Background: IL-6 trans-signaling plays a critical role in chronic inflammation and cancer.
Results: The trans-signaling inhibitor sgp130(Fc) also inhibits classic signaling depending on IL-6/sIL-6R ratios.
Conclusion: The additional function of sgp130(Fc) suggests that in vivo only low therapeutic concentrations guarantee blockade of trans-signaling but not classic signaling.
Significance: The demonstration that the trans-signaling inhibitor can also inhibit classic signaling is central for the field of IL-6 biology.
Keywords: Cytokine, Interleukin, Membrane Proteins, Signal Transduction, STAT3, Interleukin 6, Classic Signaling, sgp130, Trans-signaling
Abstract
IL-6 trans-signaling via the soluble IL-6 receptor (sIL-6R) plays a critical role in chronic inflammation and cancer. Soluble gp130 (sgp130) specifically inhibits IL-6 trans-signaling but was described to not interfere with classic signaling via the membrane-bound IL-6R. Physiological and most pathophysiological conditions are characterized by a molar excess of serum sIL-6R over IL-6 characterized by free IL-6 and IL-6 found in IL-6·sIL-6R complexes allowing both classic and trans-signaling. Surprisingly, under these conditions, sgp130 was able to trap all free IL-6 molecules in IL-6·sIL-6R·sgp130 complexes, resulting in inhibition of classic signaling. Because a significant fraction of IL-6 molecules did not form complexes with sIL-6R, our results demonstrate that compared with the anti-IL-6R antibody tocilizumab or the anti-trans-signaling monoclonal antibody 25F10, much lower concentrations of the dimeric sgp130Fc were sufficient to block trans-signaling. In vivo, sgp130Fc blocked IL-6 signaling in the colon but not in liver and lung, indicating that the colon is a prominent target of IL-6 trans-signaling. Our results point to a so far unanticipated role of sgp130 in the blockade of classic signaling and indicate that in vivo only low therapeutic concentrations of sgp130Fc guarantee blockade of IL-6 trans-signaling without affecting IL-6 classic signaling.
Introduction
Interleukin 6 is centrally involved in the development of chronic inflammatory diseases, such as rheumatoid arthritis, peritonitis, asthma, or inflammatory bowel disease. Furthermore, it is also associated with the establishment of cardiovascular diseases such as atherosclerosis and the initiation and progression of cancer (1). This broad and multifunctional spectrum makes IL-6 an important factor in understanding disease development and a promising therapeutic target. IL-6 is secreted in response to infection or induced by other stress-triggered factors including IL-1 or TNFα. Main producers of IL-6 are lymphocytes as well as fibroblasts, adipocytes, keratinocytes, and endothelial cells, resulting in homeostatic plasma levels of ∼1–10 pg/ml (2, 3). However, during infection, inflammatory diseases, or cancer development, IL-6 production is increased as part of the local tissue responses with infiltrating immune cells, and IL-6 serum levels are usually elevated up to the lower ng/ml range in serum but, with few drastic exceptions such as fatal sepsis, still lower than soluble IL-6 receptor (sIL-6R)3 serum levels (4).
The pleiotropic cytokine IL-6 belongs to a group of cytokines that share the ability to use the signal transducer molecule gp130. In classic signaling, IL-6 first binds to a nonsignaling membrane-bound IL-6 α-receptor (IL-6R, gp80, or CD126), which in turn associates with and activates the signal-transducing β-receptor chain gp130. The restricted expression of the IL-6R limits classic IL-6 signaling to only a few tissues such as the liver and some cells of the immune system. In hepatocytes, IL-6R expression is essential for the production of acute phase response proteins including C-reactive protein and fibrinogen. Being transiently expressed on lymphocytes, macrophages, and megakaryocytes, the IL-6R also orchestrates phases of the immune response (5). Furthermore, a sIL-6R is present in human sera at high concentrations (25–145 ng/ml, 0.45–2.59 nm), and these levels increase during inflammation (6–8). The sIL-6R is formed either by limited proteolysis of membrane-bound receptors, a process referred to as ectodomain shedding, or directly secreted from the cells after alternative mRNA splicing (9, 10).
Importantly, unlike other soluble cytokine receptors, the sIL-6R does not act antagonistically, thus limiting the IL-6 cytokine activity, but rather agonistically. The IL-6·sIL-6R complex or Hyper-IL-6, which is a designer cytokine in which IL-6 is connected to sIL-6R by a flexible peptide linker (11), can bind to gp130 on cell surfaces and induce signaling also on cells that do not express membrane-bound IL-6R in a process called IL-6 trans-signaling (12). Interestingly, human IL-6 can signal via human and murine IL-6R, but murine IL-6 cannot bind to the human IL-6R and therefore can only induce signaling via the murine IL-6R (13). Because gp130 is ubiquitously expressed, IL-6 trans-signaling broadens the spectrum of possible target cells for IL-6 because the complex of IL-6·sIL-6R can virtually bind to and activate all cells of the body. Compared with IL-6 classic signaling, trans-signaling shows a different spectrum of IL-6-mediated actions. In contrast to the role of IL-6 classic signaling in developmental processes, tissue homeostasis, and acute phase response, trans-signaling is mainly involved in inflammatory diseases and cancer development (5). It has recently also been linked to differentiation and activation of regulatory T cells as well as Th17 cells (14, 15). Furthermore, IL-6 trans-signaling is involved in cell migration, as well as the establishment of anti-apoptosis of T cells. Thus, sIL-6R is an important player in the development of chronic inflammatory diseases (1).
In addition, a soluble form of gp130 (sgp130) is found at concentrations between 100 and 400 ng/ml (1–4 nm) in sera of healthy humans (8), and recombinant sgp130 was shown to be a specific inhibitor of IL-6 trans-signaling (13). Because IL-6 alone does not interact with sgp130, signaling via the membrane-bound IL-6R is not inhibited by sgp130 (13). Based on these findings, endogenous sgp130 was suggested to be the natural antagonist of the IL-6·sIL-6R complex in vivo, probably to prevent systemic IL-6 trans-signaling during inflammatory diseases (13). Two extracellular parts of gp130 linked to the Fc-portions of a human IgG1 antibody (sgp130Fc) have been shown to be ∼10–100 times more effective than sgp130 alone (13). sgp130Fc also exclusively inhibited IL-6 trans-signaling by the IL-6·IL-6R complex and has no affinity of IL-6 or IL-6R alone (13). An improved sgp130Fc is currently in clinical development (16). Recently, the anti-mouse IL-6 trans-signaling mAb 25F10 was developed (17). Tocilizumab, an anti-human IL-6R neutralizing antibody (18), which prevents binding of IL-6 to the IL-6R, thereby inhibiting both classic and trans-signaling, is approved for the treatment of rheumatoid arthritis in Europe and the United States (19). Here, we present data that reveal a novel function of sgp130Fc in the inhibition of classic IL-6 signaling when the sIL-6R levels exceeded those of IL-6 on a molar level and show that the colon is a major target of IL-6 trans-signaling.
EXPERIMENTAL PROCEDURES
Cells and Reagents
Ba/F3-gp130 cells transduced with human gp130 were obtained from Immunex (Seattle, WA) (20). Ba/F3-gp130-hIL-6R cells (21) and Ba/F3-gp130-mIL-6R cells (22) have been described previously. All cells were grown in DMEM high glucose culture medium (PAA Laboratories, Cölbe, Germany) supplemented with 10% fetal bovine serum, penicillin (60 mg/liter), and streptomycin (100 mg/liter) at 37 °C with 5% CO2 in a water-saturated atmosphere. Ba/F3-gp130 cells were cultured using 10 ng/ml recombinant Hyper-IL-6, which is a fusion protein of IL-6 and the sIL-6R that mimics IL-6 trans-signaling and acts as a growth factor for Ba/F3-gp130 cells (11, 23). Hyper-IL-6 was expressed and purified as described previously (11). Ba/F3-gp130-hIL-6R cells and Ba/F3-gp130-mIL-6R cells were cultured using 10 ng/ml recombinant human IL-6 instead of Hyper-IL-6. Human IL-6 and soluble human IL-6R were expressed and purified as described previously (24). Murine IL-6 and soluble murine IL-6R were purchased from R & D Systems (Minneapolis, MN). The anti-hIL-6R mAb tocilizumab (RoACTEMRA) was obtained from Roche Applied Science. The anti-mIL-6R mAb 25F10 was described previously (17). sgp130Fc and His-tagged sgp130 were expressed in CHO cells and purified as described previously (13). Anti-phospho-STAT3 mAb (Tyr705) and anti-STAT3 mAb (124H6) were purchased from Cell Signaling Technology (Frankfurt, Germany), and anti-β-actin mAb (sc-47778) was from Santa Cruz Biotechnology (Heidelberg, Germany). The peroxidase-conjugated secondary antibodies were purchased from Pierce.
