Abstract
Bronchial biopsies of asthmatic patients show a negative correlation between desmin expression in airway smooth muscle cell (ASMC) and airway hyperresponsiveness. We previously showed that desmin is an intracellular load-bearing protein, which influences airway compliance, lung recoil, and airway contractile responsiveness (Shardonofsky, F. R., Capetanaki, Y., and Boriek, A. M. (2006) Am. J. Physiol. Lung Cell. Mol. Physiol. 290, L890–L896). These results suggest that desmin may play an important role in ASMC homeostasis. Here, we report that ASMCs of desmin null mice (ASMCsDes−/−) show hypertrophy and up-regulation microRNA-26a (miR-26a). Knockdown of miR-26a in ASMCsDes−/− inhibits hypertrophy, whereas enforced expression of miR-26a in ASMCsDes+/+ induces hypertrophy. We identify that Egr1 (early growth responsive protein-1) activates miR-26a promoter via enhanced phosphorylation of Erk1/2 in ASMCsDes−/−. We show glycogen synthase kinase-3β (GSK-3β) as a target gene of miR-26a. Moreover, induction of ASMCsDes−/− hypertrophy by the Erk-1/2/Egr-1/miR-26a/GSK-3β pathway is consistent in human recombinant ASMCs, which stably suppresses 90% endogenous desmin expression. Overall, our data demonstrate a novel role for desmin as an anti-hypertrophic protein necessary for ASMC homeostasis and identifies desmin as a novel regulator of microRNA.
Keywords: Cell Division, Gene Expression, Glycogen Synthase Kinase 3, MicroRNA, Signal Transduction, Smooth Muscle
Introduction
Asthma is a chronic lung disease with airway inflammation and abnormal airway smooth muscle contraction (bronchospasm), which is due to an intrinsic abnormality of the airway smooth muscle cells (ASMCs).2 Airway remodeling and hyperresponsiveness are characteristic features of severe asthma and chronic obstructive pulmonary disease. Clinical studies show that both hypertrophy (increase in cell size) and hyperplasia (increase in cell number) of ASMCs play key roles in airway remodeling (1–5) and hyperresponsiveness in asthmatic patients (6). Moreover, a recent study in bronchial biopsies of asthmatic patients shows a correlation between airway hyperresponsiveness, predicted percentage of forced expiratory volume in 1 s, and airway responsiveness to deep inspiration and the levels of selective expression of ASMC proteins, including desmin (7). Desmin is a cytoskeleton protein and type III intermediate filament. All types of muscle cells, including ASMCs and alveolar ducts, express desmin (8–12). Our physiologic data from desmin knock-out (Des−/−) mice show that desmin is an intracellular load-bearing protein that influences airway compliance, lung recoil, and airway contractile responsiveness (13). These results suggest that desmin may play an important role in modulating ASMC phenotype.
MicroRNAs (miRNAs) are now recognized as key regulators of gene expression and are associated with many human diseases. Recently, we have shown the role of miR-26a in the induction of hypertrophy in human ASMC (14). However, whether desmin can regulate the ASMC phenotype through a miRNA-regulated pathway is unknown. The aim of the present study is to investigate whether loss of desmin triggers induction of ASMC hypertrophy through a miR-26a-regulated pathway. Our results demonstrated that ASMCs of desmin null mice (ASMCsDes−/−) showed hypertrophy and miR-26a up-regulation. Loss of desmin in mouse ASMCs activated Erk-1/2, which up-regulated miR-26a through Egr-1 (early growth responsive protein-1). The anti-hypertrophic protein glycogen synthase kinase-3β (GSK-3β) is a target gene of miR-26a in ASMCsDes−/−. In addition, the siRNA-mediated knockdown of desmin protein in human ASMCs induced hypertrophy through the activation of the Egr-1/miR-26a/GSK-3β pathway. Overall, our data show that desmin is an essential intermediate filament for maintaining ASMC phenotype in both in vitro and in vivo systems. Our data also provide novel experimental evidence that desmin intermediate filaments are important in the regulation of microRNA.
EXPERIMENTAL PROCEDURES
Isolation and Culture of Primary Mouse Airway Smooth Muscle Cells
Des+/+ (normal C57/BL6) and Des−/− (B6.129S2/Sv-Destm1Cba/Orl) mice were obtained from Jackson Laboratory (Bar Harbor, ME) and the European Mutant Mouse Archive (Munich, Germany), respectively. Mice were maintained in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the Institutional Animal Care and Use Committee of Baylor College of Medicine approved animal protocols. Tracheal segments of Des+/+ and the age-matched Des−/− littermates (from 8 to 10 weeks old) were removed after deep anesthesia. The trachea was gently dissected from the surrounding tissues and placed in oxygenated Krebs-Ringer solution bubbled with 95% O2 and 5% CO2. The smooth muscle was carefully dissected from the adherent epithelial, connective, and parenchymal tissues and digested in 2 ml of DMEM (supplemented with 1 mm sodium pyruvate, 2 mm l-glutamine, 1:100 nonessential amino acid mixture, 50 mg/ml gentamicin, 1.5 mg/ml amphotericin B, 1 mm insulin, 5 mg/ml transferrin, 100 mm ascorbate, and 1 mg/ml bovine serum albumin) containing 3 mg/ml collagenase for 30 min at 37 °C. After partial digestion, the tissue was chopped finely and placed in an incubator until fully digested. The resulting cell suspension was centrifuged (200 × g for 5 min), washed, and seeded in supplemented DMEM containing 10% fetal calf serum at 5 × 105 in 25-cm2 culture flasks. The cells were maintained in a humidified atmosphere at 37 °C in 5% CO2 and 95% air, and the medium was replaced with fresh medium every 3 days. A portion of these cells was used for immunocytochemistry with mouse anti-α-smooth muscle actin, anti-SM22, or vimentin to confirm the existence of smooth muscle. Mouse anti-cytokeratin antibody was used to identify and eliminate epithelial cells. All experiments were conducted after the cells were serum-deprived for 24 h.
Generation of Recombinant Human Airway Smooth Muscle Cells
Recombinant human airway smooth muscle cells (reHASMCs) were generated by siRNA strategy as described earlier (15).
miRNA Microarray Analysis
Total RNA samples were isolated by TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Ten micrograms of total RNA was size-fractionated with a YM-100 Microcon centrifugal filter (Millipore, Billerica, MA). The following formulas was used to calculate percentage filtrate and retentate recovery: % retentate recovery = 100 × Wr × Cr/W0 × C0 and % filtrate recovery = 100 × Wf × Cf/W0 × C0, where Wr represents total weight of retentate before assay, Wo is the weight of starting material, Wf is the weight of filtrate, Cr is the retentate concentration, Co is the starting material concentration, and Cf is the filtrate concentration. The sample recovery was >95% of the initial samples. The recovered samples were used for miRNA expression analysis with an miRNA microarray (LC Sciences, Houston, TX).
Construction of Expression Plasmids
The pSilencer 4.1-CMV miR-26a expression construct was prepared as described previously (14). Mouse GSK-3β cDNA 3′-UTR was synthesized and cloned into pcDNA 3.1D/V5-His-TOPO vector (Invitrogen) according to the manufacturer's instructions. To generate reporter vector bearing miR-26a binding sites, a 650-bp mouse GSK-3β 3′-UTR sequence was synthesized and cloned into pmirGLO vector (Promega, Madison, WI) according to the manufacturer's instructions. Reporter vector bearing 571-bp Egr-1, 715-bp C/EBPα, or 540-bp NF-κB binding elements was synthesized from mouse Ctdsp2/miR-26a 5′-UTR DNA (as shown in Fig. 6) and cloned into pGL4.1 luciferase reporter vector (Promega). PCRs were performed to synthesize inserts with AccuPrime Pfx DNA polymerase according to the manufacturer's protocols (Invitrogen). Primer information is detailed in Table 1. Constructs were sequenced by the DNA sequence Core Facility of the Baylor College of Medicine to verify insert identities.
