Background: The 14-3-3 protein binds to and regulates the function of the regulator of G protein signaling 3 (RGS3).
Results: The 14-3-3 binding affects the structure of the Gα interaction portion of RGS3.
Conclusion: The 14-3-3 protein blocks the interaction between the RGS3 and the Gα.
Significance: This might explain the inhibitory function of 14-3-3 in the regulation of RGS3.
Keywords: Fluorescence Resonance Energy Transfer (FRET), Phosphorylation, RGS Proteins, Signal Transduction, Structural Biology, 14-3-3 Protein, H/D Exchange, SAXS, Low-resolution Structure
Abstract
Regulator of G protein signaling (RGS) proteins function as GTPase-activating proteins for the α-subunit of heterotrimeric G proteins. The function of certain RGS proteins is negatively regulated by 14-3-3 proteins, a family of highly conserved regulatory molecules expressed in all eukaryotes. In this study, we provide a structural mechanism for 14-3-3-dependent inhibition of RGS3-Gα interaction. We have used small angle x-ray scattering, hydrogen/deuterium exchange kinetics, and Förster resonance energy transfer measurements to determine the low-resolution solution structure of the 14-3-3ζ·RGS3 complex. The structure shows the RGS domain of RGS3 bound to the 14-3-3ζ dimer in an as-yet-unrecognized manner interacting with less conserved regions on the outer surface of the 14-3-3 dimer outside its central channel. Our results suggest that the 14-3-3 protein binding affects the structure of the Gα interaction portion of RGS3 as well as sterically blocks the interaction between the RGS domain and the Gα subunit of heterotrimeric G proteins.
Introduction
The amplitude and duration of G protein signaling are regulated at the G protein level by two different mechanisms. The first one involves the inhibition of Gβγ interactions with Gα-GDP and downstream effectors (1, 2), whereas the second one is based on the enhancement of the low intrinsic GTPase activity of the Gα subunit (3, 4). The second mechanism is carried out by a highly conserved regulator of G protein signaling (RGS)2 domain of RGS proteins that specifically recognizes the GTP-bound form of Gα and enhances its GTPase activity by stabilizing the transition state (5, 6). Members of RGS protein family also play additional functions based on either their ability to interact with proteins other than Gα or their nuclear localization (7, 8). The function of certain RGS proteins, including RGS3, is negatively regulated by 14-3-3 proteins by a still unknown mechanism (9–13).
The 14-3-3 proteins, a eukaryotic family of highly conserved regulatory proteins, function as scaffold molecules that modulate the activity of their binding partners by a number of different mechanisms including conformational change and relocalization (reviewed in Refs. 14–17). They fold into cup-shaped mono- or heterodimers with a large 40 Å wide channel (18, 19). This central channel contains two amphipathic grooves through which 14-3-3 proteins bind their protein ligands in both a phosphorylation-dependent and a phosphorylation-independent manner (20–23). A few currently available structures of 14-3-3 complexes with a ligand of a size that exceeds the phosphorylated binding motif suggest that the 14-3-3 dimer employs the highly conserved and rigid surface of the central channel as the main interface that permits the ligand binding in a structurally reproducible manner (24–26). However, it also appears that many interactions are isoform-specific and, thus, might involve less conserved surfaces outside the central channel.
A putative 14-3-3 binding site was first identified within the RGS domain of RGS3 and RGS7 at a conserved SYP motif (9, 10). Other studies, however, identified Ser-264 within the N-terminal part of RGS3 (outside the RGS domain) as the 14-3-3 binding site (11, 12). This residue is presumably phosphorylated by protein kinase A (PKA), and its sequence 259RRRTHSEG266 is consistent with the type II binding motif for 14-3-3 (14, 22). Recently, we have used time-resolved tryptophan fluorescence to show that the 14-3-3ζ protein binding induces significant structural changes in both the vicinity of the phosphorylation site Ser-264 and the C-terminal RGS domain of RGS3 protein (27). This suggests that both regions of RGS3 physically interact with the 14-3-3ζ protein and hence, are involved in the 14-3-3-dependent inhibition of RGS3 function.