Mice
8–12-week-old C57BL/6N wild-type mice were obtained from Charles River Laboratories (Sulzfeld, Germany). For all experiments, five male mice/group were used. All of the experiments were performed according to the German guidelines for animal care and protection. 5 μg of IL-6, 1 μg of Hyper-IL6, and 250 μg of sgp130Fc or the appropriate volume of the vehicle PBS per mouse were injected intraperitoneally. Cytokines were preincubated in vitro at 37 °C for 30 min before intraperitoneal injection. Mice were sacrificed 90 min thereafter. IL-6R (CD126)-deficient (CD126−/−) mice were described previously (14).
Calculation of Free and Complexed IL-6
To determine the amounts of free IL-6 and IL-6 complexed with sIL-6R, we used the formula described previously (26): [IL-6·sIL-6R] = 0.5[sIL-6R]i + 0.5[IL-6]i + 0.5KD1 − 0.5([sIL-6R]i2 + [IL-6]i2 + 2[IL-6]iKD1 + KD12)0.5. [sIL-6R]i and [IL-6]i represent the initially used concentrations, KD1 for the formation of IL-6·IL-6R complexes was determined to be 500 pm (27). The amount of free IL-6 was calculated by subtracting the result of the formula above from the IL-6 concentration used initially.
Proliferation Assays
Ba/F3 cells were washed three times with sterile PBS and resuspended in DMEM containing 10% FBS at a final concentration of 5 × 103 cells/well in a 96-well plate. The cells were incubated for 2 days as indicated with the cytokines and cytokine receptors in a final volume of 100 μl. For inhibitory studies, IL-6R targeting antibodies, sgp130 or sgp130Fc were added as indicated. After 2 days, cell growth was measured using the Cell Titer Blue Cell viability assay reagent (Promega, Karlsruhe, Germany) following the manufacturer's protocol. The extinction was measured using a Lambda FLUORO 320 microplate fluorescence reader (excitation filter, 530/25; emission filter, 590/35; sensitivity, 75; Software KC4; BioTek Instruments, Winooski, VT). Normalization of relative light units was achieved by subtraction of negative control values. All of the values were measured in triplicate per experiment.
Western Blotting
Organs from mice were homogenized in lysis buffer (500 mm NaCl, 50 mm Tris, pH 7.4, 0.1% SDS, 1% Nonidet P-40) containing complete protease inhibitor mixture (Roche Applied Science). 40 μg of total protein were subjected to SDS-PAGE gel electrophoresis. For cell culture experiments, ∼2 × 107 Ba/F3 cells/experiment were washed three times with sterile PBS. The cells were distributed to 2-ml tubes and starved in FBS-free medium for 4 h at 37 °C and CO2 saturation under constant shaking. The cells were stimulated with the indicated cytokines for 10 min followed by centrifugation at 4 °C and 2,000 rpm for 10 min and transfer to liquid nitrogen. 50 μl of 2.5× Laemmli buffer (92.5 mm Tris-HCl, pH 6.8, 25% glycerol, 5% SDS, 2.5% β-mercaptoethanol, and 0.025% bromphenol blue) were added to each tube, and the cells were lysed by boiling at 95 °C for 10 min. The proteins were separated by SDS-PAGE and transferred to a PVDF membrane using a Trans-Blot SD semi-dry transfer cell (Bio-Rad). The membrane was blocked in 5% low fat milk in TBS-T (10 mm Tris-HCl, pH 7.6, 150 mm NaCl, and 1% Tween 20) and probed with the primary antibody in 1% low fat milk in TBS-T (STAT3, β-actin mAb) or 5% BSA (pSTAT3-mAb) at 4 °C overnight. The blots were washed and incubated with the secondary peroxidase-conjugated antibody for 1 h before applying the ECL-plus peroxidase substrate (GE Healthcare). The Fluor ChemQ system (Cell Biosciences, Santa Clara, CA) was used for signal detection according to the manufacturer's instructions. The membranes were stripped with stripping buffer (20 ml of 10% SDS, 12.5 ml of 0.5 m Tris-HCl, pH 6.8, 67.5 ml of ultra-pure water, 0.8 ml of β-mercaptoethanol), blocked again, and probed with another primary antibody.
FACS Analysis
CD4+ T cells were stimulated with mouse IL-6 (20 ng/ml) or IL-6·sIL-6R (10 and 200 ng/ml, respectively) in the presence of 10−1–104 ng/ml sgp130Fc for 15 min. For intracellular staining of phosphorylated tyrosine residues of STAT1 and STAT3, murine T cells were fixed in 2% (w/v) paraformaldehyde at 37 °C for 15 min, followed by permeabilization of cells in 90% (v/v) methanol for 30 min on ice. The cells were then stained for CD4, CD3, and phosphorylated STAT1 (clone 4a) or STAT3 (clone 4/P-STAT3) purchased from BD Biosciences. Flow cytometry was performed on cells acquired using a CyAn ADP (Beckman Coulter), and data were analyzed using FlowJo (Tree Star, Ashland, OR) software.
Tissue Processing and Immunohistochemistry
Liver, lung, and colon tissue was fixed in 4% formaline, processed, and immunostained. Staining for STAT3 phosphorylation was carried out using anti-pSTAT3 mAb (New England Biolabs, Frankfurt am Main, Germany) diluted 1:3,000, and the signal was amplified using the tyramide signal amplification kit (PerkinElmer Life Sciences), developed as described before, differentiated in 0.5% acetic acid, rinsed in tap water, and stained with Giemsa's azur eosin methylene blue solution (Merck). STAT3 phosphorylation was quantified by counting magenta nuclear color reaction in 10 random high power fields at 20× magnification in five mice/group.
Statistical Analysis
For animal experiments, five mice/experimental group were used. The data are expressed as the mean values ± standard deviation calculated from at least three independent experiments unless otherwise stated. Statistical analysis was performed using Student's unpaired t test. A p value below 0.05 was considered statistically significant. One asterisk (*) represents a p value below 0.05, two (**) represent a p value below 0.01, and three (***) represent a p value below 0.001. Calculation of the effective dose for 50% growth reduction (IC50) was done using the GraphPad Prism 4 software.
RESULTS
The Inhibitory Profile of Anti-IL-6R mAb Tocilizumab Is Independent of Membrane-bound IL-6R Expression
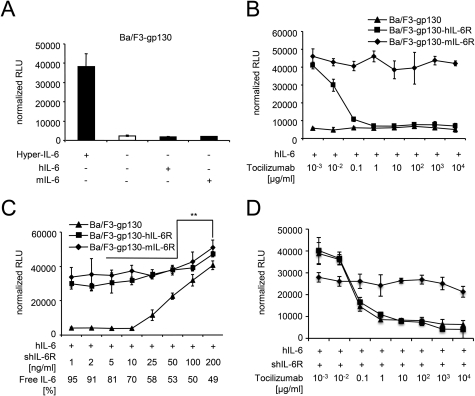
The paradigm of IL-6 trans-signaling explains how cells can be stimulated by IL-6 and soluble IL-6R. Tocilizumab, which binds to the IL-6R, inhibits both classic and trans-signaling. However, it was not investigated so far whether expression of the membrane-bound IL-6R influenced the inhibitory profile of tocilizumab under conditions promoting IL-6 trans-signaling. Because IL-6 signaling is specifically induced upon binding of IL-6 to membrane-bound IL-6R or soluble IL-6R and subsequent gp130 transmembrane complex formation, both classic and trans-signaling might operate in parallel in cells expressing gp130 and membrane-bound IL-6R. Here, we analyzed how Ba/F3 cells expressing gp130 with and without membrane-bound IL-6R react to the treatment of tocilizumab under conditions promoting IL-6 classic or trans-signaling. After stable transfection with human gp130 (referred to as Ba/F3-gp130), proliferation of Ba/F3-gp130 cells was dependent on IL-6 trans-signaling, and after additional introduction of murine or human IL-6R (referred to Ba/F3-gp130-mIL-6R and Ba/F3-gp130-hIL-6R, respectively) it was dependent on both classic and trans-signaling (Fig. 1, A and B).
FIGURE 1.
The inhibitory profile of tocilizumab is independent of IL-6R expression. A, equal numbers of Ba/F3-gp130 cells were cultured for 2 days with 10 ng/ml Hyper-IL-6, 10 ng/ml human, or 10 ng/ml murine IL-6. As a control, Ba/F3-gp130 cells were cultured without any cytokine. B, equal numbers of Ba/F3-gp130, Ba/F3-gp130-hIL-6R, or Ba/F3-gp130-mIL-6R cells were cultured for 2 days with 10 ng/ml human IL-6 and increasing concentrations of the anti-human-IL-6R antibody tocilizumab (0.001–10,000 μg/ml). C, equal numbers of Ba/F3-gp130 or Ba/F3-gp130-hIL-6R cells were cultured for 2 days with 10 ng/ml hIL-6 plus increasing concentrations of shIL-6R as indicated. D, equal numbers of Ba/F3-gp130, Ba/F3-gp130-hIL-6R, or Ba/F3-gp130-mIL-6R cells were cultured for 2 days with 10 ng/ml human IL-6 plus 200 ng/ml shIL-6R and increasing concentrations of the anti-human-IL-6R antibody tocilizumab (0.001–10,000 μg/ml). Statistical analysis was carried out using a paired Student's t test, and statistical significance was p < 0.01 (**). The proliferation measured with the lowest inhibitor concentration was about the same as without inhibitor.
