Background: The mechanism underlying incomplete dominance of KCNQ2 channel mutations, which causes epilepsy, remains unknown.
Results: We have identified a novel degradation signal with a key five-amino acid (RCXRG) motif from distal C-terminal frameshift mutations of KCNQ2.
Conclusion: The novel degradation signal accelerates degradation of surface mutant proteins through ubiquitin-independent proteasome machinery.
Significance: Our findings reveal a mechanism by which mutant KCNQ2 proteins cause epilepsy.
Keywords: Cell Surface Protein, Epilepsy, Ion Channels, Potassium Channels, Proteasome, Protein Degradation, BFNC, KCNQ2, Kv7.2
Abstract
Benign familial neonatal convulsions is an autosomal-dominant idiopathic form of epilepsy primarily caused by gene mutations of the voltage-gated Kv7.2/KCNQ2/M-channel that exert only partial dominant-negative effects. However, the mechanism underlying the incomplete dominance of channel mutations, which cause epilepsy in infancy, remains unknown. Using mutagenesis and biochemistry combined with electrophysiology, we identified a novel degradation signal derived from distal C-terminal frameshift mutations, which impairs channel function. This degradation signal, transferable to non-channel CD4, can lead to accelerated degradation of mutant proteins through ubiquitin-independent proteasome machinery but does not affect mRNA quantity and protein trafficking. Functional dissection of this signal has revealed a key five-amino acid (RCXRG) motif critical for degradation. Taken together, our findings reveal a mechanism by which proteins that carry this signal are subject to degradation, leading to M-current dysfunction, which causes epilepsy.
Introduction
Benign familial neonatal convulsions (BFNC)2 is an autosomal-dominant form of idiopathic epilepsy characterized by unprovoked partial or generalized seizures within a week after birth and remission after 3–10 weeks (1). In most patients psychomotor development is normal, but about 15% of individuals suffer from seizures later in life (2, 3). BFNC is caused by mutations in voltage-gated Kv7.2/KCNQ2 or Kv7.3/KCNQ3 potassium channel genes, which encode a low threshold, non-inactivating native M-current (4–7). M-current regulates neuronal excitability, and inhibition of M-current or loss of function of KCNQ2 leads to repetitive firing and epileptic seizures in mice (8, 9). Most known BFNC mutations have been identified in KCNQ2, but a few reside in KCNQ3 (10). Among the KCNQ2 mutations that have been tested, none exert complete dominant-negative effects on wild-type KCNQ2 (6, 11, 12). These observations have led to the conclusion that BFNC is caused by haploinsufficiency, indicating that the single functional copy of the KCNQ2 gene in BFNC patients does not produce enough functional channel protein.
Kv7.2/KCNQ2 is a member of the Kv7 family and has six transmembrane domains followed by a long cytoplasmic C terminus that contains many functional motifs and domains. It has previously been shown that most BFNC mutations are located in the C terminus of KCNQ2 (6, 13), including two distal C-terminal frameshift mutations of KCNQ2, Q2-2513del1bp (1-nucleotide deletion at 2513 bp) and Q2-2516ins5bp (5-nucleotide insertion at 2516 bp), in which there is substitution of only the last 7 or 2 residues, respectively. These two distal C-terminal frameshift mutations exhibit normal biophysical properties but show reduced current density (13–15). However, the clinical phenotype of epileptic patients carrying these two distal C-terminal frameshift mutations is no less serious than that in patients with loss-of-function mutations in the pore region that functions as the channel gate and ion selectivity filter (13, 14, 16). This raises questions as to how the two distal C-terminal frameshift mutations impair channel function and cause epilepsy.
Protein degradation is highly selective with individual protein half-lives ranging from minutes to years (17). A degradation signal is defined as a protein sequence that can reduce the half-life of the target protein and is sufficient for recognition and degradation by distinct proteolytic pathways (17, 18). An important property of degradation signals is their transferability, and genetically engineered attachment of such signal sequences leads to metabolic instability in otherwise long-lived proteins (17).
In this study we investigated the mechanism by which two C-terminal frameshift mutations that bring about the impairment of the channel function underlie BFNC. Using biochemical approaches combined with electrophysiology, we identified a novel degradation signal in the extended segments of the two frameshift mutations. This degradation signal contains a critical five-amino acid motif that leads to accelerated degradation of the mutant proteins through the ubiquitin-independent proteasome machinery but does not affect mRNA quantity and protein trafficking. Our findings demonstrate that BFNC haploinsufficiency is caused by accelerated degradation of channel proteins carrying the degradation signal, and this cellular mechanism brings about impaired channel function and causes neonatal epilepsy.
EXPERIMENTAL PROCEDURES
Molecular Biology
The human KCNQ2 and KCNQ3 cDNAs were kindly provided by Thomas J. Jentsch, and all point mutants, deletion mutants, and fusion proteins were created by PCR-based mutagenesis with LA taq (Takara). KCNQ2 and its mutants were tagged with three tandem-repeated FLAG epitopes (DYKDDDDK) at their N terminus. All CD4 and CD4 mutant constructs were tagged with HA epitope (YPYDVPDYA). CD4-GFP60aa was generated by adding 60 C-terminal residues of green fluorescent protein to the C terminus of CD4. CD4-ExtraC was constructed by appending ExtraC to the C terminus of CD4. For whole-cell patch clamp recording, cycloheximide treatment, pulse-chase assay, surface biotinylation, ubiquitination assay, and quantitative RT-PCR analysis, all cDNA clones used in these experiments were constructed and inserted into the pIRES2-EGFP vector (Clontech). For two-electrode voltage clamp recording, wild-type constructs of KCNQ2, KCNQ3, and KCNQ5 and their mutants were introduced into the KSM vector (19). KCNQ2 and Q2-2513del1bp were constructed and inserted into pEGFPC2 (Clontech) for fluorescence microscopy.
Confocal Fluorescence Microscopy
For staining of localization in the ER, HEK293 cells were co-transfected with EGFP-KCNQ2 or EGFP-fsKCNQ2 (Q2-2513del1bp) and pDsRed2-ER (Clontech) using Lipofectamine 2000 (Invitrogen). After 36 h of incubation, cells were washed in phosphate-buffered saline (PBS containing 10 mm phosphate buffer, pH 7.4, 150 mm NaCl, 0.5 mm MgCl2, and 1 mm CaC12) three times, and fixed in 4% paraformaldehyde, PBS for 15 min before washing in PBS three times. Slides were mounted with mounting medium, and images were obtained using a confocal microscope (FV1000; Olympus). For staining of localization in the Golgi apparatus, cells transfected with EGFP-KCNQ2 or EGFP-fsKCNQ2 were fixed as above. Cells were permeabilized in 0.2% Triton X-100/PBS for 10 min, and blocked in 5% goat serum/PBS for 1 h prior to incubation for 2 h at room temperature with rat antibody against GM130 (a Golgi matrix protein) at dilution of 1:150 (BD Transduction Laboratories) with serum/PBS. Cells were washed in PBS three times, and then incubated for 1 h at room temperature with TRITC-conjugated anti-rat antibody (1:250; Zhongshanjinqiao, Beijing, China). After washing three times in PBS, slides were mounted with mounting medium and images were obtained using the same confocal microscope.