In this study, we provide a structural mechanism for 14-3-3-dependent inhibition of the RGS3-Gα interaction. We have used small angle x-ray scattering, hydrogen/deuterium exchange kinetics, and Förster resonance energy transfer measurements to determine the low-resolution solution structure (hybrid structure) of the 14-3-3ζ·RGS3 complex. The structure shows the RGS domain of RGS3 bound to the 14-3-3ζ dimer in an as-yet-unrecognized manner interacting with less conserved regions on the outer surface of the 14-3-3 dimer outside of its central channel. Our results suggest that the 14-3-3 protein binding affects the structure of the Gα-interacting region of RGS3 as well as sterically blocks the interaction between the RGS domain and the Gα subunit of heterotrimeric G proteins.
EXPERIMENTAL PROCEDURES
Expression, Purification, and Phosphorylation of RGS3 Protein
Both wild type and all mutants of RGS3 (human isoform 1, sequence 255–513) were expressed, purified, and phosphorylated as described previously (27). Mutants of RGS3 protein were created using the QuikChange approach (Stratagene). All mutations were confirmed by sequencing and phosphorylation by mass spectrometry.
Labeling of RGS3 Protein Mutants by 1,5-IAEDANS
Covalent modification of RGS3 protein mutants containing a single cysteine residue (at positions 276, 285, 427, 456, or 480) with the thiol-reactive probe 1,5-IAEDANS was carried out as described elsewhere (28, 29). Briefly, the protein (50–70 μm) in buffer containing 20 mm Tris-HCl (pH 7.5), 250 mm NaCl, 1 mm EDTA, 10% (v/w) glycerol, and label was mixed at a molar ratio of 1:40 and incubated at 30 °C for 2 h and then at 4 °C overnight in the dark. The free unreacted label was removed by size exclusion chromatography. The incorporation stoichiometry was determined by comparing the peak protein absorbance at 280 nm with the absorbance of bound AEDANS measured at 336 nm using the extinction coefficient of 5700 m−1·cm−1 (Invitrogen).
Expression and Purification of 14-3-3ζ Protein
Both wild type and all mutants of the 14-3-3 protein (human ζ isoform) were expressed, purified, and phosphorylated as described previously (24, 28).
Labeling of 14-3-3ζ Protein Mutants by 5-Iodoacetamidofluorescein for FRET Measurement
Covalent modification of the 14-3-3ζ protein mutants containing a single cysteine residue with the thiol-reactive probe 5-iodoacetamidofluorescein was carried out as described elsewhere (26, 28). Briefly, we constructed two mutants containing a single cysteine residue (either at position 25 or at position 189) of human monomeric 14-3-3ζ protein (mutant S58D) (30). The protein (40 μm) in 50 mm Tris-HCl (pH 7.5), 100 mm NaCl, 1 mm EDTA, and label 5-iodoacetamidofluorescein (Invitrogen) were mixed at a molar ratio of 1:40 and incubated at room temperature for 8 h and then at 4 °C overnight in the dark. The free unreacted label was removed by size exclusion chromatography. The incorporation stoichiometry was determined by the absorbance at 492 nm using an extinction coefficient of 78,000 m−1·cm−1. The stoichiometry of fluorescein incorporation/mol of protein was found to be 99–100%.
Analytical Ultracentrifugation
Sedimentation equilibrium and sedimentation velocity experiments were performed using a ProteomeLab XL-I Beckman Coulter analytical ultracentrifuge. Sedimentation equilibrium experiments at loading concentrations of 3–15 μm were conducted at 4 °C with rotor speeds from 10,000 to 16,000 rpm. Sedimentation velocity experiments were conducted at loading concentrations of 3–18 μm, 20 °C, with rotor speeds of 42,000 or 48,000 rpm. Samples were dialyzed against the buffer containing 20 mm Tris-HCl (pH 7.5), 200 mm NaCl, and 2 mm 2-mercaptoethanol prior to analysis. All data were collected at 280 nm, and data analysis was performed with the SEDFIT and SEDPHAT packages (31, 32). Sedimentation velocity data were analyzed using a sedimentation coefficient distribution model c(s). Peaks were integrated to determine the weight-averaged sedimentation coefficients sw.
