cells were stained with FITC annexin v and PI using the Vybrant Apoptosis Assay kit (Molecular Probes) for 15 min. Stained cells were analyzed with FACSCalibur (Becton Dickinson).
Immunofluorescent Staining for Activated Caspases
Cells were stained for activated caspases 3 and 7 following the protocols of the sulforhodamine FLICA apoptosis detection kit (Immunochemistry Technologies). Nuclei were labeled by Hoechst stain for 5 min. The cells were then seeded onto slides and viewed under a fluorescent microscope (Nikon). Images were taken by a Nikon CoolSNAP camera. The number of FLICA-positive cells was quantified by counting cells from multiple fields.
ELISA Assay for TNFα
Cells treated with or without PMA were cultured for 24 h, and the culture supernatants were subsequently collected. The amount of TNFα in the media was measured using the human TNF ELISA kit II from BD Biosciences.
Western Blotting
Cells were harvested and lysed in 50 mm Tris buffer (pH 7.4) containing 1% Triton X-100 and a protease inhibitor mixture (Roche Applied Science). Protein concentrations were quantified by Bio-Rad assay. Lysates were separated by SDS-PAGE and transferred to PVDF membranes. The membranes were blocked with 5% nonfat milk and then incubated with a primary antibody, followed by an HRP-conjugated secondary antibody, and detection was accomplished using the Immobilon Chemiluminescent HRP substrate (Millipore). Human and mouse TNFR1 were detected using a rabbit monoclonal antibody from Cell Signaling, and a polyclonal antibody from Abcam, respectively. V5 was immunoblotted with a monoclonal antibody from Sigma-Aldrich. ST6Gal-I expression in mouse primary macrophages was detected with a rabbit polyclonal antibody from Santa Cruz Biotechnology, whereas ST6Gal-I expression in U937 cells was detected using a mouse monoclonal antibody from Sigma-Aldrich or a goat polyclonal antibody from R&D Systems.
SNA and MAA Precipitation
500 μg of cell lysates were incubated overnight at 4 °C with either 60 μl of SNA-conjugated agarose beads (Vector Laboratories) or MAA-conjugated agarose beads (EY Laboratories). α2-6 or α2-3-sialylated proteins bound to the beads were precipitated by centrifugation and washed extensively with lysis buffer. The sialoprotein/lectin conjugates were then resolved by SDS-PAGE and immunoblotted for TNFR1 as described previously.
PNGaseF Treatment
30 μg of cell lysates were incubated with 2 μl of Flavobacterium meningosepticum PNGaseF (New England Biolabs) at 37 °C for 1 h according to manufacturer's instructions. The treated lysates were then immunoblotted for TNFR1. To confirm whether TNFR1 sialylation is through α2-6-linked N-glycans, 250 μg of cell lysates were digested with 10 μl of PNGaseF at 37 °C for 4 h. The lysates were then precipitated by SNA lectin, and precipitates were immunoblotted for TNFR1.
ST6Gal-I Transgenic Mice and Genotyping
A rat ST6Gal-I gene engineered into a pcDNA 3.1/V5-His vector, driven by a CMV promoter, was initially obtained from Dr. Karen Colley (University of Illinois, Chicago). The CMV promoter/V5-epitope-tagged ST6Gal-I transgenic construct with BGH polyadenylation signal was excised from the plasmid backbone and injected into C57BL/6 embryos. Two strains (Line1 and Line 2) from two independent founder mice were maintained for the studies. Genotyping was used to select the mice carrying the rat ST6Gal-I gene. The sequences of the primers for the genotyping are as follows: T7 forward-CTATAGGGAGACCCAAGCTG and ST6 reverse-TCAGAGGCTAGGTAAACCAC. The product of the PCR reaction was excised from gels and sequenced to confirm integration of the ST6Gal-I transgene. Age- and gender-matched C57BL/6 wild-type mice were used as controls for experiments in this study. All experiments involving wild-type and transgenic mice were performed with prior approval from the UAB Institutional Animal Care and Use Committee (IACUC).
Isolation and Apoptosis Analysis of Peritoneal Macrophages from Mice
A 4% thioglycollate solution in distilled water (Sigma-Aldrich) was autoclaved for 15 min. To elicit an inflammatory response and macrophage emigration, 1 ml of the thioglycollate solution was injected intraperitoneally into 8–10-week-old mice. Four days later, the mice were sacrificed and peritoneal macrophages were collected by flushing the peritoneal cavity with ice-cold and serum-free DMEM. After washing with DMEM, the cells were re-suspended in 1 ml of medium. 10 μl of cells were placed into 990 μl of 2% acetic acid, and then 10 μl of cells in the acetic acid solution were placed on a hemacytometer, and cell number was quantified. The acetic acid lyses the cell membrane while keeping the nuclear membrane intact so that the cell counts represent nucleated cell numbers.
2 × 105 freshly-harvested peritoneal cells were stained with FITC annexin V and PI using the Vybrant Apoptosis Assay kit, and then evaluated for apoptosis by flow cytometry. The remaining cells were then resuspended in DMEM supplemented with 10% FBS, and seeded at a density of 5 × 105 cells/cm2. After incubating for 3 h to allow attachment, the media was replaced with fresh medium to remove non-adherent cells and attached macrophages were cultured overnight. The cells were then treated with 10 ng/ml TNFα plus 5 μm CHX for 24 h and stained for apoptosis using Vybrant Apoptosis Assay kit.