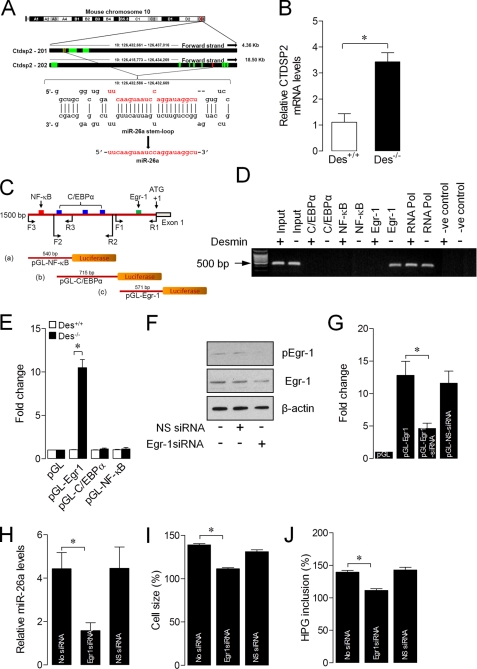
FIGURE 6.
Egr1 regulates miR-26a expression. A, schematic representation of the genomic structure of the mouse Ctdsp2 coding RNA genes in chromosome 10. The red band indicates the position of the Ctdsp2 gene on chromosome 10 or the position of miR-26a on the Ctdsp2 gene. B, total RNA was isolated from the ASMCs of Des−/− mice. Ctdsp2 mRNA levels were analyzed by qPCR. C, schematic representation of the 5′-UTR of the mouse Ctdsp2 gene (miR-26a promoter). The region between −1500 and +1 bp contains putative binding elements for Egr-1 (red), C/EBPα (blue), and NF-κB (green). A 500-bp (pGL-NF-κB), 750-bp (pGL-C/EBPα), or 500-bp (pGL-Egr-1) promoter region was synthesized and linked to the luciferase (Luc) reporter gene. D, chromatin was isolated from the ASMCs of Des+/+ and Des−/− mice and precipitated with anti-c-Egr-1, anti-C/EBPα, anti-NF-κB, anti-RNA Poly II, or nonspecific IgG. qPCRs were performed with three sets of primers, specific for the miR-26a promoter, to identify the specific transcription factor and its region of binding to the miR-26a promoter and resolved in 1% agarose gel. E, a 500-bp (pGL-Egr-1), 700-bp (pGL-C/EBPα), or 500-bp (pGL-NF-κB) promoter region was synthesized and linked to the luciferase reporter gene. ASMCs of Des−/− mice were transfected with empty vector, pGL-Egr-1, pGL-C/EBPα, or pGL-NF-κB miR-26a promoter region. Forty-eight hours after transfection, firefly luciferase activities were estimated and normalized to Renilla luciferase activities. F, ASMCs of Des+/+ mice were transfected with Egr-1 siRNA or nonspecific siRNA (NS-siRNA). After 48 h, total protein was isolated, and Egr-1 protein expression was determined by Western blot. G, ASMCs of Des+/+ and Des−/− mice were transfected with Egr-1 siRNA or its scramble form (SC-Egr-1siRNA). Forty-eight hours after RNAi transfection, cells were transfected with pGL or pGL-Egr-1 vector. After 48 h, firefly luciferase activities were estimated and normalized to Renilla luciferase activities. H–J, Des−/− ASMCs were transfected as shown in F. After 48 h of transfection, miR-26a levels (H), cell size (I), and protein synthesis (J) were estimated. Gel pictures are representative of three separate experiments. Each bar indicates mean ± S.E. (n = 3). *, p < 0.05.
TABLE 1.
Primers used in PCR
| Primer name | Sequence 5′–3′ | Underline | Purpose |
|---|---|---|---|
| miR-26a-C-F | GGATCCGTGATATCACAAGGTCCCAG | BamHI | Cloning |
| miR-26a-C-R | AAGCTTCTACATGCAAAGGGCAGGAG | HindIII | Cloning |
| GSK-3β-F1 | CACCCATTTATCATTAACCTAGCA | Cloning | |
| GSK-3β-R1 | TCTTTCCAAACGTGACCAGT | Cloning | |
| GSK-3β3′-UTR-F | GCTAGCCCCGTGACATTGTTAGCAGTT | SacI | Cloning |
| GSK-3β3′-UTR-R | CTCGAGTGCCAGTGTCTTTACGTCCA | XhoI | Cloning |
| miR-26a 5′-UTR-F1 | GGTACCCATAGACTGGGTGGCGAGTT | KpnI | Cloning |
| miR-26a 5′-UTR-R1 | GAGCTCATCGACGTAAGGCCTCTTGA | SacI | Cloning |
| miR-26a 5′-UTR-F2 | GGTACCCCTCTGGTGAAGAGCTACTGAA | KpnI | Cloning |
| miR-26a 5′-UTR-R2 | GAGCTCAGCCTGCACAAGTGTTCCTT | SacI | Cloning |
| miR-26a 5′-UTR-F3 | GGTACCGCTGGGATTTGAACTCAGGA | KpnI | Cloning |
| miR-26a 5′-UTR-R3 | GAGCTCTGACAGATGCATATGGAGCAC | SacI | Cloning |
| ChIP-F | CCCCCTGCTGTCTAGTTGAG | qPCR | |
| ChIP-R | CAGCCTCCACAGTAGCCTTC | qPCR | |
| Ctdsp2-F | GCCAAGTATGCTGACCCTGT | qPCR | |
| Ctdsp2-R | GTTTTCCTCAGGTCCCTTCC | qPCR | |
| GSK-3β-F | TTCGCCGTCCTTAACTCTTG | qPCR | |
| GSK-3β-R | CGAAGGTAGCCGAACAGAAG | qPCR | |
| GAPDH-F | TGTTCCTACCCCCAATGTGT | qPCR | |
| GAPDH-R | CCTGCTTCACCACCTTCTTG | qPCR |
Transfection and Luciferase Assays
Cells were grown in OptiMEM I medium (Invitrogen) for 24 h before transfection. Cells were transfected with 2.0 μg of expression vector bearing has-miR-26a precursor or mouse GSK-3β cDNA 3′-UTR or 2.4 μg of reporter vector bearing GSK-3β 3′-UTR, Egr-1, C/EBPα, or NF-κB binding elements using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols. The Renilla luciferase vector pGL4.74 was co-transfected with firefly luciferase vectors as a normalizer. For miR-26a inhibitor assays, cells were transfected with 400 ng of has-miR-26a miRCURY LNA knockdown probe (antagomir) or scrambled probe (Exiqon, Woburn, MA). For siRNA-mediated knockdown studies, cells were transfected with 475 pmol of siRNA specific for mouse Egr-1 or nonspecific siRNA (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). RNA transfection studies were performed with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. After 8 h, the transfection medium was replaced with the growth medium. Subsequent assays were made after 24–48-h transfection. Luciferase activity was measured with the Dual-Glo luciferase assay system (Promega) according to the manufacturer's protocol.
Solution Hybridization Detection Analysis
The expression levels of mature miRNAs were measured by solution hybridization detection method as described earlier (14).