Small Angle X-ray Scattering (SAXS)
SAXS data were collected on the European Molecular Biology Laboratory (EMBL) X33 beamline on the storage ring DORIS III (Deutsches Elektronen Synchrotron (DESY), Hamburg, Germany). The 14-3-3ζ·pRGS3 protein complex3 was measured in a concentration range of 2–8 mg·ml−1. No measurable radiation damage was detected by comparison of eight successive time frames with 15-s exposures. The data were averaged after normalization to the intensity of the transmitted beam, and the scattering of the buffer was subtracted using PRIMUS (33). The forward scattering I(0) and the radius of gyration Rg were evaluated using the Guinier approximation (34). These parameters were also computed from the entire scattering pattern using the program GNOM (35), which provides the distance distribution functions P(r) and the maximum particle dimensions Dmax. The solute apparent molecular mass (MMexp) was estimated by comparison of the forward scattering with that from reference solutions of bovine serum albumin (molecular mass 66 kDa). The excluded volume of the hydrated particle (the Porod volume, Vp) was computed as reported by Porod (36). Ab initio molecular envelopes were computed by the program DAMMIF (37), which represents the protein by an assembly of densely packed beads. Multiple iterations of DAMMIF were averaged using the program DAMAVER (38).
Hydrogen/Deuterium Exchange Kinetics Coupled to Mass Spectrometry (HDX-MS)
HDX of the 14-3-3ζ protein (25 μm), unphosphorylated RGS3 protein (25 μm), phosphorylated pRGS3 protein (25 μm), RGS3 in the presence of the 14-3-3ζ protein, and pRGS3 in the presence of the 14-3-3ζ protein was initiated by 10-fold dilution in a deuterated buffer containing 20 mm Tris-HCl (pD 7.1), 1 mm EDTA, 3 mm DTT, 200 mm NaCl, and 10% (w/v) glycerol. The molar ratio between 14-3-3ζ and RGS3 was 2:2. Aliquots (80 μl) were taken after 20 s, 1 min, 5 min, 10 min, 1 h, 3 h, and 5 h of exchange. The exchange was quenched by adding 20 μl of 0.1 m HCl and rapid freezing in liquid nitrogen. Analysis of deuterated samples (HPLC-MS) was done using a HPLC (1200 Agilent Technologies, Waldbronn, Germany) connected to an ESI Fourier transform ion cyclotron MS (9.4T APEX-Ultra, Bruker Daltonics, Billerica, MA). The analysis was started by quick thawing of the sample followed by a digestion on a pepsin column (66-μl bed volume, flow rate 100 μl·min−1). Generated peptides were desalted online using a Peptide MicroTrap (Michrom Bioresources, Auburn, CA) and separated on a C18 reversed phase column (1 × 100 mm, Jupiter, Phenomenex) using a linear gradient of 10–45% solvent B in solvent A in 20 min, where solvent A was 2% acetonitrile, 0.4% formic acid in water, and solvent B was 95% acetonitrile, 5% water, 0.4% formic acid. The injection and switching valve, pepsin column, peptide trap, and the analytical column were placed in an ice box to minimize the back exchange. Peptide identification (mapping, HPLC-MS/MS) was done using the same system as described above, and the MS/MS spectra were searched by MASCOT against a database containing sequences of 14-3-3ζ and RGS3 (the only allowed partial modification was Ser/Thr phosphorylation). Spectra of partially deuterated peptides were exported to txt files using DataAnalysis version 4.0 (Bruker Daltonics), and the centroids of the isotopic envelopes were read out using MMass version 3.11.0 (39).
Circular Dichroism Measurements
Circular dichroism (CD) measurements were carried out using a J-810 spectropolarimeter (JASCO, Tokyo, Japan) as described previously (40). The spectra were recorded at room temperature (23 °C) in a buffer containing 20 mm Tris-HCl (pH 7.5), 250 mm NaCl, 1 mm EDTA, 2 mm 2-mercaptoethanol, and 10% (w/v) glycerol. The RGS3 concentration was 3.2 μm; the 14-3-3ζ protein concentration was 6.4 μm.
