Background: Species-dependent pharmacology is an obstacle for TRPV1 antagonist development.
Results: By exchanging the pore domains TRPV1 antagonist JYL-1421, which is modality-selective in rTRPV1 can be made modality-selective in hTRPV1 and vice-versa.
Conclusion: The pore region is critical for the observed species differences.
Significance: Thus, the findings are of significance for the development of more specific and selective TRPV1 antagonists.
Keywords: Calcium, Cloning, Drug Discovery, Electrophysiology, Fluorescence, TRP Channels
Abstract
The capsaicin-, heat-, and proton-activated ion channel TRPV1, a member of the transient receptor potential cation channel family is a polymodal nociceptor. For almost a decade, TRPV1 has been explored by the pharmaceutical industry as a potential target for example for pain conditions. Antagonists which block TRPV1 activation by capsaicin, heat, and protons were developed by a number of pharmaceutical companies. The unexpected finding of hyperthermia as an on-target side effect in clinical studies using polymodal TRPV1 antagonists has prompted companies to search for ways to circumvent hyperthermia, for example by the development of modality-selective antagonists. The significant lack of consistency of the pharmacology of many TRPV1 antagonists across different species has been a further obstacle. JYL-1421 for example was shown to block capsaicin and heat responses in human and monkey TRPV1 while it was largely ineffective in blocking heat responses in rat TRPV1. These findings suggested structural dissimilarities between different TRPV1 species relevant for small compound antagonism for example of heat activation. Using a chimeric approach (human and rat TRPV1) in combination with a novel FLIPR-based heat activation assay and patch-clamp electrophysiology we have identified the pore region as being strongly linked to the observed species differences. We demonstrate that by exchanging the pore domains JYL-1421, which is modality-selective in rat can be made modality-selective in human TRPV1 and vice-versa.
Introduction
The superfamily of transient receptor potential (TRP)3 cation channels plays a major role in sensory transduction including vision, touch, hearing, thermosensation, taste sensation, osmosensation, and pain sensation (1–8).
TRPV1 is a polymodal nociceptor activated by heat, protons and the alkaloid irritant capsaicin. Over the last decade the pharmaceutical industry has developed a plethora of different TRPV1 antagonists with the aim to treat for example pain conditions. Many of these antagonists show species-dependent pharmacological profiles. More importantly, candidate antagonists exhibited species-dependent hyperthermia, an undesired on-target side effect, in vivo (9–12). Currently, it is unclear whether block of heat or proton or capsaicin activation, or combinations of them cause hyperthermia.
Molecular determinants of heat and proton activation have recently been identified in the pore region of TRPV1. In analogy to a previous approach for TRPV3, Grandl et al. performed high-throughput site-directed mutagenesis screenings leading to the identification of amino acids selectively involved in heat activation of rat TRPV1 (rTRPV1) (13–14). TRPV1 heat activation could thus be linked to three distinct amino acids in the pore region (N628K/N652T/Y653T). In another mutagenesis approach focused on the pore region of TRPV1, a position in close proximity to two of these heat-sensitive amino acids, Phe-660, has been found to lead to selective and complete loss of proton activation and potentiation when mutated (15).
Although our knowledge about molecular determinants of different TRPV1 activation modes has increased, the observed lack of consistency of the pharmacology of many TRPV1 antagonists across different species remains an obstacle. Capsazepine for instance appears to be quite ineffective as a proton blocker in rTRPV1. Heat and capsaicin responses are blocked at much lower capsazepine concentrations: >40,000 nm (pH) versus 887 nm (capsaicin) and 6 nm (heat). However, in hTRPV1 all three modes of activation are blocked with similar potency (12). Another compound, JYL-1421, was shown to block capsaicin responses with a much lower IC50 than heat responses in rTRPV1. This apparent modality selectivity in rat could however not be confirmed for human or monkey TRPV1 (9, 12). These data suggest that structural dissimilarities between different TRPV1 species can have significant functional and pharmacological consequences.
Here we show that the pore region of TRPV1 is critical for the species-dependent block of TRPV1 heat activation by small molecule antagonists such as JYL-1421. We used a chimeric approach to determine potential domain(s) responsible for the observed differences between human and rat TRPV1. By exchanging the pore domains, JYL-1421, which is modality-selective in rTRPV1 can thus be made modality-selective in hTRPV1 and vice-versa.
EXPERIMENTAL PROCEDURES
DNA Constructs, Cell Lines, Cell Culture, and Transfection
Human TRPV1 (NM_080704) was subcloned into TOPO pcDNA3.1 V5 His expression vector. Human and rat TRPV1 chimeras (cDNA chimeric variants) were generated by PCR amplification using the oligonucleotides as listed in Table 1. Internal oligonucleotides were designed to match both human and rat sequences. All constructs were verified by full-length sequence analysis. HEK293 Freestyle cells and 293fectin Transfection Reagent (Invitrogen, Breda, The Netherlands) were used for transient transfection of TRPV1 wild-type and chimeric isoforms. Transfections were performed according to manufacturer's protocol. For calcium flux experiments transfected cells were plated after 24–48 h into poly-d-lysine coated 384-well black, clear-bottom assay plates (Greiner) at a cell density of 20,000/well. For validation experiments a CHO T-Rex hTRPV1 stable cell line was used. The CHO T-Rex hTRPV1 stable cell line was cultured in T-225 cm2 flasks in Dulbecco's modified Eagle medium (DMEM) growth media (PAA Laboratories GmbH, A) containing 1× l-Glutamax, 1× Non-Essential Amino Acid (NEAA), 25 mm HEPES, supplemented with 10% tetracycline free fetal bovine serum (FBS) (PAA Laboratories GmbH, A), 10 μg/ml of blasticidin, 150 μg/ml of zeocin in 37 °C incubator under humidified atmosphere 5% CO2. Parental CHO T-Rex cells were maintained in DMEM growth media containing 1× l-Glutamax, 1× NEAA, 25 mm HEPES, supplemented with 10% tetracycline-free FBS and 10 μg/ml of blasticidin in T-225 cm2 flasks. Cells at ∼80% confluence were harvested and seeded at the appropriate seeding densities for assays.