Electrophysiology
For whole-cell patch clamp recording, currents were recorded at room temperature using the EPC 10 UBS patch clamp amplifier (HEKA). Patch electrodes were pulled from borosilicate glass and had a final resistance of 3–5 megaohms when filled with internal pipette solution that contained 130 mm KCl, 3 mm NaCl, 1 mm MgCl2, 5 mm EGTA, 10 mm HEPES, 5 mm glucose, and 3 mm Mg-ATP at pH 7.3. Cells were perfused with the external solution containing 140 mm NaCl, 4.7 mm KCl, 1.2 mm MgCl2, 1 mm CaCl2, 11 mm glucose, and 5 mm HEPES at pH 7.4. Data were acquired and analyzed using PatchMaster software (HEKA).
For two-electrode voltage clamp recording, Xenopus oocytes (stages V and VI) were selected and injected with 46 nl of solution containing 10 ng of cRNA using a microinjector (Drummond Scientific Co.). 3 days after injection, oocytes were impaled with two microelectrodes (0.5–1.0 megaohm) filled with 3 m KCl in a 40-μl recording chamber, and whole cell currents were recorded using a two-electrode voltage clamp. The chamber was constantly perfused with ND-96 recording solution containing 96 mm NaCl, 2 mm KCl, 1 mm MgCl2, 1 mm CaCl2, and 5 mm HEPES at pH 7.6. Currents were recorded in ND-96 solution at room temperature using a GeneClamp 500 amplifier (Axon Instruments) or an OC-725C amplifier (Warner Instruments). Data were acquired and analyzed using Pulse software (HEKA).
Cycloheximide Treatment and Western Blotting Assay
For cycloheximide treatment, transfected Cos-7 cells were treated with cycloheximide for various time periods and then analyzed by Western blot. For Western blot assay, protein samples were loaded on SDS-PAGE and transferred onto nitrocellulose membranes (Millipore) before blocking with a blocking buffer of 5% powdered nonfat milk in TBS-T (Tris-buffered saline with 0.05% Tween 20). The membranes were incubated overnight at 4 °C with primary antibody (1:2000 anti-HA from Roche Applied Science; 1:4000, anti-FLAG from Sigma; 1:500 anti-actin and anti-GAPDH from Santa Cruz) diluted with blocking buffer. After washing with TBS-T three times, the membranes were incubated for 1 h at room temperature with their corresponding secondary HRP-coupled antibodies (1:5000 Santa Cruz) diluted with blocking buffer. After washing three times, signal was detected by using the Immobilon Western HRP substrate (Millipore).
Cell Surface Biotinylation Assay
Confluent monolayers of HEK293 cells transfected with FLAG-KCNQ2 or FLAG-fsKCNQ2 were washed 3 times with ice-cold Ca2+/Mg2+ PBS. HEK293 cells were biotinylated with 0.5 mg/ml Sulfo-NHS-SS-biotin (Pierce) in Ca2+/Mg2+ PBS, pH 7.4, for 30 min at 4 °C. The remaining biotin was quenched by incubating the cells for an additional 10 min with 100 mm glycine in TBS. The cells were then washed with PBS and incubated in radioimmune precipitation lysis buffer containing 150 mm NaC1, 20 mm Tris, pH 8.0, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 10 mm EDTA, and protease inhibitor mixture (Roche Applied Science) at 4 °C for 30 min. One fraction of cell lysate containing 200 μg of protein was incubated with 20 μl of neutravidin beads (Pierce) for 2 h at 4 °C, and the other fraction was prepared as total protein. After incubation, the beads carrying surface proteins were washed with radioimmune precipitation lysis buffer and eluted with loading buffer. The total and surface proteins were both loaded onto 8% SDS-PAGE and assayed by Western blotting.
Pulse-Chase Assay
HEK293 cells expressing HA-tagged CD4 or CD4-ExtraC were starved of methionine and cysteine for 1 h (with DMEM lacking methionine and cysteine) and then labeled with [35S]methionine/cysteine (Easy Tag Express; PerkinElmer Life Sciences) for 1 h. Cells were washed with PBS three times and incubated in regular DMEM for various time periods (0, 1, 2, 3, and 6 h). The cells were then washed twice in PBS and solubilized with radioimmune precipitation lysis buffer. Cell lysate containing 200 μg of protein was incubated with Protein G-Sepharose (20 μl, GE Healthcare) and anti-HA antibody (1 μg, Roche Applied Science) at 4 °C overnight. After washing three times with radioimmune precipitation lysis buffer, the samples were dissolved in 30 μl of loading buffer, loaded onto 10% SDS-PAGE, and assayed by autoradiography.
Ubiquitination Assay
Cos-7 cells transfected with a combination of cDNA constructs of FLAG-KCNQ2 or FLAG-fsKCNQ2 with HA-ubiquitin (Ub) were harvested and lysed after expression for 24 h and treatment with MG132 for an additional 12 h. Cell lysate containing 200 μg of protein was incubated with Protein G-Sepharose (20 μl, GE Healthcare) and anti-FLAG antibody (1 μg, Sigma) at 4 °C overnight. Immunoprecipitates were washed 3 times in lysis buffer, dissolved in 30 μl of loading buffer, and loaded onto 6% SDS-PAGE before immunoblotting with anti-HA (1:2000, Roche Applied Science).
RNA Extraction and Quantitative RT-PCR Amplification
Total RNA was extracted from HEK293 cells using the TRIzol reagent (Invitrogen) according to manufacturer's protocol. A total of 10 μg of RNA was treated with RNase-free Turbo DNase (Ambion) for 30 min at 37 °C. RNA were extracted with phenol/chloroform and precipitated with ethanol. The purified RNA was reverse-transcribed directly using the MMLV-RT, SPCL kit (Invitrogen) according to the manufacturer's protocol. After cDNA synthesis, real-time RT-PCR was performed with the 7500 Fast Real-time PCR System (Applied Biosystems) using the SYBR Premix Ex TaqII kit (Takara) according to the manufacturer's protocol. Pairs of sequence-specific primers used in this study are as follows: human KCNQ2 forward (5′-GGCTTCAGCATCTCCCAGT-3′) and reverse (5′-GACTCTCCCTCCGCAATGTA-3′) and human GAPDH forward (5′-CCACTCCTCCACCTTTGAC-3′) and reverse (5′-ACCCTGTTGCTGTAGCCA-3′).
Statistics
All values are reported as the mean ± S.E. Statistics were performed using Prism 4.0 software. Statistical significance was assessed using the t test.