RESULTS
ST6Gal-I Prevents PMA-induced Apoptosis in U937 Cells
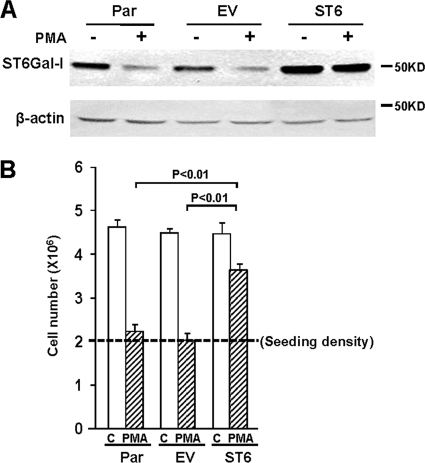
To better understand the role of ST6Gal-I in monocyte/macrophage behavior, we previously generated a stable U937 cell line that constitutively expresses ST6Gal-I (9). As shown in Fig. 1A, treatment of Par or EV U937 cells with PMA causes down-regulation of the endogenous ST6Gal-I enzyme. However, cells with forced expression of ST6 have increased basal levels of enzyme (representing the combined endogenous and exogenous ST6Gal-I), and high levels of ST6Gal-I are maintained following PMA treatment.
FIGURE 1.
ST6Gal-I inhibits PMA-induced suppression of cell growth. A, Western blot shows ST6Gal-I expression in control or PMA-treated Par, EV, or ST6Gal-I transduced (ST6) U937 cells. B, cells were suspended in media with PMA or without PMA (control, C), and then seeded into culture dishes at a density of 2 × 106 (dashed line). Cell number was quantified 24 h following this incubation. Values represent means and S.E. from three independent experiments.
PMA causes monocytic cells, including U937 cells, to exit the cell cycle, begin differentiating along the macrophage lineage, and ultimately undergo apoptosis. Accordingly, we evaluated the effects of ST6Gal-I activity on cell proliferation and survival. Cells were suspended in media with or without PMA, and then seeded into tissue culture dishes at a density of 2 × 106. Twenty-four hours later, there was a 2-fold increase in cell number for Par, EV and ST6 cells in control media, which is consistent with the known rapid doubling time of U937 cells (Fig. 1B). Treatment of Par and EV cells with PMA completely prevented this cell doubling, suggesting exit from the cell cycle and/or decreased cell survival. Remarkably, the ST6 cell population continued to expand in the presence of PMA, suggesting that ST6Gal-I-mediated sialylation may play some role in the shift between a proliferative versus differentiated phenotype.
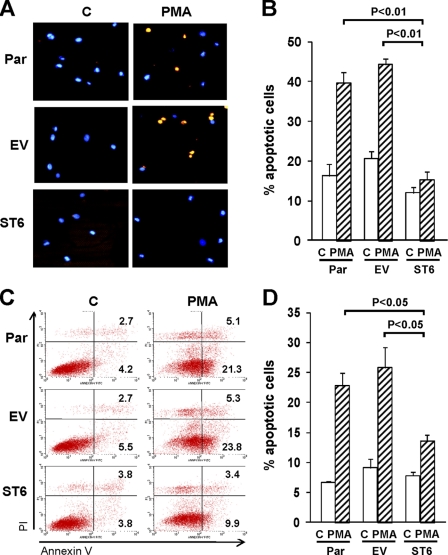
Although the results in Fig. 1 suggested that ST6Gal-I likely affects both proliferation and cell survival, we focused on apoptosis in the current study because many of the molecular events in PMA-mediated apoptosis are known. To evaluate whether ST6Gal-I activity influences cell death, we performed immunostaining to detect activated caspases 3 and 7 (Fig. 2, A and B). As compared with untreated cells, a substantially greater number of apoptotic cells was apparent in Par and EV cells treated with PMA. In contrast, no significant difference in the number of apoptotic cells was observed for untreated and PMA-treated ST6 cells.
FIGURE 2.
ST6Gal-I protects U937 cells from PMA-induced apoptosis. A, cells were treated with or without PMA (control, C) for 24 h and then stained for the apoptotic markers, activated caspases 3 and 7 (orange). Nuclei were counterstained with Hoechst (blue). B, percentage of apoptotic cells was quantified by counting cells positive for activated caspases 3 and 7 relative to total cells from multiple microscopic fields. C, cells treated with or without PMA were stained with FITC annexin V and propidium iodide (PI). Lower right quadrant, annexin V positive; Upper right quadrant, annexin V + PI positive. D, quantification of annexin-positive cells (representing combined totals from lower and upper-right quadrants). Values represent means and S.E. from three independent experiments.
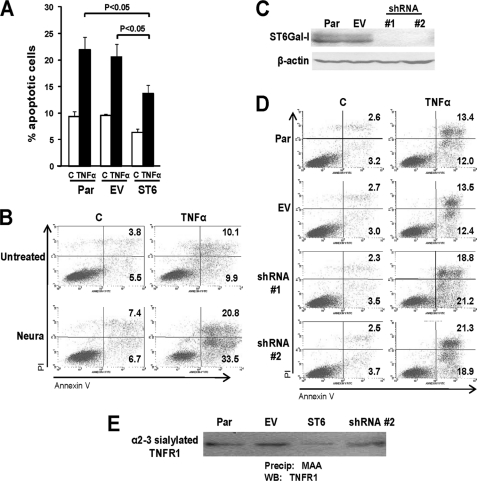
Inhibition of PMA-induced apoptosis by ST6Gal-I was further confirmed by monitoring reactivity of the cells to annexin V and propidium iodide (PI). As shown in Fig. 2, C and D, PMA treatment of Par and EV cells caused the apoptotic rates to reach ∼20–25%, whereas PMA-induced apoptosis was markedly attenuated in ST6 cells.