TABLE 1.
Human and rat TRPV1 chimeras were generated by PCR amplification using the oligonucleotides listed above
Internal oligonucleotides were designed to match both human and rat sequences. All constructs were verified by full-length sequence analysis.
| Species | Orientation | Location | Sequence of oligonucleotides (5′-3′) |
|---|---|---|---|
| Human | Sense | Start | GCCACCATGAAGAAATGGAGCAGCACAGACTTGG |
| Rat | Antisense | Stop | TTATTTCTCCCCTGGGACCATGGAATC |
| Human/rat | Sense | Internal | GTCAAGCGCATCTTCTACTTCAACTTC |
| Human/rat | Antisense | Internal | GAAGTTGAAGTAGAAGATGCGCTTGAC |
| Rat | Sense | Start | GCCACCATGGAACAACGGGGCTAGCTTAGACTCAGA |
| Human | Antisense | Stop | TCACTTCTCCCCGGAAGCGGCAGGACTCT |
| Human/rat | Sense | Internal | GAGAGCAAGAACATCTGGAAGCTGCAGAG |
| Human/rat | Antisense | Internal | CTCTGCAGCTTCCAGATGTTCTTGCTCTC |
| Human/rat | Sense | Internal | GGGCTGGACCAACATGCTCTACTA |
| Human/rat | Antisense | Internal | TAGTAGAGCATGTTGGTCCAGCCC |
FDSS Calcium Flux Assay for Agonistic Studies on TRPV1
4–5 h post-plating cells were loaded with calcium-4 dye (Molecular Devices) for 1 h at room temperature in the dark. FDSS was used for detection and analysis of capsaicin responses. Concentration response curves were analyzed using GraphPad Prism (GraphPad Software, La Jolla, CA).
FLIPR Custom-built Temperature Control Unit
Existing microplate readers (for example FLIPR or FDSS) for ion channel cell-based assays do not control temperature within individual wells. For high-throughput applications, we established a calcium mobilization, heat activation assay. To do this we collaborated with The Ideas Studio Ltd (Canterbury, Kent, UK) and designed a custom-built temperature control unit which was fitted into a FLIPR384. The temperature control unit was comprised of the custom-built heat/cold device with adjacent washing station fitted into the FLIPR384, the remote MTC-1 control tower and its adjacent chiller (Betta-Tech Controls). The custom-built apparatus for altering temperature within each individual well of the microplate was equipped with six thermoelectric heating/cooling modules that made contact on the lower side with an anodized aluminum plate that had 384 pins, which were inserted into the 384-well microplate. On the opposite side of the pins and on the top of the thermoelectric heating/cooling modules there was a hollow aluminum chamber that acted as a heat/cold sink for the surplus thermal energy. The temperature of this chamber was maintained at ∼13 °C by liquid coolant (Hexid A4, Applied Thermal Control) circulated by a temperature-stable pump. Temperature changes in the heat/cold device were communicated to the MTC-1 control tower via a PT100 temperature sensor. The MTC-1 control tower was equipped with a temperature controller that controlled the power that was delivered to the thermoelectric modules in the custom-built heat/cold device and was driven by the user directly or remotely via the iTools software package (version 7, Eurotherm).
Heat Activation Assay Using the FLIPR Custom-built Temperature Control Unit
24 h prior to the experiment, CHO-TRex cells stably expressing hTRPV1 wild type were harvested and plated in the presence or absence of induction agent (1 μg/ml doxycycline) in 384-well black, clear bottom plates (Greiner) at a density of 10,000/well. In a similar manner, transfected HEK293 Freestyle cells with human or rat TRPV1 wild-type or TRPV1 chimeric isoforms after 24 h were harvested and plated into poly-d-lysine coated 384-well black, clear-bottom assay plates (Greiner) at a cell density of 20,000/well. On the day of the experiment, cell-culture media was removed from the plates by inversion and replaced with 30 μl/well of freshly-made calcium-5 dye solution (Molecular Devices). Calcium-5 dye (1×) was prepared according to manufacturer's instructions and kept at −20 °C. On the day of the experiment the dye solution was made by diluting the calcium-5 dye (3×) in assay buffer (1× HBSS (Invitrogen) supplemented with 20 mm HEPES and 0.05% Pluronic F-127, pH 7.4) and was further supplemented with 2.5 mm water soluble probenecid (Invitrogen). Cell plates loaded with dye solution were kept at room temperature in the dark for 1 h. After addition of 10 μl of compound or assay buffer using PlateMate Plus (Matrix), the cells were left at room temperature in the dark for 15 min and then an extra 10 μl of assay buffer was added to adjust the final assay volume to 50 μl/well. Cell plates were then transferred to the modified FLIPR for measurements. The following program was applied: 22 °C for 2 min, desired temperature for 3 min, followed by 22 °C for 2 min. The applied temperature ramp was 1 °C/s. Reading time in the FLIPR was 180 s (1 image/s). The fluorescence value of the measured baseline was subtracted from all other time points in each well. The numerical sum of the fluorescence counts of the selected images, similar to area under the curve, was exported from FLIPR384 into Excel. Concentration response curves were analyzed using GraphPad Prism (GraphPad Software, La Jolla, CA). Values are represented as average of either pEC50 or pIC50 ± S.E.
