Background: Import of proteins into peroxisomes requires the PTS2 receptor Pex7p and its co-receptor Pex18p.
Results: Poly- and monoubiquitination of the co-receptor at newly identified sites triggers proteasomal degradation or receptor recycling.
Conclusion: Monoubiquitination at the conserved cysteine regulates cargo import and receptor recycling.
Significance: Pex18p export is linked to cargo translocation, which supports the idea of an export-driven import of proteins into peroxisomes.
Keywords: Cell Biology, Peroxisomes, Protein Targeting, Protein Translocation, Protein Import, Yeast, Peroxins, Peroxisome Biogenesis, Pex7p, Pex18p
Abstract
The peroxisomal matrix protein import is facilitated by cycling receptor molecules that shuttle between the cytosol and the peroxisomal membrane. In the yeast Saccharomyces cerevisiae, the import of proteins harboring a peroxisomal targeting signal of type II (PTS2) is mediated by the receptor Pex7p and its co-receptor Pex18p. Here we demonstrate that Pex18p undergoes two kinds of ubiquitin modifications. One of these ubiquitination events depends on lysines 13 and 20 and forces rapid Pex18p turnover by proteasomal degradation. A cysteine residue near the extreme Pex18p amino-terminus is required for the second type of ubiquitination. It turned out that this cysteine residue at position 6 is essential for the function of Pex18p in peroxisomal protein import but does not contribute to receptor-cargo association and binding to the peroxisomal import apparatus. However, in contrast to the wild-type protein, cysteine 6-mutated Pex18p is arrested in a membrane-protected state, whereas Pex7p is accessible in a protease protection assay. This finding indicates that Pex18p export is linked to cargo translocation, which supports the idea of an export-driven import of proteins into peroxisomes.
Introduction
The maintenance of peroxisome function depends on the formation of the peroxisomal membrane and import of both membrane and matrix proteins. Without exception, peroxisomal matrix proteins are nuclear encoded, synthesized on free ribosomes, and imported post-translationally (1). In contrast to the protein translocation systems of mitochondria, chloroplasts, and the endoplasmic reticulum that transport unfolded polypeptide chains, the peroxisomal import apparatus allows the transport of folded and even oligomeric proteins across the peroxisomal membrane. This process is facilitated by a transiently formed import pore with the receptor molecule itself as one of its constituents (2).
Based on the concept of cycling receptors (3, 4), the receptor-mediated peroxisomal import of proteins can be divided into four steps. First, proteins destined for transport into the peroxisomal matrix are recognized in the cytosol by their cognate receptor proteins Pex5p or Pex7p. This initial step in general depends on either one of the two well characterized peroxisomal targeting signals, PTS1 or PTS2, which are recognized and bound by Pex5p and Pex7p, respectively (for a review, see Ref. 5). In the second step, the cargo-loaded receptors dock to distinct proteins accessible at the surface of the peroxisomal membrane, namely Pex13p and Pex14p. These two proteins bind directly both receptors and are, together with Pex17p, established as the docking subcomplex. A second subcomplex acts downstream to the docking event and consists of the three peroxins Pex2p, Pex10p, and Pex12p. A common feature of these proteins is a RING finger domain at their extreme C termini. The RING finger subcomplex and the docking subcomplex are both linked together in a Pex8p- or Pex3p-dependent manner to form a larger complex, the so-called importomer (6, 7). In the third step of the receptor cycle, the cargo is delivered into the peroxisomal matrix, and finally, the receptor is exported in an ATP-dependent manner back to the cytosol to perform a next round of import (8).
With respect to the PTS1 receptor Pex5p, recent reports demonstrated that this final step in the receptor cycle is catalyzed by the AAA2 peroxins Pex1p and Pex6p (9, 10). The signal for the export process could be either a mono- or a polyubiquitin moiety. The polyubiquitination of Pex5p depends on Ubc4p/Ubc5p and is not a prerequisite for its function in peroxisomal protein import but might be a crucial step of a quality control system for the disposal of waste Pex5p (11–13). Pex5p monoubiquitination is facilitated by the E2 enzyme Pex4p in conjunction with the RING finger complex, which represents the corresponding E3 enzymes. In contrast to polyubiquitination, the monoubiquitin brings Pex5p back to the cytosol, where the ubiquitin is cleaved off in either a non-enzymatic manner by a nucleophilic attack of glutathione or enzyme-catalyzed by a ubiquitin hydrolase (14, 15). The PTS2 receptor shares the main features of the Pex5p-mediated import of PTS1 proteins (16). However, unlike Pex5p, the PTS2 receptor Pex7p is necessary, but not sufficient, to carry out all steps of the receptor cycle. Rather, Pex7p requires species-specific auxiliary proteins for its function in peroxisomal protein import. These are Pex18p or Pex21p in Saccharomyces cerevisiae; the orthologue Pex20p in Yarrowia lipolytica, Neurospora crassa, Hansenula polymorpha, and Pichia pastoris; or the longer of two splice isoforms of Pex5p in mammals (16).
In order to gain more insight into the PTS2 receptor cycle, we analyzed the turnover of Pex7p and its co-receptor Pex18p. We demonstrate that the half-life of Pex18p is significantly shorter than that of Pex7p and that it depends on ubiquitination on two conserved lysine residues. Comparable with Pex5p, Pex18p is also ubiquitinated on an amino-terminal cysteine residue. This modification, originally described as signal required for receptor export back to the cytosol, turns out to be required for Pex18p function in Pex7p translocation. This finding demonstrates that receptor export and cargo import are linked and thus supports the idea of a peroxisomal export-driven import of proteins.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
S. cerevisiae strains used in this study are listed in Table 1. Gene deletion and genomic tagging of the PEX14 locus was carried out as described (17, 18). Yeast complete (YPD) and minimal media (SD) have been described previously (19). YNO medium contained 0.1% oleic acid, 0.05% Tween 40, 0.1% yeast extract, and 0.67% yeast nitrogen base without amino acids, adjusted to pH 6.0. When necessary, auxotrophic requirements were added according to Ref. 20. When indicated, 20 mm N-ethylmaleimide (NEM) (Sigma) was added according to Ref. 21 to accumulate cysteine-dependent ubiquitination forms of Pex18p.
TABLE 1.