RESULTS
An Extra Segment Derived from the Distal C-terminal Frameshift Mutation of KCNQ2 Is Responsible for Current Reduction
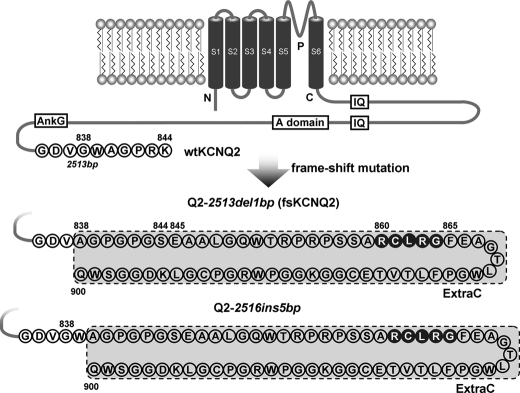
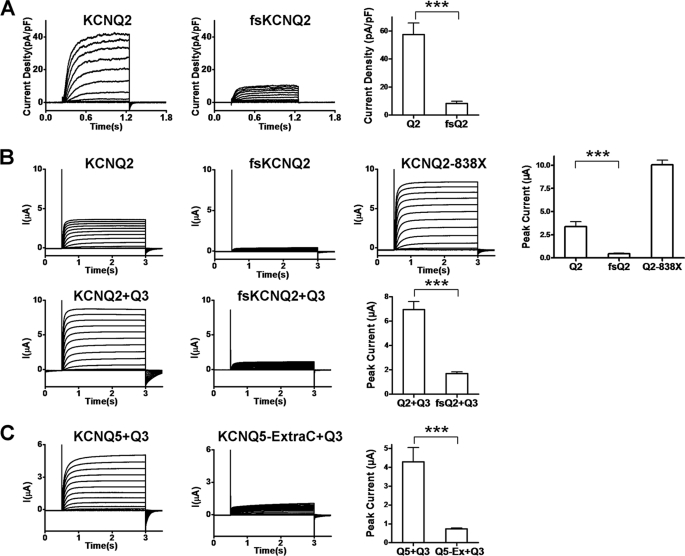
The human mutant Q2-2513del1bp studied in this paper results from one nucleotide deletion at position 2513 bp, which was designated fsKCNQ2 (frameshift mutant of KCNQ2). The fsKCNQ2 mutation results in a distal C-terminal deletion of the last seven amino acids before the stop codon, which are replaced with an extra sequence of 63 amino acids (designated ExtraC). This fsKCNQ2 mutant does not change the intrinsic motifs or domains of the channel, as compared with wild-type (WT) KCNQ2 (Fig. 1). Another distal C-terminal frameshift mutation in this study results from an insertion of five nucleotides at 2516 bp (designated Q2-2516ins5bp), leading to the generation of the same ExtraC (Fig. 1). To investigate the mechanism by which these distal frameshift mutations of KCNQ2 underlie BFNC, we began by confirming previous electrophysiological findings. Consistent with previous reports (13, 14, 16), expression of the fsKCNQ2 mutant in HEK293 cells or injection of its cRNA into oocytes resulted in small currents (Fig. 2, A and B).
FIGURE 1.
Schematic presentation of wild-type KCNQ2 and the distal C-terminal frameshift mutations in KCNQ2. The Kv7.2/KCNQ2 channel has six transmembrane domains and a long C terminus that contains important functional motifs and domains. The two distal frameshift mutants, Q2-2513del1bp and Q2-2516ins5bp, result from a 1-nucleotide deletion at 2513 bp and a 5-nucleotide insertion at 2516 bp in the human KCNQ2 gene, respectively. As a result, the fsKCNQ2 mutant (Q2-2513del1bp) is truncated and lacks seven terminal amino acids, and the Q2-2516ins5bp mutant is truncated and lacks the last five amino acids. Both mutations are replaced with a peptide of 63 amino acids at their C terminus (designated ExtraC in this study), as compared with wild-type (WT) human KCNQ2 (KCNQ2) with 844 residues. The C-terminal amino acids are represented as a one-letter symbol in the protein sequence. The italicized characters and numbers represent the mutation of the nucleotide sequence; non-italicized numbers indicate the residue in the protein sequence. IQ, calmodulin binding motif; A domain, channel assembly domain; AnkG, ankyrin-G binding motif.
FIGURE 2.
Current reduction in KCNQ channel mutations containing the ExtraC peptide. A, current traces were recorded by whole-cell patch clamp in HEK293 cells expressing KCNQ2 (left panel) or fsKCNQ2 (middle panel). Cells were held at −80 mV in response to a family of depolarizing potentials from −80 to +40 mV with 10-mV increments for 1 s. The rightmost panel shows their peak current density determined at the end of 1 s depolarization of +40 mV. Data are expressed as the mean ± S.E. (n = 5–7; ***, p < 0.001). pF, picofarads. B, representative current traces were recorded from Xenopus laevis oocytes by a two-electrode voltage clamp. The upper panels show comparison of current amplitudes of KCNQ2, fsKCNQ2, and KCNQ2–838X. The bottom panels show comparison of co-expression of KCNQ2 or fsKCNQ2 with KCNQ3. Oocytes from the same batch of cells were injected with identical amounts of cRNA and were held at −80 mV in response to a family of depolarizing potentials from −80 to +40 mV in 10-mV increments for 2.5 s. The rightmost upper and bottom panels summarize the peak current amplitudes determined at the end of 2.5-s depolarization of +40 mV. Data are expressed as the mean ± S.E. (n = 5–7; ***, p < 0.001). C, representative current traces were recorded with a two-electrode voltage clamp from X. laevis oocytes expressing KCNQ5 and KCNQ3 or expressing KCNQ5-ExtraC (the ExtraC peptide was appended to the C terminus of KCNQ5) and KCNQ3 using the same pulse protocol as in B. The rightmost panel summarizes the peak current amplitudes determined at the end of 2.5-s depolarization of +40 mV. Data are expressed as the mean ± S.E. (n = 4–7; ***, p < 0.001).
To make sure the inhibitory effect of the fsKCNQ2 mutant on channel function was specifically mediated by the extended C-terminal segment (ExtraC), we generated a truncation mutant (Q2–838X) by deleting the extended C terminus (63 amino acids) of the fsKCNQ2 mutant. In the Q2–838X mutant, the number with letter X stands for the amino acid mutated to the stop code where translation ends. The Q2–838X mutant without ExtraC expressed a robust current with gain of function (Fig. 2B), suggesting that dysfunction of the fsKCNQ2 mutant is due to ExtraC causing current reduction.