Heat Activation Assay Using Electrophysiology (Patchliner)
To further corroborate and validate the assay results we used an automated patch-clamp platform (4-channel Patchliner, Nanion). Internal solution: 50 mm CsCl, 10 mm NaCl, 60 mm CsF, 2 mm MgCl2, 20 mm EGTA,10 mm HEPES/CsOH, pH 7.2 (Osm ∼290 mOsm); external recording solution: 140 mm NaCl, 4 mm KCl, 1 mm MgCl2, 2 mm CaCl2, 5 mm d-glucose monohydrate, 10 mm HEPES/NaOH pH 7.4 (Osm ∼298 mOsm). Chips with a resistance of ∼2–3 MΩ were used for recordings. CHO-TRex cells stably expressing hTRPV1 wild type were kept in a “cell hotel” where they were periodically pipetted up and down to maintain homogeneity. Cells were viable for ∼1–2 h. Cells were automatically captured to the patch-clamp aperture using the PatchControlHT software. The PatchControlHT software was also used to obtain a good seal between glass and cell membrane (typically >1 GΩ) and to obtain the whole cell configuration. Suction was used to obtain whole cell configuration. A voltage ramp was run from −100 mV to +100 mV for 200 ms and repeated every 1 s from a holding potential of −100 mV. External recording buffer was heated up to the appropriate temperature in the pipette and then rapidly applied to the cell at a speed of 14 μl/s. The solution then rapidly cooled to room temperature, thus inactivating the channel. For compound addition, compounds were pre-incubated for at least 80 s prior to application at elevated temperature. Data recording and analysis were performed with an EPC10 Quadro amplifier, PatchMaster software package (HEKA Electronics, Lambrecht/Pfalz, Germany), Excel and Igor (WaveMetrics). The peak amplitude at +90 mV was calculated using online analysis functions in PatchMaster and these could be plotted as a function of time. Values were exported to Excel to calculate temperature activation curves. The figures were drawn and exported using Igor (WaveMetrics). Concentration response curves were constructed in Igor and a Hill equation fitted to estimate the IC50. Values are represented as average IC50 ± S.E.
Heat Activation Assay Using Patch Clamp Electrophysiology
Heat-activated currents were recorded from HEK293 cells transiently transfected with human or rat TRPV1 wild type or TRPV1 chimeric isoforms, which were identified by GFP fluorescence using the whole-cell configuration of the patch-clamp technique. Cells were voltage-clamped using a Multiclamp 700 A amplifier (Axon Instruments). The resistance of a typical patch pipette was 3–4 MΩ when filled with the intracellular solution containing (mm) 121 CsF, 1 MgCl2, 10 BAPTA, 1 MgATP, and 10 HEPES, adjusted to pH 7.3. The extracellular solution contained (mm) 150 NaCl, 5 KCl, 2 MgCl2, 1 CaCl2, 10 glucose, and 20 HEPES adjusted to pH 7.4. Series resistance was typically between 5 and 7 mΩ. Capacitive currents were cancelled and series resistance was compensated by at least 70%. Whole-cell currents were low pass filtered at 5 kHz and digitized at 20 kHz. Data were acquired using Clampex software (Axon Instruments) and analyzed using Campfit (Axon Instruments) and Origin 8.0 (OriginLab) software. Experiments were performed at room temperature (23–25 °C). Heat-evoked currents were elicited by heating solution using an SC-20 dual in-line solution heater/Cooler (Warner Instruments) and a CL-100 automatic temperature controller (Warner Instruments). Voltage-clamp experiments were performed using a voltage ramp protocol from −100 mV to +100 mV in 60 ms applied every 2 s from a holding potential of −60 mV. A heat ramp from 23 to 47 °C in 60 s was applied every 4 min. Drugs were pre-perfused for 2 min before eliciting heat-activated currents. One-way ANOVA with post-hoc Bonferroni tests was used to determine significant differences between independent groups.
RESULTS
Development and Validation of FLIPR Heat Activation Assay for TRPV1
As a consequence of the development of hyperthermia in clinical trials using polymodal TRPV1 antagonists, the pharmaceutical industry has started to search for “non-hyperthermic” antagonists. Based on the assumption that modality-specific antagonists may be devoid of hyperthermia, compounds that do not block heat activation of TRPV1 have been sought. However, compounds found to be modality-selective in certain TRPV1 species, turned out to be not modality-selective in other species. This suggests that hitherto unidentified structural differences between TRPV1 orthologues must be responsible for the observed species-specific pharmacology. To investigate multiple TRPV1 orthologues and chimeras in the heat activation mode, we constructed a custom-built temperature control unit for the FLIPR and used it to develop and validate a heat activation assay.
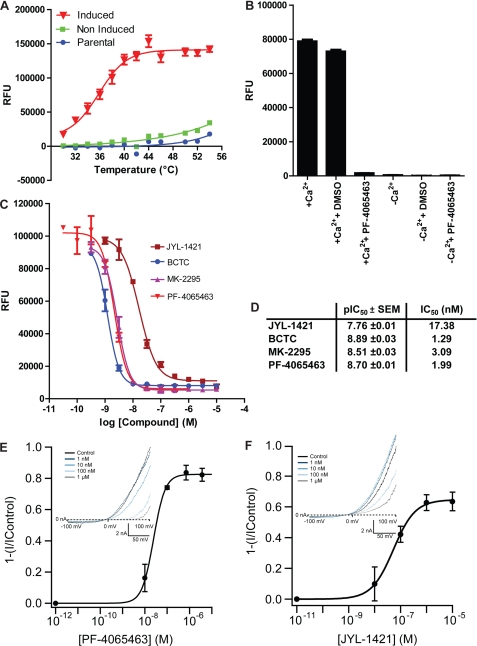
CHO-TRex cells stably expressing hTRPV1 (induced and non-induced) were exposed to a range of temperatures and a temperature response curve was obtained with maximal channel activity at 46 °C with no further fluorescence increase above this temperature (Fig. 1A). The EC50 value was calculated to be 34.76 ± 0.57 °C (mean ± S.E.). Non-induced or parental cells showed minor activation by heat only at high temperatures (>46 °C). Using a Ca2+-free assay buffer in combination with the hTRPV1-specific antagonist PF-4065463 (16), we demonstrated that the heat induced increase in fluorescence was mediated by Ca2+-influx via hTRPV1 channels (Fig. 1B).