Forced Expression of ST6Gal-I Blocks PMA-induced Loss of Sialylation from TNFR1
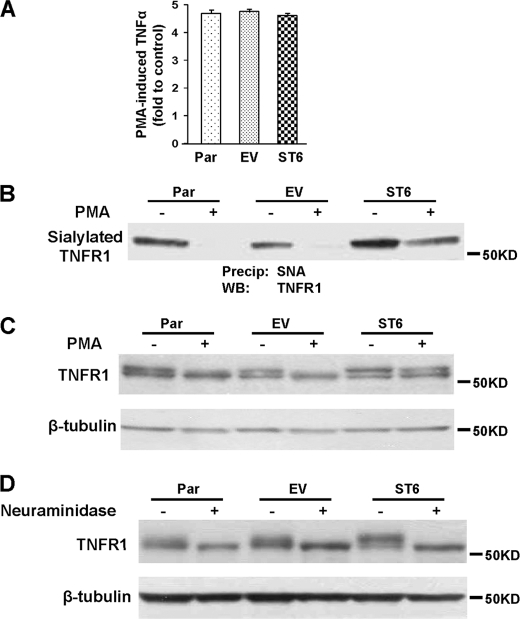
PMA-mediated apoptosis of U937 cells is thought to be due to PMA-induced synthesis and secretion of TNFα, which then directs cell death by signaling through the TNFR1 receptor (12–15). To test whether ST6Gal-I activity alters TNFα expression, cells were treated with or without PMA for 24 h, and then culture supernatants were collected and TNFα production was monitored by ELISA. As shown in Fig. 3A, TNFα production was increased by about 4.5-fold following PMA treatment in all three cell lines, with no significant difference in levels observed. These results indicate that ST6Gal-I-mediated inhibition of apoptosis is not due to changes in TNFα synthesis or release.
FIGURE 3.
PMA-induced down-regulation of ST6Gal-I is associated with loss of α2-6 sialylation from TNFR1. A, cells were treated with or without PMA for 24 h, and then the amount of TNFα secreted into culture media was measured by ELISA. Data are plotted as fold TNFα induced by PMA as compared with control samples. Values represent means and S.E. from three independent experiments. B, lysates from cells treated with or without PMA were incubated with agarose-conjugated SNA lectin. α2-6 sialylated proteins were precipitated and blotted for TNFR1. C, total TNFR1 levels were detected by blots of whole cell lysates. D, cell lysates were treated with neuraminidase at 37 °C for 1 h, and then Western blot was performed with anti-TNFR1 antibody.
We next tested whether ST6Gal-I activity caused greater α2-6 sialylation of TNFR1, leading to altered receptor signaling. To measure TNFR1 sialylation levels, cell lysates were incubated with agarose-conjugated SNA lectin, which binds specifically to α2-6 sialylated proteins. Sialylated proteins were then precipitated and immunoblotted for TNFR1. As shown in Fig. 3B, the highest expression level of α2-6 sialylated TNFR1 was observed in the untreated ST6 cells, although a considerable amount of α2-6 sialylated TNFR1 was observed in untreated Par and EV cells as well. Upon PMA treatment, levels of sialylated TNFR1 were dramatically reduced in Par and EV cells, consistent with PMA-induced down-regulation of endogenous ST6Gal-I. In contrast, TNFR1 from PMA-treated ST6 cells still retained substantial reactivity toward SNA, reflecting high α2-6 sialylation. These results suggest that TNFR1 is modified by ST6Gal-I, and that PMA induces expression of a hyposialylated TNFR1 glycoform in parental U937 cells.
Total TNFR1 levels (sialylated + unsialylated) were also monitored by Western blots of whole cell lysates (Fig. 3C). Interestingly, two bands were observed for TNFR1 in untreated Par and EV cells, and the upper band was lost in these cells following PMA treatment. These data suggested that the upper band represents the α2-6 sialylated form of TNFR1. In contrast to Par and EV cells, the upper band was retained in PMA-treated ST6 cells, consistent with the expression of α2-6 sialylated TNFR1.
To further confirm that the upper band represents an α2-6 sialylated TNFR1 isoform, Par, EV, and ST6 cell lysates were treated with neuraminidase and then Western blotted for TNFR1 (Fig. 3D). The upper band was eliminated by neuraminidase treatment in all three cell lines, confirming the identity of this band as a sialylated TNFR1 variant. These collective results are the first to identify TNFR1 as an ST6Gal-I substrate, and to show that α2-6 sialylation of TNFR1 is lost upon differentiation of monocytic cells along the macrophage lineage.
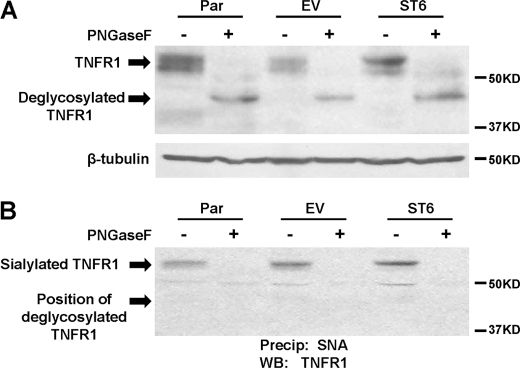
To determine whether α2-6 sialic acids were present on N-glycans, cell lysates were pretreated with PNGase F enzyme to remove N-linked glycosylation, and then TNFR1 was characterized. Immunoblots of TNFR1 in the PNGase F-treated lysates revealed a single band at ∼45kD, indicating effective cleavage by PNGase F (Fig. 4A). The PNGase F-treated and control cell lysates were then incubated with SNA-agarose to precipitate α2-6 sialylated TNFR1 (Fig. 4B). TNFR1 was precipitated by SNA from control lysates, however in contrast, no detectable TNFR1 was observed in precipitates from PNGase F-treated lysates. These results suggest that the major source of α2-6 sialic acids on TNFR1 is N-linked, rather than O-linked, glycans.