KCNQ3 that functions as an auxiliary subunit has been shown to facilitate KCNQ2 function. To test whether KCNQ3 can reverse the effect of this frameshift mutation, we co-injected the cRNA of fsKCNQ2 with KCNQ3 into Xenopus oocytes. Co-injection of the fsKCNQ2 mutant with WT KCNQ3 failed to cause an increase in current expression as compared with co-injection of WT KCNQ2 with KCNQ3, demonstrating that KCNQ3 cannot rescue the function of the fsKCNQ2 mutant (Fig. 2B). To confirm the effect of the ExtraC peptide, we appended it to the C terminus of KCNQ5, another member of the KCNQ family. Consistent with the result of co-expression of fsKCNQ2 with KCNQ3, KCNQ5 with ExtraC appended (KCNQ5-ExtraC) resulted in significant reduction of current density when coexpressed with KCNQ3 (Fig. 2C). These results show that the ExtraC derived from the C-terminal frameshift mutations of KCNQ2 is responsible for current reduction.
Accelerated Degradation of fsKCNQ2 by a Ubiquitin-independent Proteasome Pathway
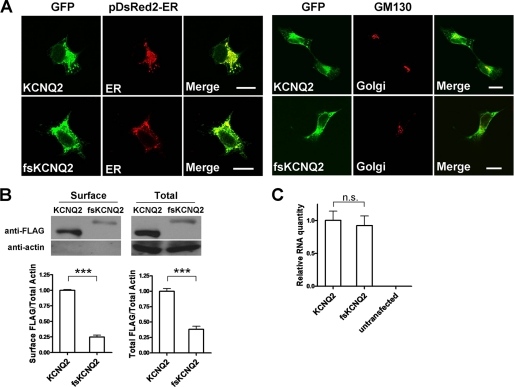
To examine whether the reduced current density of fsKCNQ2 mutant resulted from reduced surface expression, we used confocal fluorescence microscopy and biotin labeling for surface protein assay. We used the pDsRed2-ER plasmid to label the ER and the GM130 antibody (GM130) to stain the Golgi apparatus for identification of localization with wild-type KCNQ2 or the fsKCNQ2 mutant by confocal imaging.
Similar to WT KCNQ2, fsKCNQ2 proteins were localized in both the ER and Golgi apparatus (Fig. 3A), showing that the mutant can leave the ER and traffic to the Golgi apparatus. To relatively quantify surface expression, surface proteins WT KCNQ2 or fsKCNQ2 expressed in HEK293 were labeled by biotin and analyzed by Western blotting. Biotin labeling showed that both surface and total protein of the fsKCNQ2 mutant were much less than WT KCNQ2 (Fig. 3B). These results suggest that decreased surface fsKCNQ2, which underlies the reduced current density, is likely due to the reduction of the total amount of fsKCNQ2 protein (Fig. 3B). To eliminate the possibility of protein reduction caused by reduced plasmid transfection or gene transcription of fsKCNQ2, we also quantified KCNQ2 mRNA by quantitative RT-PCR. Cells transfected with WT KCNQ2 or fsKCNQ2 plasmid showed little difference in mRNA quantity (Fig. 3C), demonstrating that neither transfection nor transcription was affected. These results suggest that reduced surface expression of the mutant protein likely results from increased protein degradation.
FIGURE 3.
Reduced surface expression of fsKCNQ2 is a result of total protein reduction and is not due to ER retention. A, left panels, confocal imaging is shown of HEK293 cells co-transfected with plasmids of either EGFP-KCNQ2 or EGFP-fsKCNQ2 (green) and pDsRed2-ER for ER staining (red) after fixation with paraformaldehyde. Right panels, confocal imaging is shown of HEK293 cells transfected with plasmids of either EGFP-KCNQ2 or EGFP-fsKCNQ2 (green) for Golgi staining with GM130 antibody (red). The bar indicates 20 μm. B, shown is Western blotting analysis of biotin-labeled surface KCNQ2 and fsKCNQ2 proteins. The surface proteins from HEK293 cells transfected with FLAG-KCNQ2 or FLAG-fsKCNQ2 plasmids were labeled by biotin and segregated by avidin beads. Surface protein and total protein were separated by SDS-PAGE and Western-blotted with anti-FLAG or anti-actin antibody. In the lower panels surface FLAG-tagged KCNQ2 and fsKCNQ2 proteins over actin or total FLAG-tagged KCNQ2 and fsKCNQ2 proteins over actin were normalized to that of KCNQ2 over actin. Data are expressed as the mean ± S.E. (n = 3; ***, p < 0.001). C, quantitative analysis is shown of mRNA for KCNQ2 and fsKCNQ2 expressed in HEK293 cells lysed with TRIzol reagent. Total RNA was extracted from the lysate, treated with DNase, and quantified by quantitative RT-PCR analysis. There is no obvious difference in the quantity of mRNA in KCNQ2 and fsKCNQ2. Data are expressed as mean ± S.E. (n = 3, n.s., not significant, p > 0.05).
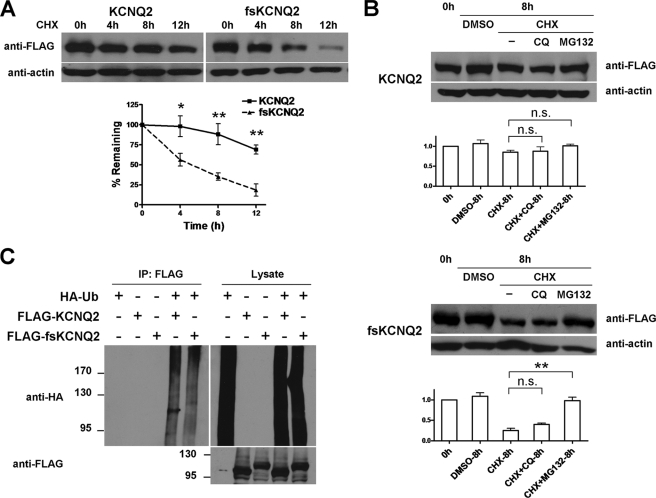
To test the stability of fsKCNQ2, we treated Cos-7 cells expressing fsKCNQ2 with the protein synthesis blocker cycloheximide (CHX) and evaluated protein stability by Western blotting of remaining proteins after degradation. After 12 h of CHX treatment, the half-life of fsKCNQ2 was about 4 h. In contrast, the half-life of WT KCNQ2 was more than 12 h (Fig. 4A). This result of this shortened half-life of these mutant proteins was also confirmed in both HEK293 and CHO cells (supplemental Fig. 1). These results indicate that fsKCNQ2 protein degradation is accelerated, leading to reduced surface expression.
FIGURE 4.