FIGURE 1.
Validation of a FLIPR heat activation assay for hTRPV1 wild-type. A, representative temperature-response curves obtained from calcium influx experiments using CHO-TRex cells stably expressing hTRPV1 (induced with 1 μg/ml doxycycline) compared with non-induced and parental cells. The experiment was performed in triplicate and repeated at least three times with similar results. B, bar diagram showing maximum response levels after heat activation of hTRPV1 channel in the presence or absence of calcium in the assay buffer. When calcium was omitted the assay buffer was supplemented with 2 mm EGTA. The experiment which was carried out in triplicate was also performed in the presence of 1 μm PF-4065463 or 0.25% DMSO (vehicle) and repeated at least three times with identical results. C, representative concentration-response curves of antagonists obtained from calcium influx experiments using CHO-TRex cells stably expressing hTRPV1 wild-type activated by heat at 46 °C. The experiment was performed in triplicate and repeated at least three times with similar results. D, summary of average pIC50 values (mean ± S.E.) calculated from experiments as described in C (n 3). E and F, concentration-response curves of PF-4065463 and JYL-1421 antagonists in heat activation mode (at 47 °C, obtained from patch-clamp experiments using Patchliner (Nanion)); insets show I-V relationships of the heat activation of hTRPV1 channels by heated solution (47 °C) and block of the heat-activated response by increasing concentrations of the two compounds.
In a subsequent validation step we showed typical hTRPV1 pharmacology in this assay format (at 46 °C) using four well-characterized hTRPV1 antagonists: PF-4065463 (16), JYL-1421 (17, 18), BCTC (18), and MK-2295 (19) (Fig. 1, C and D). All compounds blocked heat-activated hTRPV1 responses in a concentration-dependent manner (Fig. 1, C and D). The assay proved to be robust displaying a Z′ value of >0.7. As recommended by Zhang et al. (20) an assay with 0.5 ⩽ Z′ < 1, indicates an excellent assay that can be used for high-throughput screening. Recently, the development of a Ca2+-influx assay with fluorescence readout using a RT-PCR machine was reported (21). We also developed the RT-PCR heat activation assay (data presented by Papakosta et al. at FASEB Ion Channel Conference, June, 2009) but we found that our FLIPR heat activation assay was far superior in terms of assay window and reproducibility in a high-throughput format.
To further corroborate these results and validate the stable hTRPV1 cell line and compounds used in the FLIPR heat activation assay, patch-clamp experiments were performed using the Patchliner. Both compounds used in this format, PF-4065463 and JYL-1421, blocked heat activation of hTRPV1 (measured at 47 °C) in a concentration-dependent manner. Fig. 1, E and F shows an example of the heat response of hTRPV1 and the block induced by increasing concentrations of PF-4065463 or JYL-1421 (Fig. 1, E and F). The IC50 values obtained for PF-4065463 and JYL-1421 were 34 ± 16 nm (n = 3) and 59 ± 16 (n = 8), respectively, which is in good agreement with the data obtained in the FLIPR heat activation assay given the fact that different assay systems were used.
Generation of Chimeric TRPV1 Constructs and Functional Assessment in Capsaicin Activation Mode
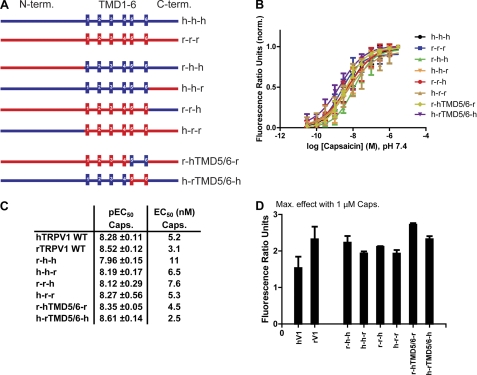
To identify candidate region(s) or domain(s) responsible for the different potencies of antagonists to block heat activation in rat and human TRPV1, we decided to take a chimeric approach. The following TRPV1 chimeras were generated by PCR cloning (N terminus-TMD1–6-C terminus): r-h-h, r-r-h, r-h-r, h-h-r, h-r-r, and h-r-h (Fig. 2A). In addition, chimeras with exchanged pore-regions (TMD5/6) were generated: r-hTMD5/6-r and h-rTMD5/6-h (Fig. 2A). After sequence verification, the chimeras were tested side-by-side with the respective TRPV1 wild-type isoforms in Ca2+-influx experiments for their respective capsaicin and heat activation profiles.
FIGURE 2.
Generation and functional characterization of chimeric TRPV1 constructs (human/rat). A, schematic representation of chimeric TRPV1 constructs. Human and rat TRPV1 are indicated by h and r, respectively. Chimeric constructs were generated by PCR cloning. Internal oligonucleotides were designed to match both human and rat sequences. B, representative capsaicin concentration-response curves obtained from calcium influx experiments using HEK293 Freestyle cells transiently transfected with human or rat TRPV1 wild-type or TRPV1 chimeric constructs as shown in A. All concentration-response measurements were performed in standard buffer solution (1× HBSS) containing 20 mm HEPES, calcium, and magnesium (1 mm, each), adjusted to pH 7.4. Calcium-4 was used as fluorescent Ca2+ indicator. The experiments were performed in triplicate and repeated at least three times with similar results. C, summary of average capsaicin pEC50 values (mean ± S.E.) calculated from experiments as described in B (n 3). D, bar diagram showing maximum response levels after application of 1 μm capsaicin. Data were obtained from at least three independent experiments, each one performed in triplicate, as described in B. Shown are mean ± S.E.
