Background: Lhx and Isl proteins contribute to genetic control in developing neurons.
Results: The Lhx3/4-binding motif in Isl2 was identified, and the structures of Lhx-Isl complexes were characterized and compared.
Conclusion: There are minor differences in the structures of Lhx3/4 binding Isl1/2 reflected by mutational and biophysical analyses.
Significance: Redundant sets of interactions conserve function in developing neurons while allowing divergence in other contexts.
Keywords: Neurodevelopment, Protein Structure, Protein-Protein Interactions, Transcription Factors, X-ray Crystallography, X-ray Scattering, LIM Homeodomain Proteins, Cell Specification
Abstract
Combinations of LIM homeodomain proteins form a transcriptional “LIM code” to direct the specification of neural cell types. Two paralogous pairs of LIM homeodomain proteins, LIM homeobox protein 3/4 (Lhx3/Lhx4) and Islet-1/2 (Isl1/Isl2), are expressed in developing ventral motor neurons. Lhx3 and Isl1 interact within a well characterized transcriptional complex that triggers motor neuron development, but it was not known whether Lhx4 and Isl2 could participate in equivalent complexes. We have identified an Lhx3-binding domain (LBD) in Isl2 based on sequence homology with the Isl1LBD and show that both Isl2LBD and Isl1LBD can bind each of Lhx3 and Lhx4. X-ray crystal- and small-angle x-ray scattering-derived solution structures of an Lhx4·Isl2 complex exhibit many similarities with that of Lhx3·Isl1; however, structural differences supported by mutagenic studies reveal differences in the mechanisms of binding. Differences in binding have implications for the mode of exchange of protein partners in transcriptional complexes and indicate a divergence in functions of Lhx3/4 and Isl1/2. The formation of weaker Lhx·Isl complexes would likely be masked by the availability of the other Lhx·Isl complexes in postmitotic motor neurons.
Introduction
LIM domains, named for the first three proteins in which the motif was identified (Lin-11/Islet-1/Mec-3), are zinc fingers that coordinate two zinc ions and mediate protein-protein interactions (reviewed in Ref. 1). Two related families of LIM-containing proteins, LIM homeodomain and LIM-only proteins, are expressed in a combinatorial manner along with their protein-binding partners to form the “LIM code,” a transcriptional code that helps specify cell type during the development of the central nervous system and many other tissues and organs (e.g. Refs. 2 and 3). The 12 mammalian LIM homeodomain proteins comprise six pairs of paralogues (4), which often have overlapping expression patterns and functions. LIM homeobox protein 3 and 4 (Lhx3 and Lhx4)5 and Islet-1 and -2 (Isl1 and Isl2) are two such paralogous pairs. All four proteins are functional in developing motor neurons (3, 5). Mice in which these genes have been individually disrupted are all embryonic or perinatal lethal, but the phenotypes differ, showing that the pairs of paralogues are not genetically redundant. For example, both Lhx3−/− and Lhx4−/− mice are perinatal lethal but show primary defects in pituitary and lung development, respectively (6, 7). Isl1−/− mice die about halfway through embryonic development (embryonic day 11.5) with abnormal heart and pancreas development and an absence of motor neurons (8), whereas Isl2−/− mice are perinatal lethal, probably due to disrupted breathing resulting from defects in motor neuron differentiation in the thoracic levels of the spinal cord (9).
In differentiating motor neurons, the paralogues show some redundancy. For example, during chick ventral motor neuron development, Lhx3 and Lhx4 must both be knocked out to specify dorsal rather than ventral motor neurons (5). In mice and zebrafish, exogenous Isl1 or Isl2 can trigger motor neuron differentiation, but both must be expressed to maintain proper motor neuron cell fate (9–11).
Lhx3 and Isl1 form a simple and well characterized example of the LIM code associated with the development of adjacent cell types (V2 interneurons and motor neurons) in the ventral spinal cord. Lhx3 is expressed in developing V2 interneurons, whereas both Lhx3 and Isl1 are expressed in postmitotic motor neurons. These LIM homeodomain proteins are strong drivers of cell specification. The addition of Lhx3 to dorsal spinal cord cells results in the formation of V2 interneurons, whereas further addition of Isl1 switches their identity to motor neuron cells (12).
Lhx3 and Isl1 form cell-specific transcriptional complexes that also contain LIM domain-binding protein 1 (Ldb1), a widely expressed transcriptional regulator that binds to, and is essential for the activity of, members of the LIM homeodomain and LIM-only families (13–15). Lhx3 binds Ldb1 to generate an active binary complex in V2 interneurons (Fig. 1A). In motor neurons, Isl1 disrupts this interaction to form a ternary complex in which Isl1 displaces Lhx3 as the binding partner of Ldb1 and forms additional contacts with Lhx3 (Fig. 1B) (12). The contacts between Lhx3 or Isl1 and Ldb1 are made by the tandem LIM domains from the LIM homeodomain proteins and the ∼30-residue LIM interaction domain from Ldb1 (Ldb1LID) (Fig. 1C) (13, 14, 16). An essentially identical interaction is formed between the LIM domains from Lhx3 and an Ldb1LID-like sequence near the C terminus of Isl1, the Lhx3-binding domain (Isl1LBD) (17).
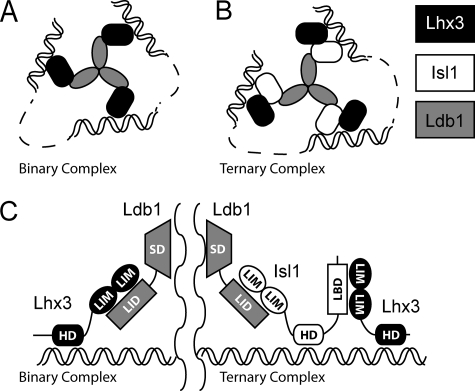
FIGURE 1.
LIM homeodomain complexes in development. A, the proteins Ldb1 (gray) and Lhx3 (black) assemble into a binary complex. B, with the addition of Isl1 (white), the ternary complex is preferentially formed (12). Note that the self-association domain of Ldb1 forms trimers in vitro (21) and that the homeodomains from Lhx3 and Isl1 bind DNA. The spacing between the homeodomain-binding sites (dashed lines) is not defined and may represent close or distant binding sites. C, details of domain contacts within the binary and ternary complexes. The LIM interaction domain (LID) from Ldb1 makes contacts with the LIM domains (LIM) from Lhx3 in the binary complex, but in the ternary complex, Ldb1LID contacts the LIM domains of Isl1, and the LBD from Isl1 binds the LIM domains from Lhx3. SD, self-association domain; HD, homeodomain.