Accelerated degradation of fsKCNQ2 mediated by the ubiquitin-independent proteasome pathway. A, shown is a CHX treatment assay of KCNQ2 and fsKCNQ2 in Cos-7 cells. Cells were transfected with plasmids FLAG-KCNQ2 or FLAG-fsKCNQ2 and treated with protein synthesis inhibitor CHX (75 μg/ml) for the indicated time period (0, 4, 8, or 12 h). After degradation, the remaining proteins were separated by SDS-PAGE and analyzed by Western blotting using anti-FLAG or anti-actin antibody. Data are expressed as the mean ± S.E. (n = 4; *, p < 0.05; **, p < 0.01). The half-life of KCNQ2 was more than 12 h, and the half-life of fsKCNQ2 was about 4 h. B, shown is a Western blotting analysis of KCNQ2 and fsKCNQ2 in Cos-7 cells treated with CHX (75 μg/ml) and chloroquine (CQ, 50 μm) or MG132 (15 μg/ml). Samples were loaded in lanes from the left in the following order: control without any treatment, DMSO treatment for 8 h, CHX for 8 h, CHX with chloroquine for 8 h, CHX with MG132 for 8 h. Data were normalized to that of control without any treatment and are expressed as the mean ± S.E. (n = 3, **, p < 0.01, n.s., not significant, p > 0.05). C, Cos-7 cells were transfected with combinations of FLAG-KCNQ2 or FLAG-fsKCNQ2 and HA-ubiquitin plasmids as indicated with a plus sign and treated with MG132 for 12 h. Cell lysates were subjected to immunoprecipitation (IP) by anti-FLAG antibody before Western blotting with anti-HA antibody. As a loading control, lysates were directly Western-blotted with anti-HA or anti-FLAG antibody.
Proteasome and lysosome pathways are widely considered to be mechanisms for protein degradation. Using the proteasome-specific inhibitor MG132 or the lysosome-specific inhibitor chloroquine, we sought to determine whether fsKCNQ2 protein degradation is mediated by these two pathways. In WT KCNQ2 expressing Cos-7 cells treated with CHX, KCNQ2 protein showed no obvious difference in degradation in the presence or absence of either chloroquine or MG132 (Fig. 4B, top panels). In contrast, Cos-7 cells expressing the fsKCNQ2 mutant that were treated only with CHX resulted in significant protein degradation that was inhibited upon treatment with MG132, as compared with the chloroquine group (Fig. 4B, bottom panels). These results show that the proteasome pathway, and not the lysosome pathway, underlies the destabilization of fsKCNQ2 mutant protein.
We next investigated whether accelerated proteasome-dependent degradation of fsKCNQ2 is due to ubiquitination of this mutant. To test for ubiquitination of the fsKCNQ2 mutant, Cos-7 cells expressing different combinations of HA-Ub, FLAG-KCNQ2, and FLAG-fsKCNQ2 were treated with MG132 for 12 h. Proteins were subject to immunoprecipitation with anti-FLAG antibody and Western-blotted with anti-HA antibody. Ubiquitination of a target protein resulted in smeared bands at a high molecular weight (Fig. 4C). As a control, WT KCNQ2 was subjected to ubiquitination, as previously demonstrated in a similar assay (20). Similar to WT KCNQ2, the fsKCNQ2 mutant showed no further ubiquitination when it was co-expressed with HA-Ub proteins (Fig. 4C). This result indicates that the proteasome-dependent degradation of fsKCNQ2 does not rely on ubiquitination.
The ExtraC Peptide Serves as a Degradation Signal That Is Transferable
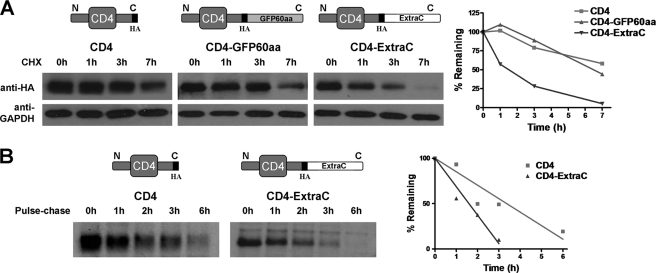
To test whether the degradation effect of the ExtraC peptide can be transferred to another unrelated protein, we fused the ExtraC to the C terminus of CD4 (CD4-ExtraC). As a control, we attached the C terminus of CD4 to GFP60aa (C-terminal 60 residues of green fluorescent protein). GFP60aa contains the same number of residues as ExtraC. After CHX treatment for 7 h in HEK293 cells, CD4-ExtraC was dramatically degraded with a half-life of about 1.5 h, as compared with WT CD4 or CD4-GFPC60aa that had a half-life of more than 6 h, showing that the function of ExtraC, which is specific for protein degradation, is transferable (Fig. 5A).
FIGURE 5.
The degradation function of ExtraC is transferable to CD4 protein. A, HEK293 cells transfected with HA-tagged plasmids of CD4 (left panels, as a blank control), CD4-GFP60aa (middle left panels, as a negative control), or CD4-ExtraC (middle right panels) were treated with CHX for the indicated time period (0, 1, 3, or 7 h). Proteins were separated by SDS-PAGE and Western-blotted with anti-HA or anti-GAPDH antibody. CD4-GFP60aa is a chimera of CD4 with 60 residues from the C-terminal sequence of GFP appended, and the number of residues is approximately the same as ExtraC. The rightmost plot shows a summary of protein degradation over time. Both the half-lives of CD4 and CD4-GFP60aa are more than 6 h, and that of CD4-ExtraC is about 1.5 h. B, pulse-chase analysis of protein degradation for CD4 (left panel) and CD4-ExtraC proteins (middle panel) is shown. Cells transfected with CD4 or CD4-ExtraC plasmids were labeled with [35S]Met/Cys culture medium for 1 h followed by a chase with “cold” Met/Cys culture medium for the indicated time periods (0, 1, 2, 3, or 6 h). The same amount of product from chase incubations was immunoprecipitated using anti-HA antibody and separated by SDS-PAGE for analysis by autoradiography. In the rightmost panel, the plot summarizes the protein degradation over time, and it is fitted with linear regression.
To further confirm this result, we used the metabolism inhibitor-free pulse-chase assay in which newly synthesized proteins were labeled by 32S and then evaluated for isotope-labeled CD4 and CD4-ExtraC proteins over a period of 6 h. Appending the ExtraC to the C terminus of CD4 resulted in fast degradation (Fig. 5B), further demonstrating the transferability of ExtraC, which was identified as a degradation signal.