Real-time Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Expression Analysis
Real-time RT-PCR assays were performed as described earlier (14). The amount of amplified transcripts (2 − ΔΔCT) was estimated by the comparative CT (ΔCT) method and normalized to an endogenous reference (GAPDH) relative to a calibrator. All PCR products were verified on agarose gel stained with ethidium bromide to discriminate between the correct amplification products and the potential primer dimers.
Western Blot
Cell lysates were isolated by using NE-PER nuclear and cytoplasmic extraction reagents (Pierce) according to the manufacturer's instructions. Sixty micrograms of protein was resolved by SDS-PAGE and transferred to nitrocellulose membrane. Membrane was blocked with 5% fat-free milk for 1 h and probed with mouse anti-α-smooth muscle actin, anti-SM22, anti-smMHC (myosin heavy chain), anti-GSK-3β, anti-Egr-1, anti-Erk-1/2, anti-p38, anti-JNK, or anti-β-actin. Antibody binding was detected with a peroxidase-conjugated goat anti-mouse IgG and chemiluminescence (Pierce).
DNA and Protein Synthesis Analyses
Click-iT EdU (5-ethynyl-2′-deoxyuridine, a thymidine analog) and Clic-iT HPG (l-homopropargylglycine, a glycine analog) kits were used to estimate DNA and protein synthesis, respectively, as described previously (14).
Cell Size Analysis
Cells were stained for α-actin fibers, and cell size was determined by computer-assisted planimetry. 100–200 cells in 20–30 fields were examined in each experiment.
Statistical Analysis
The results are expressed as means ± S.E. of at least three independent experiments. The comparison among different groups was performed by one-way analysis of variance followed by Bonferroni's test using SigmaStat 3.5 software. Paired data were evaluated by Student's t test. p < 0.05 was considered statistically significant.
RESULTS
ASMCsDes−/− Are Hypertrophic Phenotype
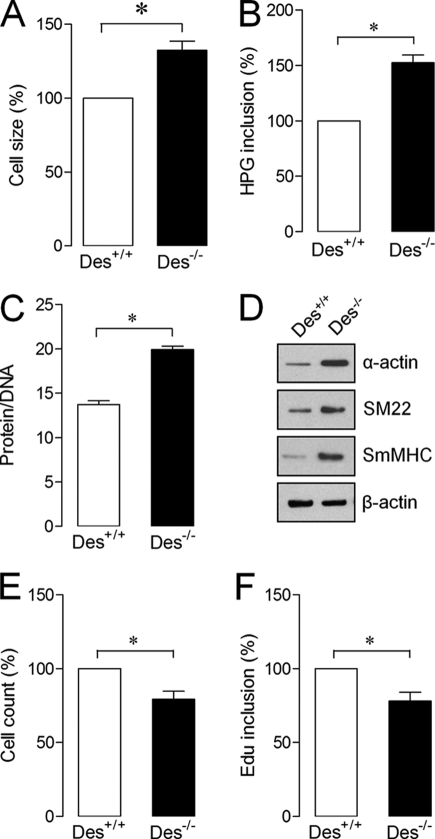
To examine whether desmin can involve in the regulation of ASMC phenotype, we isolated and cultured tracheal ASMCs from Des−/− mice and age-matched Des+/+ littermates (control). Our data showed that ASMCsDes−/− displayed hypertrophic phenotype, as evidenced by increased cell size (Fig. 1A), protein synthesis (Fig. 1B), and DNA/protein ratio (Fig. 1C). Expression of hypertrophic contractile proteins, such as α-actin, SM22, and SmMHC, was also high in ASMCsDes−/− (Fig. 1D). In contrast, loss of desmin in mouse ASMCs decreased proliferation, as evidenced by reduced cell numbers (Fig. 1E) and DNA synthesis (Fig. 1F). These results indicate that desmin is essential for maintaining ASMC phenotype.
FIGURE 1.
ASMCs isolated from of Des−/− mice display hypertrophy. ASMCs from Des+/+ and Des−/− mice were isolated and cultured to determine cell size (A), protein synthesis (B), protein/DNA ratio (C), smooth muscle-specific contractile proteins (D), proliferation (E), and DNA synthesis (F). Gel pictures are representative of three separate experiments. Each bar indicates mean ± S.E. (error bars) (n = 3). *, p < 0.05. Edu inclusion, DNA synthesis.
Loss of Desmin Alters miRNA Expression Profile in ASMC
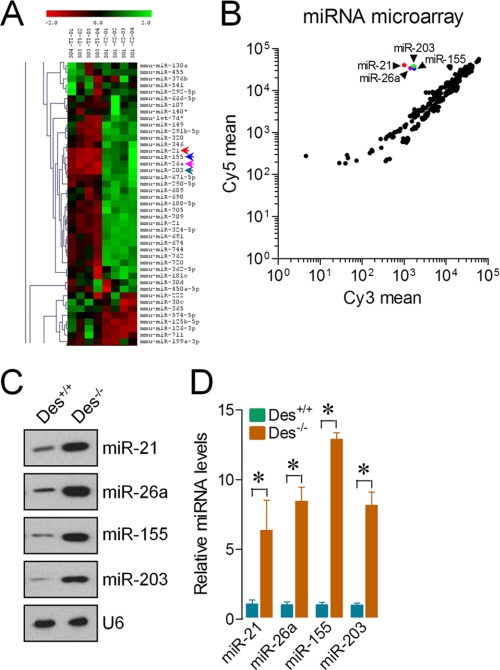
To explore whether desmin is necessary for the regulation of miRNAs in ASMCs, we performed an miRNA array screening using total RNA isolated from ASMCsDes+/+ and ASMCsDes−/−. The array uncovered the induction of 25 up-regulated and 26 down-regulated miRNAs in ASMCsDes−/−. The highly up-regulated miRNAs were miR-21, miR-26a, miR-155, and miR-203 (Fig. 2, A and B). Consistent with the microarray findings, solution hybridization and qPCR assays confirmed the miR-21, miR-26a, miR-155, and miR-203 up-regulations in ASMCsDes−/− (Fig. 2, C and D). The small nuclear RNA U6, a control and normalizer for miRNAs, was relatively unchanged, which excluded the possibility of artifactual changes in miRNA recovery. These results indicate that desmin plays a key role in the regulation of miRNA expression in mouse ASMCs.
FIGURE 2.
ASMCs of Des−/− mice show altered micro-RNA expression profiles. Total RNA was isolated from the ASMCs of Des+/+ and Des−/− mice to determine expression patterns of miRNA clusters. A, heat map diagram of the two-way hierarchical clustering showing relative miRNA expression levels between the ASMCs of Des+/+ and Des−/− mice. B, data presented on a scatter plot show log10-transformed signal intensities for each probe on both channels for the Cy3-labeled (Des+/+) and Cy5-labeled (Des−/−) samples. Each dot represents one miRNA probe. C and D, RNA used in A was analyzed by a solution hybridization technique with 5′-biotin-labeled miR-21, miR-26a, miR-155, miR-203, and small nuclear RNA U6 (C) and in a separate experiment by qPCR to assay expression of miR-21, miR-26a, miR-155, miR-203, and U6 under the same conditions (D). U6 served as both loading control and normalizer. Gel pictures are representative of three separate experiments. Each bar indicates mean ± S.E. (error bars) (n = 3). *, p < 0.05.