FIGURE 4.
N-glycans are the major source of α2-6 sialic acids on TNFR1. A, cell lysates were treated with or without PNGaseF, and then TNFR1 immunoblots were performed. B, cell lysates were treated with or without PNGaseF, and then lysates were incubated with agarose-conjugated SNA lectin. α2-6 sialylated proteins were precipitated and blotted for TNFR1.
α2-6 Sialylation of TNFR1 Controls TNFα-directed Apoptosis
To evaluate whether altered α2-6 sialylation of TNFR1 regulates apoptotic signaling, Par, EV, and ST6 cells were treated with TNFα and then monitored for annexin V reactivity. As shown in Fig. 5A, ST6 cells exhibited significantly less TNFα-induced apoptosis than Par or EV cells. Notably, α2-6 sialylation is present on TNFR1 expressed by Par and EV (in the absence of PMA treatment), however the level of TNFR1 α2-6 sialylation in ST6 cells is clearly higher (see Fig. 3B). These results suggest that α2-6 sialylation of TNFR1 has a protective effect on cell death.
FIGURE 5.
Decreased α2-6 sialylation of TNFR1 promotes U937 cell apoptosis. A, cells were treated with or without TNFα plus CHX for 24 h (C, control). Cells were then analyzed by flow cytometry for annexin V and PI. Values represent means and S.E. for three independent experiments. B, annexin V and PI reactivity were analyzed in Par cells treated with neuraminidase at 37 °C for 1 h, followed by treatment with TNFα plus CHX for 24 h. C, Par cells were transduced with two different shRNA sequences (#1 and #2) against ST6Gal-I. Western blotting was performed to confirm knockdown. D, Par, EV, and shRNA-transduced cells were treated with TNFα plus CHX for 24 h, and then evaluated for annexin V and PI reactivity. E, lysates from Par, EV, ST6, or shRNA (sequence #2) cells were incubated with MAA-agarose and α2-3 sialylated proteins were precipitated by centrifugation. Precipitates were immunoblotted for TNFR1.
We next examined whether the forced loss of α2-6 sialylation from TNFR1 would sensitize cells to TNFα-mediated apoptosis. To this end, Par cells were treated with neuraminidase to remove sialic acids, and apoptotic rates were analyzed by annexin-V reactivity. TNFα-induced apoptosis was substantially increased in the cells treated with neuraminidase, indicating that reduced α2-6 sialylation of TNFR1 makes cells more susceptible to apoptosis (Fig. 5B).
To more directly investigate whether loss of α2-6 sialylation from TNFR1 promotes cell apoptosis, endogenous ST6Gal-I was knocked down in parental cells using shRNA. Two different shRNA sequences were tested, and for each a pooled population of stable clones was generated. As shown in Fig. 5C, ST6Gal-I levels were greatly reduced in both of the shRNA-transduced cells indicating effective knockdown. Par, EV, and the two ST6Gal-I knock-down cell lines were then evaluated for apoptosis (Fig. 5D). Annexin-V labeling experiments revealed that forced down-regulation of ST6Gal-I dramatically increased the amount of TNFα-induced apoptosis. Hence, loss of α2-6 sialylation from TNFR1 sensitizes cells to TNFα-mediated apoptosis, whereas elevated ST6Gal-I-mediated α2-6 sialylation of TNFR1 conversely provides protection against TNFα-induced cell death. To further assess the functional importance of α2-6 sialylation, we screened for potential changes in α2-3 sialylation that may have occurred as a consequence of indirect effects of ST6Gal-I knockdown. Lysates from Par, EV, ST6, or shRNA cell lines were subjected to precipitation with the α2-3 sialic acid-selective lectin, MAA, and then blotted for TNFR1. Comparable levels of α2-3-sialylated TNFR1 were observed in parental and shRNA-transduced cells, indicating that the effects of ST6Gal-I knockdown on apoptosis are not due to any compensatory changes in α2-3 sialylation (Fig. 5E).
TNFR1 Has Increased α2-6 Sialylation in Cells from Overexpressing ST6Gal-I Transgenic Mice
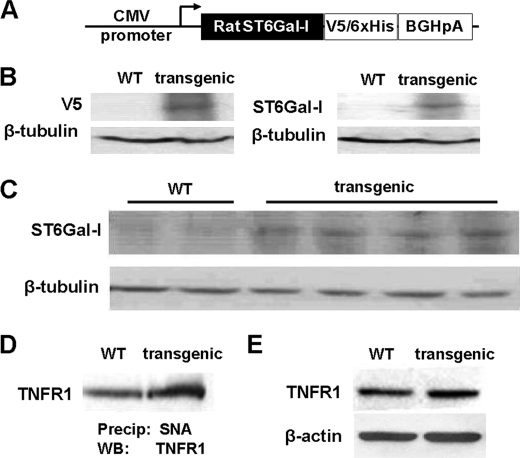
To elucidate the physiologic importance of ST6Gal-I in regulating TNFR1-mediated apoptosis, we generated transgenic mice that constitutively express ST6Gal-I. Briefly, a V5-tagged rat ST6Gal-I cDNA, cloned into the pcDNA 3.1/V5 vector that contains the CMV promoter (Fig. 6A), was injected into fertilized C57BL/6 eggs. Two independent transgenic lines were identified and studied (Line 1 and Line 2). Expression of the transgene was evaluated by immunoblotting. As shown in Fig. 6B, splenic homogenates from transgenic mice were positive for both V5 and ST6Gal-I, verifying protein expression of the transgene. The transgenic mice were fertile and did not present with any gross phenotypic abnormalities. Also, histologic analyses of multiple organs from older (>9 months) transgenic animals did not reveal any obvious differences from non-transgenic littermate controls.