S. cerevisiae strains used in this study
| S. cerevisiae strain | Description | Source or reference |
|---|---|---|
| UTL-7A (wild type) | MATα leu2-3, 112 ura3-52 trp1 | Ref. 19 |
| pex3Δ | MATα leu2-3, 112 ura3-52 trp1, pex3::loxP | Ref. 60 |
| pex4Δ | MATα leu2-3, 112 ura3-52 trp1, pex4::LEU2 | Ref. 61 |
| pex5Δ | MATα leu2-3, 112 ura3-52 trp1, pex5::loxP | Ref. 28 |
| pex7Δ | MATα leu2-3, 112 ura3-52 trp1, pex7::LEU2 | Ref. 4 |
| pex8Δ | MATα leu2-3, 112 ura3-52 trp1, pex8::LEU2 | Ref. 62 |
| pex12Δ | MATα leu2-3, 112 ura3-52 trp1, pex12::LEU2 | Ref. 27 |
| pex1Δ6Δ | MATα leu2-3, 112 ura3-52 trp1, pex1::loxP, pex6::loxP | Ref. 9 |
| pex18Δpex21Δ | MATα leu2-3, 112 ura3-52 trp1, pex18::loxP, pex21::loxP | Ref. 33 |
| fox3Δ | MATα leu2-3, 112 ura3-52 trp1, fox3::loxP | Ref. 33 |
| cim5-1 | cim5-1, ura3-52, leu2Δ1, his3Δ200 | Ref. 38 |
| UTL-7A-Pex14p-TEV-ProtA | MATα leu2-3, 112 ura3-52 trp1, PEX14-TEV-ProtA-kanMX6 | Ref. 6 |
| pex4Δ-Pex14p-TEV-ProtA | MATα leu2-3, 112 ura3-52 trp1, pex4::loxP, PEX14-TEV-ProtA-kanMX6 | This study |
| pex6Δ-Pex14p-TEV-ProtA | MATα leu2-3, 112 ura3-52 trp1, pex6::loxP, PEX14-TEV-ProtA-kanMX6 | This study |
| pex8Δ-Pex14p-TEV-ProtA | MATα leu2-3, 112 ura3-52 trp1, pex8::loxP, PEX14-TEV-ProtA-kanMX6 | Ref. 6 |
| pex4Δ18Δ21Δ-Pex14p-TEV-ProtA | MATα leu2-3, 112 ura3-52 trp1, pex4::loxP, pex18::loxP, pex21::loxP,PEX14-TEV-ProtA-kanMX6 | This study |
Plasmid Constructions
Plasmids and oligonucleotides used are listed in Tables 2 and 3, respectively. PEX18, including 300 bp of the 5′- and 100 bp of the 3′-non-coding region, was amplified by PCR from genomic DNA of S. cerevisiae using primer pair KU1204/KU1205. Introduced BamHI/HindIII sites were used to subclone the PCR product into equally digested pRS416 (22), resulting in vector pRSPEX18. For construction of Pex18p variants, mutations were introduced by PCR using the QuikChange® site-directed mutagenesis kit (Stratagene), with plasmid pRSPEX18 as template.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or reference | Oligonucleotides |
|---|---|---|---|
| pRSPEX18 | PEX18 | This study | KU1204/KU1205 |
| pAHe1 | PEX18(K13R) | This study | RE2548/RE2549 |
| pAHe2 | PEX18(K20R) | This study | RE2550/RE2551 |
| pAHe3 | PEX18(K13R/K20R) | This study | |
| pAHe4 | PEX18(C6S) | This study | RE2546/RE2547 |
| pAHe5 | PEX18(C6S, K13R/K20R) | This study | |
| Yep96 | Ub | Ref. 40 | |
| Yep105 | mycUb | Ref. 40 |
TABLE 3.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| KU1204 | 5′-CACAAGCTTCTAGTATAATCAGGTATGTAAGG-3′ |
| KU1205 | 5′-GCGGATCCGAACGCAGTATGTAATTTAATA-3′ |
| RE2546 | 5′-ATAAAATACAAACAATGAATAGTAACCGAAGCCAAACGAATGAGGT-3′ |
| RE2547 | 5′-ACCTCATTCGTTTGGCTTCGGTTACTATTCATTGTTTGTATTTTAT-3′ |
| RE2548 | 5′-ATGCCAAACGAATGAGGTGAATAGATTTATTAGTAGTACAGAAAAGG-3′ |
| RE2549 | 5′-CCTTTTCTGTACTACTAATAAATCTATTCACCTCATTCGTTTGGCAT-3′ |
| RE2550 | 5′-GGTGAATAAATTTATTAGTAGTACAGAAAGGGGGCCTTTTACGGG-3′ |
| RE2551 | 5′-CCCGTAAAAGGCCCCCTTTCTGTACTACTAATAAATTTATTCACC-3′ |
Protease Protection Assay
Protease protection assays were performed according to Ref. 23. Equal portions of the organelle pellets were incubated with constant amounts of proteinase K (20 μg/50 μg of lysate). After defined time points, the proteinase K was inhibited by the addition of 2 mm PMSF, and samples were immediately precipitated with TCA and processed for SDS-PAGE (23).
Antibodies and Immunoblotting
Immunoblot analyses were performed according to standard protocols (24). Immunoblots were incubated with polyclonal rabbit antibodies raised against Pex7p (25), Pex18p, Pex5p (26), Pex12p (27), Pex13p (28), Pex14p (26), and Fbp1p (29). Anti-rabbit coupled HRP (Sigma-Aldrich) was used as second antibody, and blots were developed using the ECL system (Amersham Biosciences). Alternatively, primary antibody was detected with IRDye 800CW goat anti-rabbit IgG secondary antibody (LI-COR Bioscience, Bad Homburg, Germany) followed by a detection using the “Infrarot Imaging System” (LI-COR Bioscience).
Miscellaneous Methods
Immunopurification of native Pex14p complexes from yeast cells using IgG-Sepharose was performed according to Ref. 6. Protein turnover was monitored by the addition of 15 μg/ml cycloheximide (Sigma-Aldrich) to the cell culture prior sample collection. Whole cell yeast extracts were prepared from oleic acid-induced cells as described earlier (30). dsRed-tagged proteins were monitored by life cell imaging with a Zeiss Axioplan fluorescence microscope and AxioVision 4.4 software (Zeiss, Jena, Germany).
RESULTS
Defects in the Peroxisomal Receptor Cycle Affect the Relative Abundance of Pex18p
The peroxisomal matrix protein import of PTS2-proteins is facilitated by the import receptor Pex7p assisted by auxiliary proteins also referred to as co-receptors (31). In S. cerevisiae, this function is carried out by Pex18p and Pex21p, two proteins that partially overlap in function (32). Upon oleic acid induction, Pex18p turned out to be most important for the import of the PTS2-carrying thiolase (Fox3p) (32, 33). Pex18p was demonstrated to be ubiquitinated and constitutively degraded, most likely by the ubiquitin-proteasome system (34).
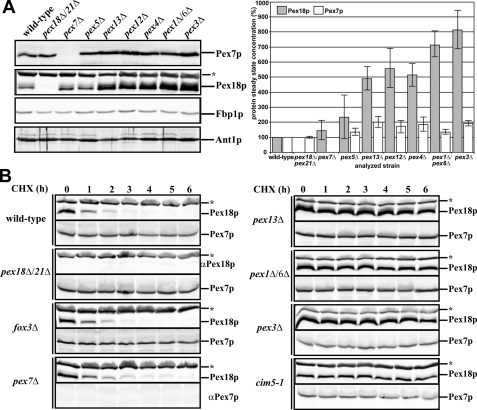
In order to understand these processes in greater detail, we compared the relative abundance of the auxiliary Pex18p and the PTS2 receptor Pex7p in wild-type and selected PEX deletion strains. Whole cell lysates of the oleic acid-induced strains were prepared and subjected to SDS-PAGE and immunoblot analysis. In samples derived from either pex7Δ or pex5Δ cells, the Pex18p abundance was similar to those obtained from wild-type cells (Fig. 1A). However, a significant increase of Pex18p abundance was visible when mutants with an affected matrix protein import were analyzed, whereas Pex7p levels only slightly increased. In fact, deletion of components of the receptor docking complex (Pex13p), the RING finger complex (Pex12p), or the receptor export apparatus (Pex4p and Pex1p/Pex6p) raised the steady-state concentration of Pex18p up to 8-fold compared with wild-type strain (Fig. 1A). Such an increase was also observed when pex3Δ cells were analyzed. To exclude the possibility that our finding was caused by uneven sample loading, we probed for the cytosolic marker fructose-1,6-bisphosphate phosphatase (Fbp1p) (35) and for the peroxisomal ATP transporter Ant1p, which both turned out to be equal in all samples (Fig. 1A). Thus, our results demonstrate that Pex18p abundance is increased in strains affected in receptor binding or recycling, which supplements previous findings (34). Because Pex3p is required for the targeting of peroxisomal membrane proteins and its deletion leads to a loss of peroxisomal membranes (36), our results moreover indicate that functional peroxisomes are required for the maintenance of the wild-type concentration of Pex18p.
FIGURE 1.