miR-26a Involves in the Initiation of ASMCDes−/− Hypertrophy
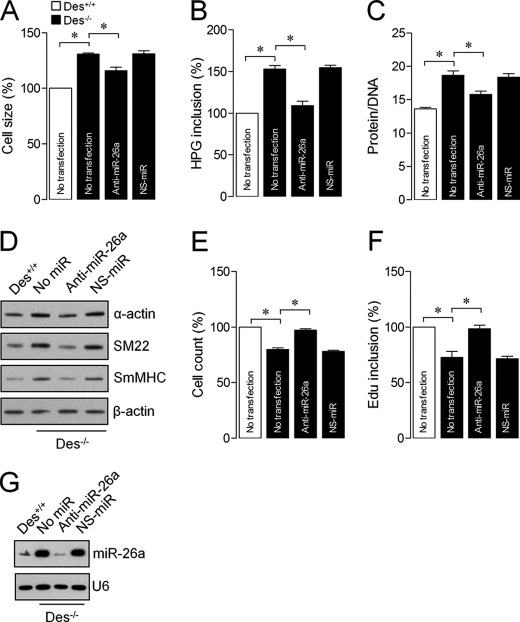
Previously, we showed miR-26a as a regulator of human ASMC hypertrophy (14). To evaluate whether miR-26a up-regulation in ASMCsDes−/− induces hypertrophy, we determined the growth activity of ASMCsDes−/− after miR-26a or nonspecific antagomir (nonspecific miRNA; NS-miR) transfection. Knockdown of miR-26a in ASMCsDes−/− significantly inhibited the loss of desmin-induced hypertrophy, as evidenced by decreased cell size (Fig. 3A), protein synthesis (Fig. 3B), protein/DNA ratio (Fig. 3C), and contractile protein expression (Fig. 3D). Interestingly, knockdown of miR-26a increased ASMCDes−/− proliferation, as evidenced by increased cell number (Fig. 3E) and DNA synthesis (Fig. 3F). Transfection of ASMCsDes−/− with NS-miR had no effect on cell phenotype. These results indicate the dynamic nature of miR-26a on ASMCDes−/− phenotype. We examined endogenous miR-26a expression using solution hybridization after transfection of ASMCsDes−/− with miR-26a antagomir. As expected, miR-26a antagomir decreased the endogenous miR-26a levels of expression, suggesting that particular antagomir is specific to miR-26a (Fig. 3G).
FIGURE 3.
miR-26a is responsible for the induction of ASMC hypertrophy in Des−/− mice. ASMCs of Des−/− mice were transfected with anti-miR-26a or NS-miR. Forty-eight hours after transfection, cell size (A), protein synthesis (B), protein/DNA ratio (C), smooth muscle-specific contractile proteins (D), proliferation (E), and DNA synthesis (F) were determined. G, total RNA was isolated and analyzed by a solution hybridization technique with 5′-biotin-labeled miR-26a and small nuclear RNA U6 to confirm the effect of anti-miR-26a on miR-26a expression. Gel pictures are representative of three separate experiments. Each bar indicates mean ± S.E. (error bars) (n = 3). *, p < 0.05.
Overexpression of miR-26a in ASMCDes+/+ Induces Hypertrophy
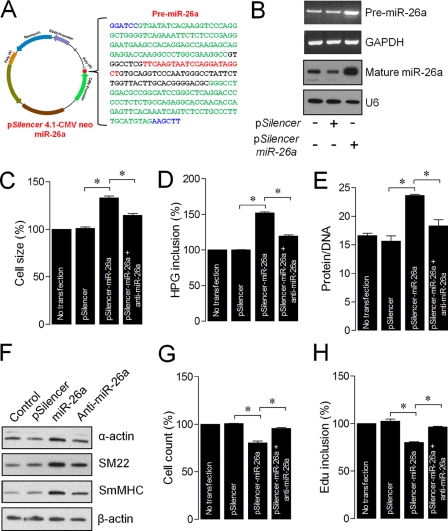
To investigate whether the miR-26a-induced ASMC hypertrophy occurs only in response to loss of desmin or independent of desmin, we transfected ASMCsDes+/+ with miR-26a expression vector (Fig. 4A) and confirmed the endogenous pre-miR-26a and mature miR-26a levels (Fig. 4B). The results showed that miR-26a overexpression in ASMCsDes+/+ induced hypertrophy (Fig. 4, C–F). Introduction of miR-26a-antagomir reversed the miR-26a-induced hypertrophic effects. Overexpression of miR-26a also inhibited ASMCsDes+/+ proliferation, and introduction of miR-26a-antagomir reversed the scenario (Fig. 4, G and H). These results indicate that miR-26a-induced-hypertrophy or inhibition of proliferation is independent of desmin expression.
FIGURE 4.
Enforced expression of miR-26a transforms normal ASMCs to hypertrophic phenotype. A, structure of the pSilencer-miR-26a construct contains pre-miR-26a, cytomegalovirus promoter (CMV), simian virus 40 (SV40), and neomycin. The blue letters represent two restriction sites, BamHI and HindIII. The red letters indicate 22 bases of mature miR-26a sequence. The underlined letters represent the 77-bp stem-loop sequence. The green letters indicate the 100-bp native flank sequence to both upstream and downstream of the stem loop sequence. B, ASMCs of Des+/+ mice were transfected with pSilencer or pSilencer-miR-26a and/or anti-miR-26a. Thirty-six hours after transfection, the overexpression of pre-miR-26a and mature miR-26a were confirmed by solution hybridization methods. Cell size (C), protein synthesis (D), protein/DNA ratio (E), smooth muscle-specific contractile proteins (F), proliferation (G), and DNA synthesis (H) were determined 72 h after transfection. Gel pictures are representative of three separate experiments. Each bar indicates mean ± S.E. (error bars) (n = 3). *, p < 0.05.
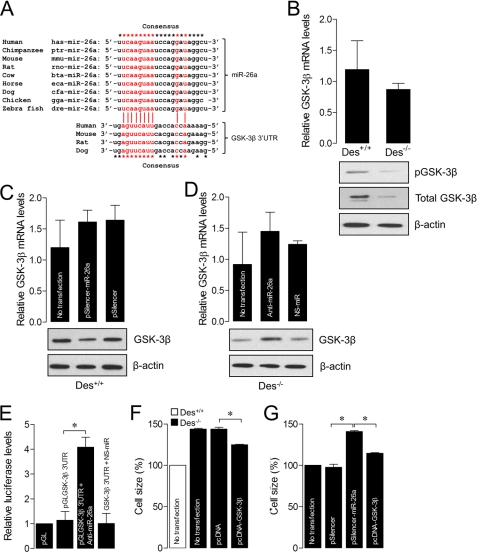
GSK-3β Is a Target mRNA of miR-26a
Studies show that GSK-3β is an anti-hypertrophic protein, which negatively regulates ASMC hypertrophy (14, 16, 17). Furthermore, GSK-3β has a conservative miR-26a seed sequence in its 3′-UTR (Fig. 5A). In agreement with this, our results showed that the levels of total and phosphorylated GSK-3β protein expression were lower in ASMCsDes−/− than ASMCsDes+/+ (Fig. 5B). Enforced expression of miR-26a significantly decreased GSK-3β protein expression in ASMCsDes+/+ (Fig. 5C). In contrast, miR-26a knockdown in ASMCsDes−/− significantly increased GSK-3β protein expression (Fig. 5D). However, in both experiments, there was no significant change in GSK-3β mRNA levels, suggesting that miR-26a predominantly inhibits GSK-3β translation. Subsequently, we confirmed the effect of miR-26a on GSK-3β translation. To determine whether miR-26a indeed binds on GSK-3β 3′-UTR, we transfected ASMCsDes−/− with a reporter construct containing the luciferase gene fused to the GSK-3β 3′-UTRβ (luc-GSK-3β3′UTR). As shown in Fig. 5E, cells transfected with luc-GSK-3β-3′UTR had less luciferase activity, and introduction of anti-miR-26a significantly increased the luciferase activity. NS-miR-26a antagomir had no effect on the luciferase activity. To further study whether GSK-3β itself inhibits ASMC hypertrophy, we overexpressed GSK-3β in ASMCsDes−/−. GSK-3β overexpression in ASMCsDes−/− inhibited hypertrophy (Fig. 5F). In contrast, ectopic expression of miR-26a in ASMCsDes+/+ induced hypertrophy, and GSK-3β overexpression reversed miR-26a-induced hypertrophy (Fig. 5G). Overall, our data support a model in which loss of desmin in ASMCs up-regulates miR-26a that inhibits GSK-3β protein expression, which in turn induces hypertrophy by increasing the expression of smooth muscle-specific markers.