Identification of Key Motif/Residues within ExtraC Critical for Degradation
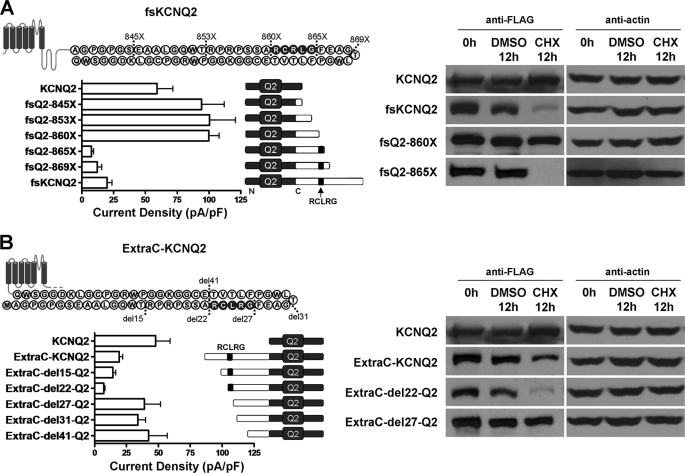
To identify the key motif/residues within ExtraC that mediate degradation, we constructed several deletions of fsKCNQ2. Serial C-terminal deletions of fsKCNQ2 were designated as fsQ2–869X, -865X, -860X, -853X, and -845X, in which the three digit numbers represent the position of the amino acid mutated to the stop codon that is designated as X. Whole-cell patch clamp recordings of HEK293 cells expressing either fsKCNQ2 or truncated mutants fsQ2–869X and fsQ2–865X resulted in significant reduction of current density as compared with the robust current expression of WT KCNQ2 or truncated mutants fsQ2–860X, fsQ2–853X, and fsQ2–845X (Fig. 6A, left panel). Comparison of fsQ2–860X and fsQ2–865X reveals the motif RCLRG containing five residues that distinguishes the two mutants (Fig. 6A). fsKCNQ2 and two truncated mutants that all carry this RCLRG motif showed a small amount of current, which was in contrast with WT KCNQ2 and mutants without the motif that showed robust current expression (Fig. 6A, left panel). To investigate whether reduced currents were due to degradation, we transfected WT KCNQ2 and the three mutants fsQ2–865X, fsQ2–860X, and fsKCNQ2 into Cos-7 cells treated with or without CHX (75 μg/ml) for 12 h. Blocking protein synthesis with CHX revealed that both fsQ2–865X truncation and fsKCNQ2 mutant proteins showed fast degradation, as compared with either fsQ2–860X or WT KCNQ2 (Fig. 6A, right panel).
FIGURE 6.
Functional identification of the RCLRG motif critical for current reduction and accelerated degradation. A, left panels, shown are current densities of HEK293 cells expressing KCNQ2, fsKCNQ2, or fsKCNQ2 truncation mutants: fsQ2–845X, -853X, -860X, -865X, and -869X (the number indicates amino acid sequence number of fsKCNQ2 that was mutated to a stop codon). Currents were recorded by whole-cell patch clamp in cells held at −80 mV and depolarized to +40 mV for 1 s. Data are expressed as the mean ± S.E. (n = 4–7). In the right panels, Western blotting analysis of proteins from transfected Cos-7 cells treated with CHX (75 μg/ml) for 0 or 12 h or with vehicle (0.75‰ DMSO) for 12 h is shown. Proteins were extracted, separated by SDS-PAGE, and Western-blotted with anti-FLAG or anti-actin antibody. pF, picofarads. B, left panels, shown are current densities of HEK293 cells expressing KCNQ2 or ExtraC-KCNQ2 chimeras in which the ExtraC peptide and its truncations were fused to the N terminus of KCNQ2. ExtraC truncations were generated from N-terminal deletions of the ExtraC-KCNQ2 chimera, leading to ExtraC-del15-Q2, ExtraC-del22-Q2, ExtraC-del27-Q2, ExtraC-del31-Q2, and ExtraC-del41-Q2 mutants. Currents were recorded by whole-cell patch clamp in cells held at −80 mV and depolarized to +40 mV for 1 s. Data are expressed as the mean ± S.E. (n = 4–6). Right panels, shown is Western blotting analysis of proteins from transfected Cos-7 cells treated with CHX (75 μg/ml) for 0 or 12 h or vehicle for 12 h as above in A.
Degradation signals can transfer their function when they are attached to either the N or C terminus (17, 21, 22). To test whether the function of ExtraC can be transferred to the N terminus, we attached ExtraC to the N terminus of WT KCNQ2 (ExtraC-KCNQ2) and made several N-terminal truncation mutants based on ExtraC-KCNQ2 designated as ExtraC-del15-Q2, ExtraC-del22-Q2, ExtraC-del27-Q2, ExtraC-del31-Q2, and ExtraC-del41-Q2 (Fig. 6B, left panel). Whole-cell recordings of HEK293 cells expressing ExtraC-KCNQ2 or the two N-terminal deletion mutants, ExtraC-del15-Q2 and ExtraC-del22-Q2, showed dramatic reduction of current as compared with robust current expression associated with either WT KCNQ2 or its counterparts ExtraC-del27, -del31, and -del41-Q2, in which the RCLRG motif was removed (Fig. 6B, left panel). To further test the effect of the RCLRG motif on protein degradation, we transfected WT KCNQ2 and three mutants, ExtraC-KCNQ2, ExtraC-del22-Q2, and ExtraC-del27-Q2, into Cos-7 cells. Consistent with current recordings, protein degradation of either ExtraC-del22-Q2 or ExtraC-KCNQ2 was faster than that of WT KCNQ2 or ExtraC-del27-Q2 without the RCLRG motif (Fig. 6B, right panel). These results demonstrate that the degradation function of ExtraC with the RCLRG motif can be conferred upon both N and C termini of channel proteins.
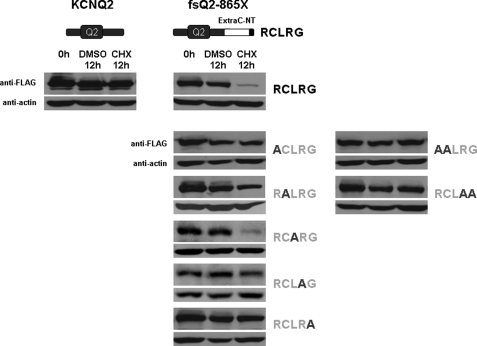
To further identify which individual residues are critical in mediating degradation, we mutated each of the five residues within the RCLRG motif into alanine. As shown in Fig. 7, the fsQ2–865X (fsQ2–860-RCLRG) mutant showed rapid degradation (middle panels) as compared with WT KCNQ2 (left panels). Mutating the first two individual residues of the RCLRG motif to alanine in either the N or C terminus abolished accelerated degradation. In contrast, the fsQ2–860-RCARG mutant in which the middle residue leucine was mutated had no affect on degradation (Fig. 7, middle panels). To further confirm the two flanking residues of the RCLRG motif were critical for degradation, we generated two double mutations. As shown in Fig. 7, right panels, the two double mutations prevented degradation, further confirming the two flanking residues necessary for degradation. These results indicate that the four residues of the RCXRG motif play a critical role in mediating accelerated degradation effect of the ExtraC in fsKCNQ2 mutant (Fig. 7).
FIGURE 7.