Pex18p is subjected to a rapid turnover in wild-type cells. A, whole cell lysates of oleic acid-induced strains as indicated were analyzed for the amount of Pex18p and Pex7p by immunological detection. Cytosolic Fbp1p and the peroxisomal ATP transporter Ant1p serve as control for equal loading (left). Signal intensities of Pex7p and Pex18p of two independent experiments were estimated by densitometry. Error bars, S.E. (right). B, the indicated strains were grown on oleic acid medium for 14 h and then treated with 15 μg/ml cycloheximide. At defined time points, whole cell lysates were prepared and analyzed by immunoblotting for Pex7p and Pex18p. *, cross-reactive band labeled with anti-Pex18p antibodies.
Pex18p Is Subjected to a Rapid Turnover in Wild-type Cells
Cellular protein concentration is regulated by the balance of protein synthesis and degradation. To discriminate whether the observed increase in the steady-state concentration of Pex18p is due to a higher expression or a lower degradation rate of Pex18p, we performed cycloheximide chase experiments. Cycloheximide is an inhibitor of protein biosynthesis in eukaryotic organisms because it blocks translational elongation (37). Thus, the analysis of proteins after application of this inhibitor allows determination of their turnover rate. Cycloheximide was added to the medium of oleic acid-induced cells, and the stability of Pex18p and Pex7p was analyzed. In wild-type cells, Pex18p was below the detection level already after 3 h of cycloheximide chase, indicating its rapid turnover by proteasomal degradation (Fig. 1B (left) and supplemental Table S1). To corroborate these data, we monitored Pex18p level in a strain affected in CIM5, a gene that encodes a regulatory subunit of the 26 S proteasome (38). In this strain, the level of Pex18p showed no discernible decline even 6 h after application of cycloheximide, which demonstrates that the Pex18p turnover is caused by proteasomal degradation. The turnover rate of Pex18p in a fox3Δ as well as pex7Δ strain was comparable with the wild type, indicating that neither the PTS2 receptor nor its cargo protein thiolase is required for Pex18p delivery to the proteasome. In contrast, Pex18p remained stable in strains with general matrix protein import defect (Fig. 1B (right) and supplemental Table S1, pex13Δ, pex1Δ/6Δ, and pex3Δ). Thus, a lacking receptor cycle does not enhance Pex18p expression but avoids its proteasomal degradation. Again, Pex7p behaved quite different from its co-receptor. In all tested strains, the PTS2 receptor remained stable during the measured time period of 6 h.
Similar Ubiquitination Features of the PTS1 Receptor Pex5p and Pex18p
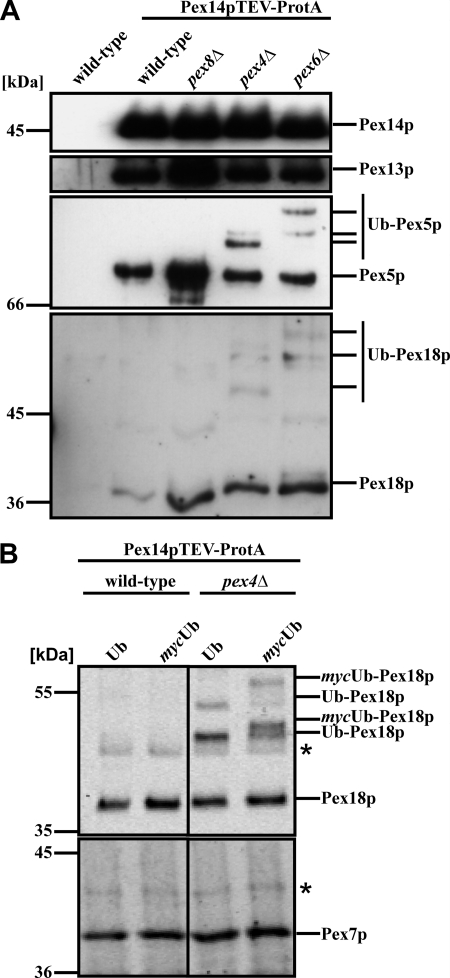
Pex5p as well as Pex18p were demonstrated to be modified by ubiquitination (11, 13, 34). Although much progress has been made analyzing ubiquitination of Pex5p, including elucidation of the corresponding enzymatic cascade and identification of the target residues for ubiquitin (Ub) conjunction, our knowledge of Pex18p ubiquitination remained scarce. To gain more insight into the functional role of Pex18p, we investigated ubiquitination of this co-receptor in more detail. Hampered by cross-reactive proteins when using Pex18p-specific antibodies, our strategy was to enrich for membrane-bound Pex18p by isolation of the Pex14p complex of the peroxisomal protein import that contains both PTS receptors (26, 39) as well as Pex18p (33). To this end, we applied wild-type or selected mutant strains that expressed Pex14p genomically tagged with protein A and with a tobacco etch virus (TEV) cleavage site between the protein A tag and its fusion partner protein. Purified complexes were subjected to SDS-PAGE and immunoblot analysis to monitor for the presence of the bait protein Pex14p, the docking protein Pex13p, and the PTS1 import receptor Pex5p and PTS2-co-receptor Pex18p. When isolated from wild-type cells, Pex13p, Pex5p, and Pex18p were present in the Pex14p complex (Fig. 2A, lane 2), which is in line with previous findings (6, 33). The same was true for the complex isolated from a pex8Δ, pex4Δ, or pex6Δ strain. However, although both Pex5p and Pex18p were detected as native and thus unmodified proteins when derived from wild-type or pex8Δ cells, immunoreactive signals with lower mobility appeared when the receptor molecules were isolated from either a strain defective in the E2 enzyme Pex4p or the AAA protein Pex6p. With respect to Pex5p, these higher molecular weight signals are known to represent polyubiquitinated Pex5p species (11, 13). The modification pattern of Pex5p differs between strains that are affected either in Pex4p or its membrane anchor protein Pex22p or in one of the AAA proteins Pex1p and Pex6p or their membrane anchor Pex15p (Fig. 2A) (11, 13). Interestingly, Pex18p also exhibits Pex5p-like modification patterns in a pex4Δ and pex6Δ strain (Fig. 2A), indicative of ubiquitinated Pex18p. In order to investigate whether the higher molecular weight bands of Pex18p that appeared in the mutant strains indeed represent ubiquitinated species of the PTS2 co-receptor, we analyzed yeast strains expressing either the wild-type Ub or the mycUb fusion gene (40). The mycUb variant is about 1.5 kDa larger than wild-type Ub but is indistinguishable from wild-type Ub in its ability to be enzymatically conjugated to and cleaved from acceptor molecules (40). In vivo substitution of the Ub with the larger mycUb should decrease the electrophoretic mobility of higher molecular weight bands labeled with Pex18p-specific antibodies when these represent Ub conjugates. Therefore, we isolated the Pex14p complex from a wild-type strain as well as the pex4Δ strain expressing either Ub or mycUb. Obtained eluate fractions were separated by SDS-PAGE and processed for immunoblotting. Again, high molecular weight bands were not detected in the wild-type sample but were present when Pex18p was isolated from pex4Δ cells (Fig. 2B). Comparison of pex4Δ samples upon expression of either Ub or mycUb showed that the putative ubiquitinated species of Pex18p were replaced by new bands with a decreased electrophoretic mobility (Fig. 2B). These data show that overexpression of mycUb was accompanied by an increase in size of all higher molecular mass Pex18p species. Thus, our data demonstrated that these bands represent ubiquitinated species of the PTS2 co-receptor Pex18p. It is worth noting that in contrast to Pex18p, Pex7p exhibited no obvious modification in pex4Δ cells, as shown in Fig. 2B.
FIGURE 2.