This study explores the possibility that Isl2 and Lhx4 may be capable of substituting for Isl1 and Lhx3, respectively, in the ternary complex. We identify a new Lhx3-binding domain in Isl2 (Isl2LBD), which binds both Lhx3 and Lhx4, and show that Isl1LBD also binds Lhx4. The x-ray crystal structure of Lhx4LIM1+2·Isl2LBD,6 combined with biophysical and mutagenic analysis of the series of pairwise interactions, shows that although the structures of Lhx3/4·Isl1/2 pairwise complexes are very similar, there are differences in the nature of the binding interactions between these LIM homeodomain family proteins that likely contribute to diverging biological functions.
EXPERIMENTAL PROCEDURES
Cloning, Mutagenesis, and Protein Expression
The numbering of residues from the proteins refers to the following National Center for Biotechnology Information (NCBI) entries for mouse proteins: Lhx3, P50481; Lhx4, NP_034842; Isl1, NP_067434; Isl2, NP_081673; Ldb1, NP_034827.1. Lhx3LIM1+2 refers to residues 28–153, Lhx4LIM1+2 refers to residues 24–149, Isl1LBD refers to residues 262–291, Isl2LBD refers to residues 272–301, and Ldb1LID refers to residues 300–330. All clones and mutants were generated by PCR and sequenced to confirm identity (Sydney University Prince Alfred Macromolecular Analysis Centre (SUPAMAC), Royal Prince Alfred Hospital, Sydney, Australia). Constructs of the tethered complexes were generated as described previously (18, 19). All proteins were expressed with a GST tag using pGEX-2T (GE Healthcare) in Escherichia coli BL21 (DE3) at 20 °C for 16–20 h. Proteins were purified on glutathione-Sepharose 4B resin (GE Healthcare) and eluted by proteolytic cleavage of the tag with thrombin (Sigma-Aldrich). The proteins were additionally purified by size-exclusion chromatography using a Superdex 200 16/60 column (GE Healthcare).
Protein Characterization
Far-UV CD spectra were collected for samples in 10 mm Tris base (pH 8.5), 150 mm NaF, and 0.5 mm tris(2-carboxyethyl) phosphine at 20 °C as described previously (20). Multi-angle laser light scattering data were collected on samples eluted from a Superose 12 10/30 (GE Healthcare) size-exclusion column as described previously (21).
X-ray Structure Determination and Refinement
The crystallization, collection, and processing of data for Lhx4-Isl2 were described previously (19). The program SOLVE (22) was used with anomalous data to 3.0 Å resolution to identify the zinc atom positions and generate initial phases. Statistical density modification and local pattern matching were carried out using RESOLVE (23). The model was built and revised manually with Coot (24). Refinement of the diffraction data was carried out to 2.16 Å using REFMAC5 and PHENIX with TLS refinement (25–27). The final model was refined using PHENIX. The MOLPROBITY server was used to identify steric clashes and unconventional geometry for validation of the structure (28).
Small-angle X-ray Scattering (SAXS) Data Acquisition and Analysis
Solution SAXS data (I(q) versus q, where q = (4πsinθ)/λ, 2θ is the scattering angle, and λ the x-ray wavelength) were measured at 10 °C using a SAXSess (Anton Paar) line collimation instrument (10-mm slit width; Ref. 29). For details of sample environments, see supplemental data 3. Data reduction included corrections for beam geometry, sample absorbance, and detector sensitivity as described previously (29). Data were placed on an absolute scale, and the forward scattering at zero angle (I(0)) was used to evaluate the molecular weight of the scattering particles (30) using contrast values (ΔρM) and partial specific volumes (υ) derived from the program MULCh (31). Guinier analysis of the desmeared data was performed using PRIMUS (32). GIFT (31) was used to calculate probable atom pair distance distributions (P(r) versus r) from which the maximum dimension (Dmax), radius of gyration (Rg), and I(0) of each tethered complex were determined. Ab initio shape restorations of the molecular envelopes for each complex were performed using DAMMIF with the outputs from GIFT. Further rigid body modeling was performed using BUNCH (33) (see supplemental data 3 for details). The fits of the refined BUNCH models against the SAXS data were assessed using CRYSOL, which also was used to calculate scattering profiles and fits of the original crystal structures to the data. The P(r) versus r of the crystal or BUNCH structures were determined as described in Ref. 34 using GNOM.
Yeast Two-hybrid Analysis
Inserts were cloned into pGAD10 and modified pGBT9 plasmids and co-transformed into AH109 cells (Clontech), as described previously (35). Transformed cells were selected by growth on leucine- and tryptophan-deficient media. For the detection of an interaction, the following selection media were used as indicated (all selection media lacked leucine and tryptophan). For low stringency (−H + X-α-gal), the medium was additionally deficient in histidine (−H) and supplemented with 40 μg ml−1 X-α-gal; for moderate stringency (−H + X-α-gal + 3-AT), the medium was as for low stringency but further supplemented with 1 mm 3-amino-1,2,4-triazole (3-AT); for high stringency, the medium was deficient in both histidine and adenine (−H-A). Numbers of yeast cells were normalized such that A600 nm = 0.2 and were deposited in 2-μl drops at the dilutions indicated.
Chemical Denaturation
Stability studies using guanidine hydrochloride as the denaturant were conducted as described previously (36). Tryptophan fluorescence following excitation at 295 nm was recorded in the range 320–380 nm. The fluorescence emission wavelength maximum (λmax) was used as an indication of the unfolding of the tethered protein complex. The λmax was obtained by fitting of a Gaussian curve through the emission spectra using Origin 6.1 (OriginLab).
RESULTS
Identification of the Lhx3-binding Domain in Isl2
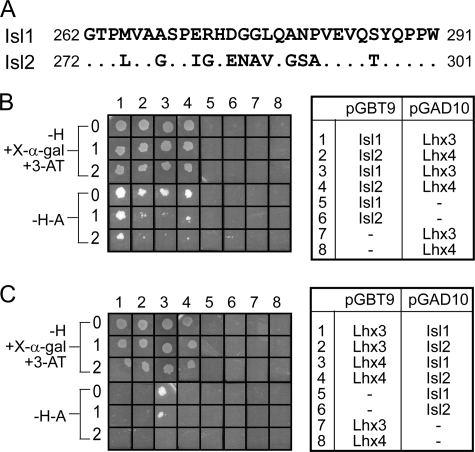
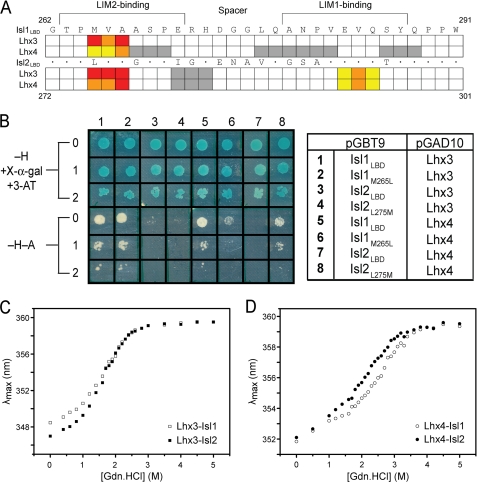
Isl1LBD spans residues 262–291 (Fig. 2A) (17). Comparison of the sequences of Isl1 and Isl2 suggested that a similar region in Isl2, residues 272–301 (Isl2LBD), might mediate binding to Lhx3 (Fig. 2A). We used yeast two-hybrid analysis to test the ability of Isl1LBD and the putative Isl2LBD to interact with the LIM domains of Lhx3 and Lhx4. When the IslLBDs were fused to the GAL4 DNA-binding domain (GAL4DBD), all pairs of proteins exhibited growth on high stringency (−H-A) selection plates, indicating that all four pairwise interactions among these proteins take place (Fig. 2B). When the Lhx3/4LIM1+2 constructs were fused to the GAL4DBD, the Lhx3/4LIM1+2·Isl1/2LBD interactions were all evident under moderate stringency conditions (−H + X-α-gal + 3-AT), and only the interaction between Lhx4LIM1+2 and Isl1LBD was detected under high stringency conditions (Fig. 2C). Thus, Isl2LBD does mediate interactions with Lhx3, and both Lhx3 and Lhx4 can interact with equivalent domains in Isl1 and Isl2.