The IC50 for tocilizumab-mediated inhibition of Ba/F3-gp130-IL-6R cell proliferation stimulated with 10 ng/ml hIL-6 was 13.5 ± 2.2 ng/ml (Fig. 1B and Table 1). Ba/F3-gp130 cells expressing no membrane-bound IL-6R served as negative control (Fig. 1B).
TABLE 1.
Inhibitory profile of tocilizumab, 25F10, and sgp130Fc
The data were obtained with 10 ng/ml Hyper-IL-6 or 10 ng/ml IL-6 plus 200 ng/ml sIL-6R. The IC50 values were calculated from three individual experiments and correspond to tocilizumab, 25F10 mAb, and sgp130Fc, respectively.
| Inhibitor | Ba/F3 cell receptor composition | Cytokine stimulation | Classic signaling | Trans-signaling | IC50 |
|
|---|---|---|---|---|---|---|
| ng/ml | nm | |||||
| Tocilizumab (classic and trans-signaling) | gp130 | hIL-6/shIL-6R | No | Yes | 39.7 ± 2.8 | 0.27 ± 0.02 |
| gp130 + hIL-6R | hIL-6 | Yes | No | 13.5 ± 2.2 | 0.09 ± 0.015 | |
| hIL-6/shIL-6R | Yes | Yes | 67 ± 11.4 | 0.45 ± 0.08 | ||
| gp130 + mIL-6R | hIL-6 | Yes | No | No inhibition | ||
| hIL-6/shIL-6R | Yes | Yes | No inhibition | |||
| 25F10 mAb (selective for trans-signaling) | gp130 | mIL-6/smIL-6R | No | Yes | 967.1 ± 222.7 | 6.5 ± 1.5 |
| gp130 + hIL-6R | mIL-6/smIL-6R | No | Yes | 1226.2 ± 254.5 | 7.6 ± 1.7 | |
| gp130 + mIL-6R | mIL-6 | Yes | No | No inhibition | ||
| mIL-6/smIL-6R | Yes | Yes | 1032.4 ± 249 | 7 ± 1.7 | ||
| sgp130Fc (selective for trans-signaling) | gp130 | Hyper-IL-6 | No | Yes | 12.5 ± 2 | 0.067 ± 0.010 |
| hIL-6/shIL-6R | No | Yes | 0.5 ± 0.05 | 0.003 ± 0.001 | ||
| mIL-6/smIL-6R | No | Yes | 0.1 ± 0.09 | 0.001 ± 0.001 | ||
| gp130 + hIL-6R | Hyper-IL-6 | No | Yes | 9.7 ± 1.5 | 0.05 ± 0.008 | |
| hIL-6 | Yes | No | No inhibition | |||
| hIL-6/shIL-6R | Yes | Yes | 155.2 ± 20.1 | 0.83 ± 0.11 | ||
| mIL-6 | No | No | No proliferation | |||
| mIL-6/smIL-6R | No | Yes | 0.07 ± 0.01 | 0.001 ± 0.001 | ||
| gp130 + mIL-6R | hIL-6 | Yes | No | No inhibition | ||
| hIL-6/shIL-6R | Yes | Yes | 15.4 ± 4.1 | 0.08 ± 0.02 | ||
| mIL-6 | Yes | No | No inhibition | |||
| mIL-6/smIL-6R | Yes | Yes | 177.2 ± 64.1 | 0.95 ± 0.34 | ||
Next, a dose-response analysis with 10 ng/ml IL-6 and increasing amounts of sIL-6R (1–200 ng/ml) revealed that 50–200 ng/ml sIL-6R induced IL-6 trans-signaling in Ba/F3-gp130 cells lacking IL-6R expression. Importantly, IL-6-induced proliferation of Ba/F3-gp130-IL-6R cells expressing membrane-bound IL-6R was significantly enhanced by the addition of 200 ng/ml sIL-6R, indicating that both signaling pathways, classic and trans-signaling, can act in parallel (Fig. 1C). The same has been shown for HepG2 cells, which express gp130 and IL-6R (24).
Based on the KD of IL-6 to IL-6R of ∼500 pm (27) and using the equation (26) described under “Experimental Procedures,” we have calculated that even if the sIL-6R is present in molar excess to IL-6, the equilibrium of free IL-6 versus trapped IL-6 in IL-6·sIL-6R complexes will not completely eliminate all free IL-6 molecules, supporting our hypothesis that IL-6 classic and trans-signaling can act in parallel (Fig. 1C). For example, a 10-fold molar excess of sIL-6R over IL-6 (10 ng/ml IL-6 and 200 ng/ml sIL-6R, molecular mass of ∼20 kDa for IL-6 and ∼40 kDa for sIL-6R) should result in ∼28% free IL-6 molecules and 72% IL-6 molecules trapped in biologically active IL-6·sIL-6R complexes. Importantly, these concentrations reflect average IL-6·sIL-6R levels found under inflammatory conditions in human serum (29) and were used to analyze the inhibitory profile of tocilizumab, the anti-trans-signaling mAb 25F10 and sgp130Fc.
The concentrations of tocilizumab needed for 50% growth reduction of Ba/F3-gp130 and Ba/F3-gp130-hIL-6R stimulated with 10 ng/ml hIL-6 and 200 ng/ml shIL-6R were 39.7 ± 2.8 and 57 ± 4.5 ng/ml, respectively (Fig. 1D and Table 1) and revealed no statistically significant differences. As a control, tocilizumab did not inhibit proliferation of Ba/F3-gp130-mIL-6R cells stimulated with IL-6 and sIL-6R (Fig. 1D). We conclude from these experiments that under conditions in which classic- and trans-signaling occur in parallel, tocilizumab inhibits IL-6 classic and trans-signaling with comparable kinetics, irrespective of the expression of membrane-bound IL-6R on the target cell.
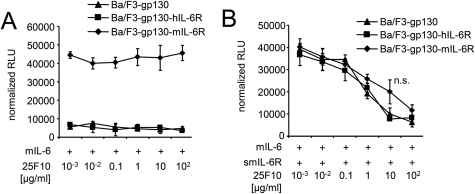
The Inhibitory Profile of the Anti-trans-signaling mAb 25F10 Is Independent of Membrane-bound IL-6R Expression
Recently, the anti-murine IL-6 trans-signaling mAb 25F10 was described (17). Here, the inhibitory properties of 25F10 were analyzed using Ba/F3-gp130, Ba/F3-gp130-hIL-6R, and Ba/F3-gp130-mIL-6R cells. First, we confirmed that 25F10 did not inhibit IL-6 classic signaling on Ba/F3-gp130-mIL-6R cells stimulated with murine IL-6 (Fig. 2A). 25F10 inhibited IL-6 signaling on Ba/F3-gp130, Ba/F3-gp130-hIL-6R, and BaF/3-gp130-mIL-6R cells stimulated with 10 ng/ml mIL-6 and 200 ng/ml smIL-6R in a dose-dependent manner with a IC50 values of 961.1 ± 222.7, 1226.4 ± 254.5, and 1032.4 ± 249 ng/ml, respectively (Fig. 2B and Table 1). Surprisingly, 25F10 inhibited proliferation of Ba/F3-gp130-mIL-6R only slightly less effectively as compared with Ba/F3-gp130 cells (IC50 Ba/F3-gp130-mIL-6R = 1032.4 ± 249 ng/ml versus IC50 Ba/F3-gp130 = 961.1 ± 222.7 ng/ml). We conclude from these experiments that 25F10 actively promotes the formation of IL-6·sIL-6R/25F10 complexes, because 25F10 was also shown to bind to sIL-6R in the absence of IL-6 (17).
FIGURE 2.
The inhibitory profile of the anti-trans-signaling antibody 25F10 is independent of IL-6R expression. A, equal numbers of Ba/F3-gp130, Ba/F3-gp130-hIL-6R, or Ba/F3-gp130-mIL-6R cells were cultured for 2 days with 10 ng/ml mIL-6 and with different concentrations of the anti-murine-IL-6R trans-signaling antibody 25F10 (0.001–100 μg/ml). B, equal numbers of Ba/F3-gp130, Ba/F3-gp130-hIL-6R, or Ba/F3-gp130-mIL-6R cells were cultured for 2 days with 10 ng/ml mIL-6 plus 200 ng/ml smIL-6R and with different concentrations of the anti-murine-IL-6R trans-signaling antibody 25F10 (0.001–100 μg/ml). Statistical analysis was carried out using a paired Student's t test. n.s., not significant. The proliferation measured with the lowest inhibitor concentration was about the same as without inhibitor.