Ubiquitination of Pex18p. Pex14p-TEV-ProtA-complexes were isolated by affinity chromatography from membrane fractions of the indicated strains. A, eluate fractions were analyzed by immunoblotting for the presence of Pex14p, Pex13p, and Pex18p as well as the PTS1 receptor Pex5p. Higher molecular weight species of Pex18p were detected in strains deficient in either PEX4 or PEX6, which resembled the appearance of ubiquitinated Pex5p (Ub-Pex5p) in these strains. B, Pex14p-TEV-ProtA complexes were isolated from wild-type and pex4Δ cells expressing either plasmid-encoded wild-type Ub or mycUb. In pex4Δ-cells, Pex18p modifications with a higher molecular weight were visible, which shifted to protein species of even lower electrophoretic mobility upon expression of mycUb, demonstrating that these modifications represent ubiquitinated Pex18p.
A Conserved Cysteine Residue of Pex18p Is Required for Its Function
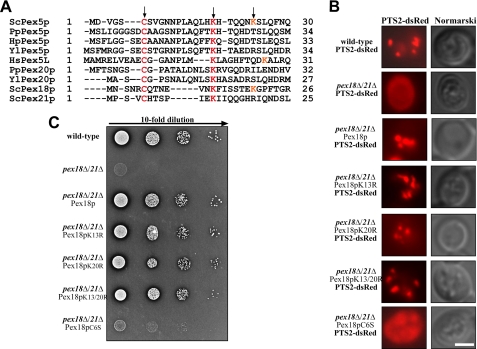
It has been demonstrated that Pex18p can functionally replace the N-terminal half of Pex5p in transporting PTS1 proteins into peroxisomes (41). Despite low overall similarities between the primary sequences of Pex18p and the PTS1 receptor, one common feature is obvious. Both Pex18p and Pex5p contain a cysteine and at least one lysine residue at similar positions (Fig. 3A). These residues are conserved among species and thus present in Pex5p from different organisms as well as Pex21p and the PTS2 co-receptor Pex20p from P. pastoris and Y. lipolytica. It is worth noting that Pex18p contains a second lysine residue adjacent to the first one, which is also present in Pex5p from bakers' yeast and mammals. Either one or both of these lysine residues were demonstrated to serve as a target for polyubiquitination of S. cerevsiae and H. polymorpha Pex5p as well as P. pastoris Pex20p (42–44). The conserved cysteine turned out to be essential for the recycling of P. pastoris Pex20p and rat Pex5p and has been shown to be required for Pex5p monoubiquitination (42, 45–47).
FIGURE 3.
Functional relevance of a conserved cysteine residue of Pex18p. A, primary sequence alignment of N-terminal regions of Pex5p and Pex20p of different species and S. cerevisiae Pex18p and Pex21p. The conserved cysteine and first lysine residue present in all (co-)receptor molecules as well as a second lysine residue specific for S. cerevisiae Pex5p and Pex18p are indicated with an arrow and displayed in red. B, the PTS2 marker protein PTS2-dsRed was transformed in wild-type, pex18Δ21Δ, and pex18Δ21Δ cells expressing the indicated Pex18p variants. The transformed strains were grown on oleic acid plates for 2 days and examined by fluorescence microscopy. Structural integrity of the cells is documented by bright field microscopy. In contrast to pex18Δ21Δ, cells that are impaired in PTS2-dependent matrix protein import and mislocalize the marker protein to the cytosol, the wild type exhibits a punctate fluorescence staining. pex18Δ21Δ cells expressing either wild-type Pex18p or lysine residue substitutions of the PTS2-receptor displayed similar staining for the synthetic PTS2 marker protein. The strain expressing Pex18pC6S displays cytosolic staining of the marker protein, indicating an import defect for PTS2 proteins and, thus, a loss of function of this Pex18p mutant. Scale bar, 5 μm. C, growth analysis of wild-type and pex18Δ/21Δ cells. The indicated strains were spotted as a series of 10-fold dilutions on media containing oleic acid as the sole carbon source. The plates were incubated for 3–5 days at 30 °C and scored for the appearance of colonies and halo formation. In comparison with wild-type cells, pex18Δ/21Δ as well as pex18Δ/21Δ cells expressing Pex18pC6S showed no growth on oleate as the sole carbon source, corroborating the observed protein import defect.
In order to elucidate the role of the conserved residues for Pex18p function in more detail, we generated Pex18p variants with substitutions of the first (Lys-13) or the second (Lys-20) lysine to arginine, the conserved cysteine (Cys-6) to serine, or a combination of these variations. Wild-type and mutated Pex18p were expressed from a plasmid in pex18Δpex21Δ cells. Untransformed and transformed oleic acid-induced live cells were monitored by fluorescence microscopy for PTS2-dependent matrix protein import. The synthetic marker protein PTS2-dsRed revealed a punctate staining pattern in the wild-type strain that is typical for peroxisomal labeling (Fig. 3B). In contrast, the marker was cytosolic in pex18Δpex21Δ cells (Fig. 3B), which is in line with the PTS2 import deficiencies of these cells (32). This import defect was restored by expression of wild-type Pex18p. Likewise, expression of Pex18p variants with substitution of either one or both lysine residues led to a punctate pattern of PTS2-dsRed. This finding indicates that the conserved lysine residues within the N terminus of Pex18p are not required for functional PTS2-dependent protein import into peroxisomes. However, when Pex18p(C6S) was expressed in pex18Δpex21Δ cells, a cytosolic distribution of the synthetic PTS2 protein was observed, comparable with the staining revealed from untransformed mutant cells. Because expression of Pex18p(C6S) was the same as for wild-type Pex18p (Fig. 4B and supplemental Table S2), the results demonstrate a loss of function of this Pex18p variant. To corroborate this finding, we performed growth test on oleic acid as single carbon source. Peroxisomes are required for growth of bakers' yeast on this carbon source, and a growth defect indicates peroxisomal dysfunction. In contrast to wild-type cells, cells deficient in PEX18/PEX21 were unable to grow on oleic acid (Fig. 3C), as typical for S. cerevisiae mutant strains that are defective in peroxisome biogenesis (19). The mutant strain expressing either wild-type Pex18p or Pex18p variants with substitution of conserved lysine residues grew normally. In contrast, no growth was observed for pex18Δpex21Δ cells expressing Pex18p(C6S) (Fig. 3C). From this we conclude that the conserved cysteine at position 6 is essential for Pex18p function in PTS2-dependent matrix protein import.
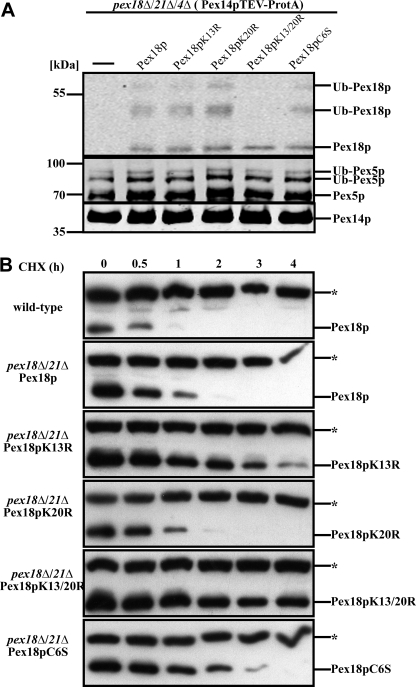
FIGURE 4.
Lysines 13 and 20 of Pex18p are required for co-receptor polyubiquitination and proteasomal degradation. A, Pex14p complexes were isolated from pex18Δ/21Δ/4Δ and pex18Δ/21Δ/4Δ cells expressing the indicated Pex18p variants. Eluate fractions were inspected for the presence of Pex14p, Pex18p, and Pex5p by immunoblot analysis. In all samples, Pex5p displayed the ubiquitination pattern typical for pex4Δ cells. Although different Pex18p variants exhibited a normal modification pattern, no ubiquitinated Pex18p species were detected when both lysine 13 and 20 were changed to arginine (Pex18pK13R/K20R). This finding identifies both lysine residues as targets for ubiquitin attachment. B, wild-type as well as pex18Δ21Δ cells expressing the indicated Pex18p variants were grown on oleic acid for 14 h and treated with 15 μg/ml cycloheximide. At the indicated time points, samples were taken and analyzed for Pex18p by immunoblotting. *, cross-reactive band. Although wild-type Pex18p was below the detection level after 1 h, Pex18pK13R/K20R was almost stable along the whole time period. These data indicate that the conserved lysine residues are required for proteasomal degradation of Pex18p and support the notion that they provide the target residues for polyubiquitination.