No significant differences in capsaicin activation could be found in these experiments between wild-type channels and chimeras. Average capsaicin EC50 values for rTRPV1 and hTRPV1 wild-type isoforms were 3.1 and 5.2 nm, respectively. Average capsaicin EC50 values of all chimeras were in the range of 2.5 - 11 nm (Fig. 2, B and C). Also, average maximum effect values for capsaicin (1 μm) activation were comparable between wild-type isoforms and chimeras (Fig. 2D).
Antagonist Assays Using Chimeric TRPV1 Constructs Reveal That the Pore Region Is Critical for Species-dependent Block of Heat Activation
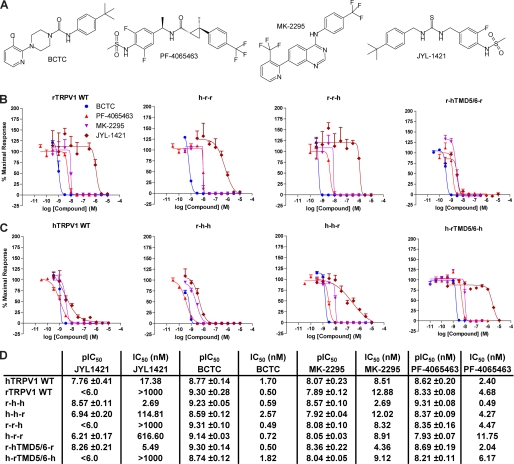
HEK293 cells transiently expressing TRPV1 wild-type or mutant isoforms were activated with capsaicin or heat in the presence of the four different TRPV1 antagonists previously used for assay validation: JYL-1421 (17, 18), MK-2295 (19), PF-4065463 (16), and BCTC (18) (Fig. 3A). As expected rTRPV1 wild-type was blocked by the three polymodal antagonists MK-2295, PF-4065463, and BCTC while JYL-1421 demonstrated weak antagonism; potential lack of pharmacological selectivity precluded use of concentrations greater than 10 μm (Fig. 3B). In contrast, cells overexpressing hTRPV1 wild-type showed block of heat activation by all four compounds with similar IC50 values (Fig. 3C). Experiments with the chimeric channels revealed that exchanging the pore domains between rat and human TRPV1 (r-hTMD5/6-r and h-rTMD5/6-h) reversed the expected phenotypes (Fig. 3, B–D). Human TRPV1 with a rat pore domain (h-rTMD5/6-h) thus showed an antagonist profile normally obtained with rTRPV1 wild-type and vice versa. Data from JYL-1421 antagonism were analyzed using one-way ANOVA with posthoc Bonferroni pairwise comparisons. ANOVA was found to be significant [F(3,11) = 20.61, p < 0.001] and posthoc tests revealed significant differences when comparing hTRPV1WT versus rTRPV1WT (p < 0.05), hTRPV1WT versus h-rTMD5/6-h (p < 0.01), rTRPV1WT versus r-hTMD5/6-r (p < 0.05) and r-hTMD5/6-r versus h-rTMD5/6-h (p < 0.001).
FIGURE 3.
Effect of TRPV1 antagonists on heat-activated TRPV1 chimeric and wild-type isoforms. A, depicted are the chemical structures of the four TRPV1 antagonists used in the studies. B and C, representative antagonist concentration-response curves obtained from calcium influx experiments using HEK293 Freestyle cells transiently transfected with human or rat TRPV1 wild-type or TRPV1 chimeric constructs. The experiments were performed in triplicate and repeated at least three times with similar results. D, summary of average antagonist mean pIC50 ± S.E. values calculated from experiments as described in B and C (n 3).
A shift of the mean IC50 of JYL-1421 (17.4 nm versus 114.8 nm) was also observed when exchanging the human with the rat C terminus (h-h-r). Replacement of the rat with the human C terminus (r-r-h) however, did not affect the lack of a significant JYL-1421 block of heat responses in rTRPV1 (Fig. 3, B–D).
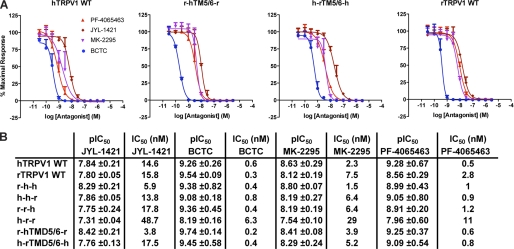
Block of capsaicin activation on the other hand did not show any significant differences between wild-type isoforms and chimeras. All four antagonists showed similar IC50s values for wild-type isoforms compared with chimeras (Fig. 4, A and B).
FIGURE 4.
Effect of TRPV1 antagonists on capsaicin-activated TRPV1 chimeric and wild-type isoforms. A, shown are antagonist concentration-response curves obtained from calcium mobilization experiments using HEK293 Freestyle cells transiently transfected with human or rat TRPV1 wild-type as well as TRPV1 pore chimeras. Concentration-response measurements were performed in standard buffer solution (1× HBSS) containing 20 mm HEPES, calcium, and magnesium (1 mm, each), adjusted to pH 7.4. Calcium-4 was used as fluorescent Ca2+ indicator. Cells were incubated at room temperature for 15 min after application of antagonist and subsequently stimulated with respective average capsaicin EC80 concentrations obtained from experiments described in Fig. 2. The experiments were performed in triplicate and repeated at least three times with similar results. B, summary of average antagonist pIC50 ± S.E. values calculated from experiments as described in A (n 3).