FIGURE 6.
ST6Gal-I activity causes increased α2-6 sialylation of TNFR1 in vivo. A, construct used to generate ST6Gal-I-expressing transgenic mice. B, splenic homogenates were prepared from wild-type (WT) or transgenic mice, and immunoblotting was performed with anti-V5 antibody and anti-ST6Gal-I antibody. C, peritoneal cells collected 96 h following thioglycollate injection were immunoblotted for ST6Gal-I. D, α2-6 sialylated proteins from peritoneal cell lysates were precipitated with SNA-agarose and immunoblotted for TNFR1. E, cell lysates from wild-type and transgenic mice were immunoblotted for TNFR1.
To assess expression of ST6Gal-I in immune cells of transgenic mice, animals were injected intraperitoneally with 4% thioglycollate, and infiltrating cells were collected by lavage 96 h following injection. At 96 h, cells of the monocyte/macrophage lineage represent the predominate cell type within peritoneal infiltrates (19, 20). Peritoneal cells collected from 4 different transgenic mice exhibited elevated ST6Gal-I relative to wild-type mice (Fig. 6C), although the level of ST6Gal-I up-regulation was relatively modest in this cell type. It is possible that this may reflect cell-type specific differences in the efficacy of the CMV promoter, and it should also be noted that transgene expression is hemiallelic. Nonetheless, SNA analyses (Fig. 6D) revealed that the TNFR1 expressed by transgenic cells had increased levels of α2-6 sialylation. Interestingly, immunoblots of TNFR1 expressed by mouse macrophages revealed only one band for the receptor (Fig. 6E), whereas two bands were observed in lysates from human U937 cells, with the upper band representing the sialylated form. We hypothesize that the size of variantly glycosylated TNFR1 isoforms in transgenic mouse cells may be too small to resolve by SDS-PAGE, because of more modest changes in the degree of sialylation.
Macrophages from Transgenic ST6Gal-I Mice Are Resistant to TNFα-induced Apoptosis
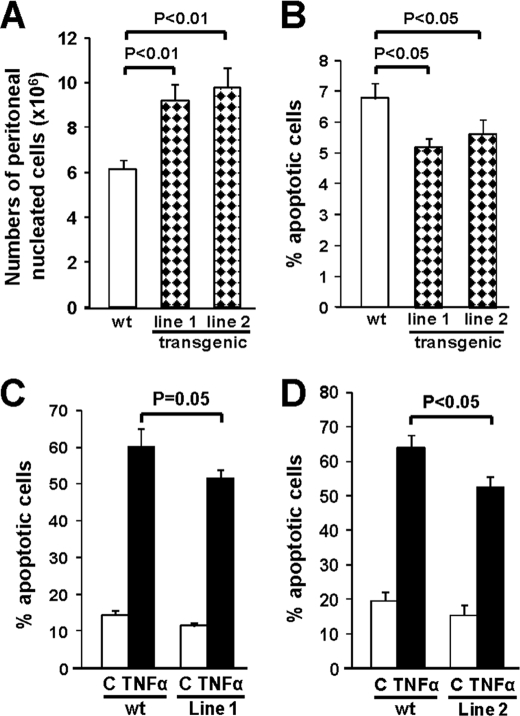
To examine whether forced ST6Gal-I-mediated sialylation of TNFR1 prevents monocyte/macrophage apoptosis in vivo, peritoneal cells were collected at 96 h following thioglycollate injection and evaluated for annexin reactivity. Intriguingly, an increased number of peritoneal cells was observed for each of the two independent transgenic mouse lines (Fig. 7A), potentially reflecting greater survival of these cells (although other mechanisms are also possible). Furthermore, cells from the transgenic animals displayed decreased annexin V reactivity (Fig. 7B), which may be due to reduced apoptosis induced by endogenous TNFα or other factors within the inflammatory milieu (e.g. galectins). To more directly assess TNFR1-mediated apoptosis in macrophages, peritoneal cells were cultured overnight, and then cells were treated with TNFα (Fig. 7, C and D). These experiments revealed that TNFα-induced apoptosis was also significantly attenuated in macrophages harvested from transgenic mice. These results provide strong evidence that ST6Gal-I mediated α2-6 sialylation of TNFR1 plays a critical role in regulating apoptotic signaling from this receptor.
FIGURE 7.
ST6Gal-I inhibits apoptosis of macrophages from ST6Gal-I transgenic mice. A, peritoneal cells were harvested at 96 h following thioglycollate injection and nucleated cell counts were obtained using a hemacytometer (n = 10 for each group of animals). B, peritoneal cells were stained with annexin V and PI, and the apoptosis rate was analyzed by flow cytometry (n = 11 WT; n = 10 line 1; n = 9 line 2). C and D, peritoneal cells were cultured overnight, and the adherent fraction (representing macrophages) was treated with TNFα plus CHX for 24 h. Annexin V and PI staining revealed significantly lower levels of apoptosis in each of the two transgenic lines. (For panel C, n = 5 for each group; for panel D, n = 8 for each group.)