Lys-13/Lys-20-dependent Polyubiquitination and Degradation of Pex18p
Next we addressed whether substitution of conserved amino acid residues interferes with the ubiquitination status of Pex18p. A pex18Δpex21Δ strain with genomically expressed Pex14p-TEV-ProtA for complex isolation and additional deletion of PEX4 was applied. pex4Δ cells were applied because they are not affected in the formation of the peroxisomal importomer (6) but accumulate polyubiquitinated Pex5p and Pex18p, which here could be analyzed. To this end, protein complexes isolated from these mutant cells expressing different Pex18p mutants were probed for the bait protein Pex14p as well as for Pex18p and Pex5p. In all complexes, Pex5p was present at similar amounts and with the ubiquitination pattern typical for PEX4-deficient cells (Fig. 4A) (11, 13). Associated Pex18p with substitution of either one or both lysine residues or the cysteine residue (Pex18pK13R, -K20R, -K13R/K20R, and -C6S) exhibited the same protein pattern and intensity as wild-type Pex18p. However, Pex18pK13R/K20R, which contained replacement of both lysine residues by arginine, still associated with Pex14p but was not anymore ubiquitinated. This observation strongly argues that the lysines at position 13 and 20 represent the target residues for co-receptor polyubiquitination. Moreover, the data demonstrated that the conserved cysteine that proved to be essential for protein function is not required for binding of Pex18p to the docking complex of the peroxisomal membrane (Figs. 3 (B and C) and 4A).
Next we investigated whether mutation of the target residues for ubiquitination of Pex18p has an influence on the turnover rate of the protein. Cells were grown on oleic acid medium, cycloheximide was added, and at different time points whole cell lysates were prepared and analyzed. Wild-type Pex18p was completely degraded within 2 h after the addition of cycloheximide (Fig. 4B and supplemental Table S2). The substitution of K20R did not affect the half-life of Pex18p because the same turnover was observed for the mutated proteins as for plasmid-encoded wild-type Pex18p. Substitution of K13R did slightly increase the stability. The highest degree of stabilization, however, was observed for Pex18p harboring substitution of both K13R and K20R residues. This is in agreement with the role of these lysine residues in polyubiquitination and direction of Pex18p for proteasomal degradation. Interestingly, the biologically inactive Pex18pC6S exhibited only a slight increase in stability, which is similar to the stabilization observed for the K13R substitution. However, in contrast to the K13R substitution, replacement of the cysteine results in a complete loss of function of Pex18p, pointing to a special role of the conserved cysteine.
Cysteine-dependent Ubiquitination of Pex18p
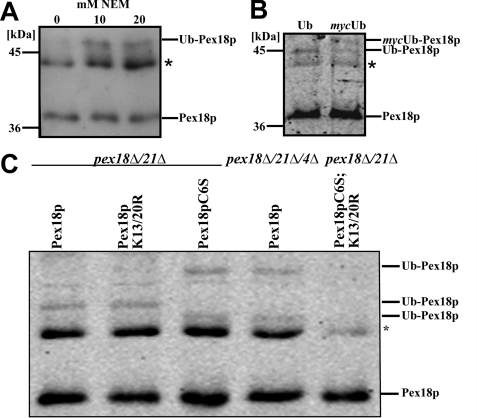
Because Pex18p shares some features with the N-terminal domain of the PTS1 receptor Pex5p, which is unmodified under wild-type conditions but monoubiquitinated when isopeptidase inhibitor NEM was used (48), we inspected Pex18p under these conditions. Oleic acid-induced cells expressing Pex14p-TEV-ProtA were treated with NEM, and Pex14p complexes were isolated and probed by immunoblotting with Pex18p-specific antibodies. The appearance of an additional high molecular weight species of Pex18p was observed when NEM was added to the preparation (Fig. 5A). In order to verify whether the higher molecular weight species represents ubiquitinated Pex18p, the protein was isolated from strains overexpressing plasmid-encoded Ub or mycUb. Upon expression of mycUb, the electrophoretic mobility of the Pex18p modification decreased, which demonstrated its nature as a Ub conjugate (Fig. 5B).
FIGURE 5.
Pex18p is ubiquitinated at a conserved cysteine residue. Pex14p-TEV-ProtA was isolated from oleic acid-induced wild-type cells in the absence or presence of NEM (A), upon expressing either plasmid-encoded Ub or mycUb in the presence of 20 mm NEM (B), or from pex18Δ21Δ expressing either wild-type Pex18p or mutated Pex18p as indicated as well as pex18Δ21Δ4Δ expressing wild-type Pex18p (C). Equal portions of the obtained eluate fractions were analyzed by immunoblotting for the presence of Pex18p and its modified species. *, cross-reactive band.
In Pex5p, the NEM-protected monoubiquitination takes place at the conserved cysteine residue at the extreme N terminus of the PTS1 receptor. Because this residue is also conserved in Pex18p (Fig. 3A), we analyzed whether it also represents a target residue for Pex18p ubiquitination. Wild-type Pex18p or mutated Pex18p variants were expressed in pex18Δ21Δ cells; the Pex14p complex was isolated in the presence of NEM and probed for Pex18p. As shown before, monoubiquitinated Pex18p was visible under these conditions (Fig. 5C, lane 1). The modification pattern did not change upon substitution of the two conserved lysine residues with arginine, demonstrating that these residues are dispensable for Pex18p monoubiquitination (Fig. 5C, lane 2). Upon replacement of the conserved cysteine residue at position 6 of Pex18p by serine, the ubiquitination pattern of Pex18p changed (Fig. 5C, lane 3). The monoubiquitinated form disappeared, and instead a pattern resembling the polyubiquitinated Pex18p in the pex4Δ appeared (Fig. 5C, lane 4). This pattern was also observed in pex4Δ even in the absence of NEM (not shown). When, in addition to cysteine 6, the two conserved lysine residues were substituted (Fig. 5C, lane 5), no ubiquitination of Pex18p was visible anymore. These findings indicate that Pex18p undergoes a cysteine-dependent NEM-protected ubiquitination. Moreover, replacement of the cysteine leads to polyubiquitination of Pex18p at the conserved lysine residues.
Pex7p Import Requires the Export of Its Co-receptor Pex18p
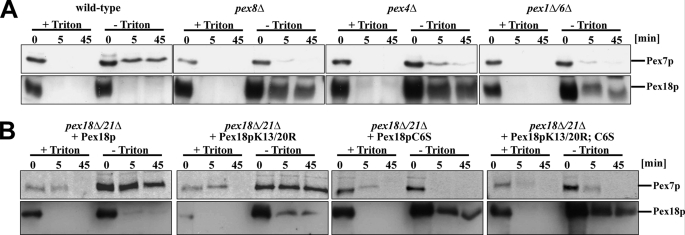
Pex18p has a dual localization with a major cytosolic fraction and smaller portion localized at the outer surface of the peroxisomal membrane (32). In order to disclose changes in the topology of Pex18p during the protein import cascade, we performed protease protection assays with wild-type as well as selected pex mutants affected at different stages of the receptor cycle. Organellar fractions derived from wild-type, pex8Δ, pex4Δ, or pex1Δ/6Δ cells were prepared and incubated with proteinase K in the presence or absence of Triton X-100. In line with previous results, Pex18p of wild-type cells was accessible to exogenously added protease even in the absence of detergent Triton X-100, which destroys membrane integrity (Fig. 6A) (32). Under these conditions, however, a portion of the Pex18p-binding partner Pex7p was protected against degradation, indicating its peroxisomal localization. Interestingly, a different topology for receptor and co-receptor is observed in the mutant strains. Although Pex7p was completely accessible to proteinase K even in the absence of detergent, a significant portion of Pex18p was protease-protected (Fig. 6A). This result indicates that Pex7p has not been translocated across the peroxisomal membrane and that Pex18p is halted in a protease-protected form. Because this result was obtained for all tested mutants strains, it seems to be a common phenotype for a defect in components of the peroxisomal matrix protein import machinery.