Time-resolved Fluorescence Measurements
Fluorescence intensity and anisotropy decays were measured on a time-correlated single photon counting apparatus, as described previously (28, 41). The fluorescence decays have been acquired under the “magic angle” conditions when the measured intensity decay, I(t), is independent of the rotational diffusion of the chromophore. Samples were placed in a thermostatic holder, and all experiments were performed at 22 °C in a buffer containing 20 mm Tris-HCl (pH 7.5), 250 mm NaCl, 1 mm EDTA, 3 mm 2-mercaptoethanol, and 10% (w/v) glycerol. The RGS3 concentration was 10 μm; the 14-3-3ζ protein concentration was 20 μm. Dansyl fluorescence was excited at 315 nm by a picosecond dye laser (Spectra Physics). Fluorescence was measured at 480 nm using a monochromator with a 400-nm cut-off filter placed in front of its input slit. Fluorescence was assumed to decay multiexponentially according to the formula,
where τi and αi are the fluorescence lifetimes and the corresponding amplitudes, respectively. I(t) was analyzed by the singular value decomposition maximum entropy method (42). The program yields amplitudes, αi, that represent the lifetime distribution in the decay. We have chosen 100 lifetimes equidistantly spaced in the logarithmic scale and covering the range from 20 ps to 20 ns. The mean lifetime was calculated as follows,
where fi is the fractional intensity of the ith lifetime component. The fluorescence anisotropy decays r(t) were obtained from the parallel I‖(t) and perpendicular I⊥(t) decay components. Data were analyzed by a model independent singular value decomposition maximum entropy method approach that does not set prior limits on the shape of the distribution (42). The anisotropies r(t) were analyzed for a series of exponentials,
where the amplitudes βi represent the distribution of the correlation times φi and are related to the initial anisotropy r0 by the formula,
we used 100 correlation times φi equidistantly spaced in the logarithmic scale and ranging from 100 ps to 500 ns.
Fluorescence Resonance Energy Transfer Analysis
The fluorescence resonance energy transfer data were measured and analyzed as described previously (26, 28). The Förster critical distance, R0, was calculated as follows,
where n is the refractive index of the medium, QD is the emission quantum yield of the donor in the absence of the acceptor, κ2 is the orientation factor, and J(λ) is the spectral overlap integral of donor fluorescence and acceptor absorption. The quantum yield of the donor, which is the AEDANS moiety attached at Cys-456 within the flexible region of the C-terminal RGS domain of the RGS3 molecule, was calculated relative to the quantum yield of quinine sulfate in 0.1 m H2SO4 (43). The spectral overlap J(λ) between AEDANS and fluorescein was obtained by numerical integration of a product of an area-normalized emission spectrum of AEDANS-labeled pRGS3 and the absorption spectrum of fluorescein-labeled 14-3-3ζ.
RESULTS
Biophysical Characterization of the 14-3-3ζ·pRGS3 Protein Complex
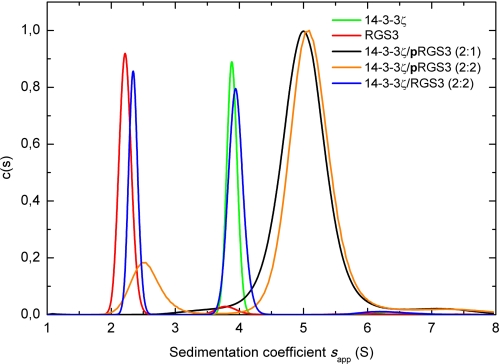
The analytical ultracentrifugation was used for the basic biophysical characterization of the 14-3-3ζ·pRGS3 complex. Fig. 1 shows the continuous sedimentation coefficient distribution c(s) obtained from the sedimentation velocity experiments. Analysis of this distribution reveals that the phosphorylated RGS3 and the 14-3-3ζ protein form a stable complex with a weight-averaged sedimentation coefficient (sw) of 5.0 s, although the unphosphorylated RGS3 shows no interaction with 14-3-3ζ. The 14-3-3ζ protein and RGS3 alone show single peaks with sw values of 3.8 and 2.2 s, respectively. The value of sw for the complex corresponds to a molecular mass of ∼84 kDa, hence suggesting a 2:1 stoichiometry (a dimer of 14-3-3ζ binds one molecule of pRGS3).
FIGURE 1.