FIGURE 2.
Identification of the Isl2 Lhx3-binding domain, Isl2LBD. A, sequence alignment of Isl1LBD and Isl2272–301. Dots are identical residues. B and C, pairwise interactions between Isl1/Isl2 and Lhx3/Lhx4 identified by yeast two-hybrid analysis. AH109 yeast cells co-transformed with pGBT9/pGAD10 vectors shown on the right were tested for growth under different selection conditions (−H + X-α-gal + 3-AT) or (−H-A); 0 indicates no dilution of yeast cells (A600 nm = 0.2), 1 indicates a 1:10 dilution (A600 nm = 0.02), and 2 indicates a 1:100 dilution (A600 nm = 0.002). pGBT9 encodes the GAL4DBD and pGAD10 encodes the GAL4AD.
Generation of Stable Lhx-Isl Constructs
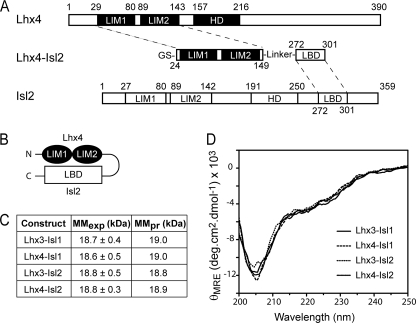
When expressed in bacteria, the LIM domains from Lhx3 and Lhx4, like those from other LIM-only/LIM homeodomain proteins, tend to aggregate and are largely insoluble (37, 38). Tethering an interaction partner such as Isl1LBD or Ldb1LID to the LIM domains protects them from aggregation, allowing soluble tethered complexes to be produced (e.g. Refs. 18, 38, and 39). As shown for Lhx4·Isl2 in Fig. 3, A and B, we generated constructs of the form Lhx3/4LIM1+2-linker-Isl1/2LBD, where the 11-residue linker was designed to be flexible by the inclusion of glycine and serine residues (18, 19). These constructs are subsequently referred to as Lhx3-Isl1, Lhx3-Isl2, Lhx4-Isl1, and Lhx4-Isl2.
FIGURE 3.
Construction and characterization of the tethered Lhx3/4-Isl1/2 complexes. A and B, schematics illustrating the generation of the tethered constructs (A) and arrangement of domains (B) in the constructs, using Lhx4-Isl2 as an example. C, experimental (MMexp) and predicted (MMpr) molecular masses of the complexes as determined by multi-angle laser light scattering. D, far-UV CD spectra of the tethered complexes.
All four tethered complexes were monomeric as determined by multi-angle laser light scattering (Fig. 3C and supplemental Fig. S1) (17). Far-UV CD spectra of each of the complexes show that the proteins are folded and have similar secondary structure compositions (16–18% helix, 23–26% β-turn, 17–19% β-sheet, and 36–41% coil; Fig. 3D).
Crystal Structure of Lhx4-Isl2
The protein Lhx4-Isl2 was crystallized as described previously (19). The structure of Lhx4-Isl2 was determined using single-wavelength anomalous dispersion data recorded at the zinc absorption edge (λ = 1.2819 Å) to a resolution of 2.16 Å (Table 1). The Rwork and Rfree are consistent with an x-ray crystal structure of this resolution with some disordered regions (e.g. the linker between Lhx4LIM1+2 and Isl2LBD).
TABLE 1.
Refinement statistics for Lhx4-Isl2
Values for the highest resolution shell are given in parentheses.
| Resolution limit (Å) | 2.16 (2.21-2.16) |
| Rworka | 0.235 (0.294) |
| Rfreeb | 0.265 (0.314) |
| No. of reflections used in refinement | 39,191 (2407) |
| No. of reflections in the test set | 2022 (92) |
| Protein atoms (including zinc) | 2201 |
| Solvent molecules | 35 |
| r.m.s.d. bond length (Å) | 0.003 |
| r.m.s.d. bond angles (°) | 0.66 |
| Wilson B factor (Å2) | 53 |
| Mean protein B factor (Å2) | 58 |
| Mean solvent B factor (Å2) | 53 |
| Ramachandran plot, residues in | |
| Favored regions (%) | 95.3 |
| Allowed regions (%) | 4.1 |
| Disallowed regions (%) | 0.7 |
| Missing side chainsc | |
| Common to both chain A and chain B | 17 |
| In chain A only | 10 |
| In chain B only | 4 |
a Rwork = Σ|Fobs − Fcalc|/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure amplitudes, respectively.
b Rfree is Rwork for the 5% validation set.
c Lists of residues for which there was overall insufficient electron density or that had missing side chains are presented in supplemental Table S2A and supplemental Table S2B, respectively.
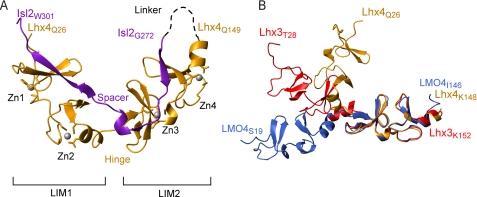
Lhx4 contains two typical LIM domains, each comprising two zinc-coordinating modules made up of two sequential β-hairpins and a short segment of α-helix (Fig. 4A, orange ribbon). Isl2LBD (purple ribbon) binds across the length of both LIM domains, forming a “head-to-tail” complex. Isl2LBD forms small stretches of β-strand structure along the interface, packing against the second β-hairpin of all four zinc-coordinating modules to make small, three-stranded antiparallel β-sheets.
FIGURE 4.