FIGURE 6.
Pex18p arrests in a protease-protected form when the receptor cycle is blocked at late stages. Organelle pellets of wild-type and selected pex mutant strains (A) or pex18Δ/21Δ cells expressing different variants of Pex18p (B) were isolated and subjected to a protease protection assay with proteinase K in the absence or presence of Triton X-100. At the indicated time points, the protease was inhibited by the addition of PMSF. Equal portions of the samples were subjected to SDS-PAGE and immunoblot analysis for the presence of Pex7p and Pex18p.
Ubiquitination of the import receptor Pex5p was shown to be required for its export back to the cytosol (43). In the yeast S. cerevisiae, a block of receptor export by either deletion of components of the export machinery or mutation of amino acids of Pex5p that prevent receptor ubiquitination leads to Pex5p accumulation at the peroxisomal membrane (11, 13, 43). Here, we observed a similar behavior for Pex18p as a higher amount of the co-receptor associates with Pex14p in pex deletion strains (Fig. 2A). This finding raised the question of whether this effect could be resembled by amino acid substitution of the Ub target residues of Pex18p. To address this question, we analyzed the topology of Pex18p variants and especially the consequence for Pex7p localization. The biologically active Pex18pK13R/K20R substitutions that are not any more polyubiquitinated behaved almost like wild-type Pex18p with only a small fraction of Pex18p but the majority of Pex7p being protease-protected by the peroxisomal membrane (Fig. 6B). In contrast, substitution of cysteine 6 of Pex18p by serine arrested a significant portion of Pex18p in a protease-protected form, whereas Pex7p was protease-sensitive even in the absence of detergent. The same was observed when both conserved lysine and cysteine residues were substituted (Fig. 6B). Accordingly, blocking of the Cys-6-dependent monoubiquitination results in an arrest of Pex18p in a membrane protected form with Pex7p still facing the cytosol.
Thus, the cysteine-dependent ubiquitination, which triggers receptor export as shown for Pex5p, is a prerequisite for cargo translocation. This supports the idea that export triggers import, as postulated in the export-driven import model (49).
DISCUSSION
Pex18p acts as a co-receptor for Pex7p and is required for the import of PTS2 proteins into the peroxisomal matrix. In this report, we demonstrate that Pex18p is ubiquitinated in two different ways and identified the target residues for this protein modification. Lysine-dependent ubiquitination is not required for Pex18p function but for its rapid degradation via the proteasome. In contrast, modification of the conserved cysteine residue at the extreme N terminus of Pex18p leads to a biologically inactive protein. This protein still associates with Pex7p and binds to the peroxisomal docking machinery but is unable to facilitate receptor translocation. Moreover, cysteine substitution arrests Pex18p in a membrane-protected state, whereas Pex7p faces the cytosol, supporting the idea of an export-driven import of peroxisomal proteins.
Ubiquitination of Pex18p
Ubiquitination of peroxisomal proteins was first observed for the PTS1-receptor Pex5p, and this has been a matter of extensive investigation (31). For ubiquitination within the PTS2-pathway, it was observed that the PTS2 co-receptors Pex18p of S. cerevisiae and Pex20p of P. pastoris are ubiquitinated, and evidence was provided for a ubiquitin-dependent regulated turnover of these proteins (34, 42). Here, we show that the two lysine residues of Pex18p that correspond to the ubiquitination sites in S. cerevisiae Pex5p also function as targets for Pex18p polyubiquitination. Blocking polyubiquitination of Pex18p by mutation of these lysines resulted in a striking extension of the half-life of the protein. However, the mutant Pex18pK13R/K20R variant still associated with the peroxisomal docking machinery (Fig. 4A) and was still functional because it maintained the capability to restore the import defect of pex18Δ21Δ cells (Fig. 3, B and C). Thus, polyubiquitination of Pex18p proved not to be required for its function as co-receptor in PTS2-dependent protein import into peroxisomes. In this respect, Pex18p behaves like Pex5p from S. cerevisiae. For Pex5p, the lysine-dependent polyubiquitination turned out to be dispensable for the function of the protein as import receptor; however, polyubiquitination turned out to be required for degradation via the ubiquitin-proteasome system in a process termed receptor accumulation and degradation in the absence of recycling (RADAR) (11, 13, 50). RADAR was originally described for the regulated degradation of Pex20p, the P. pastoris counterpart of Pex18p (51). Interestingly, substitution of the conserved cysteine of Pex20p blocks recycling of the co-receptor from the peroxisome to the cytosol. Under these conditions, a lysine-dependent polyubiquitination of Pex20p is responsible for release of the co-receptor and its rapid degradation via the ubiquitin-proteasome system (42). It is worth noting that Pex20p as well as Pex5p of P. pastoris share similar regulation and dynamics during the import cycle (51). Defects in the late steps of protein import preventing Pex5p recycling also affect Pex20p localization and stability (50).
We identified a second type of ubiquitination of Pex18p, which occurred in wild-type cells but was only visible when NEM was used (Fig. 5). NEM is a known inhibitor of deubiquitinating enzymes but also inhibits AAA proteins like the NEM-sensitive fusion protein NSF (52, 53). The NEM-protected ubiquitination occurs on a conserved cysteine residue of Pex18p, similar to Pex5p monoubiquitination (Fig. 5) (48). At first glance, these data seem to oppose a previous report describing ubiquitinated forms of Pex18p in wild-type cells even without NEM (34). However, these studies were performed with FLAG-tagged Pex18p, and the authors stated that no modification was seen with the untagged co-receptor. One could assume that the tagging influenced the performance of the receptor, which then is directed to ubiquitination and RADAR. In support of this assumption, the ubiquitination pattern observed for FLAG-tagged Pex18p (34) resembles the ubiquitination pattern for Pex18p in strains affected for Pex6p (Fig. 2A).
When the conserved cysteine 6 of Pex18p was replaced by serine, Pex18pC6S still associated with the peroxisomal import apparatus, and the half-life of the protein was only slightly enhanced in comparison with wild-type Pex18p (Figs. 4 and 5). In this respect, the functional relevance of the conserved cysteine seems to be the same for Pex20p and Pex18p. In both cases, the cysteine is required for protein function, most likely co-receptor export back to the cytosol. However, the consequence for the fate of the protein harboring mutation of the crucial cysteine is different. Pex20p is rapidly degraded, whereas Pex18p is arrested in a membrane-protected state (Fig. 6).
Different behavior of Pex7p and Pex18p
Pex7p is guided to the peroxisomal membrane in a Pex18p- and Pex21p-dependent manner. Although two co-receptors are present, the phenotype of the single deletion strains is more drastic for pex18Δ than for pex21Δ (32), indicating a more prominent function of Pex18p under oleic acid conditions. Peroxisomal targeting of Pex7p is almost abolished when Pex18p is missing, and Pex7p remains almost completely in the cytosol, unable to associate with peroxisomes (32). In fact, the functional ternary receptor-cargo complex consists of Pex7p, which binds the cargo, and Pex18p, which binds the cargo-loaded Pex7p. In this respect, Pex18p highly resembles the function of the N-terminal domain of the PTS1 receptor Pex5p (54), whereas Pex7p resembles the Pex5p cargo-binding C-terminal domain. In fact, a chimerical protein of Pex18p and the PTS1-binding domain of Pex5p proved to be functional in PTS1 protein import (41). Consequently, the architecture of the Pex7p-Pex18p receptor complex, consisting of two proteins that can be separately monitored, allowed us to distinguish the fates of the two functional elements or “domains” of the receptor complex upon different steps of the import process.