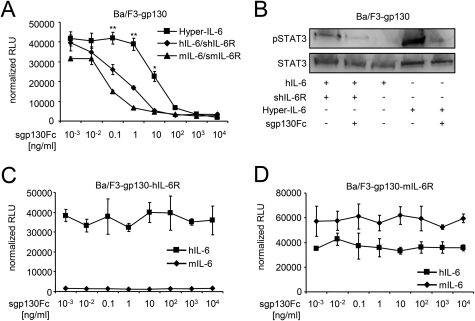
Inhibition of Classic and Trans-signaling by sgp130Fc Is Controlled by IL-6/sIL-6R Ratios
Murine and human IL-6·sIL-6R-induced as well as Hyper-IL-6-induced proliferation was inhibited by sgp130Fc in a dose-dependent manner on Ba/F3-gp130 cells (IC50 hIL-6·hsIL-6R = 0.5 ± 0.05 ng/ml; IC50 mIL-6·msIL-6R = 0.1 ± 0.09 ng/ml; IC50 Hyper IL-6 = 12.5 ± 2 ng/ml]) (Fig. 3A and Table 1). Accordingly, high concentrations of sgp130Fc (10 μg/ml) were able to completely block hIL-6·shIL-6R and Hyper-IL-6-induced STAT3 phosphorylation of Ba/F3-gp130 cells (Fig. 3B). As a control, we verified that sgp130Fc was not able to inhibit IL-6 classic signaling on Ba/F3-gp130-hIL6R (Fig. 3C) or Ba/F3-gp130-mIL6R cells (Fig. 3D) stimulated with human or murine IL-6 alone. mIL-6 failed to induce proliferation of Ba/F3-gp130-hIL6R, because mIL-6 cannot bind to the hIL-6R (13). Inhibition of Hyper-IL-6-induced proliferation needed higher sgp130Fc concentrations compared with IL-6 plus sIL-6R, because all Hyper-IL-6 molecules are biologically active to induce trans-signaling, whereas in the stimulation with IL-6 and sIL-6R, only a minority of molecules are biologically active in IL-6·sIL-6R complexes, with the majority present as free IL-6 and sIL-6R.
FIGURE 3.
sgp130Fc specifically inhibits IL-6 trans-signaling on cells lacking membrane-bound IL-6R. A, equal numbers of Ba/F3-gp130 cells were cultured for 2 days with 10 ng/ml Hyper-IL-6, 10 ng/ml hIL-6 plus 200 ng/ml shIL-6R, or 10 ng/ml mIL-6 plus 200 ng/ml smIL-6R. Inhibition of growth was achieved by increasing concentrations of sgp130Fc (0.001–10,000 ng/ml). B, equal numbers of Ba/F3-gp130 cells were incubated with the indicated cytokines for 30 min after 6 h of serum starvation. Western blot analysis of cell lysates was performed with an antibody against phospho-STAT3 and against STAT3 as loading control. C and D, equal numbers of Ba/F3-gp130-hIL-6R or Ba/F3-gp130-mIL-6R cells were cultured for 2 days with either 10 ng/ml human or murine IL-6 plus increasing concentrations of sgp130Fc (0.001–10,000 ng/ml). Statistical analysis was carried out using a paired Student's t test, and statistical significance was p < 0.05 (*) or p < 0.01 (**). The proliferation measured with the lowest inhibitor concentration was about the same as without inhibitor.
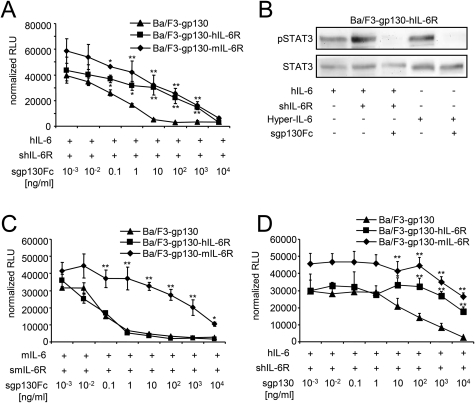
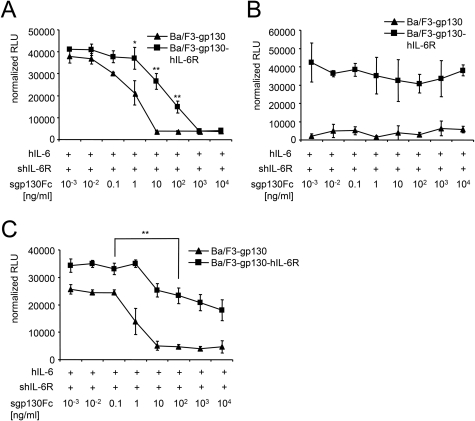
hIL-6·shIL-6R-induced (10 and 200 ng/ml) proliferation of Ba/F3-gp130 was inhibited by sgp130Fc (IC50 = 0.5 ± 0.05 ng/ml). The proliferation of Ba/F3-gp130-hIL-6R and BaF/3-gp130-mIL-6R was also completely inhibited by sgp130Fc (IC50 hIL-6R = 155.2 ± 20.1 ng/ml and IC50 mIL-6R = 15.4 ± 4.1 ng/ml) (Fig. 4A and Table 1), albeit 300-fold and 30-fold less efficient as compared with Ba/F3-gp130 cells, respectively. In line with these findings, high concentrations of sgp130Fc (10 μg/ml) were able to block hIL-6·shIL-6R and Hyper-IL-6-induced STAT3 phosphorylation of Ba/F3-gp130-hIL-6R cells (Fig. 4B), which express ∼2.5-fold more membrane-bound IL-6R than gp130 (30). Because all in vivo experiments were performed with human sgp130Fc in mice, we validated our results with 10 ng/ml murine IL-6 and 200 ng/ml murine sIL-6R. mIL-6·smIL-6R-induced proliferation of Ba/F3-gp130, Ba/F3-gp130-hIL-6R, and Ba/F3-gp130-mIL-6R cells was inhibited by sgp130Fc with IC50 values of 0.01 ± 0.09, 0.07 ± 0.01, and 15.4 ± 4.1 ng/ml, respectively (Fig. 4C and Table 1), revealing that Ba/F3-mIL-6R cells needed ∼200-fold higher sgp130Fc protein concentrations for 50% growth inhibition as compared with Ba/F3-gp130 cells. This result supports the data obtained with human IL-6. The IC50 sgp130Fc of mIL-6·smIL-6R was the same for Ba/F3-gp130 and Ba/F3-gp130-hIL-6R cells because mIL-6 cannot bind to human IL-6R (13). On these cells, mIL-6-induced proliferation was solely dependent on IL-6 trans-signaling (Fig. 4C). Compared with the anti-IL-6 trans-signaling mAb 25F10, sgp130Fc inhibited IL-6 trans-signaling on Ba/F3-gp130 cells at a ∼10,000-fold lower concentration (IC50 25F10 = 967.1 ± 222.7 ng/ml and IC50 sgp130Fc = 0.1 ± 0.09 ng/ml).
FIGURE 4.
Inhibition of classic signaling by sgp130 on Ba/F3-gp130-IL-6R cells. A, equal numbers of Ba/F3-gp130, Ba/F3-gp130-hIL-6R, or Ba/F3-gp130-mIL-6R cells were cultured for 2 days with 10 ng/ml hIL-6 plus 200 ng/ml shIL-6R and with increasing concentrations of sgp130Fc (0.001–10,000 ng/ml). B, equal numbers of Ba/F3-gp130-hIL-6R cells were incubated with the indicated cytokines for 30 min after 6 h of serum starvation. Western blot analysis of cell lysates was performed with an antibody against phospho-STAT3 and against STAT3 as loading control. C, equal numbers of Ba/F3-gp130, Ba/F3-gp130-hIL-6R, or Ba/F3-gp130-mIL-6R cells were cultured for 2 days with 10 ng/ml hIL-6 plus 200 ng/ml shIL-6R and with increasing concentrations of sgp130Fc (0.001–10,000 ng/ml). D, equal numbers of Ba/F3-gp130, Ba/F3-gp130-hIL-6R, or Ba/F3-gp130-mIL-6R cells were cultured for 2 days with 10 ng/ml hIL-6 plus 200 ng/ml shIL-6R and with increasing concentrations of sgp130 (0.001–10,000 ng/ml). Statistical analysis was carried out using a paired Student's t test, and statistical significance was p < 0.05 (*) or p < 0.01 (**). The proliferation measured with the lowest inhibitor concentration was about the same as without inhibitor.
Next, hIL-6·shIL-6R-induced proliferation of Ba/F3-gp130 cells was inhibited by the naturally occurring sgp130, which is not dimerized by an Fc part, with an IC50 of 50.13 ± 42.95 ng/ml (Fig. 4D), which was ∼100 times lower compared with sgp130Fc and is in good agreement with previous reports (14). However, the highest concentration of sgp130 of 10 μg/ml was not sufficient to completely block IL-6·sIL-6R-induced proliferation, again showing that higher concentrations of sgp130 irrespective of the Fc part are needed to block IL-6·sIL-6R signaling on cells expressing IL-6R compared with cells lacking IL-6R expression.