The crystal structure of Lhx4-Isl2. A, ribbon diagram of chain B of the Lhx4-Isl2 complex. Lhx4 is shown in orange, and Isl2 is in purple. The position of the linker is shown as a black dashed line. B, overall shape comparison of tethered LIM homeodomain and LIM-only complexes. Molecules are aligned over the backbone residues of the LIM2 domains of Lhx4-Isl2 (3MMK), Lhx3-Isl1 (2RGT), and LMO4(LIM-only protein 4)LIM1+2-Ldb1LID (1RUT) complexes. For clarity, only the LIM domains are shown; Lhx4 is in orange, Lhx3 is in red, and LMO4 is in blue. Images were prepared and alignment of molecules was performed using MolMol (46).
The two molecules in the asymmetric unit (chains A and B, chain B is shown in Fig. 4A) are highly similar (root mean square deviation ∼0.28 Å for the backbone atoms). Fourteen residues are missing from both chains, an additional residue is found only in chain A, and another four are found only in chain B. The missing residues are from the linker and termini of the protein (supplemental Table S2A). Of the residues modeled, ∼20% have missing side chains (see supplemental Table S2B), most commonly in the region between the two LIM domains (the “hinge”) and the adjacent region in Isl2LBD (the “spacer”). In both chains, residue Asp-66 has poorly defined electron density and lies in the disallowed region of the Ramachandran plot. Apart from these minor differences, both molecules have the same basic structure.
The overall conformation of the structure is unexpectedly compact when compared with previously determined structures of Lhx3-Isl1 and a related LIM-only complex (e.g. Fig. 4B) (17, 35). The Lhx4-Isl2 complex has a sharp bend of ∼100° at the hinge/spacer region between the two domains. As a result, the two LIM domains of Lhx4 lie almost perpendicular to one another.
Solution Properties of Lhx4-Isl2
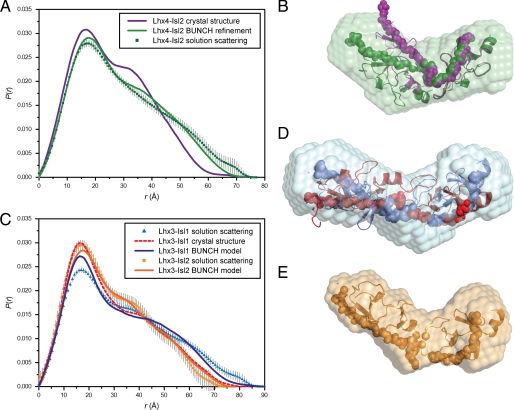
SAXS data were measured to probe the structure of Lhx4-Isl2 in solution (supplemental Fig. S3). The linearity of the Guinier plot and the molecular weight of Lhx4-Isl2 estimated from the extrapolated zero-angle scattering (I(0)) indicate that under these conditions, the solution contains non-interacting, monodisperse Lhx4-Isl2 molecules (Table 2 and supplemental Fig. S3). The calculated scattering curve from CRYSOL (40) using the coordinates of the Lhx4-Isl2 crystal structure shows deviations from the shape of the experimental curve in the very low-q and mid-q regimes (supplemental Fig. S3). These differences become more pronounced when comparing the interatomic distance distribution functions, P(r), which are very sensitive to symmetry and domain structure and can provide more intuitive information about the shape of a molecule (Fig. 5A). The larger radius of gyration, Rg, and longer maximum dimension, Dmax, of the molecule for the solution data (Rg = 23.5 Å; Dmax = 75–80 Å) when compared with the crystal structure (Rg = 20.5 Å; Dmax = 70–75 Å) suggest that the solution structure is more elongated than that in the crystal form.
TABLE 2.
Structural parameters from small-angle X-ray scattering data
| Molecule | Lhx4-Isl2 | Lhx3-Isl1 | Lhx3-Isl2 |
|---|---|---|---|
| Dmax (Å) | 77 | 89 | 75 |
| [protein] (mg ml−1) | 3.10 ± 0.08a | 5.00 ± 0.11a | 2.57 ± 0.07a |
| I(0) (cm−1) | 0.0517 (± 0.0003)b | 0.0698 (± 0.0003)b | 0.0400 (± 0.0003)b |
| υ (cm3 g−1) | 0.719 | 0.718 | 0.720 |
| ΔρM (× 1010 cm−2) | 2.23 | 2.24 | 2.22 |
| Rg (Å) | 23.5 (± 0.3)b | 26.9 (± 0.2)b | 22.6 (± 0.4)b |
| MMexpc (kDa) | 20.2 ± 0.5 | 16.8 ± 0.4 | 19.0 ± 0.6 |
| MMpr (kDa) | 18.9 | 19.0 | 18.8 |
| MMexp/MMpr | 1.07 | 0.88 | 1.01 |
a Error is 1 S.D.
b I(0) and Rg were derived from P(r) using GIFT. The errors for these values were obtained by analyzing the Guinier region of the scattering in PRIMUS. GIFT does not produce errors for these values.
c MMexp = I(0)NA/[protein]ΔρM2.
FIGURE 5.
The solution structures of Lhx3/4-Isl1/2. A, P(r) profiles from experimental scattering data for Lhx4-Isl2 (green squares) and calculated scattering profiles from the Lhx4-Isl2 crystal structure (purple line) and the generated BUNCH model (green line). B, alignment using the atoms of Lhx4LIM2 of the Lhx4-Isl2 crystal structure (purple) with the best-fit BUNCH model (green) and ab initio DAMMIF reconstruction (transparent green surface). C, P(r) profiles from experimental scattering data for Lhx3-Isl1 (blue triangles) and Lhx3-Isl2 (orange squares) and calculated scattering profiles from the Lhx3-Isl1 crystal structure (red dashed line), the Lhx3-Isl1 BUNCH model (dark blue line), and the Lhx3-Isl2 BUNCH model (dark orange line). D, alignment using the atoms of Lhx3LIM2 of the Lhx3-Isl1 crystal structure (red) with the best-fit BUNCH model (blue) and ab initio DAMMIF reconstruction (transparent blue surface). E, the best-fit BUNCH model (orange) and ab initio DAMMIF reconstruction (transparent orange surface) for Lhx3-Isl2. LIM domains are shown as ribbons, and LBDs as are shown as spheres.
This type of tandem LIM-binding peptide complex is thought to have some flexibility at the hinge/spacer (17, 41). A rigid body refinement using the crystal structure and the SAXS data was carried out in which the two LIM peptide halves of the crystal structure were allowed to flex at the hinge/spacer. The BUNCH models support a more elongated conformation in solution with an average angle at the hinge/spacer of ∼120° (Fig. 5B). The more extreme bend in the Lhx4-Isl2 crystal structure likely represents a rarely populated conformation that had a preference for crystallization under the conditions used.