Our findings demonstrate that Pex7p exhibits a half-life longer than 6 h and thus is much more stable than Pex18p (Fig. 1). The different turnover rates of receptor and co-receptor imply a disassembly of this functional unit during the receptor cycle, at the latest prior to the proteasomal degradation of Pex18p. A model of the early steps of the PTS2-driven import postulates the binding of the PTS2 cargo protein by its receptor Pex7p in the cytosol (4, 55). Subsequently, Pex18p joins the receptor-cargo complex and mediates docking of this trimeric complex to the peroxisomal membrane (33). This scenario excludes a dissociation prior to membrane contact.
So far, no ubiquitination has been reported for the PTS2 receptor Pex7p. Thus, Pex7p might be still in conjunction with Pex18p upon export, the complex might rapidly dissociate afterward, and Pex18p is degraded while Pex7p is recycled. Alternatively, Pex7p export may be independent of Pex18p and thus require specific export machinery. In this case, ubiquitin might not serve as export signal for Pex7p.
Pex18p Export Triggers Pex7p Translocation
In wild-type cells, Pex7p is partially protected by the peroxisomal membrane, whereas Pex18p is completely accessible to exogenously added protease (32, 56) (Fig. 6A). This observation led to the idea that Pex18p is a peripheral membrane protein facing the cytosol (32). Pex5p and Pex7p have been reported to reach into the peroxisomal lumen during the PTS1 and PTS2 protein import cascade (57, 58), in agreement with the idea of an extended shuttle, which proposes that the cargo-loaded receptor would accompany the cargo into the peroxisomal lumen, where cargo is released and the receptors are recycled (57, 58). Our finding that Pex18p is arrested in a protease-protected state when the receptor cycle is halted indicated that also this protein inserts into the peroxisomal membrane or reaches the luminal site during the import cascade (Fig. 6). A protease-protected form of Pex20p has also been described for Y. lipolytica mutants lacking Pex8p (59). Pex8p is an intraperoxisomal peroxin, which has been related to cargo release (63).
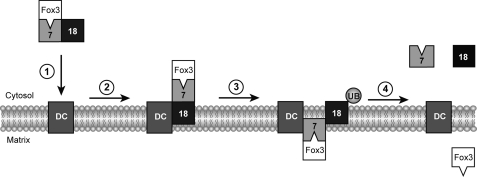
Incorporation of our results into the current model leads to the following emerging picture of yeast PTS2-dependent protein import into peroxisomes (Fig. 7). Pex7p binds its cargo in the cytosol, followed by the recruitment of Pex18p (33). This trimeric complex associates with the peroxisomal docking complex. In analogy to Pex5p, Pex18p is supposed to insert into the membrane into a protease-protective state, whereas Pex7p remains on the cytosolic face of the peroxisomal compartment. This might be a brief intermediate because it is not visible in wild-type background but accumulates when receptor cycling is blocked (Fig. 6). Next, Pex18p and Pex7p change their topology. Whereas Pex18p becomes accessible to externally added proteases, Pex7p gains a protected state, which might indicate translocation of the cargo-loaded receptor across the membrane (Fig. 6). Interestingly, this topology change depends on modification of the conserved cysteine residue at the extreme N terminus of Pex18p. The counterpart of Pex18p in the PTS1 pathway is the import receptor Pex5p, which undergoes monoubiquitination on a cysteine at the same position. The monoubiquitination at the conserved cysteine is required for Pex5p function and export (43, 48). The importance of the cysteine for recycling has also been demonstrated for P. pastoris Pex20p (42), and the evidence is convincing that cysteine 6 has the same function for Pex18p. Accordingly, mutation of the cysteine results in the loss of Pex18p cycling, causing the observed block of Pex7p translocation across the peroxisomal membrane (Fig. 6). Thus, export of Pex18p is supposed to be a prerequisite for Pex7p import. An explanation may provide the model of export-driven import (49), which describes how receptor export might be mechanically coupled to cargo import. Accordingly, the mechanical coupling of Pex18p export would drive peroxisomal import of cargo-loaded Pex7p.
FIGURE 7.
Model of yeast PTS2-dependent protein import into peroxisomes. The Fox3p (thiolase)-loaded PTS2-receptor complex consisting of Pex7p and Pex18p associates with the peroxisomal docking complex (1). Pex18p is supposed to insert into the membrane, whereas Pex7p remains on the cytosolic face of the peroxisomal compartment (2). Next, Pex18p as well as Pex7p change their topology. While Pex18p becomes accessible to externally added proteases, Pex7p gains a protected state, which might indicate translocation of the cargo-loaded receptor across the membrane. This change in topology depends on ubiquitination of the conserved cysteine residue in the N-terminal domain of Pex18p (3). Finally, after the cargo has been released into the peroxisomal lumen, Pex7p as well as Pex18p are transported back to the cytosol (4).
Supplementary Material
Acknowledgments
We are grateful to Ulrike Freimann for technical assistance and to Wolfgang Schliebs for reading of the manuscript.
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB642.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- AAA
- ATPases associated with diverse cellular activities
- NEM
- N-ethylmaleimide
- Ub
- ubiquitin
- mycUb
- ubiquitin fused to a myc tag
- TEV
- tobacco etch virus
- RADAR
- receptor accumulation and degradation in the absence of recycling.