To reduce the amount of free IL-6 molecules, we used a ∼150-fold molar excess of sIL-6R over IL-6. Again, the same Ba/F3 cell lines were used and stimulated with 3.3 ng/ml hIL-6 and 1000 ng/ml shIL-6R. hIL-6·shIL-6R-induced proliferation of Ba/F3-gp130 and Ba/F3-gp130-hIL-6R cells was inhibited by sgp130Fc with IC50 values of 0.6 ± 0.3 and of 15.2 ± 7.3 ng/ml, respectively (Fig. 5A). In this scenario, sgp130Fc was ∼25-fold more efficient to inhibit proliferation of cells expressing only gp130 compared with cells expressing gp130 and IL-6R.
FIGURE 5.
Inhibition of classic signaling by sgp130Fc is dependent on the ratio of IL-6 to sIL-6R on Ba/F3-gp130-IL-6R cells. A, equal numbers of Ba/F3-gp130 or Ba/F3-gp130-hIL-6R cells were cultured for 2 days with 3.3 ng/ml hIL-6 plus 1000 ng/ml shIL-6R and with increasing concentrations of sgp130Fc (0.001–10,000 ng/ml). B, equal numbers of Ba/F3-gp130 or Ba/F3-gp130-hIL-6R cells were cultured for 2 days with 10 ng/ml hIL-6 plus 10 ng/ml shIL-6R and with increasing concentrations of sgp130Fc (0.001–10,000 ng/ml). C, equal numbers of Ba/F3-gp130 or Ba/F3-gp130-hIL-6R cells were cultured for 2 days with 10 ng/ml hIL-6 plus 50 ng/ml shIL-6R and with increasing concentrations of sgp130Fc (0.001–10,000 ng/ml). Statistical analysis was carried out using a paired Student's t test, and statistical significance was p < 0.05 (*) or p < 0.01 (**). The proliferation measured with the lowest inhibitor concentration was about the same as without inhibitor.
Next, we stimulated Ba/F3-gp130 and Ba/F3-gp130-hIL-6R cells with 10 ng/ml IL-6 and 10 or 50 ng/ml sIL-6R, respectively. As shown in Fig. 1C, 10 ng/ml IL-6 and 50 ng/ml sIL-6R were barely sufficient to induce IL-6 trans-signaling on Ba/F3-gp130 cells, whereas 10 ng/ml sIL-6R was not. Accordingly, proliferation of Ba/F3-gp130-hIL-6R cells stimulated with 10 ng/ml IL-6 and 10 ng/ml sIL-6R was not inhibited by sgp130Fc, because under these conditions, cells only proliferate in response to classic signaling, and sIL-6R levels only trapped a minority of the free IL-6 molecules in IL-6·sIL-6R·sgp130Fc complexes (Fig. 5B). In contrast, proliferation of Ba/F3-gp130 cells induced with 10 ng/ml IL-6 and 50 ng/ml sIL-6R was inhibited by sgp130Fc in a dose-dependent manner (IC50 = 0.4 ± 0.3 ng/ml; Fig. 5C). Proliferation of Ba/F3-gp130-IL-6R cells was inhibited by sgp130Fc with an IC50 of 5.2 ± 2.5 ng/ml but only by ∼50%, indicating that sIL-6R levels were also not sufficient to trap all IL-6 molecules in IL-6·sIL-6R·sgp130Fc complexes (Fig. 5C). However, the IC50 for Ba/F3-gp130-hIL-6R was ∼14-fold higher than for Ba/F3-gp130 cells, which express no membrane-bound IL-6R.
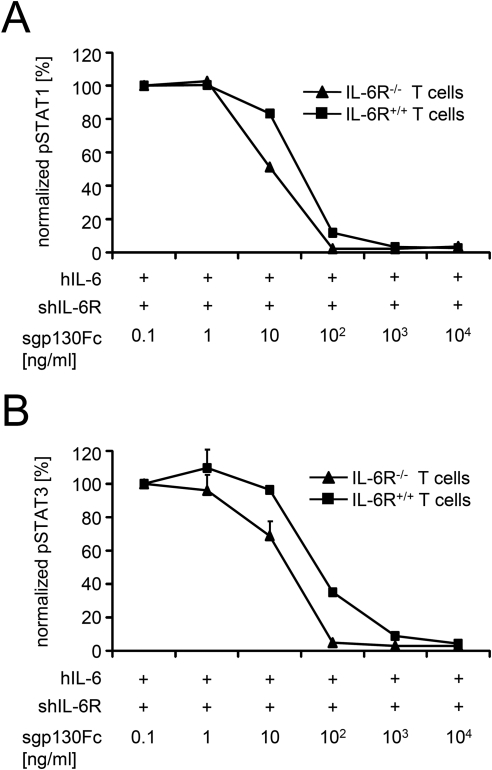
To confirm our results generated in in vitro experiments with overexpression of gp130 and IL-6R, we made use of the recently developed IL-6R-deficient (CD126−/−) mice, which were generated by disruption of exons 4, 5, and 6 that encode the IL-6 recognition sites of the IL-6R (14). Splenic CD4+ T cells from IL-6R-deficient mice, but not from wild-type mice, did not respond to murine IL-6 alone and showed no increase in STAT1 or STAT3 phosphorylation following direct stimulation with 20 ng/ml IL-6 (supplemental Fig. S1), indicating that T cells of wild-type mice but not of IL-6R-deficient mice express IL-6R. Phosphorylation of STAT1 and STAT3 was only observed in T cells from IL-6R-deficient mice when IL-6 trans-signaling was reconstituted using 10 ng/ml IL-6 and 200 ng/ml sIL-6R (Fig. 6 and supplemental Figs. S2 and S3). Phosphorylation of STAT1 and STAT3 of T cells from wild-type mice and from IL-6R-deficient mice was also completely inhibited by sgp130Fc (IC50 STAT1/wild type = 26.8 ± 1.7 ng/ml, IC50 STAT3/wild type = 52.3 ± 1.7 ng/ml, and IC50 STAT1/IL-6R-deficient = 9.3 ± 0.4 ng/ml, IC50 STAT3/IL-6R-deficient = 17.1 ± 5 ng/ml) (Fig. 6). Importantly, even on primary T cells that express less IL-6R than gp130, inhibition of STAT1/STAT3 phosphorylation was at least ∼3-fold less efficient on IL-6R-deficient T cells as compared with T cells from wild-type mice expressing membrane-bound IL-6R.
FIGURE 6.
Inhibition of classic signaling by sgp130Fc on primary T cells from wild-type and IL-6R-deficient mice. A, CD4+ T cells were stimulated with a combination of IL-6 (10 ng/ml) and sIL-6R (200 ng/ml) for 15 min in the presence of 10−1–104 ng/ml sgp130Fc. STAT1 activation was determined by flow cytometry and normalized. B, CD4+ T cells were stimulated with a combination of IL-6 (10 ng/ml) and sIL-6R (200 ng/ml) for 15 min in the presence of 10−1–104 ng/ml sgp130Fc. STAT3 activation was determined by flow cytometry and normalized. The proliferation measured with the lowest inhibitor concentration was about the same as without inhibitor.
We conclude from these experiments that increasing concentrations of sgp130(Fc) affect the equilibrium of free IL-6 versus IL-6 trapped in IL-6·sIL-6R complexes, and with increasing concentrations, sgp130(Fc) is able to trap all free IL-6 molecules in biologically inactive IL-6·sIL-6R·sgp130Fc complexes. Our results show for the first time that sgp130(Fc) can inhibit both classic and trans-signaling when sIL-6R levels exceed those of IL-6 on a molar level.
Physiological Levels of sIL-6R Are Sufficient to Induce IL-6 Trans-signaling
Endogenous sgp130 was suggested to be the natural inhibitor of IL-6 trans-signaling in vivo to prevent systemic activation by IL-6 trans-signaling by buffering IL-6·sIL-6R in biologically inactive IL-6·sIL-6R·sgp130 complexes. If this assumption is correct, a simple injection of IL-6 into mice should lead to the following situation: (i) IL-6 will form complexes with endogenous sIL-6R and thereafter with sgp130, which blocks IL-6 trans-signaling, and (ii) free IL-6 (molar excess of injected IL-6 over endogenous sIL-6R) will induce IL-6 classic signaling in cells such as hepatocytes that express membrane-bound IL-6R (31). If this assumption is incorrect, injection of IL-6 will also lead to trans-signaling in classic signaling unresponsive cells, which in turn should be inhibited by preventive injection of recombinant sgp130Fc.