DISCUSSION
The TNFα/TNFR signaling axis plays a critical, but complex, role in the pathogenesis of chronic inflammatory and autoimmune disorders. The TNFR1 receptor can initiate both proliferative and apoptotic signaling cascades, and elucidation of mechanisms regulating these divergent pathways is of obvious importance for developing effective clinical treatments. Most studies of pathogenic TNFR1-dependent signaling have focused on changes in the expression level of TNFR1 or associated signaling molecules (21–23) whereas limited attention has been paid to potential mechanisms that control TNFR1 activity independently of variant protein expression, for example, regulation via post-translational modifications such as glycosylation. To date there is very little information concerning the glycosylation of TNFR1. Analyses of the secreted form of TNFR1 present in human urine suggest that TNFR1 is elaborated with both N- and O-linked glycans (24), and a soluble chimeric TNFR1-IgG protein was also found to be N-glycosylated (25). However, to our knowledge, the glycan profile on the native transmembrane form of TNFR1 has not been previously characterized. There are three consensus sequences for N-glycosylation on the human TNFR1 molecule (25, 26), but it is not known which of these sites are filled, nor is anything known regarding oligosaccharide composition. In the current study we show for the first time that TNFR1 is α2-6 sialylated by ST6Gal-I, and that the degree of TNFR1 α2-6 sialylation changes as a consequence of cell differentiation status. Induction of macrophage differentiation in U937 monocytic cells causes down-regulation of ST6Gal-I, leading to a loss of α2-6 sialylation on TNFR1, and an associated increase in cell death. Preventing TNFR1 hyposialylation through forced overexpression of ST6Gal-I protects against apoptosis, whereas forced down-regulation of ST6Gal-I in undifferentiated U937 cells (with high endogenous ST6Gal-I) correspondingly enhances apoptosis stimulated by TNFα. Further confirming a role for α2-6 sialylation in TNFR1 activity, macrophages from ST6Gal-I-overexpressing transgenic animals are protected against TNFα-induced apoptosis. These collective results highlight a completely new mechanism for regulation of TNFR1. Additional experiments will be required to decipher the molecular mechanism by which α2-6 sialylation alters TNFR1 structure/function, however studies of other ST6Gal-I substrates suggest that α2-6 sialylation can affect receptor conformation, clustering and/or internalization, depending upon the specific receptor. For example, ST6Gal-I-mediated α2-6 sialylation of the β1 integrin modulates receptor conformation (9), whereas α2-6 sialylation alternatively regulates clustering of the CD45 tyrosine phosphatase on T cells (27), and both clustering and internalization of the PECAM molecule on endothelial cells (28).
In general, research centered on the role of α2-6 sialylation in regulating the activity of specific receptors has been relatively limited, which is surprising given that ST6Gal-I expression changes dramatically in association with many immune cell phenotypic alterations. In addition to down-regulation of ST6Gal-I during macrophage differentiation (7–10), ST6Gal-I levels are markedly decreased as a consequence of T cell activation (29, 30), as well as dendritic cell maturation (31, 32). One known critical function of ST6Gal-I-directed α2-6 sialylation is to inhibit the binding of galectins to cell surface oligosaccharides (33). Galectins are β-galactoside-binding lectins that regulate multiple cell behaviors including induction of apoptosis, and the addition of α2-6 sialic acid to the galactosyl structure blocks galectin binding (27, 34–37). In an elegant study by Toscano et al. (38), ST6Gal-I levels were shown to be significantly lower in Th1 and Th17 cells as compared with Th2 cells, and consequently, the lower levels of ST6Gal-I-mediated α2-6 sialylation on Th1 and Th17 cells allowed greater galectin-1-induced apoptosis. Thus, reduced expression of ST6Gal-I facilitated the selective killing of Th1 and Th17 cells. The results presented in the current study now highlight another major molecular pathway by which ST6Gal-I-mediated sialylation regulates cell death; specifically, inactivation of TNFR1 through α2-6 sialylation. Given the multiplicity of apoptotic pathways regulated by ST6Gal-I, this enzyme may serve as a major regulator of the lifespan of a subset of activated and/or differentiated immune cell populations. Finally, it is intriguing that loss of sialylation has recently been identified as an cell surface marker for ingestion of apoptotic cells by macrophages (39). These data suggest that diminished α2-6 sialylation may couple the induction of apoptotic mechanisms to the process of apoptotic cell elimination by phagocytes.
The finding that α2-6 sialylation of TNFR1 modulates apoptotic signaling adds to an emerging body of literature suggesting that variant glycosylation represents an important mechanism for regulating death receptor function. The first evidence supporting this concept was provided by studies of sialylation on the Fas death receptor, which is another member of the TNFR superfamily. Peter et al. reported that treatment of T cells with neuraminidase to remove sialic acids (both α2-3 and α2-6-linked)-sensitized cells to Fas-mediated apoptosis (40). Consistent with these results, Fas-mediated apoptosis was enhanced in hyposialylated subclones of human B lymphoma cells as compared with highly sialylated clones (α2-3, and α2-6 sialic acids were not distinguished in this study) (41). More recently our group established that Fas is a substrate for ST6Gal-I in colon carcinoma cells, and determined that α2-6 sialylation of Fas prevented apoptosis by blocking receptor internalization as well as the binding of FADD to Fas cytosolic tails (42); this latter event is crucial for the formation of the Death Inducing Signaling Complex (DISC). However, we also reported in this same study that ST6Gal-I activity does not alter TRAIL-mediated apoptosis, suggesting that α2-6 sialylation has a selective effect on signaling through the Fas receptor, but not DR5. Of note, DR5 does not contain any consensus sequences for N-glycosylation. As a fascinating counterpoint to these studies, Wagner et al. reported that a specific type of O-linked glycan on DR5 controls sensitivity to TRAIL, but does not affect signaling through Fas (43), and importantly, the consensus sequences for the functionally-relevant O-glycans identified by this group are not conserved in either TNFR1 or Fas (43). Although minimal attention has been directed at understanding the role of glycosylation in death receptor function, these landmark studies strongly point to the functional importance of variant glycosylation, and also highlight the concept that there is specificity in the molecular pathways that control the glycosylation-dependent regulation of distinct death receptors.