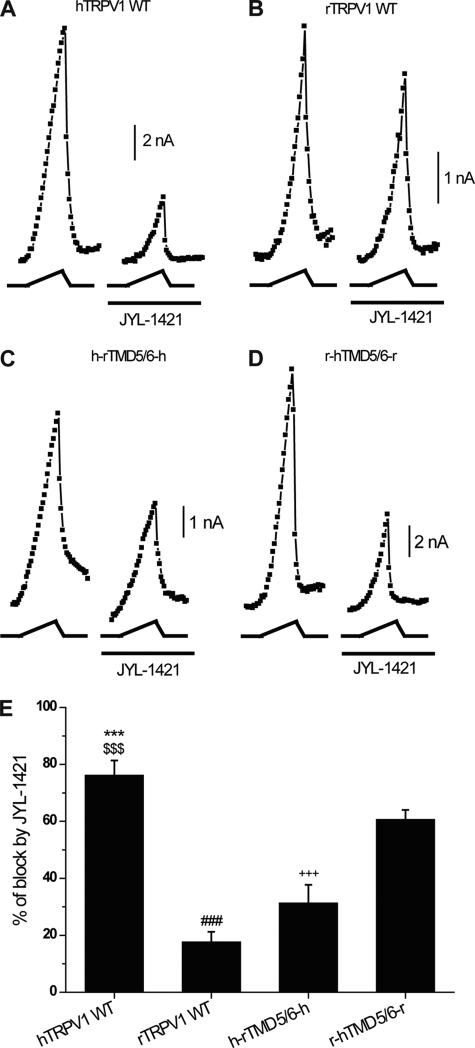
To further corroborate these data, we performed manual patch-clamp electrophysiological recordings (Fig. 5, A–E). Both, JYL-1421 and PF-4065463 (330 nm), were tested on heat-activated (47 °C) cells transiently transfected with hTRPV1, rTRPV1 wild-type isoforms and h-rTMD5/6-h, r-hTMD5/6-r chimeras. JYL-1421 clearly blocked the current of heat-activated hTRPV1, while it had only a weak effect on the current of heat-activated rTRPV1 (76 ± 5.1% and 17 ± 3.5% respectively). JYL-1421 blocked the current of the heat activated h-rTMD5/6-h chimera by 31 ± 6.4% and of r-hTMD5/6-r chimera by 60 ± 3.3%. Significant changes were detected (ANOVA [F(3,24) = 32.8, p < 0.001], posthoc Bonferroni tests (all p < 0.001) *** hTRPV1 versus rTRPV1,$$$ hTRPV1 versus h-rTMD5/6-h, ### rTRPV1 versus r-hTMD5/6-r, +++ h-rTMD5/6-h versus r-hTMD5/6-r). PF-4065463 completely abolished the current of heat-activated hTRPV1, rTRPV1 wild-type isoforms and h-rTMD5/6-h, r-hTMD5/6-r chimeras (data not shown). The patch-clamp data were in agreement with those of the heat assay performed on the FLIPR (Fig. 3, B–D).
FIGURE 5.
Effect of TRPV1 antagonists on heat-activated TRPV1 chimeric and wild-type isoforms using manual patch-clamp electrophysiology. A–D, representative plots of heat-activated current measured at +100 mV during heat ramp in HEK293 cells transfected with hTRPV1WT, rTRPV1WT, h-rTMD5/6-h, r-hTMD5/6-r in the absence and presence of JYL-1421 (0.33 μm). Heat ramp is depicted under each current plot (heat ramp from 23 °C to 47 °C). The compound was pre-applied for 2 min before heat activation. E, summary of effect of JYL-1421 on heat activated hTRPV1WT, rTRPV1WT, h-rTMD5/6-h, r-hTMD5/6-r channels. Each bar is the mean ± S.E. (n = 5 for hTRPV1WT, n = 7 for rTRPV1WT, n = 6 for h-rTMD5/6-h, and n = 7 for r-hTMD5/6-r).
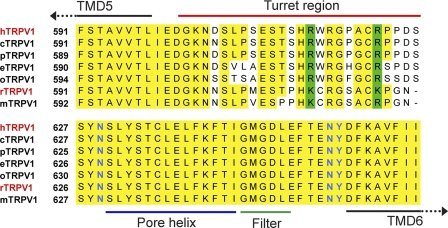
In summary, these results demonstrate that the species-specific pharmacology of compound JYL-1421 which potently blocks heat activation of human but not rat TRPV1 can be overcome by exchanging the orthologous pore domains. An amino acid sequence alignment of rat and human TRPV1 pore domains revealed nine amino acid differences and one deletion between TMD5 and TMD6 (Fig. 6). All amino acid differences in this region are found in the pore turret domain (highlighted in red, Fig. 6). Generally, the pore turret domain which stretches from shortly after TMD5 to the beginning of the pore helix is much less conserved across different species than the rest of the pore region which shows almost no cross-species amino acid differences. Interestingly, this is also the region that Yang et al. (22) have recently proposed as being involved in heat activation. In addition, Grandl et al. (13) identified an amino acid, Asn-628 just downstream of this motif that affects the heat activation of the TRPV1 channel, suggesting that cross-species differences in or near that region may be able to interfere with TRPV1 activation and inhibition mechanisms.
FIGURE 6.
Amino acid alignment of part of the TRPV1 pore domain in different TRPV1 species (h, homo sapiens, r, rattus norvegicus, o, oryctolagus cuniculus, p, pongo pygmaeus, e, equus caballus, m, mus musculus, c, cynomologus). The pore turret domain (marked by a red line) is significantly less conserved than the rest of the pore region. Residues specifically involved in heat activation of TRPV1 are highlighted in blue.
DISCUSSION
The differential compound pharmacology and modality selectivity across different species led us to use a chimeric approach to identify the underlying molecular determinants. To test a broad set of chimeras in the presence of a number of different TRPV1 antagonists in a heat activation assay, an automated platform capable of accurately delivering heat to individual wells of a cell plate was required. However, this platform did not exist commercially and therefore a custom built temperature control unit was designed and fitted to the FLIPR.