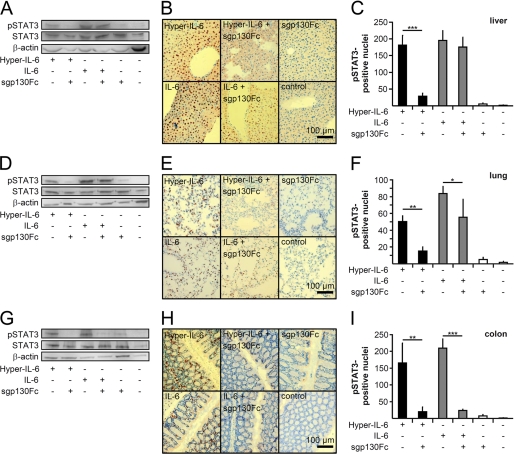
The inhibitory capacity of endogenous sgp130 and of recombinant sgp130Fc to inhibit IL-6 trans-signaling was determined for liver, lung, and colon. Therefore, 5 μg of IL-6 and, as a control, 1 μg of Hyper-IL-6 with and without 250 μg of sgp130Fc were injected intraperitoneally in 8–12-week-old C57Bl/6N mice. The majority of injected human IL-6 has a very short serum half-life of ∼3 min with ∼80% found in the liver of rats (32), which likely explains why comparably high doses of IL-6 were needed to obtain sustained IL-6 responses in mice (33). Previously, we demonstrated that intraperitoneal injection of sgp130Fc results in amounts of approximately one-fifth to one-tenth of the totally injected sgp130Fc in the serum (34).
STAT3 phosphorylation was detected in liver, lung, and colon by Western blotting and immunohistochemistry 90 min after cytokine injection (Fig. 7). As expected, Hyper-IL-6-induced STAT3 phosphorylation was significantly inhibited by sgp130Fc in all organs examined, indicating that the amount of sgp130Fc was sufficient to competitively block Hyper-IL-6-induced trans-signaling. However, sgp130Fc did not inhibit IL-6-induced STAT3 phosphorylation in liver and only partially in lung (Fig. 7, A–C and D–F, respectively), suggesting that IL-6-induced STAT3 phosphorylation in the liver and lung is mainly mediated by IL-6 classic signaling via the endogenous membrane-bound IL-6R and not via the soluble IL-6R. In the colon, both IL-6- and Hyper-IL-6-induced STAT3 phosphorylation was strongly inhibited by the sgp130Fc protein (Fig. 7, G–I), suggesting that the colon is a target of IL-6 trans-signaling via the endogenous soluble IL-6R. Moreover, at least under our experimental conditions, the level of endogenous sgp130 protein was not sufficient to prevent IL-6 trans-signaling responses in the colon. It is, however, conceivable that endogenous sgp130 might inhibit IL-6 trans-signaling at lower IL-6 concentrations in the pg/ml range.
FIGURE 7.
Physiological levels of endogenous sIL-6R induce IL-6 trans-signaling in colon but not in liver or lung. A, Western blot analysis of STAT3 phosphorylation in the liver after IL-6 and Hyper-IL-6 injection with or without the addition of sgp130Fc. A representative example of five individual experiments is shown. B, immunohistochemical pSTAT3 staining of representative photomicrographs of liver sections from corresponding mice treated as described in A. Scale bar, 100 μm. C, quantification of STAT3 phosphorylation from 10 random high power fields of five mice/group (magnification, 20×). D, Western blot analysis of STAT3 phosphorylation in the lung after IL-6 and Hyper-IL-6 injection with or without the addition of sgp130Fc. A representative example of five individual experiments is shown. E, immunohistochemical pSTAT3 staining of representative photomicrographs of lung sections from corresponding mice treated as described in D. Scale bar, 100 μm. F, quantification of STAT3 phosphorylation from 10 random high power fields of five mice/group (magnification, 20×). G, Western blot analysis of STAT3 phosphorylation in the colon after IL-6 and Hyper-IL-6 injection with or without the addition of sgp130Fc. A representative example of five individual experiments is shown. H, immunohistochemical pSTAT3 staining of representative photomicrographs of colon sections from corresponding mice treated as described in G. Scale bar, 100 μm. I, quantification of STAT3 phosphorylation from 10 random high power fields of five mice/group (magnification, 20×). Statistical analysis was carried out using a paired Student's t test, and statistical significance was p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***).
DISCUSSION
In the present study, we show that the colon, but not liver or lung, is a major target of IL-6 trans-signaling in vivo. IL-6 trans-signaling was induced by injection of IL-6 into mice. Circulating IL-6 formed IL-6·sIL-6R complexes with endogenous sIL-6R. Our findings are in line with previous reports showing IL-6R expression in the lung and liver (33, 35, 36). Importantly, the colon seemed to be a target of IL-6 trans-signaling under our conditions, even though it has been reported that intestinal epithelial cells can express membrane-bound IL-6R (37). This is of particular importance, because a recent report suggested a role for IL-6 in the regeneration of intestinal epithelial cells in a dextran/sodium/sulfate-induced colitis (38). Previously, it has been reported that circulating IL-6 is rapidly removed from the circulation via the liver. Our finding might be explained by a higher level of local sIL-6R in the gut or the relatively low amounts of IL-6 that reach intestinal epithelial cells. Moreover, the endogenous sgp130 in mice cannot inhibit IL-6 trans-signaling when IL-6 levels exceed physiological sIL-6R levels. Inhibition of IL-6 trans-signaling was, however, achieved by preventive injection of sgp130Fc.
Because the sIL-6R serum levels exceed the levels of IL-6 in normal physiology and also in many pathophysiological situations (29, 34), we decided to mimic these conditions in a cell-based assay to test the inhibitory properties of sgp130Fc on cells lacking or expressing membrane-bound IL-6R. In cells lacking IL-6 receptor expression, trans-signaling was inhibited at comparably low concentrations of sgp130Fc. For Ba/F3-gp130 cells stimulated with 10 ng/ml Hyper-IL6 or a mixture of 10 ng/ml IL-6 and 200 ng/ml sIL-6R, the molar ratio for Hyper-IL-6:sgp130Fc to achieve 50% inhibition of proliferation was 2.5:1, but for hIL-6·shIL-6R:sgp130Fc, it was 55:1. The molar ratios of Hyper-IL-6 and IL-6·sIL-6R versus sgp130 were calculated for the dimeric sgp130Fc, which can bind one or two IL-6·IL-6R complexes (39). Compared with a mixture of IL-6 and sIL-6R proteins, which contained free IL-6, free sIL-6R, and biologically active IL-6·sIL-6R complexes, Hyper-IL-6 mimics a situation when all IL-6 molecules are trapped. Calculations showed that a 5-fold molar excess of sIL-6R over IL-6 leads to ∼50% of free IL-6 and 50% of IL-6 in IL-6·sIL-6R complexes. Our experimental data (Fig. 3A), however, indicate that the total amount of free IL-6 was above 90%, and only 10% or less formed biologically active IL-6·sIL-6R complexes. The discrepancy between the calculated and the experimentally determined IL-6·sIL-6R levels might be explained by the nonstatic experimental situation. During the experiment, IL-6 might become thermally or chemically inactivated, degraded by secreted proteases, or depleted by cellular internalization, thereby reducing the amount of IL-6, which can build IL-6·sIL-6R complexes. In conclusion, the high amount of free IL-6 in relation to biologically active IL-6·sIL-6R complexes explains why ∼22-fold lower sgp130Fc concentrations were needed to inhibit the biological activity of a mixture of IL-6 and sIL-6R molecules as compared with Hyper-IL-6.
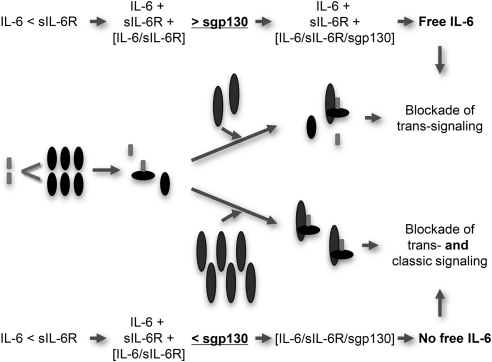
Using cells expressing the IL-6R, we demonstrated that sgp130Fc can inhibit IL-6 classic signaling on Ba/F3-gp130-IL-6R and IL-6R-expressing primary T cells, if sIL-6R levels exceed IL-6 levels. sgp130Fc is able to shift the balance of free IL-6 molecules and IL-6·sIL-6R-complexes to the side of biologically inactive IL-6·sIL-6R·sgp130Fc complexes. sgp130 has no intrinsic affinity to IL-6 or sIL-6R alone, but the affinity of sgp130 to IL-6·sIL-6R is ∼100 times higher than the affinity of IL-6 to IL-6R (27). Therefore, once an IL-6·sIL-6R complex has formed, it can be trapped by sgp130Fc and is thereby eliminated from the first equilibrium of free IL-6 and IL-6·sIL-6R complexes. Elimination of one IL-6·sIL-6R complex from the first equilibrium subsequently favors the formation of a compensatory IL-6·sIL-6R complex to reestablish the initial balance of free IL-6 and IL-6·sIL-6R complexes. Finally, trapping of IL-6·sIL-6R complexes by sgp130Fc may result in a complete elimination of free IL-6 molecules and indirect inhibition of classic signaling. The molar ratio of hIL-6·shIL-6R to sgp130Fc for 50% inhibition of proliferation of Ba/F3-gp130-hIL6R cells stimulated with 10 ng/ml IL-6 and 200 ng/ml sIL-6R was 0.19:1. Compared with cells lacking IL-6R expression, ∼300-fold more sgp130Fc was thus needed to inhibit IL-6 signaling in cells expressing membrane-bound IL-6R, indicating that only high amounts of sgp130Fc were able to shift the balance of free IL-6 and IL-6·sIL-6R complexes (Fig. 8).