Taken together, results presented in the current study elucidate a novel glycosylation-dependent mechanism for regulating immune cell survival. The loss of α2-6 sialylation from selected cell surface receptors, due to down-regulation of ST6Gal-I during the activation/maturation of monocytes, T cells and dendritic cells, would be expected to sensitize cells to several apoptotic stimuli including TNFα, FasL, and galectins, thus limiting immune cell lifespan. Conversely, overexpression of ST6Gal-I, which is observed in both leukemia and epithelial cancers (33), confers protection against apoptosis. One anticipates that manipulating ST6Gal-I expression to control death receptor signaling, and therefore immune cell apoptosis, could provide a novel therapeutic tool for treating inflammatory and autoimmune disorders.
Acknowledgments
We thank the UAB Arthritis and Musculoskeletal Center Analytic and Preparative Core Facility for assistance with flow cytometry, and the UAB Transgenic Mouse Facility for assistance in generating the ST6Gal-I transgenic mice.
This work was supported, in whole or in part, by Grants R01 CA84248 (to S. L. B.) and P30AR48311 from the National Institutes of Health, the UAB Rheumatic Diseases Core Center, and the American Heart Association (Grant-in-Aid to S. L. B.).
- ST6Gal-I
- β-galactoside α2-6-sialyltransferase I
- TNFR1
- tumor necrosis factor receptor I
- PMA
- phorbol-12-myristate-13-acetate
- MAA
- Maackia amurensis agglutinin
- SNA
- Sambucus nigra agglutinin
- PNGaseF
- peptide:N-glycosidase F
- EV
- empty-vector transduced
- Par
- parental
- ST6
- ST6Gal-I.
REFERENCES
- 1. Takashima S. (2008) Biosci. Biotechnol. Biochem. 72, 1155–1167 [DOI] [PubMed] [Google Scholar]
- 2. Harduin-Lepers A., Vallejo-Ruiz V., Krzewinski-Recchi M. A., Samyn-Petit B., Julien S., Delannoy P. (2001) Biochimie 83, 727–737 [DOI] [PubMed] [Google Scholar]
- 3. Dall'Olio F. (2000) Glycoconj J. 17, 669–676 [DOI] [PubMed] [Google Scholar]
- 4. Marino J. H., Tan C., Davis B., Han E. S., Hickey M., Naukam R., Taylor A., Miller K. S., Van De Wiele C. J., Teague T. K. (2008) Glycobiology 18, 719–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nasirikenari M., Segal B. H., Ostberg J. R., Urbasic A., Lau J. T. (2006) Blood 108, 3397–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hennet T., Chui D., Paulson J. C., Marth J. D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4504–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Semel A. C., Seales E. C., Singhal A., Eklund E. A., Colley K. J., Bellis S. L. (2002) J. Biol. Chem. 277, 32830–32836 [DOI] [PubMed] [Google Scholar]
- 8. Seales E. C., Shaikh F. M., Woodard-Grice A. V., Aggarwal P., McBrayer A. C., Hennessy K. M., Bellis S. L. (2005) J. Biol. Chem. 280, 37610–37615 [DOI] [PubMed] [Google Scholar]
- 9. Woodard-Grice A. V., McBrayer A. C., Wakefield J. K., Zhuo Y., Bellis S. L. (2008) J. Biol. Chem. 283, 26364–26373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taniguchi A., Higai K., Hasegawa Y., Utsumi K., Matsumoto K. (1998) FEBS Lett. 441, 191–194 [DOI] [PubMed] [Google Scholar]
- 11. de Vente J., Kiley S., Garris T., Bryant W., Hooker J., Posekany K., Parker P., Cook P., Fletcher D., Ways D. K. (1995) Cell Growth Differ. 6, 371–382 [PubMed] [Google Scholar]
- 12. Rahmani M., Dai Y., Grant S. (2002) Exp. Cell Res. 277, 31–47 [DOI] [PubMed] [Google Scholar]
- 13. Takada Y., Hachiya M., Osawa Y., Hasegawa Y., Ando K., Kobayashi Y., Akashi M. (1999) J. Biol. Chem. 274, 28286–28292 [DOI] [PubMed] [Google Scholar]
- 14. Cartee L., Smith R., Dai Y., Rahmani M., Rosato R., Almenara J., Dent P., Grant S. (2002) Mol. Pharmacol. 61, 1313–1321 [DOI] [PubMed] [Google Scholar]
- 15. O'Sullivan A. W., Wang J. H., Redmond H. P. (2009) J. Surg Res. 151, 138–144 [DOI] [PubMed] [Google Scholar]
- 16. Gonzalez-Guerrico A. M., Kazanietz M. G. (2005) J. Biol. Chem. 280, 38982–38991 [DOI] [PubMed] [Google Scholar]
- 17. Thorburn A. (2004) Cell Signal. 16, 139–144 [DOI] [PubMed] [Google Scholar]
- 18. Guicciardi M. E., Gores G. J. (2009) FASEB J. 23, 1625–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chan J., Leenen P. J., Bertoncello I., Nishikawa S. I., Hamilton J. A. (1998) Blood 92, 1423–1431 [PubMed] [Google Scholar]