It was found that the pore region of TRPV1 is critical for the species-dependent block of TRPV1 heat activation by small molecule antagonists such as JYL-1421. Specifically, exchanging the pore domain between human and rat TRPV1 changed JYL-1421, which is normally a weak antagonist of rTRPV1 into a potent antagonist of this channel. In a reciprocal fashion, it was also possible to weaken the potency of JYL-1421 at hTRPV1 by incorporation of the rat pore domain. The differential impact of JYL-1421 on heat activation in rat and human TRPV1 by exchanging the pore domain (with the pore turret domain showing the only significant amino acid differences), suggests that a change to the overall conformation in that region can generally affect the ability to block heat activation.
Yang et al. (22) have recently suggested that the pore turret domain of thermal TRP channels is part of the temperature-sensing apparatus. They replaced a 14-residue region of the turret of TRPV1 (amino acids 613–626) with a glycine-based fragment of equal length and found that it inhibited the thermal response. This interpretation was however disputed recently by Yao et al. (23). They had previously studied the same region in TRPV1 (one residue longer, Thre-612—Ser-626) and found that it was functionally redundant (24). Yao et al. also argued that TRPV3, a homologous heat-activated channel, does not contain the region studied by Yang et al. (22). It was therefore speculated that the contradiction between the data published by Ryu et al. (24) and those presented by Yang et al. may have occurred because the substitution by Yang et al. exerted an unintended effect on channel structure. The data presented here do neither support nor refute the hypothesis that parts of the pore turret domain may be involved in temperature activation as postulated by Yang et al. They only suggest that amino acid differences in the pore turret domain can explain the different pharmacological profiles of compounds such as JYL-1421 as seen for example in rat and human TRPV1.
JYL-1421 has been shown previously to be a competitive antagonist of capsaicin and resiniferatoxin (RTX) which bind to TRPV1 between TMD3 and TMD4 (25, 26). JYL1421 inhibited [3H]resiniferatoxin binding to rTRPV1 with an affinity of 53.5 ± 6.5 nm and antagonized capsaicin-induced calcium uptake with an EC50 of 9.2 ± 1.6 nm (26). The portion of the pore turret domain suggested here to underlie the observed species-specific antagonist profiles is therefore expected not to be involved in direct JYL-1421 binding. Nevertheless, these amino acid variations appear to be sufficient to differentially impact TRPV1 function and pharmacology (for example block of heat activation) in different species. Our data suggest that compounds that competitively inhibit capsaicin binding i.e. bind to the same site on the TRPV1 channel may have entirely different effects from each other on heat activation of the channel. The mechanisms by which this may be achieved are obviously allosteric in nature but more studies are required to understand this phenomenon. The finding has profound implications for the development of modality specific TRPV1 antagonists suggesting the need for the incorporation of high throughput screening of alternative modalities for the different species orthologues, such as the heated FLIPR system described herein, to truly understand the species specific in vitro pharmacology of compounds interacting with TRPV1 and how these interactions lead to pharmacodynamic effects such as hyperthermia.
In addition to the species differences in the pore region, the chimera experiments described here also revealed a difference between rat and human TRPV1 block of heat activation relating to the C terminus, i.e. replacing the human with the rat C terminus (h-h-r) reduced the ability of JYL-1421 to block heat responses when compared with hTRPV1 wild-type. In contrast, replacement of the rat with the human C terminus (r-r-h) did not significantly affect the JYL-1421 block of heat responses which may point to the C terminus playing a unique role in temperature regulation of rat but not human TRPV1. Brauchi et al. have recently reported that the C terminus of rTRPV1 may be involved in heat activation (27). Again, the data presented here do neither support nor refute this hypothesis, but they do suggest that amino acid differences in the C terminus may have an impact on antagonist pharmacology in different species, although much less pronounced compared with the pore region differences.
Different TRPV1 mutations have been described in the literature to cause selective loss of heat, proton or capsaicin activation. TRPV1 mutants Y511A and S512Y (in the intracellular loop between TMD2 and TMD3) have lost their capsaicin sensitivity while maintaining normal heat and proton activation profiles (28). The triple mutant TRPV1 (N628K/N652T/Y653T) displays a selective loss of heat activation (13). Mutations in the extracellular loop between TMD3 and TMD4 (V538L) and in the pore region between TMD5 and TMD6 abrogate proton activation (E648Q, T633A) while preserving capsaicin and heat responsiveness or affect potentiation of heat and capsaicin responses by protons (E600) (29). Finally, TRPV1 (F660) mutants (in TMD6) show a lack of both activation and potentiation by protons while activation by heat or capsaicin is preserved (15). The literature therefore demonstrates that heat, proton and capsaicin activation of TRPV1 can be relatively well separated from each other at the molecular level. Furthermore it appears that amino acids involved in heat and proton activation reside in very close proximity to each other in the pore domain. Regardless of the mechanism by which TRPV1 antagonism causes hyperthermia for example by block of proton activation as postulated recently by Garami et al. (30) or by block of heat activation or both, it will be critical to the successful development of novel, safe TRPV1 antagonists that inhibition of such activation modes is avoided.
In conclusion, by using a chimeric approach (human and rat TRPV1) we have identified the pore region as being strongly linked to observed species differences in TRPV1 pharmacology. Furthermore we demonstrate that by exchanging the pore domains JYL-1421, which is modality-selective in rat can be made modality-selective in human TRPV1 and vice-versa.
Acknowledgments
We thank Dr. James Turner (Pfizer Global R&D) for technical support. We further thank Mark Watson, Mark Callaghan, and Steve McDonald for support in designing and constructing the custom-built temperature control unit for the FLIPR.
This work was supported by Pfizer Ltd. Global R&D.