Identification of residues within the RCLRG motif critical for protein degradation by alanine scanning mutagenesis. Shown are Cos-7 cells expressing wild-type KCNQ2, fsQ2–865X, or different forms of the fsQ2–865X mutant in which each residue of the RCLRG motif was mutated to alanine (-ACLRG, -RALRG, -RCARG, -RCLAG, -RCLRA) or two residues were mutated to alanine (-AALRG and -RCLAA). The fsQ2–865X mutant has 864 residues (see Fig. 1) and carries the RCLRG motif before the stop codon as indicated by the letter X. Cells were treated with CHX (75 μg/ml) for 0 or 12 h or with vehicle (0.75 % DMSO) for 12 h. All extracted proteins (left panels for KCNQ2, middle panels for fsQ2–865X and individual mutations, and right panels for double mutations of the fsQ2–865X mutant) were separated by SDS-PAGE and Western-blotted with anti-FLAG or anti-actin antibody.
DISCUSSION
Most human BFNC mutations that have been identified are located in the C terminus of the Kv7.2/KCNQ2 channel. However, none of the mutations that have been tested show their dominant-negative effects on wild-type subunits (5, 6, 11, 12). This indicates that BFNC is caused by haploinsufficiency, suggesting the Kv7.2/KCNQ2 mutant protein produced by the cell is not sufficient to function normally.
To better understand the mechanism by which channel mutations that show partial dominant-negative effects underlie epilepsy, we studied the two distal C-terminal frameshift mutations of KCNQ2 that exhibit normal biophysical properties of channel function but have reduced current density (13, 14, 16). Based on their functional properties, we reasoned that the frameshift KCNQ2 mutations can fold properly and be transported to the plasma membrane. To verify this concept we carried out staining of one of the mutations tagged with GFP in HEK293 cells, and the results show that those C-terminal mutations can be translocated across from ER to the Golgi and plasma membrane and distributed in the same fashion as WT KCNQ2 (Fig. 3, A and B). This is consistent with previous reports which have noted that there is little difference in the distribution in the axon and the axon initial segment in neurons and translocation across Golgi and plasma membranes (15, 16), suggesting that the fsKCNQ2 mutant channel is correctly folded. In contrast, the frameshift mutation Q2-2043del1bp with a much larger frameshift region (219 amino acids) cannot form functional channels and cannot be transported from the ER to the Golgi. This mutant is thought to be degraded through the ER-associated degradation pathway, which degrades misfolded proteins (16, 23, 24). Based on the findings of this study, we propose a novel degradation mechanism that removes frameshift mutant proteins with normal folding that carry a degradation signal.
Is the degradation signal within the extended C terminus specific for channel degradation? To address this question, we constructed another frameshift mutant, Q2-2513del2bp, in which two nucleotides are deleted as compared with the fsKCNQ2 mutant (Q2-2513del1bp), where only one nucleotide is deleted. This Q2-2513del2bp mutation introduces another extra C-terminal sequence with 39 amino acids that does not contain the RCXRG motif. However, this frameshift mutant reveals no difference in degradation as compared with WT KCNQ2 (supplemental Fig. 2), indicating that random frameshift mutations without the RCXRG motif have no effect on protein stability. Regarding the role of the RCXRG motif within ExtraC as a general signal for degradation (Figs. 5 and 6B), we propose that any mutant protein with the RCXRG motif within a degradation signal will be subject to accelerated breakdown.
Why is it so easy to create a degradation signal in such a short extra region? Do native proteins with longer sequences contain degradation signals? Correctly folded proteins may carry numerous degradation signals. However, these signals are likely sequestered or enfolded in structured proteins or assembled with auxiliary subunits, in which case they may not be functional (17). When correct folding goes awry due to truncation or mutation at key sites, these signals may become exposed and functional (17, 24, 25). Based on our findings, we suspect that these distal frameshift mutations may have minor changes of their original sequence as most parts of mutant proteins can be folded correctly in the manner of wild-type proteins. When mutations cause random rearrangement of codons after site mutation, random sequences that are not properly structured as native proteins selected by evolution can be created (27–29), thus generating signals that are exposed resulting in protein degradation. Without specific structural information, we speculate that the ExtraC polypeptide of the fsKCNQ2 mutant may be highly disordered or unstructured due to its random codon rearrangement (27–29). This property of ExtraC may facilitate RCXRG motif recognition by degradation-related molecules (30, 31). On the other hand, the disordered ExtraC may provide necessary unstructured initials for proteasome digestion (32, 33).
Does the degradation signal from the ExtraC have any implications for actual physiological degradation because the signal is generated from a random mutagenesis? We performed literature and data base search to look into whether the RCXRG motif exists as a physiological degradation signal in native proteins. We have found that many native proteins contain the RCXRG motif, but there is no report that native proteins carrying the RCXRG motif are subject to physiological degradation. This suggests that the RCXRG motif/ExtraC may not have any physiological implications. However, the motif/degradation signal can be hidden or become non-functional when native proteins with the degradation signal have structures (17). When unstructured in an environment such as oxidative stress, native proteins can be subject to degradation (34, 35). It has also been shown that a random distal C-terminal frameshift mutation of aquaporin-2 (water channel protein) can result in generating an extra region that contains several signal motifs for basolateral sorting or retention (36). Those signal motifs from aquaporin-2 mutation have been demonstrated in other native proteins (for instance, low density lipoprotein receptors), functioning as physiological signals for basolateral sorting and retention (26, 37–39).
In summary, we have identified a novel degradation signal that is derived from distal C-terminal frameshift mutations of KCNQ2, which causes the impairment of channel function leading to epilepsy. This degradation signal contains a primary critical motif of five amino acids (RCXRG) that can be transferred to non-channel proteins for degradation. Our findings demonstrate that the distal C-terminal frameshift mutations of the Kv7.2/KCNQ2/M-channel are subject to degradation through the ubiquitin-independent proteasome pathway, a cellular mechanism by which incomplete dominant-negative mutations of Kv7.2/KCNQ2 cause epilepsy of infancy. Here, we propose a novel degradation mechanism that removes mutant proteins that carry the degradation signal, serving as a general mechanism for clearance of numerous mutant proteins in the cell. Moreover, identification and removal of this degradation signal may potentially help production of functional recombinant proteins that are of importance for pharmaceutical manufacture.
Supplementary Material
Acknowledgments
We thank our laboratory members Yanxin Lu, Dr. Ping Liang, Ka Lu, and Wen Liu for assistance and comments on this work. We also thank Dr. Michael A. McNutt for evaluation and the correction of English in this manuscript. K. W. W. thanks J. M. Wang for consistent support during this research.