Solution Properties of Lhx3-Isl1 and Lhx3-Isl2
SAXS data were also collected for Lhx3-Isl1, Lhx3-Isl2, and Lhx4-Isl1. The Lhx4-Isl1 sample showed signs of aggregation, and data were not further analyzed (data not shown), but Lhx3-Isl1 and Lhx3-Isl2 were monomers in solution as evidenced by molecular mass estimates from I(0) (Table 2). Lhx3-Isl1 and Lhx3-Isl2 have similar asymmetric P(r) profiles, indicating that the complexes adopted extended conformations in solution (Fig. 5C). However, the Rg and Dmax for Lhx3-Isl2 are shorter (Rg = 22.3 Å; Dmax ∼75 Å) than Lhx3-Isl1(Rg = 26.9 Å; Dmax ∼90 Å), indicating that Lhx3-Isl2 is more compact than its Lhx3-Isl1 counterpart, as seen in the BUNCH models (Fig. 5, D and E). The solution structural parameters are essentially identical for Lhx3-Isl2 and Lhx4-Isl2 (Fig. 5, B and E, and supplemental Table S3), suggesting that binding to Isl2LBD causes both Lhx3 and Lhx4 to adopt a more compact structure than does binding to Isl1LBD.
Overall these solution scattering studies (see supplemental data 3 for a detailed analysis) indicate that tethered Lhx3/4-Isl1/2 complexes are likely to exist as an ensemble of elongated complexes with some flexibility at the hinge/spacer. Although the molecular details of each LIM domain peptide module are unlikely to be affected by flexion, details concerning the hinge/spacer regions will likely vary between members of the ensembles, and analyses of high resolution structure should be careful not to overinterpret details from these regions.
Identifying Binding Hotspots in Lhx3/4·Isl1/2 Complexes
We used yeast two-hybrid analysis to compare the ability of the individual LIM domains from Lhx3 and Lhx4 to interact with Isl1LBD, Isl2LBD, and Ldb1LID (supplemental Table S4) (17). The LIM2 domains from Lhx3 and Lhx4 were both capable of independently interacting with Isl1LBD and Ldb1LID, whereas the LIM1 domains could bind weakly to Ldb1LID (some yeast growth was evident only under low stringency selection conditions), but not to Isl1LBD. No individual LIM domain from either Lhx3 or Lhx4 showed evidence of an interaction with Isl2LBD in this assay.
The Lhx3-binding domains of Isl1 and Isl2 were subjected to mutagenesis to identify the key residues in those domains that mediate binding with Lhx3 and Lhx4. Residues were mutated to alanine (or glycine if the native residue was alanine). Initially sequential sets of three consecutive residues were mutated and tested for their ability to interact with the LIM domains from Lhx3 and Lhx4 using yeast two-hybrid analysis. For those triple mutants that showed significantly reduced levels of binding, a second round of single point mutants was generated and tested in the same manner. The results from these mutagenic screens are reported in supplemental data 4 and are summarized in Fig. 6A. Finally, two point mutants, Isl1M265L and Isl2L275M, were used to test binding specificity (Fig. 6B and supplemental data 4).
FIGURE 6.
Binding and apparent stability in Lhx3/4·Isl1/2 complexes. A, summary of alanine mutagenic screening assayed by yeast two-hybrid analysis from supplemental Table S4. The sequence of Isl1LBD (262–291) is shown. The sequence of Isl2LBD (272–301) shows where residues are conserved (*) or different. Colored boxes indicate where mutation had a strong (red), moderate (orange), or minor effect (yellow). White boxes indicate residues mutated in triple-alanine constructs that had a minor effect on growth of yeast. Gray boxes indicate no effect on yeast growth. B, comparison of wild-type and mutant LBDs from Isl1/Isl2 binding to the LIM domains of Lhx3/Lhx4 by yeast two-hybrid analysis. AH109 yeast cells co-transformed with pGBT9/pGAD10 vectors shown on the right were tested for growth under different selection conditions (−H + X-α-gal + 3-AT) or (−H−A); 0 indicates no dilution of yeast cells (A600 nm = 0.2), 1 indicates a 1:10 dilution (A600 nm = 0.02), and 2 indicates a 1:100 dilution (A600 nm = 0.002). C, resistance of Lhx3-Isl1/2 complexes to denaturation by guanidine hydrochloride (Gdn.HCl). □, Lhx3-Isl1; ■, Lhx3-Isl2. D, resistance of Lhx4-Isl1/2 complexes to denaturation by guanidine hydrochloride. ○, Lhx4-Isl1; ●, Lhx4-Isl2. For C and D, λmax reports maximum emission wavelength in the range 320–380 nm with excitation at 295 nm.
Mutations in the N-terminal (and not the C-terminal) half of Isl1LBD significantly disrupted binding to both Lhx3 and Lhx4 (Fig. 6A) (17), which is consistent with only the LIM2 domains from the Lhx proteins being able to independently bind Isl1LBD. In contrast, mutations in both the N-terminal and the C-terminal halves of Isl2LBD abrogated binding to Lhx3 and Lhx4 (Fig. 6A), consistent with both LIM domains of the Lhx proteins being required to mediate an interaction with Isl2LBD.
Among the residues identified as being most important for the interactions, Isl1265–267, and Isl2275–277/293–295, the only sequence difference between the two paralogues is a conservative switch from Met-265 in Isl1 to Leu-275 in Isl2. Interaction data for Isl1M265L and Isl2L275M suggest that although Lhx3 has no apparent preference for either residue at Isl1265/Isl2275, Lhx4 binds with higher affinity when there is a methionine at this position (Fig. 6B, supplemental Fig. S4, and supplemental Table S4).
Relative Stabilities of Lhx-Isl Complexes
We used an indirect approach to probe the relative binding affinities of Lhx3 and Lhx4 for each of Isl1 and Isl2. Stable forms of isolated LIM domains from Lhx3 and Lhx4 cannot be produced in a recombinant fashion, preventing the use of standard binding experiments (such as ELISA, isothermal titration calorimetry, or surface plasmon resonance). We instead compared the relative resistance of the Lhx3/4-Isl1/2 complexes to chemical denaturation, an approach previously used to gauge the relative stability of tethered Lhx3-Isl1 and Lhx3LIM1+2-Ldb1LID complexes (17). The LBD/LID peptides are largely unstructured, and if we only compare identical folded LIM domains, the differences in resistance to chemical denaturation should report on differences in binding affinity between the LIM domains and the peptides. As the sequences of Lhx3LIM1+2 and Lhx4LIM1+2 differ, we only compared the apparent stability of Lhx3-Isl1 with Lhx3-Isl2, or Lhx4-Isl1 with Lhx4-Isl2, and did not compare Lhx3 constructs with Lhx4 constructs. Note that these proteins do not exhibit reversible unfolding, meaning that the data cannot be used to calculate differences in free energy of unfolding and/or actual binding affinities.