FIGURE 5.
GSK-3β is a target of miR-26a. A, sequence alignment of putative miR-26a, and its targeting site on the 3′-UTR of GSK-3β shows a high level of complementarity and sequence conservation in vertebrates. B, total RNA and protein were isolated from the ASMCs of Des+/+ and Des−/− mice. GSK-3β mRNA levels were determined by qPCR. Total and phosphorylated (p) GSK-3β protein levels were determined by Western blot. C, ASMCs of Des+/+ mice were transfected with pSilencer or pSilencer-miR-26a expression construct. GSK-3β mRNA levels were determined 36 h after transfection by qPCR, and total GSK-3β protein levels were determined 48 h after transfection by Western blot. D, ASMCs of Des−/− mice were transfected with anti-miR-26a or NS-miR. GSK-3β mRNA levels were determined 36 h after transfection by qPCR, and total GSK-3β protein levels were determined 48 h after transfection by Western blot. E, ASMCs of Des−/− mice were transfected with the GSK-3β 3′-UTR-luciferase construct (luc-GSK-3β-3′UTR) with or without anti-miR-26a or NS-miR. Forty-eight hours after transfection, cells were collected, and then firefly luciferase activities were estimated and normalized to Renilla luciferase activities. F, ASMCs of Des−/− mice were transfected with pcDNA or pcDNA-GSK-3β expression construct. Forty-eight hours after transfection, cell size was determined. G, ASMCs of Des+/+ mice were transfected with pSilencer or pSilencer-miR-26a expression construct along with or without pcDNA-GSK-3β expression construct. Cell size was determined 48 h after transfection. Gel pictures are representative of three separate experiments. Each bar indicates mean ± S.E. (error bars) (n = 3). *, p < 0.05.
miR-26a Is under the Control of Egr-1
In the mouse genome, location of miR-26a is in the fifth intron of the C-terminal domain RNA polymerase II polypeptide A small phosphatase like-201 (Ctdspl-201) gene at chromosome 9 and in the fifth intron of the Ctdsp2–202 gene at chromosome 10 (Fig. 6A). To understand whether miR-26a transcription occurs through the activation of Ctdspl and/or Ctdps2 promoter, we performed qPCR with primers specific for Ctdspl or Ctdsp2. Our data showed that the mRNA level of Ctdsp2 was 3.4-fold higher in ASMCsDes−/− than that in ASMCsDes+/+ (Fig. 6B). These findings provide evidence that miR-26a transcription in ASMCsDes−/− occurs via the activation of the Ctdsp2 promoter. To determine the specific transcription factor that involves in the transcription of miR-26a in ASMCsDes−/−, we analyzed the miR-26a promoter region using the public software PATCH (available on-line). A scan of a 1.5-kb genomic sequence located upstream of the ATG of the mouse Ctdsp2 gene identified putative consensus binding sites of C/EBPα, Egr-1, and NF-κB (Fig. 6C), which play important roles in airway remodeling (14, 18–20). Using ChIP assays, we found that the activation of miR-26a promoter in ASMCsDes−/− was completely dependent on the Egr-1 binding sites (Fig. 6D). In contrast, C/EBPα and NF-κB had no activities on the miR-26a promoter in ASMCsDes−/−.
To study whether Egr-1 indeed influences the promoter activity of miR-26a, ASMCsDes−/− were transfected with pGL-C/EBPα, pGL-Egr-1, and pGL-NF-κB luciferase reporter construct for 48 h. ASMCsDes−/− transfected with the pGL-Egr-1 construct had higher luciferase activities than pGL-C/EBPα and pGL-NF-κB constructs. However, these constructs had no luciferase activities in ASMCsDes+/+ (Fig. 6E). To study the effect of endogenous Egr-1 on miR-26a promoter activity, ASMCsDes−/− were transfected with Egr-1 siRNA for 36 h, which led to a decrease in the endogenous Egr-1 expression both in total and phosphorylated states (Fig. 6F). Transfections of the pGL-Egr-1 promoter construct into Egr-1-knocked down ASMCsDes−/− abolished miR-26 promoter activity (Fig. 6G). Furthermore, knockdown of Egr-1 decreased miR-26a expression (Fig. 6H) followed by inhibition of hypertrophy in ASMCDes−/− (Fig. 6, I and J). Taken together, these data indicate that miR-26a is a direct transcriptional target of Egr-1 in the loss of desmin-induced ASMC hypertrophy.
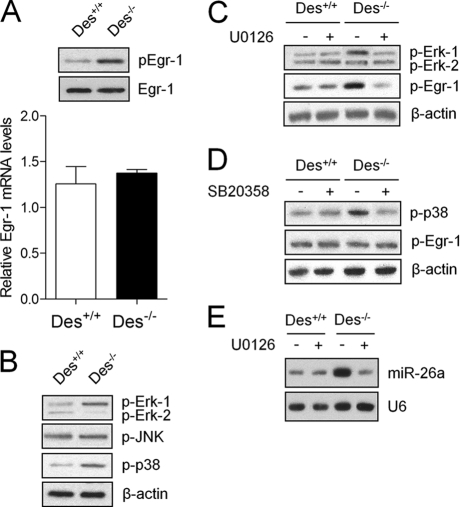
Erk-1/2 Is an Upstream Target of Egr-1 in ASMCDes−/−
To explore how loss of desmin activates Egr-1 in mouse ASMCs, we determined the activation of Erk-1/2, p38, and JNK MAPKs. Loss of desmin increased Egr-1 activation at the post-transcriptional level (Fig. 7A). Erk-1/2 and p38, but not JNK, were higher in ASMCsDes−/− than in ASMCsDes+/+ (Fig. 7B). Interestingly, treatment of ASMCsDes−/− with U0126 (Erk-1/2 inhibitor) but not SB20358 (p38 inhibitor) reduced Egr-1 activation (Fig. 7, C and D). Treatment of ASMCsDes−/− with U0126 also decreased miR-26a expression Fig. 7E). These results indicate that loss of desmin activates Erk-1/2 that post-transcriptionally activates Egr-1, which in turn up-regulates miR-26a in ASMCsDes−/−.
FIGURE 7.
Erk-1/2 is the upstream signaling protein of Egr-1 in ASMCs of Des−/− mice. Total RNA and protein were isolated from the ASMCs of Des+/+ and Des−/− mice. A, Egr-1 mRNA levels were determined by qPCR, and total and phosphorylated Egr-1 proteins were determined by Western blot. B, phosphorylated Erk-1/2, JNK, and p38 were determined by Western blot. C–E, ASMCs of Des+/+ and Des−/− mice were pretreated with either 50 μm Erk-1/2 inhibitor (U0126) or 20 μm p38 inhibitor (SB203580) for 30 min. Total protein was isolated, and phosphorylated Erk-1/2 (C), p38 (D), and Egr-1 (C and D) were determined by Western blot. Total RNA was isolated from U0126-treated cells to determine miR-26 expression by solution hybridization (E). Gel pictures are representative of three separate experiments. Each bar indicates mean ± S.E. (n = 3).