Sedimentation velocity analysis. The continuous distribution of sedimentation coefficients c(s) for 14-3-3ζ alone (green), RGS3 alone (red), 14-3-3ζ, and pRGS3 mixed in the molar ratio 2:1 (black), 14-3-3ζ and pRGS3 mixed in the molar ratio 2:2 (orange), and 14-3-3ζ and unphosphorylated RGS3 mixed in the molar ratio 2:2 (blue) is shown. The weight-averaged sedimentation coefficient (sw) of 5.0 s for the 14-3-3ζ·pRGS3 complex corresponds to the molecular mass of ∼84 kDa, indicating a 2:1 stoichiometry (dimer of 14-3-3ζ binds one molecule of pRGS3). Values of sw were obtained by integration of c(s) curve.
The sedimentation equilibrium measurements confirmed that at a 2:1 mixing molar ratio of 14-3-3ζ to pRGS3, a complex with 2:1 stoichiometry is exclusively formed. The apparent equilibrium dissociation constant (KD) of this complex was determined to be ∼20 nm. At a 2:2 mixing ratio, a complex with 2:2 stoichiometry is formed (a dimer of 14-3-3ζ binds two molecules of pRGS3). However, this complex is unstable and dissociates to the 2:1 complex with the apparent dissociation constant of ∼20 μm. Furthermore, the sedimentation equilibrium experiments revealed, in agreement with the sedimentation velocity measurements, that unphosphorylated RGS3 does not interact with the 14-3-3ζ protein.
Low-resolution Structure of the 14-3-3ζ·pRGS3 Protein Complex
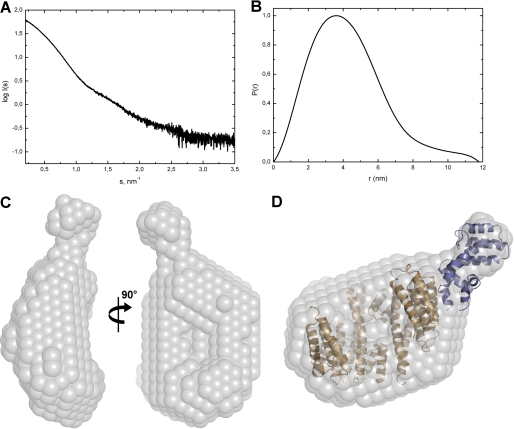
To obtain the low-resolution structure of the 14-3-3ζ·pRGS3 complex directly in solution, we carried out SAXS experiments. The experimental SAXS curve from the 14-3-3ζ·pRGS3 complex is shown in Fig. 2A. The apparent molecular mass of the 14-3-3ζ·pRGS3 complex was estimated by comparison of the forward scattering intensity I(0) with that from reference solutions of bovine serum albumin. The estimated mass of ∼78 kDa corresponds well to 2:1 stoichiometry in agreement with the results from analytical ultracentrifugation. The excluded volume of the hydrated particle (the Porod volume, Vp) was found to be 156 × 103 Å3, further corroborating the 2:1 stoichiometry of the complex (for globular proteins, the hydrated volume in Å3 should numerically be about twice the molecular mass in daltons). Other parameters that can be obtained directly from the scattering data in a model-independent manner are the radius of gyration (Rg) and the maximum particle distance (Dmax). The values of Rg and Dmax were determined to be 34 and 118 Å, respectively. The shape of the calculated distance distribution function (P(r) function) suggests that the complex is significantly asymmetric (Fig. 2B).
FIGURE 2.
SAXS scattering data and the low-resolution structure of the 14-3-3ζ·pRGS3 complex. A, solution scattering pattern for the 14-3-3ζ·pRGS3 complex. X-ray scattering was measured at three different protein concentrations (2, 4, and 8 mg·ml−1), and no concentration dependence was observed. Scattering intensity I(s) is plotted in relation to the scattering vector s (s = 4πsin(θ)/λ, where 2θ is the scattering angle and λ is the wavelength). B, plot of the distance distribution functions P(r) with the maximum particle distance (Dmax) of 11.8 nm. C, average low-resolution ab initio shape envelope (spheres around the dummy residues) of the 14-3-3ζ·pRGS3 complex as determined from solution scattering experiments using the program DAMMIF (37). D, overlay of the rigid body model of the 14-3-3ζ·pRGS3 complex with the ab initio shape envelope. The envelope is shown in gray, the 14-3-3ζ protein is shown in light brown, and the RGS domain of RGS3 is shown in blue. The rigid body model was prepared using the crystal structure of 14-3-3ζ (21) and the RGS domain of RGS3 (27).