The resistance of the constructs to denaturation was measured by monitoring the change in λmax of tryptophan fluorescence as a function of guanidine hydrochloride concentration. Lhx3-Isl1 and Lhx3-Isl2 tethered complexes had virtually identical unfolding curves, with a midpoint of unfolding at ∼2 m guanidine hydrochloride, suggesting that the Lhx3-Isl1 and Lhx3-Isl2 complexes have the same apparent stability (Fig. 6C). In contrast, Lhx4-Isl1 showed a midpoint of unfolding at ∼2.5 m when compared with ∼2 m for Lhx4-Isl2, suggesting that the Lhx4-Isl1 tethered complex is probably more stable than the Lhx4-Isl2 complex (Fig. 6D).
DISCUSSION
We have now shown that an Lhx3-binding domain exists in Isl2 as well as Isl1 and that both of these domains can bind to each of Lhx3 and Lhx4. Overall Lhx3 and Lhx4 share 66% sequence identity, whereas Isl1 and Isl2 share 74% sequence identity, which is highest in the homeodomains (95 and 98% sequence identity, respectively) and LIM domains (83 and 82% identity, respectively). Isl1LBD and Isl2LBD have lower sequence identity (60%). The structure of the Lhx4-Isl2 complex reported here is most divergent in sequence from the existing Lhx3-Isl1 complex structure (17), allowing a structure-guided interpretation of sequence, binding, and mutational data for Lhx3/Lhx4 and Isl1/Isl2 interactions.
The Lhx3-Isl1 and Lhx4-Isl2 Structures Have Similar Overall Structures
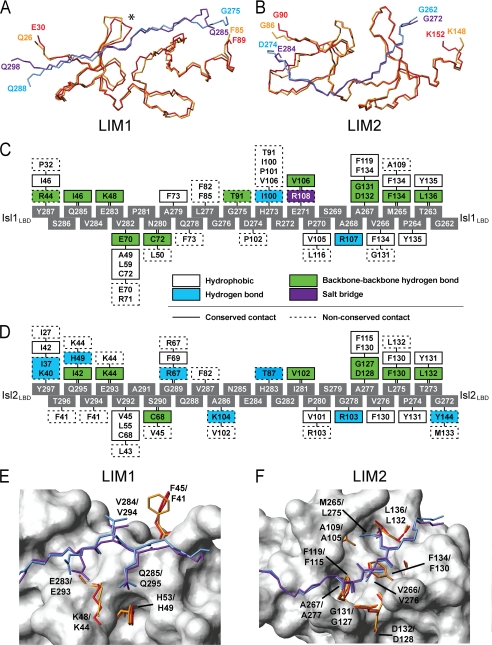
The overall shapes of Lhx3-Isl1 and Lhx4-Isl2 from the crystal structures differ by the angles at the hinge/spacer (∼100° for Lhx4-Isl2 and ∼165° for Lhx3-Isl1; Fig. 4B). The solution structural parameters derived from SAXS data indicate that, although this difference is exaggerated in the crystal forms, in solution there is a difference in the hinge/spacer angles between the two complexes such that Lhx4-Isl2 and Lhx3-Isl2 are more compact than Lhx3-Isl1 (have a smaller angle). At the domain level, the folds of the LIM domains of Lhx3 and Lhx4 are highly conserved. An alignment of the backbone atoms from chain B of the Lhx4-Isl2 structure and the Lhx3-Isl1 structure (Protein Data Bank (PDB) accession code 2RGT, chain B; Ref. 17) gives an r.m.s.d. of 0.77 Å for the LIM1 domains (Fig. 7A) and 0.59 Å for the LIM2 domains (Fig. 7B).
FIGURE 7.
Comparison of Lhx3/4·Isl1/2 interaction interfaces. A and B, backbone alignment of Lhx3 (red) and Lhx4 (orange) using the LIM1 (A) and LIM2 (B) domains. The backbones of Isl1 (blue) and Isl2 (purple) are also shown. An asterisk indicates loops in Lhx3/Lhx4 that show the most variation in the backbone alignment. C and D, interaction maps indicating the residues from Lhx3-Isl1 (C) and Lhx4-Isl2 (D) that make contacts as identified by LIGPLOT (47). Gray boxes are residues of Isl1LBD or Isl2LBD. Colored boxes for the Lhx residues define the type of interaction as indicated; solid lines represent conserved contacts between the two complexes, and dashed lines indicate non-conserved contacts between the two complexes. Note that the numbering between Lhx3 and Lhx4 differs by four (e.g. Lhx4F130 corresponds to Lhx3F134) and by 10 between Isl1 and Isl2 (e.g. Isl1M265 corresponds to Isl2L275). E and F, the key residues in the two complexes from Lhx3 and Lhx4 LIM1 domains (E) and Lhx3 and Lhx4 LIM2 domains (F). The side chains of critical residues from Isl1 (blue) and Isl2 (purple) identified using yeast two-hybrid analysis and the residues they contact are shown (Lhx3 in red and Lhx4 in orange), as well as the backbone atoms of non-critical residues from Isl1LBD and Isl2LBD. Non-Isl-binding residues from Lhx4LIM1 and Lhx4LIM2 are shown in surface representation (white). The residues Isl1M265 and Isl2L275 adopt equivalent rotamers, mtp and mt, respectively.
Isl1LBD and Isl2LBD have a backbone r.m.s.d. of 8.7 Å, which is not surprising given the different shapes of the complexes. If we consider the LIM1- and LIM2-binding regions of the LBDs separately, the r.m.s.d. drops to 0.56 Å for Isl1279–287/Isl2289–297 and 0.38 Å for Isl1262–273/Isl2272–283. Isl2LBD forms four well defined β-strands at the interface with Lhx4, and Isl1LBD shows an almost identical binding topology, with three well defined strands and evidence of a fourth short β-strand from two interdomain backbone-backbone hydrogen bonds over the third zinc-coordinating module of Lhx3. The program PISA (42) reveals similar surface areas buried at the interfaces of the two complexes, ∼1490 Å2 for Lhx3LIM1+2·Isl1LBD and ∼1540 Å2 for Lhx4LIM1+2·Isl2LBD.
Structure-guided Interpretation of Binding Data
A comparison of the interfaces in the two structures shows that there is a high sequence similarity in the stretches of the LBDs that contact each LIM domain (the binding motifs) but poor sequence conservation in and around the spacer (Fig. 6A). The specific intermolecular contacts of the two complexes are almost identical at the LIM2/N-terminal LBD interfaces but are more varied at the LIM1/C-terminal LBD interfaces (Fig. 7, C and D). In the latter region, there are minor variations in the side-chain conformations of some residues at the interface when compared with those at the LIM2/N-terminal LBD interfaces (Fig. 7, E and F). These differences in structure are reflected by different contributions to binding of the C-terminal halves of Isl1LBD and Isl2LBD and the effects of mutation on binding.