FIGURE 8.
Inhibition of classic and trans-signaling by sgp130Fc is controlled by the IL-6·sIL-6R ratio. At low concentrations, sgp130Fc specifically inhibits IL-6 trans-signaling on cells lacking IL-6R expression. sgp130Fc does not interfere with classic IL-6 signaling on cells expressing membrane-bound IL-6R, because at low sgp130Fc concentrations, free IL-6 molecules are not eliminated by forced IL-6·sIL-6R·sgp130Fc complex formation. Only at high concentrations, sgp130Fc additionally inhibits classic signaling by forced IL-6·sIL-6R·sgp130Fc complex formation.
Moreover, when cells were stimulated with 10 ng/ml IL-6 and 200 ng/ml sIL-6R, proliferation of Ba/F3-gp130 cells lacking membrane-bound IL-6R expression stimulated with hIL-6·shIL-6R was ∼90-fold less efficiently inhibited by tocilizumab compared with sgp130Fc. This difference was not apparent on cells expressing membrane-bound IL-6R. Here, comparable molar concentrations of sgp130Fc and tocilizumab were needed to inhibit IL-6 signaling.
How can the 90-fold IC50 difference between sgp130Fc and tocilizumab on cells lacking IL-6R be explained? Tocilizumab inhibits global IL-6 signaling irrespective of sIL-6R/IL-6 ratios. Tocilizumab binds to the so-called cytokine-binding module of soluble and membrane-bound IL-6R equally well, thereby inhibiting the binding of IL-6R to the so-called site I binding interface of IL-6. Under our experimental conditions, tocilizumab has to bind to all membrane-bound IL-6R or sIL-6R molecules to inhibit IL-6 signaling, whereas sgp130Fc only has to bind to the small amount of biological active IL-6·sIL-6R complexes to inhibit IL-6 trans-signaling and does not recognize free IL-6 and free sIL-6R.
The anti-trans-signaling mAb 25F10 also inhibits IL-6 signaling on cells expressing membrane-bound IL-6R, if a molar excess of sIL-6R over IL-6 is present. Unlike sgp130Fc, 25F10 has an intrinsic affinity to the sIL-6R alone, and thereby 25F10 actively favors the formation of IL-6·sIL-6R/25F10 complexes via a preformed 25F10·IL-6R complex and eliminates free IL-6 in a one-step equilibrium process, whereas trapping of IL-6 by sgp130Fc is a two-step equilibrium process. The first step is the binding of IL-6 to the sIL-6R, and the second step is the trapping of this complex by sgp130Fc. The induced IL-6·sIL-6R·25F10 complex formation might explain why 25F10 inhibits almost equally well IL-6 signaling on cells expressing or lacking membrane-bound IL-6R. The differences in IC50 between 25F10 and sgp130Fc are explained by the 10–100-fold lower affinity of 25F10 compared with sgp130Fc (17, 40).
Our revised view of the mechanism of action of sgp130Fc including its influence on classic signaling might have direct implications for therapeutic application of sgp130Fc in IL-6-dependent diseases with documented molar excesses of sIL-6R over IL-6 (29, 34). In retrospect, it might be difficult to decide whether amelioration of disease signs in mice by injection of high doses of sgp130Fc (>250–500 μg/mouse) was dependent on inhibition of trans-signaling alone or on both classic and trans-signaling (41–43). On the other hand, we provide a mechanistic explanation of why low doses of sgp130Fc (<2–50 μg/mouse) were as effective as high doses in vivo (25, 34, 40, 44, 45).
In a polymicrobial sepsis model, 45% of the challenged mice survived. Application of a low dose of sgp130Fc (12.5 μg/mouse) led to the survival of 100% compared with 80% for the high dose of sgp130Fc (250 μg/mouse) (28). Importantly, blocking of classic and trans-signaling by an anti-IL-6 mAb did not improve survival, indicating that blocking of IL-6 trans-signaling was able to prevent sepsis-induced lethality and suggesting beneficial effects of classic IL-6 signaling in this context (28).
Even though many reports demonstrated a molar excess of serum sIL-6R over serum IL-6 under inflammatory conditions, it is uncertain whether such ratios are likely at local sites of inflammation. The opposite might be true as shown for the air-pouch model of acute inflammation, where local production of IL-6 exceeds sIL-6R generation, resulting in a molar excess of IL-6 over sIL-6R (34). With a molar excess of IL-6 over sIL-6R, sgp130Fc would only be able to block trans-signaling, because free IL-6 will not or will only partially be trapped in IL-6·sIL-6R·sgp130Fc complexes. However, trapping of some IL-6 molecules (as a function of the amount of sIL-6R molecules) might reduce but not abolish classic signaling as shown by induction of acute phase response by injection of IL-6 into mice, which was not inhibited by sgp130Fc (24).
In conclusion, we show that sgp130 and sgp130Fc are able to block classic and trans-signaling, whereas comparably low doses of sgp130Fc are able and sufficient to inhibit trans-signaling exclusively on cells lacking IL-6R but do not interfere with classic and trans-signaling on cells expressing membrane-bound IL-6R.
Supplementary Material
Acknowledgments
We thank Steffi Schnell and Elsbeth Schulz (Institute of Biochemistry, University of Kiel) for excellent technical assistance. We thank Mark Baker (NovImmune) for critical discussions.
This work was funded by Deutsche Forschungsgemeinschaft Grant SFB877 of Projects A1 and A2 and by the Cluster of Excellence “Inflammation at Interfaces.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- sIL-6R
- soluble IL-6 receptor
- gp130
- glycoprotein 130
- sgp130
- soluble glycoprotein 130
- Fc
- crystallizable fragment of a human IgG1 antibody.