- TRP
- transient receptor potential
- TMD
- transmembrane domain
- CRC
- concentration-response curve
- RFU
- relative fluorescence unit
- FLIPR
- Fluorescence Imaging Plate Reader
- FDSS
- Functional Drug Screening System.
REFERENCES
- 1. Clapham D. E. (2003) Nature 426, 517–524 [DOI] [PubMed] [Google Scholar]
- 2. Clapham D. E., Montell C., Schultz G., Julius D. (2003) Pharmacol. Rev. 55, 591–596 [DOI] [PubMed] [Google Scholar]
- 3. Dhaka A., Viswanath V., Patapoutian A. (2006) Annu. Rev. Neurosci. 29, 135–161 [DOI] [PubMed] [Google Scholar]
- 4. Ramsey I. S., Delling M., Clapham D. E. (2006) Annu. Rev. Physiol. 68, 619–647 [DOI] [PubMed] [Google Scholar]
- 5. Damann N., Voets T., Nilius B. (2008) Curr. Biol. 18, R880–889 [DOI] [PubMed] [Google Scholar]
- 6. Gees M., Colsoul B., Nilius B. (2010) Cold Spring Harb. Perspect Biol. 2, a003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baez-Nieto D., Castillo J. P., Dragicevic C., Alvarez O., Latorre R. (2011) Exp. Med. Biol. 704, 469–490 [DOI] [PubMed] [Google Scholar]
- 8. Chung M. K., Jung S. J., Oh S. B. (2011) Adv. Exp. Med. Biol. 704, 615–636 [DOI] [PubMed] [Google Scholar]
- 9. Gavva N. R., Bannon A. W., Surapaneni S., Hovland D. N., Jr., Lehto S. G., Gore A., Juan T., Deng H., Han B., Klionsky L., Kuang R., Le A., Tamir R., Wang J., Youngblood B., Zhu D., Norman M. H., Magal E., Treanor J. J., Louis J. C. (2007) J. Neurosci. 27, 3366–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Szallasi A., Cortright D. N., Blum C. A., Eid S. R. (2007) Nat. Rev. Drug Discov. 6, 357–372 [DOI] [PubMed] [Google Scholar]
- 11. Khairatkar-Joshi N., Szallasi A. (2009) Trends Mol. Med. 15, 14–22 [DOI] [PubMed] [Google Scholar]
- 12. Lázár J., Gharat L., Khairathkar-Joshi N., Blumberg P. M., Szallasi A. (2009) Exp. Opin. Drug Disc. 4, 159–180 [DOI] [PubMed] [Google Scholar]
- 13. Grandl J., Kim S. E., Uzzell V., Bursulaya B., Petrus M., Bandell M., Patapoutian A. (2010) Nat. Neurosci. 13, 708–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grandl J., Hu H., Bandell M., Bursulaya B., Schmidt M., Petrus M., Patapoutian A. (2008) Nat. Neurosci. 11, 1007–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aneiros E., Cao L., Papakosta M., Stevens E. B., Phillips S., Grimm C. (2011) EMBO J. 30, 994–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hanazawa T., Hirano M., Inoue T., Nagayama S., Nakao K., Shishido Y., Tanaka H. (2006) PCT Int. Appl., CODEN: PIXXD2 WO 2006097817 [Google Scholar]
- 17. Jakab B., Helyes Z., Varga A., Bölcskei K., Szabó A., Sándor K., Elekes K., Börzsei R., Keszthelyi D., Pintér E., Petho G., Németh J., Szolcsányi J. (2005) Eur. J. Pharmacol. 517, 35–44 [DOI] [PubMed] [Google Scholar]
- 18. Wong G. Y., Gavva N. R. (2009) Brain Res. Rev. 60, 267–277 [DOI] [PubMed] [Google Scholar]
- 19. Kym P. R., Kort M. E., Hutchins C. W. (2009) Biochem. Pharmacol. 78, 211–216 [DOI] [PubMed] [Google Scholar]
- 20. Zhang J. H., Chung T. D., Oldenburg K. R. (1999) J. Biomol. Screen. 4, 67–73 [DOI] [PubMed] [Google Scholar]
- 21. Reubish D., Emerlin D., DeFalco J., Steiger D., Victoria C., Vincent F. (2009) BioTechniques 47, iii-ix [DOI] [PubMed] [Google Scholar]
- 22. Yang F., Cui Y., Wang K., Zheng J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 7083–7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yao J., Liu B., Qin F. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, E125; author reply E126–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ryu S., Liu B., Yao J., Fu Q., Qin F. (2007) J. Neurosci. 27, 12797–12807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gavva N. R., Klionsky L., Qu Y., Shi L., Tamir R., Edenson S., Zhang T. J., Viswanadhan V. N., Toth A., Pearce L. V., Vanderah T. W., Porreca F., Blumberg P. M., Lile J., Sun Y., Wild K., Louis J. C., Treanor J. J. (2004) J. Biol. Chem. 279, 20283–20295 [DOI] [PubMed] [Google Scholar]
- 26. Wang Y., Szabo T., Welter J. D., Toth A., Tran R., Lee J., Kang S. U., Suh Y. G., Blumberg P. M., Lee J. (2002) Mol. Pharmacol. 62, 947–956 [DOI] [PubMed] [Google Scholar]
- 27. Brauchi S., Orta G., Salazar M., Rosenmann E., Latorre R. (2006) J. Neurosci. 26, 4835–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jordt S. E., Julius D. (2002) Cell 108, 421–430 [DOI] [PubMed] [Google Scholar]
- 29. Jordt S. E., Tominaga M., Julius D. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8134–8139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garami A., Shimansky Y. P., Pakai E., Oliveira D. L., Gavva N. R., Romanovsky A. A. (2010) J. Neurosci. 30, 1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]