Isl2E293A/V294A/Q295A abrogated binding to both Lhx3 and Lhx4 (Fig. 6A, supplemental Fig. S4, and supplemental Table S4). Isl2E293-Q295 is equivalent to Isl1E283-Q285 in Isl1, but the side chains of those residues form predominantly different sets of contacts in the two complexes (Fig. 7, C and D), and the backbone alignment of Lhx3 and Lhx4 in this region shows more deviation than elsewhere in the molecules (Fig. 7A, marked by an asterisk). The side-chain conformations of Isl2E293 and Isl2Q295 and the residues that they contact, Lhx4F41 and Lhx4K44, also differ from those of the corresponding residues in the Lhx3-Isl1 structure (Fig. 7E). Although these differences are mostly subtle, the changes appear significant when compared with the N-terminal halves of the Isl1LBD and Isl2LBD, where sets of contacts and side-chain conformations are almost identical (e.g. Fig. 7, C, D, and F), even in non-conserved residues.
The essentially identical solution parameters for Lhx3-Isl2 and Lhx4-Isl2, coupled with the same patterns of binding of Isl2LBD for Lhx3 and Lhx4 (which differ from the Isl1LBD patterns for the same targets), imply that the binding of Isl2LBD results in a different conformation in Lhx3 and Lhx4 than does Isl1LBD. That is, although the mutation to alanine of residues in the LBD spacers does not appear to significantly affect binding, sequence variation in the spacers and the minor differences in the binding motifs induce different angles at the hinge/spacer. These differences might influence the formation of higher order complexes involving these protein pairs to regulate different activities in cells by creating or obscuring binding sites.
The chemical denaturation data for Lhx4 tethered complexes indicate that Lhx3-Isl1 and Lhx3-Isl2 have the same resistance to denaturation, but Lhx4-Isl2 is less resistant than Lhx4-Isl1. This difference may reflect an increased binding affinity of Lhx4·Isl1 over Lhx4·Isl2; however, thermodynamic binding data, which have thus far not been possible to obtain, would be required to establish whether this is the case. A small increase in the apparent affinity of Lhx4 (but not Lhx3) for the equivalent Isl1 residue and a decreased apparent affinity for the Isl2 residue at position Isl1M265/Isl2L275 (Fig. 6B, supplemental Fig. S4, and supplemental Table S4) implies that although Lhx3·Isl1 and Lhx3·Isl2 are interchangeable in terms of binding specificity, Lhx4 appears to have a small preference for binding Isl1 over Isl2.
Redundancy and Divergence of LIM Homeodomain Proteins
Paralogous pairs of genes are often thought to have arisen via genome duplication events. As demonstrated by their different knock-out phenotypes in mice, the pairs Lhx3/Lhx4 and Isl1/Isl2 have diverged in some of their functions, but all four proteins are expressed in postmitotic ventral motor neurons (43). Lhx3 and Lhx4 are apparently redundant in those cells as mice embryos in which either Lhx3 or Lhx4 is knocked out have normal ventral motor neurons (5). In addition, human Lhx3 mutations that cause combined pituitary hormone deficiency syndrome do not cause motor neuron abnormalities (44). It is only when both Lhx3 and Lhx4 are knocked out that ventral motor neurons fail to develop (5). Although at least some expression of Isl2 is required for all of the motor neuron subtypes to develop, Isl2 appears to act by increasing levels of Islet activity rather than providing a separate function (9, 11). Indeed, either Isl1 or Isl2 can effectively promote ectopic motor neuron differentiation when co-expressed with Lhx3, suggesting that the Islet proteins do have the same molecular function in this context (9–12). If Lhx4 does bind Isl2 less strongly than it does Isl1, co-expression of Isl2 with Lhx4 rather than Lhx3 should show a reduction in the efficiency of motor neuron development.
It seems likely that conservation of function of Lhx3/Lhx4 and Isl1/Isl2 in motor neurons represents a co-evolutionary process; high levels of Lhx3/Lhx4 must be matched by high levels of Islet proteins to achieve the correct balance of cell-specific transcription complexes. However, the presence of four possible pairwise complexes indicates a highly redundant system that could accommodate evolutionary divergence; simultaneous disruptions to both Lhx3 and Isl1 would probably be required to severely compromise motor neuron development. Teleost fish have higher numbers of LIM homeodomain (and many other) genes than other vertebrates, most likely from additional genome duplication events. The sequences of LBDs from the Islet proteins of these fish suggest the presence of a single Isl1 paralogue, with two or more proteins that are Isl2-like (or more divergent in sequence), suggesting that the duplicated genes are acquiring more diverse functions.
Divergence is also evident in apparent differences in the way that Isl1 and Isl2 mediate binding to the Lhx3 and Lhx4 targets. Solution scattering data indicate that Isl2 induces a more compact structure (smaller angle at the hinge/spacer) in Lhx3 and Lhx4 than does Isl1 (Fig. 5), probably due to differences in contacts between hinge and spacer residues (Fig. 7, C and D). The C-terminal half of Isl2LBD appears to make a larger contribution to binding than the same region of Isl1. Despite these different binding patterns, Isl1LBD and Isl2LBD appear to have equivalent affinities for Lhx3 (Fig. 6B), suggesting that the N-terminal half of Isl2 makes a correspondingly weaker contribution to binding. However, although these complexes combine modular interactions between the LIM domains and cognate peptide sequences to result in high affinity complexes, the interaction affinities of the modules may not be additive, as suggested by data for LIM-only protein 2 (41, 45). Differences in binding at the two domains could play a role in the kinetics of exchange of partners between different transcription complexes that contain LIM homeodomain and LIM-only proteins (17).
In conclusion, our studies have shown that although the pairwise interactions between the paralogous pairs of LIM homeodomain proteins Lhx3/Lhx4 and Isl1/Isl2 have many similarities that explain high levels of redundancy in motor neuron development, minor differences are evident that reflect evolutionary divergence for these proteins in other developmental pathways.
Supplementary Material
Acknowledgment
We thank Don Parkin for collection and preliminary evaluation of SAXS data.
This work was supported by grants from the Australian Research Council.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Figs. S1–S4, and Tables S1–S4.
The atomic coordinates and structure factors (code 3MMK) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Throughout this study, the format Lhx4·Isl2, Lhx3·Isl1, and so forth is used to represent a non-tethered complex or interaction, whereas the format Lhx4-Isl2, Lhx3-Isl1, and so forth is used to represent a tethered protein complex.