REFERENCES
- 1. Scheller J., Chalaris A., Schmidt-Arras D., Rose-John S. (2011) Biochim. Biophys. Acta 1813, 878–888 [DOI] [PubMed] [Google Scholar]
- 2. Akira S., Taga T., Kishimoto T. (1993) Adv. Immunol. 54, 1–78 [DOI] [PubMed] [Google Scholar]
- 3. Kishimoto T., Akira S., Narazaki M., Taga T. (1995) Blood 86, 1243–1254 [PubMed] [Google Scholar]
- 4. Jones S. A. (2005) J. Immunol. 175, 3463–3468 [DOI] [PubMed] [Google Scholar]
- 5. Rose-John S., Scheller J., Elson G., Jones S. A. (2006) J. Leukocyte Biol. 80, 227–236 [DOI] [PubMed] [Google Scholar]
- 6. Montero-Julian F. A. (2001) Cell. Mol. Biol. 47, 583–597 [PubMed] [Google Scholar]
- 7. Mitsuyama K., Toyonaga A., Sasaki E., Ishida O., Ikeda H., Tsuruta O., Harada K., Tateishi H., Nishiyama T., Tanikawa K. (1995) Gut 36, 45–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gaillard J. P., Bataille R., Brailly H., Zuber C., Yasukawa K., Attal M., Maruo N., Taga T., Kishimoto T., Klein B. (1993) Eur. J. Immunol. 23, 820–824 [DOI] [PubMed] [Google Scholar]
- 9. Müllberg J., Schooltink H., Stoyan T., Günther M., Graeve L., Buse G., Mackiewicz A., Heinrich P. C., Rose-John S. (1993) Eur. J. Immunol. 23, 473–480 [DOI] [PubMed] [Google Scholar]
- 10. Müllberg J., Dittrich E., Graeve L., Gerhartz C., Yasukawa K., Taga T., Kishimoto T., Heinrich P. C., Rose-John S. (1993) FEBS Lett. 332, 174–178 [DOI] [PubMed] [Google Scholar]
- 11. Fischer M., Goldschmitt J., Peschel C., Brakenhoff J. P., Kallen K. J., Wollmer A., Grötzinger J., Rose-John S. (1997) Nat. Biotechnol. 15, 142–145 [DOI] [PubMed] [Google Scholar]
- 12. Rose-John S., Heinrich P. C. (1994) Biochem. J. 300, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jostock T., Müllberg J., Ozbek S., Atreya R., Blinn G., Voltz N., Fischer M., Neurath M. F., Rose-John S. (2001) Eur. J. Biochem. 268, 160–167 [DOI] [PubMed] [Google Scholar]
- 14. Jones G. W., McLoughlin R. M., Hammond V. J., Parker C. R., Williams J. D., Malhotra R., Scheller J., Williams A. S., Rose-John S., Topley N., Jones S. A. (2010) J. Immunol. 184, 2130–2139 [DOI] [PubMed] [Google Scholar]
- 15. Dominitzki S., Fantini M. C., Neufert C., Nikolaev A., Galle P. R., Scheller J., Monteleone G., Rose-John S., Neurath M. F., Becker C. (2007) J. Immunol. 179, 2041–2045 [DOI] [PubMed] [Google Scholar]
- 16. Melton L., Coombs A. (2008) Nat. Biotechnol. 26, 957–959 [DOI] [PubMed] [Google Scholar]
- 17. Lissilaa R., Buatois V., Magistrelli G., Williams A. S., Jones G. W., Herren S., Shang L., Malinge P., Guilhot F., Chatel L., Hatterer E., Jones S. A., Kosco-Vilbois M. H., Ferlin W. G. (2010) J. Immunol. 185, 5512–5521 [DOI] [PubMed] [Google Scholar]
- 18. Sato K., Tsuchiya M., Saldanha J., Koishihara Y., Ohsugi Y., Kishimoto T., Bendig M. M. (1993) Cancer Res. 53, 851–856 [PubMed] [Google Scholar]
- 19. Yokota S., Imagawa T., Mori M., Miyamae T., Aihara Y., Takei S., Iwata N., Umebayashi H., Murata T., Miyoshi M., Tomiita M., Nishimoto N., Kishimoto T. (2008) Lancet 371, 998–1006 [DOI] [PubMed] [Google Scholar]
- 20. Gearing A. J., Beckett P., Christodoulou M., Churchill M., Clements J., Davidson A. H., Drummond A. H., Galloway W. A., Gilbert R., Gordon J. L., Leber T. M., Mangan M., Miller K., Nayee P., Owen K., Patel S., Thomas W., Wells G., Wood L. M., Woolley K. (1994) Nature 370, 555–557 [DOI] [PubMed] [Google Scholar]
- 21. Chalaris A., Rabe B., Paliga K., Lange H., Laskay T., Fielding C. A., Jones S. A., Rose-John S., Scheller J. (2007) Blood 110, 1748–1755 [DOI] [PubMed] [Google Scholar]
- 22. Garbers C., Jänner N., Chalaris A., Moss M. L., Floss D. M., Meyer D., Koch-Nolte F., Rose-John S., Scheller J. (2011) J. Biol. Chem. 286, 14804–14811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schroers A., Hecht O., Kallen K. J., Pachta M., Rose-John S., Grötzinger J. (2005) Protein Sci. 14, 783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mackiewicz A., Schooltink H., Heinrich P. C., Rose-John S. (1992) J. Immunol. 149, 2021–2027 [PubMed] [Google Scholar]
- 25. Hurst S. M., Wilkinson T. S., McLoughlin R. M., Jones S., Horiuchi S., Yamamoto N., Rose-John S., Fuller G. M., Topley N., Jones S. A. (2001) Immunity 14, 705–714 [DOI] [PubMed] [Google Scholar]
- 26. Müller-Newen G., Küster A., Hemmann U., Keul R., Horsten U., Martens A., Graeve L., Wijdenes J., Heinrich P. C. (1998) J. Immunol. 161, 6347–6355 [PubMed] [Google Scholar]
- 27. Zohlnhöfer D., Graeve L., Rose-John S., Schooltink H., Dittrich E., Heinrich P. C. (1992) FEBS Lett. 306, 219–222 [DOI] [PubMed] [Google Scholar]
- 28. Barkhausen T., Tschernig T., Rosenstiel P., van Griensven M., Vonberg R. P., Dorsch M., Mueller-Heine A., Chalaris A., Scheller J., Rose-John S., Seegert D., Krettek C., Waetzig G. H. (2011) Crit. Care Med. 39, 1407–1413 [DOI] [PubMed] [Google Scholar]
- 29. Gaillard J., Pugnière M., Tresca J., Mani J., Klein B., Brochier J. (1999) Eur. Cytokine Netw. 10, 337–344 [PubMed] [Google Scholar]
- 30. Kallen K. J., Grötzinger J., Lelièvre E., Vollmer P., Aasland D., Renné C., Müllberg J., Myer zum Büschenfelde K. H., Gascan H., Rose-John S. (1999) J. Biol. Chem. 274, 11859–11867 [DOI] [PubMed] [Google Scholar]
- 31. Peters M., Schirmacher P., Goldschmitt J., Odenthal M., Peschel C., Fattori E., Ciliberto G., Dienes H. P., Meyer zum Büschenfelde K. H., Rose-John S. (1997) J. Exp. Med. 185, 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Castell J. V., Geiger T., Gross V., Andus T., Walter E., Hirano T., Kishimoto T., Heinrich P. C. (1988) Eur. J. Biochem. 177, 357–361 [DOI] [PubMed] [Google Scholar]
- 33. Peters M., Blinn G., Solem F., Fischer M., Meyer zum Büschenfelde K. H., Rose-John S. (1998) J. Immunol. 161, 3575–3581 [PubMed] [Google Scholar]
- 34. Rabe B., Chalaris A., May U., Waetzig G. H., Seegert D., Williams A. S., Jones S. A., Rose-John S., Scheller J. (2008) Blood 111, 1021–1028 [DOI] [PubMed] [Google Scholar]
- 35. Doganci A., Eigenbrod T., Krug N., De Sanctis G. T., Hausding M., Erpenbeck V. J., El-Bdaoui H., Lehr H. A., Schmitt E., Bopp T., Kallen K. J., Herz U., Schmitt S., Luft C., Hecht O., Hohlfeld M., Ito H., Nishimoto N., Yoshizaki K., Kishimoto T., Rose-John S., Renz H., Neurath M. F., Galle P. R., Finotto S. (2005) J. Clin. Invest. 115, 313–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ellingsgaard H., Ehses J. A., Hammar E. B., Van Lommel L., Quintens R., Martens G., Kerr-Conte J., Pattou F., Berney T., Pipeleers D., Halban P. A., Schuit F. C., Donath M. Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13163–13168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Becker C., Fantini M. C., Schramm C., Lehr H. A., Wirtz S., Nikolaev A., Burg J., Strand S., Kiesslich R., Huber S., Ito H., Nishimoto N., Yoshizaki K., Kishimoto T., Galle P. R., Blessing M., Rose-John S., Neurath M. F. (2004) Immunity 21, 491–501 [DOI] [PubMed] [Google Scholar]
- 38. Grivennikov S., Karin E., Terzic J., Mucida D., Yu G. Y., Vallabhapurapu S., Scheller J., Rose-John S., Cheroutre H., Eckmann L., Karin M. (2009) Cancer Cell 15, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boulanger M. J., Chow D. C., Brevnova E. E., Garcia K. C. (2003) Science 300, 2101–2104 [DOI] [PubMed] [Google Scholar]
- 40. Tenhumberg S., Waetzig G. H., Chalaris A., Rabe B., Seegert D., Scheller J., Rose-John S., Grötzinger J. (2008) J. Biol. Chem. 283, 27200–27207 [DOI] [PubMed] [Google Scholar]
- 41. Atreya R., Mudter J., Finotto S., Müllberg J., Jostock T., Wirtz S., Schütz M., Bartsch B., Holtmann M., Becker C., Strand D., Czaja J., Schlaak J. F., Lehr H. A., Autschbach F., Schürmann G., Nishimoto N., Yoshizaki K., Ito H., Kishimoto T., Galle P. R., Rose-John S., Neurath M. F. (2000) Nat. Med. 6, 583–588 [DOI] [PubMed] [Google Scholar]
- 42. Fujimoto M., Serada S., Mihara M., Uchiyama Y., Yoshida H., Koike N., Ohsugi Y., Nishikawa T., Ripley B., Kimura A., Kishimoto T., Naka T. (2008) Arthritis Rheum. 58, 3710–3719 [DOI] [PubMed] [Google Scholar]
- 43. Serada S., Fujimoto M., Mihara M., Koike N., Ohsugi Y., Nomura S., Yoshida H., Nishikawa T., Terabe F., Ohkawara T., Takahashi T., Ripley B., Kimura A., Kishimoto T., Naka T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9041–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matsumoto S., Hara T., Mitsuyama K., Yamamoto M., Tsuruta O., Sata M., Scheller J., Rose-John S., Kado S., Takada T. (2010) J. Immunol. 184, 1543–1551 [DOI] [PubMed] [Google Scholar]
- 45. Mitsuyama K., Matsumoto S., Rose-John S., Suzuki A., Hara T., Tomiyasu N., Handa K., Tsuruta O., Funabashi H., Scheller J., Toyonaga A., Sata M. (2006) Gut 55, 1263–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.