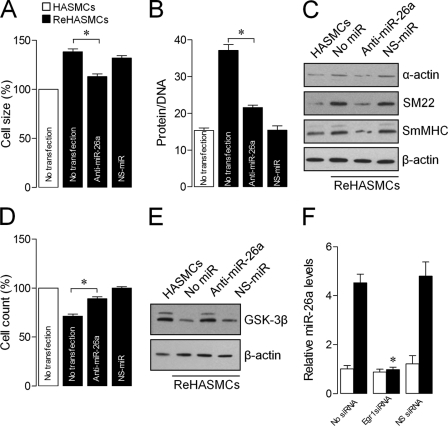
Knockdown of Desmin in HASMCs Induces Hypertrophy through miR-26a-GSK-3β Pathway
To study whether loss of desmin-induced ASMC hypertrophy by miR-26a is conserved in humans, desmin gene was knocked down in HASMCs using an siRNA strategy, as described earlier (15). siRNA-mediated knockdown of desmin in HASMCs increased cell size, protein/DNA ratio, and smooth muscle-specific gene expression. (Fig. 8, A–C) and inhibited proliferation (Fig. 8D), similar to ASMCsDes−/−. Knockdown of desmin in HASMCs also decreased GSK-3β protein expression, and transfection of these cells with miR-26a-antagomir increased GSK-3β protein expression (Fig. 8E). The Egr-1 RNAi strategy corroborates Egr-1-induced-miR-26a up-regulation in desmin-knocked down HASMCs (Fig. 8F). These data indicate that the loss of desmin-induced ASMC hypertrophy is mediated through a conserved signaling pathway in mammals at least in mice and humans.
FIGURE 8.
siRNA-mediated knockdown of desmin induces hypertrophy through the Egr-1/miR-26a/GSK-3β pathway in human ASMCs. Normal HASMCs and reHASMCs were transfected with anti-miR-26a or NS-miR. A–E, 48 h after transfection, cell size (A), protein/DNA ratio (B), smooth muscle-specific contractile proteins (C), proliferation (D), and GSK-3β protein levels (E) were determined. F, normal HASMCs and reHASMCs were transfected with Egr-1 siRNA or Egr-1 nonspecific siRNA (Egr-1NS-siRNA). miR-26a levels were determined by qPCR. Gel pictures are representative of three separate experiments. Each bar indicates mean ± S.E. (n = 3). *, p < 0.05.
DISCUSSION
In the present study, we demonstrate that desmin is an important intermediate filament that maintains ASMC phenotype through an miRNA-regulated pathway. ASMCs isolated from desmin null mice were hypertrophic phenotype and showed miRNA dysregulation. Loss and gain functional analyses of miR-26a showed that miR-26a up-regulation in ASMCsDes−/− induced hypertrophy through the suppression of the anti-hypertrophic protein GSK-3β. Furthermore, overactivation of Erk-1/2 in ASMCsDes−/− increased Egr-1 phosphorylation, which in turn up-regulated miR-26a expression. The loss of desmin-induced-ASMC hypertrophy by Erk-1/2-Egr-1/miR-26a/GSK-3β pathway is consistent in recombinant HASMCs, which stably suppress 90% of endogenous desmin expression. Overall, for the first time, our study uncovers that desmin is an anti-hypertrophic protein important for ASMC homeostasis and provides a new molecular link between intermediate filament and miRNA regulation.
Desmin is one of the structural and intermediate proteins expressed in all muscle tissues, including ASMCs. Structural changes to airway smooth muscle due to hypertrophy and/or hyperplasia are a well known feature of chronic airway diseases, such as asthma, chronic obstructive pulmonary disease, and cystic fibrosis (1–5). In addition, in the last few years, miRNAs have been recognized as potent regulators of gene expression and as a key modulator of a variety of biological processes (21, 22). Under normal physiological conditions, miRNAs remain at a steady level, and this level is altered during pathogenesis of airway diseases (23), thereby leading to changes in the protein coding gene expression. We have recently shown that miR-26a is a mechanosensitive hypertrophic miRNA in HASMC (14). Moreover, studies show that mechanical stretch increases desmin and contractile protein expression in canine ASMCs (24) and lung undifferentiated mesenchymal cells (25), suggesting a link between desmin and mechanical regulation. However, the key question of whether desmin can regulate ASMC phenotype by altering miRNA expression profile remains to be answered. Our microarray and qPCR data showed that ASMCsDes−/− up-regulated miR-21, miR-26a, miR-155, and miR-203 expressions.
Our results also showed that ASMCsDes−/− or reHASMCs were hypertrophic phenotype as evidenced by increased cell size, protein synthesis, protein-DNA ratio, and contractile protein expression, indicating a negative correlation between desmin and ASMC hypertrophy. This is in agreement with the previous study, which showed a negative correlation between the level of selective expression of ASMC proteins, including desmin and airway hyperresponsiveness in asthmatic patients (7). It is well known that airway hyperresponsiveness is directly associated with ASMC hypertrophy and/or hyperplasia (6). The negative correlation between the level of desmin expression and airway hyperresponsiveness in asthmatic patients may be due to ASMC hypertrophy rather than hyperplasia. However, our previous study showed that airways of Des−/− mice had hyporesponsiveness to cholinergic stimulation (13). This may be due to a reduced number of ASMCs in the airways of Des−/− mice. It is well documented that cell hypertrophy occurs after cell cycle arrest. In agreement with this, the present study and our previous study (14) showed that miR-26a up-regulation either by loss of desmin, enforced expression, or stretch inhibited cell proliferation and then transformed ASMCs to hypertrophic phenotype. Further studies are necessary to investigate how miR-26a inhibits ASMC proliferation.
Gene expression-modulating miRNAs are encoded in diverse genomic locations, including intergenic regions, introns of protein-coding genes, and introns/exons of non-coding RNA genes (26). Most mammalian miRNAs lie between protein-coding genes, whereas about one-third are within the introns of annotated mRNAs (27, 28). In the human and mouse genomes, two distinct genes, Ctdspl and Ctdsp2, encode different pri-miR-26a (miR-26a-1 and miR-26a-2), leading to the generation of identical mature miR-26a. In mice, miR-26a-1 is located in an intron of Ctdspl at chromosome 9, and miR-26a-2 is located in an intron of Ctdsp2 at chromosome 10. It is likely that the intronic miRNAs are processed from the same primary transcript as the precursor mRNAs, and thus, their expression levels are regulated by the expression of the host mRNA (29). We examined the correlation between the miR-26a expression profile and the expression profile of the host genes by qPCR. Interestingly, ASMCsDes−/− overexpressed Ctdsp2 but not Ctdspl mRNA (data not shown). These results indicate that processing of most of the miR-26a in ASMCsDes−/− is from the same primary transcript as its host gene, Ctdsp2. This suggests the existence of a strong correlation between the expression of miR-26a and Ctdsp2 mRNA, consistent with previous findings (30). We determined the specific transcription factor that up-regulates pri-miR-26a in ASMCsDes−/−. Using a ChIP assay, we found that activation of miR-26a promoter in ASMCsDes−/− was completely dependent on the Egr-1 binding sites. The luciferase reporter assays demonstrated that the activation of the miR-26a promoter in ASMCsDes−/− was effective only after the transfection of cells with pGL-Egr-1 construct, which contains one Egr-1 binding site. Moreover, the siRNA-mediated knockdown of Egr-1 confirmed the requirement of Egr-1 for the activation of miR-26a promoter in ASMCsDes−/−. Egr-1 is a zinc finger transcription factor expressed in a variety of cell types, including smooth muscle cells, by a large number of growth factors, cytokines, and injurious stimuli (31). Studies show that administration of epidermal growth factor in mouse lung tissue (32) and the lungs of fetal and neonatal transgenic mice expressing TGF-α (18) trigger overexpression of Egr-1 protein. Moreover, Egr-1 promotes both growth and inhibition of tumor cells (33–38). Importantly, mouse remodeling airways and vessels after TGF-α induction rapidly up-regulate Egr-1 (18). These results suggest the role of Egr-1 in cell growth. Furthermore, we found that loss of desmin in ASMCs activated Erk-1/2 but not p38 and JNK, which in turn up-regulated miR-26a through Egr1 in ASMCsDes−/−.