- Lhx3
- LIM homeobox protein 3
- Lhx4
- LIM homeobox protein 4
- Isl1
- Islet-1
- Isl2
- Islet-2
- Ldb1
- LIM domain-binding protein 1
- LBD
- Lhx3-binding domain
- LIM1+2
- tandem LIM domains
- LID
- LIM interaction domain
- 3-AT
- 3-amino-1,2,4-triazole
- λmax
- fluorescence emission wavelength maximum
- SAXS
- small-angle X-ray scattering
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Matthews J. M., Bhati M., Lehtomaki E., Mansfield R. E., Cubeddu L., Mackay J. P. (2009) Curr. Pharm. Des. 15, 3681–3696 [DOI] [PubMed] [Google Scholar]
- 2. Bachy I., Failli V., Rétaux S. (2002) Neuroreport 13, A23–A27 [DOI] [PubMed] [Google Scholar]
- 3. Tsuchida T., Ensini M., Morton S. B., Baldassare M., Edlund T., Jessell T. M., Pfaff S. L. (1994) Cell 79, 957–970 [DOI] [PubMed] [Google Scholar]
- 4. Failli V., Rogard M., Mattei M. G., Vernier P., Rétaux S. (2000) Genomics 64, 307–317 [DOI] [PubMed] [Google Scholar]
- 5. Sharma K., Sheng H. Z., Lettieri K., Li H., Karavanov A., Potter S., Westphal H., Pfaff S. L. (1998) Cell 95, 817–828 [DOI] [PubMed] [Google Scholar]
- 6. Li H., Witte D. P., Branford W. W., Aronow B. J., Weinstein M., Kaur S., Wert S., Singh G., Schreiner C. M., Whitsett J. A., et al. (1994) EMBO J. 13, 2876–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sheng H. Z., Zhadanov A. B., Mosinger B., Jr., Fujii T., Bertuzzi S., Grinberg A., Lee E. J., Huang S. P., Mahon K. A., Westphal H. (1996) Science 272, 1004–1007 [DOI] [PubMed] [Google Scholar]
- 8. Pfaff S. L., Mendelsohn M., Stewart C. L., Edlund T., Jessell T. M. (1996) Cell 84, 309–320 [DOI] [PubMed] [Google Scholar]
- 9. Thaler J. P., Koo S. J., Kania A., Lettieri K., Andrews S., Cox C., Jessell T. M., Pfaff S. L. (2004) Neuron 41, 337–350 [DOI] [PubMed] [Google Scholar]
- 10. Hutchinson S. A., Eisen J. S. (2006) Development 133, 2137–2147 [DOI] [PubMed] [Google Scholar]
- 11. Song M. R., Sun Y., Bryson A., Gill G. N., Evans S. M., Pfaff S. L. (2009) Development 136, 2923–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thaler J. P., Lee S. K., Jurata L. W., Gill G. N., Pfaff S. L. (2002) Cell 110, 237–249 [DOI] [PubMed] [Google Scholar]
- 13. Jurata L. W., Gill G. N. (1997) Mol. Cell. Biol. 17, 5688–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jurata L. W., Kenny D. A., Gill G. N. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11693–11698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Meyel D. J., O'Keefe D. D., Jurata L. W., Thor S., Gill G. N., Thomas J. B. (1999) Mol Cell 4, 259–265 [DOI] [PubMed] [Google Scholar]
- 16. Agulnick A. D., Taira M., Breen J. J., Tanaka T., Dawid I. B., Westphal H. (1996) Nature 384, 270–272 [DOI] [PubMed] [Google Scholar]
- 17. Bhati M., Lee C., Nancarrow A. L., Lee M., Craig V. J., Bach I., Guss J. M., Mackay J. P., Matthews J. M. (2008) EMBO J. 27, 2018–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bhati M., Lee M., Nancarrow A. L., Bach I., Guss J. M., Matthews J. M. (2008) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 297–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gadd M. S., Langley D. B., Guss J. M., Matthews J. M. (2009) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 151–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deane J. E., Mackay J. P., Kwan A. H., Sum E. Y., Visvader J. E., Matthews J. M. (2003) EMBO J. 22, 2224–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cross A. J., Jeffries C. M., Trewhella J., Matthews J. M. (2010) J. Mol. Biol. 399, 133–144 [DOI] [PubMed] [Google Scholar]
- 22. Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D. Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Terwilliger T. C. (2000) Acta Crystallogr. D. Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 26. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 27. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 28. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 29. Jeffries C. M., Whitten A. E., Harris S. P., Trewhella J. (2008) J. Mol. Biol. 377, 1186–1199 [DOI] [PubMed] [Google Scholar]
- 30. Orthaber D., Bergmann A., Glatter O. (2000) J. Appl. Crystallogr. 33, 218–225 [Google Scholar]
- 31. Whitten A. E., Cai S. Z., Trewhella J. (2008) J. Appl. Crystallogr. 41, 222–226 [Google Scholar]
- 32. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 33. Petoukhov M. V., Svergun D. I. (2005) Biophys. J. 89, 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Svergun D. I. (1992) J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 35. Deane J. E., Ryan D. P., Sunde M., Maher M. J., Guss J. M., Visvader J. E., Matthews J. M. (2004) EMBO J. 23, 3589–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jeffries C. M., Graham S. C., Stokes P. H., Collyer C. A., Guss J. M., Matthews J. M. (2006) Protein Sci. 15, 2612–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deane J. E., Sum E., Mackay J. P., Lindeman G. J., Visvader J. E., Matthews J. M. (2001) Protein Eng. 14, 493–499 [DOI] [PubMed] [Google Scholar]
- 38. Lee C., Nancarrow A. L., Bach I., Mackay J. P., Matthews J. M. (2005) J. Biomol. NMR 33, 198. [DOI] [PubMed] [Google Scholar]
- 39. Deane J. E., Maher M. J., Langley D. B., Graham S. C., Visvader J. E., Guss J. M., Matthews J. M. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1484–1486 [DOI] [PubMed] [Google Scholar]
- 40. Svergun D., Barberato C., Koch M. H. (1995) J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 41. El Omari K., Hoosdally S. J., Tuladhar K., Karia D., Vyas P., Patient R., Porcher C., Mancini E. J. (2011) Blood 117, 2146–2156 [DOI] [PubMed] [Google Scholar]
- 42. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 43. Tanabe Y., William C., Jessell T. M. (1998) Cell 95, 67–80 [DOI] [PubMed] [Google Scholar]
- 44. Netchine I., Sobrier M. L., Krude H., Schnabel D., Maghnie M., Marcos E., Duriez B., Cacheux V., Moers A., Goossens M., Grüters A., Amselem S. (2000) Nat. Genet. 25, 182–186 [DOI] [PubMed] [Google Scholar]
- 45. Ryan D. P., Sunde M., Kwan A. H., Marianayagam N. J., Nancarrow A. L., Vanden Hoven R. N., Thompson L. S., Baca M., Mackay J. P., Visvader J. E., Matthews J. M. (2006) J. Mol. Biol. 359, 66–75 [DOI] [PubMed] [Google Scholar]
- 46. Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 47. Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.