This work was supported by National Science Foundation of China Research Grants 30630017 and 30970919, the Ministry of Science Technology of China “973 Program” 2007CB512100, and the Ministry of Education of China, 111 Project China (B07001) (to K. W. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- BFNC
- benign familial neonatal convulsions
- CHX
- cycloheximide
- Ub
- ubiquitin
- EGFP
- enhanced green fluorescent protein
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Jentsch T. J., Schroeder B. C., Kubisch C., Friedrich T., Stein V. (2000) Epilepsia 41, 1068–1069 [DOI] [PubMed] [Google Scholar]
- 2. Psenka T. M., Holden K. R. (1996) Seizure 5, 243–245 [DOI] [PubMed] [Google Scholar]
- 3. Ronen G. M., Rosales T. O., Connolly M., Anderson V. E., Leppert M. (1993) Neurology 43, 1355–1360 [DOI] [PubMed] [Google Scholar]
- 4. Charlier C., Singh N. A., Ryan S. G., Lewis T. B., Reus B. E., Leach R. J., Leppert M. (1998) Nat. Genet. 18, 53–55 [DOI] [PubMed] [Google Scholar]
- 5. Biervert C., Schroeder B. C., Kubisch C., Berkovic S. F., Propping P., Jentsch T. J., Steinlein O. K. (1998) Science 279, 403–406 [DOI] [PubMed] [Google Scholar]
- 6. Schroeder B. C., Kubisch C., Stein V., Jentsch T. J. (1998) Nature 396, 687–690 [DOI] [PubMed] [Google Scholar]
- 7. Wang H. S., Pan Z., Shi W., Brown B. S., Wymore R. S., Cohen I. S., Dixon J. E., McKinnon D. (1998) Science 282, 1890–1893 [DOI] [PubMed] [Google Scholar]
- 8. Watanabe H., Nagata E., Kosakai A., Nakamura M., Yokoyama M., Tanaka K., Sasai H. (2000) J. Neurochem. 75, 28–33 [DOI] [PubMed] [Google Scholar]
- 9. Peters H. C., Hu H., Pongs O., Storm J. F., Isbrandt D. (2005) Nat. Neurosci. 8, 51–60 [DOI] [PubMed] [Google Scholar]
- 10. Volkers L., Rook M. B., Das J. H., Verbeek N. E., Groenewegen W. A., van Kempen M. J., Lindhout D., Koeleman B. P. C. (2009) Neurosci. Lett. 462, 24–29 [DOI] [PubMed] [Google Scholar]
- 11. Richards M. C., Heron S. E., Spendlove H. E., Scheffer I. E., Grinton B., Berkovic S. F., Mulley J. C., Davy A. (2004) J. Med. Genet. 41, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peroz D., Rodriguez N., Choveau F., Baró I., Mérot J., Loussouarn G. (2008) J. Physiol. 586, 1785–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Singh N. A., Westenskow P., Charlier C., Pappas C., Leslie J., Dillon J., Anderson V. E., Sanguinetti M. C., Leppert M. F. (2003) Brain 126, 2726–2737 [DOI] [PubMed] [Google Scholar]
- 14. Lerche H., Biervert C., Alekov A. K., Schleithoff L., Lindner M., Klinger W., Bretschneider F., Mitrovic N., Jurkat-Rott K., Bode H., Lehmann-Horn F., Steinlein O. K. (1999) Ann. Neurol. 46, 305–312 [DOI] [PubMed] [Google Scholar]
- 15. Chung H. J., Jan Y. N., Jan L. Y. (2006) Proc. Natl. Acad. Sci. U. S. A. 103, 8870–8875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Soldovieri M. V., Castaldo P., Iodice L., Miceli F., Barrese V., Bellini G., Miraglia del Giudice E., Pascotto A., Bonatti S., Annunziato L., Taglialatela M. (2006) J. Biol. Chem. 281, 418–428 [DOI] [PubMed] [Google Scholar]
- 17. Ravid T., Hochstrasser M. (2008) Nat. Rev. Mol. Cell Biol. 9, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schrader E. K., Harstad K. G., Matouschek A. (2009) Nat. Chem. Biol. 5, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jow F., Wang K. (2000) Brain Res. Mol. Brain Res. 80, 269–278 [DOI] [PubMed] [Google Scholar]
- 20. Ekberg J., Schuetz F., Boase N. A., Conroy S. J., Manning J., Kumar S., Poronnik P., Adams D. J. (2007) J. Biol. Chem. 282, 12135–12142 [DOI] [PubMed] [Google Scholar]
- 21. Tasaki T., Kwon Y. T. (2007) Trends Biochem. Sci. 32, 520–528 [DOI] [PubMed] [Google Scholar]
- 22. Li X., Coffino P. (1993) Mol. Cell. Biol. 13, 2377–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 24. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen H., MacDonald A., Coffino P. (2002) J. Biol. Chem. 277, 45957–45961 [DOI] [PubMed] [Google Scholar]
- 26. Fölsch H., Ohno H., Bonifacino J. S., Mellman I. (1999) Cell 99, 189–198 [DOI] [PubMed] [Google Scholar]
- 27. Yamauchi A., Yomo T., Tanaka F., Prijambada I. D., Ohhashi S., Yamamoto K., Shima Y., Ogasahara K., Yutani K., Kataoka M., Urabe I. (1998) FEBS Lett 421, 147–151 [DOI] [PubMed] [Google Scholar]
- 28. Doi N., Yanagawa H. (1998) Cell. Mol. Life Sci. 54, 394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Doi N., Kakukawa K., Oishi Y., Yanagawa H. (2005) Protein Eng. Des. Sel. 18, 279–284 [DOI] [PubMed] [Google Scholar]
- 30. Fuxreiter M., Tompa P., Simon I. (2007) Bioinformatics 23, 950–956 [DOI] [PubMed] [Google Scholar]
- 31. Dyson H. J., Wright P. E. (2005) Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 32. Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., Matouschek A. (2004) Nat. Struct. Mol. Biol. 11, 830–837 [DOI] [PubMed] [Google Scholar]
- 33. Komaru K., Ishida Y., Amaya Y., Goseki-Sone M., Orimo H., Oda K. (2005) FEBS J. 272, 1704–1717 [DOI] [PubMed] [Google Scholar]
- 34. Ferrington D. A., Sun H., Murray K. K., Costa J., Williams T. D., Bigelow D. J., Squier T. C. (2001) J. Biol. Chem. 276, 937–943 [DOI] [PubMed] [Google Scholar]
- 35. Kurepa J., Smalle J. A. (2008) Plant Signal. Behav. 3, 386–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kamsteeg E. J., Bichet D. G., Konings I. B., Nivet H., Lonergan M., Arthus M. F., van Os C. H., Deen P. M. (2003) J. Cell Biol. 163, 1099–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ohno H., Fournier M. C., Poy G., Bonifacino J. S. (1996) J. Biol. Chem. 271, 29009–29015 [DOI] [PubMed] [Google Scholar]
- 38. Nesterov A., Carter R. E., Sorkina T., Gill G. N., Sorkin A. (1999) EMBO J. 18, 2489–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hunziker W., Harter C., Matter K., Mellman I. (1991) Cell 66, 907–920 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.