The molecular mechanism underlying ASMC hypertrophy involves a change in the gene expression profile, mostly including the up-regulation of translation, transcription, and survival-related genes (6). Like miR-26a, many proteins emerged to regulate hypertrophy and promote disease upon their dysregulated expression. Among those, one of the potential proteins that regulate hypertrophy is GSK-3β. GSK-3β is constitutively active in non-stimulated cells and inactivated upon phosphorylation at Ser9 (39). Although initially described as an inhibitor of glycogen synthesis through phosphorylation of glycogen synthase (40), GSK-3β was later revealed as a key signaling molecule regulating many aspects of cellular function, including protein synthesis, cytoskeletal integrity, and gene expression (41). More importantly, GSK-3β negatively regulates cardiac (42–46), skeletal (47, 48), and ASMC (16, 17) hypertrophy, as evidenced by the finding that GSK-3β overexpression inhibits the induction of hypertrophy in these cells by blocking global protein synthesis that eventually reduced the expression of hypertrophic genes. These studies suggest the post-transcriptional role of GSK-3β on hypertrophy. Interestingly, our results showed that GSK-3β phosphorylation in ASMCsDes−/− was decreased due to a reduction in the total protein production, and, which induced ASMCDes−/− hypertrophy. Our experiment identified GSK-3β as a downstream target of miR-26a, which inhibits GSK-3β protein translation. A similar finding has been reported in HASMCs, in which inhibition of GSK-3β by RNAi increases cell size and protein synthesis (17). In addition, we found that the miR-26a-GSK-3β-mediated signaling pathway did not regulate Des−/− ASMC proliferation. This is also consistent with the previous finding demonstrating that GSK-3β does not participate in HASMC proliferation (17). Moreover, our search for GSK-3β targets provided only five miRNAs (miR-26a, miR-26b, miR-199a, 199b, and 302a*). Because loss of desmin in ASMC did not induce any of these miRNAs except for miR-26a in ASMC, we cannot rule out the possibility that these miRNAs are not upstream targets of GSK-3β. It is well recognized that miRNAs may function according to a “combinational circuitry model,” whereby a single miRNA targets multiple mRNAs and several miRNAs may target a single mRNA (49). Thus, further studies are required to elucidate specifically whether hypertrophic stimuli can induce the expression of these predicted miRNAs and whether miR-26a can down-regulate hypertrophic proteins of ASMCs other than GSK-3β.
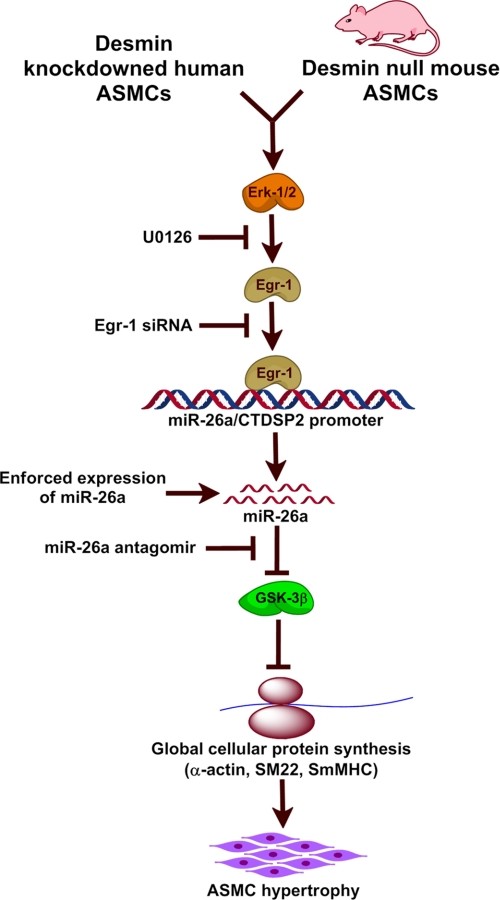
In summary, our results provide experimental evidence demonstrating that desmin is an anti-hypertrophic protein, which induces ASMC hypertrophy as a result of its down-regulation. Loss of desmin in ASMC activates Erk-1/2, which up-regulates miR-26a through Egr-1 activation. Molecular functional analyses indicate that GSK-3β is a target gene of miR-26a, which inhibits GSK-3β protein production. The Erk-1/2/Egr-1/miR-26a/GSK-3β pathway in the induction of hypertrophy is conserved in human ASMCs (Fig. 9).
FIGURE 9.
miR-26a pathway in the initiation of ASMC hypertrophy. Up-regulation of miR-26a in ASMCsDes−/− and reHASMCs, which stably suppress endogenous desmin expression, increases global cellular protein synthesis through inhibition of GSK-3β protein expression. As a result, ASMC hypertrophic marker proteins (α-actin, SM22, and MHC) are up-regulated, which in turn initiates hypertrophy.
This work was supported by the National Science Foundation and the Keck Foundation.
- ASMC
- airway smooth muscle cell
- HASMC
- human airway smooth muscle cell
- reHASMC
- recombinant human airway smooth muscle cell
- miRNA
- microRNA
- NS-miR
- nonspecific miRNA
- COPD
- chronic obstructive pulmonary disease
- qPCR
- quantitative PCR.