- 20. Greenlee M. C., Sullivan S. A., Bohlson S. S. (2009) Inflamm. Res. 58, 909–919 [DOI] [PubMed] [Google Scholar]
- 21. Parameswaran N., Patial S. (2010) Crit. Rev. Eukaryot Gene Expr. 20, 87–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang L., Connelly J. J., Peppel K., Brian L., Shah S. H., Nelson S., Crosslin D. R., Wang T., Allen A., Kraus W. E., Gregory S. G., Hauser E. R., Freedman N. J. (2010) Hum. Mol. Genet. 19, 2754–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Al-Lamki R. S., Wang J., Vandenabeele P., Bradley J. A., Thiru S., Luo D., Min W., Pober J. S., Bradley J. R. (2005) FASEB J. 19, 1637–1645 [DOI] [PubMed] [Google Scholar]
- 24. Corti A., Merli S., Bagnasco L., D'Ambrosio F., Marino M., Cassani G. (1995) J. Interferon Cytokine Res 15, 143–152 [DOI] [PubMed] [Google Scholar]
- 25. Köhne C., Johnson A., Tom S., Peers D. H., Gehant R. L., Hotaling T. A., Brousseau D., Ryll T., Fox J. A., Chamow S. M., Berman P. W. (1999) J. Cell. Biochem. 75, 446–461 [PubMed] [Google Scholar]
- 26. Zhang J., Hawari F. I., Shamburek R. D., Adamik B., Kaler M., Islam A., Liao D. W., Rouhani F. N., Ingham M., Levine S. J. (2008) Biochem. Biophys. Res. Commun. 366, 579–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Amano M., Galvan M., He J., Baum L. G. (2003) J. Biol. Chem. 278, 7469–7475 [DOI] [PubMed] [Google Scholar]
- 28. Kitazume S., Imamaki R., Ogawa K., Komi Y., Futakawa S., Kojima S., Hashimoto Y., Marth J. D., Paulson J. C., Taniguchi N. (2010) J. Biol. Chem. 285, 6515–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Comelli E. M., Sutton-Smith M., Yan Q., Amado M., Panico M., Gilmartin T., Whisenant T., Lanigan C. M., Head S. R., Goldberg D., Morris H. R., Dell A., Paulson J. C. (2006) J. Immunol. 177, 2431–2440 [DOI] [PubMed] [Google Scholar]
- 30. Kaech S. M., Hemby S., Kersh E., Ahmed R. (2002) Cell 111, 837–851 [DOI] [PubMed] [Google Scholar]
- 31. Jenner J., Kerst G., Handgretinger R., Müller I. (2006) Exp. Hematol. 34, 1212–1218 [DOI] [PubMed] [Google Scholar]
- 32. Videira P. A., Amado I. F., Crespo H. J., Algueró M. C., Dall'Olio F., Cabral M. G., Trindade H. (2008) Glycoconj J. 25, 259–268 [DOI] [PubMed] [Google Scholar]
- 33. Zhuo Y., Bellis S. L. (2011) J. Biol. Chem. 286, 5935–5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hirabayashi J., Hashidate T., Arata Y., Nishi N., Nakamura T., Hirashima M., Urashima T., Oka T., Futai M., Muller W. E., Yagi F., Kasai K. (2002) Biochim. Biophys. Acta 1572, 232–254 [DOI] [PubMed] [Google Scholar]
- 35. Leppänen A., Stowell S., Blixt O., Cummings R. D. (2005) J. Biol. Chem. 280, 5549–5562 [DOI] [PubMed] [Google Scholar]
- 36. Stowell S. R., Arthur C. M., Mehta P., Slanina K. A., Blixt O., Leffler H., Smith D. F., Cummings R. D. (2008) J. Biol. Chem. 283, 10109–10123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhuo Y., Chammas R., Bellis S. L. (2008) J. Biol. Chem. 283, 22177–22185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Toscano M. A., Bianco G. A., Ilarregui J. M., Croci D. O., Correale J., Hernandez J. D., Zwirner N. W., Poirier F., Riley E. M., Baum L. G., Rabinovich G. A. (2007) Nat. Immunol. 8, 825–834 [DOI] [PubMed] [Google Scholar]
- 39. Meesmann H. M., Fehr E. M., Kierschke S., Herrmann M., Bilyy R., Heyder P., Blank N., Krienke S., Lorenz H. M., Schiller M. (2010) J. Cell Sci. 123, 3347–3356 [DOI] [PubMed] [Google Scholar]
- 40. Peter M. E., Hellbardt S., Schwartz-Albiez R., Westendorp M. O., Walczak H., Moldenhauer G., Grell M., Krammer P. H. (1995) Cell Death Differ. 2, 163–171 [PubMed] [Google Scholar]
- 41. Keppler O. T., Peter M. E., Hinderlich S., Moldenhauer G., Stehling P., Schmitz I., Schwartz-Albiez R., Reutter W., Pawlita M. (1999) Glycobiology 9, 557–569 [DOI] [PubMed] [Google Scholar]
- 42. Swindall A. F., Bellis S. L. (2011) J. Biol. Chem. 286, 22982–22990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wagner K. W., Punnoose E. A., Januario T., Lawrence D. A., Pitti R. M., Lancaster K., Lee D., von Goetz M., Yee S. F., Totpal K., Huw L., Katta V., Cavet G., Hymowitz S. G., Amler L., Ashkenazi A. (2007) Nat. Med. 13, 1070–1077 [DOI] [PubMed] [Google Scholar]