REFERENCES
- 1. Lazarow P. B., Fujiki Y. (1985) Annu. Rev. Cell Biol. 1, 489–530 [DOI] [PubMed] [Google Scholar]
- 2. Meinecke M., Cizmowski C., Schliebs W., Krüger V., Beck S., Wagner R., Erdmann R. (2010) Nat. Cell Biol. 12, 273–277 [DOI] [PubMed] [Google Scholar]
- 3. Dodt G., Gould S. J. (1996) J. Cell Biol. 135, 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marzioch M., Erdmann R., Veenhuis M., Kunau W. H. (1994) EMBO J. 13, 4908–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown L. A., Baker A. (2008) Mol. Membr. Biol. 25, 363–375 [DOI] [PubMed] [Google Scholar]
- 6. Agne B., Meindl N. M., Niederhoff K., Einwächter H., Rehling P., Sickmann A., Meyer H. E., Girzalsky W., Kunau W. H. (2003) Mol. Cell 11, 635–646 [DOI] [PubMed] [Google Scholar]
- 7. Hazra P. P., Suriapranata I., Snyder W. B., Subramani S. (2002) Traffic 3, 560–574 [DOI] [PubMed] [Google Scholar]
- 8. Girzalsky W., Platta H. W., Erdmann R. (2009) Biol. Chem. 390, 745–751 [DOI] [PubMed] [Google Scholar]
- 9. Platta H. W., Grunau S., Rosenkranz K., Girzalsky W., Erdmann R. (2005) Nat. Cell Biol. 7, 817–822 [DOI] [PubMed] [Google Scholar]
- 10. Miyata N., Fujiki Y. (2005) Mol. Cell Biol. 25, 10822–10832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kiel J. A., Emmrich K., Meyer H. E., Kunau W. H. (2005) J. Biol. Chem. 280, 1921–1930 [DOI] [PubMed] [Google Scholar]
- 12. Erdmann R., Schliebs W. (2005) Nat. Rev. Mol. Cell Biol. 6, 738–742 [DOI] [PubMed] [Google Scholar]
- 13. Platta H. W., Girzalsky W., Erdmann R. (2004) Biochem. J. 384, 37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grou C. P., Carvalho A. F., Pinto M. P., Huybrechts S. J., Sá-Miranda C., Fransen M., Azevedo J. E. (2009) J. Biol. Chem. 284, 10504–10513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Debelyy M. O., Platta H. W., Saffian D., Hensel A., Thoms S., Meyer H. E., Warscheid B., Girzalsky W., Erdmann R. (2011) J. Biol. Chem. 286, 28223–28234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schliebs W., Kunau W. H. (2006) Biochim. Biophys. Acta 1763, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 17. Güldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H. (1996) Nucleic Acids Res. 24, 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. (1999) Yeast 15, 963–972 [DOI] [PubMed] [Google Scholar]
- 19. Erdmann R., Veenhuis M., Mertens D., Kunau W. H. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5419–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ausubel F. J., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (1992) Current Protocols in Molecular Biology, Greene Publishing Associates, New York [Google Scholar]
- 21. Platta H. W., El Magraoui F., Bäumer B. E., Schlee D., Girzalsky W., Erdmann R. (2009) Mol. Cell Biol. 29, 5505–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klein A. T., van den Berg M., Bottger G., Tabak H. F., Distel B. (2002) J. Biol. Chem. 277, 25011–25019 [DOI] [PubMed] [Google Scholar]
- 24. Harlow E., Lane D. (1988) Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 25. Stein K., Schell-Steven A., Erdmann R., Rottensteiner H. (2002) Mol. Cell Biol. 22, 6056–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Albertini M., Rehling P., Erdmann R., Girzalsky W., Kiel J. A., Veenhuis M., Kunau W. H. (1997) Cell 89, 83–92 [DOI] [PubMed] [Google Scholar]
- 27. Albertini M., Girzalsky W., Veenhuis M., Kunau W. H. (2001) Eur. J. Cell Biol. 80, 257–270 [DOI] [PubMed] [Google Scholar]
- 28. Girzalsky W., Rehling P., Stein K., Kipper J., Blank L., Kunau W. H., Erdmann R. (1999) J. Cell Biol. 144, 1151–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bigl M., Eschrich K. (1994) Biol. Chem. 375, 153–160 [DOI] [PubMed] [Google Scholar]
- 30. Girzalsky W., Hoffmann L. S., Schemenewitz A., Nolte A., Kunau W. H., Erdmann R. (2006) J. Biol. Chem. 281, 19417–19425 [DOI] [PubMed] [Google Scholar]
- 31. Lanyon-Hogg T., Warriner S. L., Baker A. (2010) Biol. Cell 102, 245–263 [DOI] [PubMed] [Google Scholar]
- 32. Purdue P. E., Yang X., Lazarow P. B. (1998) J. Cell Biol. 143, 1859–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grunau S., Schliebs W., Linnepe R., Neufeld C., Cizmowski C., Reinartz B., Meyer H. E., Warscheid B., Girzalsky W., Erdmann R. (2009) Traffic 10, 451–460 [DOI] [PubMed] [Google Scholar]
- 34. Purdue P. E., Lazarow P. B. (2001) J. Biol. Chem. 276, 47684–47689 [DOI] [PubMed] [Google Scholar]
- 35. Entian K. D., Vogel R. F., Rose M., Hofmann L., Mecke D. (1988) FEBS Lett. 236, 195–200 [DOI] [PubMed] [Google Scholar]
- 36. Fujiki Y., Matsuzono Y., Matsuzaki T., Fransen M. (2006) Biochim. Biophys. Acta 1763, 1639–1646 [DOI] [PubMed] [Google Scholar]
- 37. Obrig T. G., Culp W. J., McKeehan W. L., Hardesty B. (1971) J. Biol. Chem. 246, 174–181 [PubMed] [Google Scholar]
- 38. Ghislain M., Udvardy A., Mann C. (1993) Nature 366, 358–362 [DOI] [PubMed] [Google Scholar]
- 39. Brocard C., Lametschwandtner G., Koudelka R., Hartig A. (1997) EMBO J. 16, 5491–5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ellison M. J., Hochstrasser M. (1991) J. Biol. Chem. 266, 21150–21157 [PubMed] [Google Scholar]
- 41. Schäfer A., Kerssen D., Veenhuis M., Kunau W. H., Schliebs W. (2004) Mol. Cell Biol. 24, 8895–8906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Léon S., Subramani S. (2007) J. Biol. Chem. 282, 7424–7430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Platta H. W., El Magraoui F., Schlee D., Grunau S., Girzalsky W., Erdmann R. (2007) J. Cell Biol. 177, 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kiel J. A., Otzen M., Veenhuis M., van der Klei I. J. (2005) Biochim. Biophys. Acta 1745, 176–186 [DOI] [PubMed] [Google Scholar]
- 45. Williams C., van den Berg M., Sprenger R. R., Distel B. (2007) J. Biol. Chem. 282, 22534–22543 [DOI] [PubMed] [Google Scholar]
- 46. Carvalho A. F., Grou C. P., Pinto M. P., Alencastre I. S., Costa-Rodrigues J., Fransen M., Sá-Miranda C., Azevedo J. E. (2007) Biochim. Biophys. Acta 1773, 1141–1148 [DOI] [PubMed] [Google Scholar]
- 47. Okumoto K., Misono S., Miyata N., Matsumoto Y., Mukai S., Fujiki Y. (2011) Traffic 12, 1067–1083 [DOI] [PubMed] [Google Scholar]
- 48. Kragt A., Voorn-Brouwer T., van den Berg M., Distel B. (2005) J. Biol. Chem. 280, 7867–7874 [DOI] [PubMed] [Google Scholar]
- 49. Schliebs W., Girzalsky W., Erdmann R. (2010) Nat. Rev. Mol. Cell Biol. 11, 885–890 [DOI] [PubMed] [Google Scholar]
- 50. Léon S., Goodman J. M., Subramani S. (2006) Biochim. Biophys. Acta 1763, 1552–1564 [DOI] [PubMed] [Google Scholar]
- 51. Léon S., Zhang L., McDonald W. H., Yates J., 3rd, Cregg J. M., Subramani S. (2006) J. Cell Biol. 172, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fröhlich K. U., Fries H. W., Peters J. M., Mecke D. (1995) Biochim. Biophys. Acta 1253, 25–32 [DOI] [PubMed] [Google Scholar]
- 53. Borodovsky A., Kessler B. M., Casagrande R., Overkleeft H. S., Wilkinson K. D., Ploegh H. L. (2001) EMBO J. 20, 5187–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kiel J. A., Veenhuis M., van der Klei I. J. (2006) Traffic 7, 1291–1303 [DOI] [PubMed] [Google Scholar]
- 55. Rehling P., Marzioch M., Niesen F., Wittke E., Veenhuis M., Kunau W. H. (1996) EMBO J. 15, 2901–2913 [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang J. W., Lazarow P. B. (1995) J. Cell Biol. 129, 65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dammai V., Subramani S. (2001) Cell 105, 187–196 [DOI] [PubMed] [Google Scholar]
- 58. Nair D. M., Purdue P. E., Lazarow P. B. (2004) J. Cell Biol. 167, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Smith J. J., Rachubinski R. A. (2001) J. Biol. Chem. 276, 1618–1625 [DOI] [PubMed] [Google Scholar]
- 60. Halbach A., Rucktäschel R., Rottensteiner H., Erdmann R. (2009) J. Biol. Chem. 284, 3906–3916 [DOI] [PubMed] [Google Scholar]
- 61. Wiebel F. F., Kunau W. H. (1992) Nature 359, 73–76 [DOI] [PubMed] [Google Scholar]
- 62. Rehling P., Skaletz-Rorowski A., Girzalsky W., Voorn-Brouwer T., Franse M. M., Distel B., Veenhuis M., Kunau W. H., Erdmann R. (2000) J. Biol. Chem. 275, 3593–3602 [DOI] [PubMed] [Google Scholar]
- 63. Wang D., Visser N. V., Veenhuis M., van der Klei I. J. (2003) J. Biol. Chem. 278, 43340–43345 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.