REFERENCES
- 1. Benayoun L., Druilhe A., Dombret M. C., Aubier M., Pretolani M. (2003) Am. J. Respir. Crit. Care Med. 167, 1360–1368 [DOI] [PubMed] [Google Scholar]
- 2. Ebina M., Takahashi T., Chiba T., Motomiya M. (1993) Am. Rev. Respir. Dis. 148, 720–726 [DOI] [PubMed] [Google Scholar]
- 3. Hays S. R., Ferrando R. E., Carter R., Wong H. H., Woodruff P. G. (2005) Thorax 60, 226–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jeffery P. K. (2001) Am. J. Respir. Crit. Care Med. 164, S28–38 [DOI] [PubMed] [Google Scholar]
- 5. Regamey N., Ochs M., Hilliard T. N., Mühlfeld C., Cornish N., Fleming L., Saglani S., Alton E. W., Bush A., Jeffery P. K., Davies J. C. (2008) Am. J. Respir. Crit. Care Med. 177, 837–843 [DOI] [PubMed] [Google Scholar]
- 6. Terzano C., Laurendi G., Capoccetta G., Rapisarda M., Pacilio R., Petroianni A. (2003) Eur. Rev. Med. Pharmacol. Sci. 7, 9–26 [PubMed] [Google Scholar]
- 7. Slats A. M., Janssen K., van Schadewijk A., van der Plas D. T., Schot R., van den Aardweg J. G., de Jongste J. C., Hiemstra P. S., Mauad T., Rabe K. F., Sterk P. J. (2008) J. Allergy Clin. Immunol. 121, 1196–1202 [DOI] [PubMed] [Google Scholar]
- 8. Costa M. L., Escaleira R., Cataldo A., Oliveira F., Mermelstein C. S. (2004) Braz. J. Med. Biol. Res. 37, 1819–1830 [DOI] [PubMed] [Google Scholar]
- 9. Halayko A. J., Salari H., Ma X., Stephens N. L. (1996) Am. J. Physiol. 270, L1040–L1051 [DOI] [PubMed] [Google Scholar]
- 10. Kapanci Y., Ribaux C., Chaponnier C., Gabbiani G. (1992) J. Histochem. Cytochem. 40, 1955–1963 [DOI] [PubMed] [Google Scholar]
- 11. Park S., Rasmussen H. (1986) J. Biol. Chem. 261, 15734–15739 [PubMed] [Google Scholar]
- 12. Yamada M., Kurihara H., Kinoshita K., Sakai T. (2005) J. Histochem. Cytochem. 53, 735–744 [DOI] [PubMed] [Google Scholar]
- 13. Shardonofsky F. R., Capetanaki Y., Boriek A. M. (2006) Am. J. Physiol. Lung Cell. Mol. Physiol. 290, L890–L896 [DOI] [PubMed] [Google Scholar]
- 14. Mohamed J. S., Lopez M. A., Boriek A. M. (2010) J. Biol. Chem. 285, 29336–29347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mohamed J. S., Boriek A. M. (2011) FASEB J., in press [Google Scholar]
- 16. Bentley J. K., Deng H., Linn M. J., Lei J., Dokshin G. A., Fingar D. C., Bitar K. N., Henderson W. R., Jr., Hershenson M. B. (2009) Am. J. Physiol. Lung Cell. Mol. Physiol. 296, L176–L184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deng H., Dokshin G. A., Lei J., Goldsmith A. M., Bitar K. N., Fingar D. C., Hershenson M. B., Bentley J. K. (2008) J. Biol. Chem. 283, 10198–10207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kramer E. L., Mushaben E. M., Pastura P. A., Acciani T. H., Deutsch G. H., Khurana Hershey G. K., Korfhagen T. R., Hardie W. D., Whitsett J. A., Le Cras T. D. (2009) Am. J. Respir. Cell Mol. Biol. 41, 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roth M., Johnson P. R., Borger P., Bihl M. P., Rüdiger J. J., King G. G., Ge Q., Hostettler K., Burgess J. K., Black J. L., Tamm M. (2004) N. Engl. J. Med. 351, 560–574 [DOI] [PubMed] [Google Scholar]
- 20. Ogawa H., Azuma M., Muto S., Nishioka Y., Honjo A., Tezuka T., Uehara H., Izumi K., Itai A., Sone S. (2011) Clin. Exp. Allergy 41, 104–115 [DOI] [PubMed] [Google Scholar]
- 21. Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 22. Plasterk R. H. (2006) Cell 124, 877–881 [DOI] [PubMed] [Google Scholar]
- 23. Williams A. E., Larner-Svensson H., Perry M. M., Campbell G. A., Herrick S. E., Adcock I. M., Erjefalt J. S., Chung K. F., Lindsay M. A. (2009) PLoS ONE 4, e5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu M., Tanswell A. K., Post M. (1999) Am. J. Physiol. 277, L667–L683 [DOI] [PubMed] [Google Scholar]
- 25. Yang Y., Beqaj S., Kemp P., Ariel I., Schuger L. (2000) J. Clin. Invest. 106, 1321–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Griffiths-Jones S., Saini H. K., van Dongen S., Enright A. J. (2008) Nucleic Acids Res. 36, D154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rodriguez A., Griffiths-Jones S., Ashurst J. L., Bradley A. (2004) Genome Res. 14, 1902–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saini H. K., Griffiths-Jones S., Enright A. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17719–17724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cullen B. R. (2004) Mol. Cell 16, 861–865 [DOI] [PubMed] [Google Scholar]
- 30. Cai X., Hagedorn C. H., Cullen B. R. (2004) RNA 10, 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Silverman E. S., Collins T. (1999) Am. J. Pathol. 154, 665–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu L., Tsai J. C., Aird W. C. (2000) Blood 96, 1772–1781 [PubMed] [Google Scholar]
- 33. Calogero A., Arcella A., De Gregorio G., Porcellini A., Mercola D., Liu C., Lombari V., Zani M., Giannini G., Gagliardi F. M., Caruso R., Gulino A., Frati L., Ragona G. (2001) Clin. Cancer Res. 7, 2788–2796 [PubMed] [Google Scholar]
- 34. Baron V., De Gregorio G., Krones-Herzig A., Virolle T., Calogero A., Urcis R., Mercola D. (2003) Oncogene 22, 4194–4204 [DOI] [PubMed] [Google Scholar]
- 35. Huang R. P., Fan Y., de Belle I., Niemeyer C., Gottardis M. M., Mercola D., Adamson E. D. (1997) Int. J. Cancer 72, 102–109 [DOI] [PubMed] [Google Scholar]
- 36. Levin W. J., Casey G., Ramos J. C., Arboleda M. J., Reissmann P. T., Slamon D. J. (1994) Chest 106, 372S–376S [PubMed] [Google Scholar]
- 37. Liu C., Calogero A., Ragona G., Adamson E., Mercola D. (1996) Crit. Rev. Oncogene 7, 101–125 [PubMed] [Google Scholar]
- 38. Rauscher F. J., 3rd (1993) Adv. Exp. Med. Biol. 348, 23–29 [DOI] [PubMed] [Google Scholar]
- 39. Cohen P., Frame S. (2001) Nat. Rev. Mol. Cell Biol. 2, 769–776 [DOI] [PubMed] [Google Scholar]
- 40. Parker P. J., Caudwell F. B., Cohen P. (1983) Eur. J. Biochem. 130, 227–234 [DOI] [PubMed] [Google Scholar]
- 41. Grimes C. A., Jope R. S. (2001) Prog. Neurobiol. 65, 391–426 [DOI] [PubMed] [Google Scholar]
- 42. Antos C. L., McKinsey T. A., Frey N., Kutschke W., McAnally J., Shelton J. M., Richardson J. A., Hill J. A., Olson E. N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 907–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Badorff C., Ruetten H., Mueller S., Stahmer M., Gehring D., Jung F., Ihling C., Zeiher A. M., Dimmeler S. (2002) J. Clin. Invest. 109, 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haq S., Choukroun G., Kang Z. B., Ranu H., Matsui T., Rosenzweig A., Molkentin J. D., Alessandrini A., Woodgett J., Hajjar R., Michael A., Force T. (2000) J. Cell Biol. 151, 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hardt S. E., Sadoshima J. (2002) Circ. Res. 90, 1055–1063 [DOI] [PubMed] [Google Scholar]
- 46. Morisco C., Seta K., Hardt S. E., Lee Y., Vatner S. F., Sadoshima J. (2001) J. Biol. Chem. 276, 28586–28597 [DOI] [PubMed] [Google Scholar]
- 47. Rochat A., Fernandez A., Vandromme M., Molès J. P., Bouschet T., Carnac G., Lamb N. J. (2004) Mol. Biol. Cell 15, 4544–4555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vyas D. R., Spangenburg E. E., Abraha T. W., Childs T. E., Booth F. W. (2002) Am. J. Physiol. Cell Physiol. 283, C545–C551 [DOI] [PubMed] [Google Scholar]
- 49. He H., Jazdzewski K., Li W., Liyanarachchi S., Nagy R., Volinia S., Calin G. A., Liu C. G., Franssila K., Suster S., Kloos R. T., Croce C. M., de la Chapelle A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 19075–19080 [DOI] [PMC free article] [PubMed] [Google Scholar]