The program DAMMIF was used to produce multiple low-resolution ab initio envelopes for the 14-3-3ζ·pRGS3 complex that were further averaged using the program DAMAVER (Fig. 2C) (38). The envelope reproduces correctly the cup-like shape of the 14-3-3 dimer and suggests that a significant part of the RGS3 molecule is located outside the central channel of 14-3-3. We assume it is the C-terminal RGS domain (residues 384–513) because it possesses well ordered three-dimensional structure (27), whereas the N-terminal portion of RGS3 (residues 255–380) is, according to the theoretical prediction, likely disordered (supplemental Fig. S1). Furthermore, the N-terminal part of RGS3 contains a phosphorylated 14-3-3 binding motif (sequence RRRTHpS264EGS), which is presumably docked in the ligand binding groove of 14-3-3, as has been observed in other structures of 14-3-3 protein complexes (21, 22, 24, 25, 44). Therefore, the rigid body modeling of the 14-3-3ζ·pRGS3 complex was performed using the crystal structures of 14-3-3ζ (21) and RGS domain of RGS3 (27) (Protein Data Bank (PDB) codes 1qjb and 2oj4, respectively). The rigid body model displayed good agreement with the low-resolution ab initio shape (Fig. 2D). In addition, the radius of gyration (Rg) and the maximum particle dimension (Dmax) calculated for the suggested structural model (34.9 and 125 Å, respectively) are in excellent agreement with those determined by the program GNOM from the scattering data. The model of the complex suggests that the RGS domain interacts with less conserved regions on the outer surface of the 14-3-3ζ dimer outside its central channel involving helices H6 and H8. However, it is necessary to keep in mind that the structural model shown in Fig. 2D contains only approximately one-half of the RGS3 construct (the C-terminal RGS domain, residues 384–513) used for the SAXS experiments as no experimental structural data are available for the unstructured N-terminal part of RGS3.
Mapping of the Interactions between 14-3-3ζ and pRGS3 Using HDX-MS
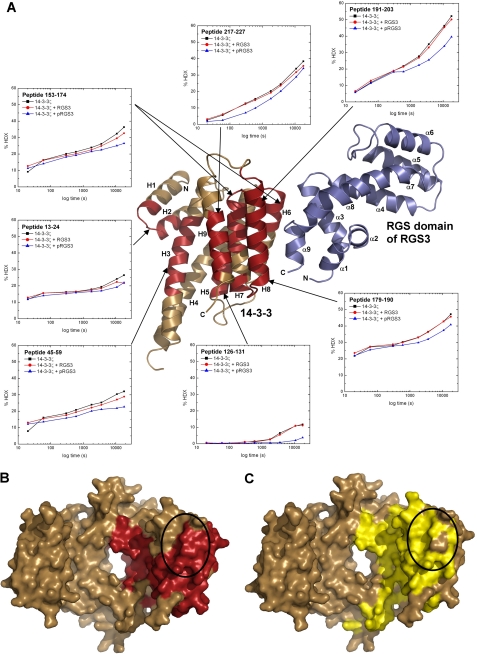
HDX-MS was used to validate the low-resolution structural model obtained from SAXS and to further map the interactions between 14-3-3ζ and pRGS3. Therefore, the HDX kinetics of the 14-3-3ζ·RGS3 and 14-3-3ζ·pRGS3 complexes as well as free 14-3-3ζ, RGS3, and pRGS3 proteins were measured. The exchange kinetics of 14-3-3ζ regions were followed on 20 peptides from the pepsin digest, together covering 98% of the sequence (supplemental Fig. S2). Upon the deuterium incorporation, the isotopic distributions of 14-3-3ζ peptides are shifted to the higher masses, and for several peptides, sequences 13–24, 45–59, 126–131, 153–174, 179–190, 191–203, and 217–227 (Fig. 3A), the incorporation is slower in the presence of phosphorylated pRGS3 when compared with unphosphorylated RGS3 or 14-3-3ζ alone. This slower deuterium incorporation can be directly interpreted as decreased accessibility to the solvent following the complex formation, thus suggesting that protected regions of 14-3-3ζ (or their parts) form the binding interface. These regions map not only to the surface of the ligand binding groove formed by helices H3, H5, H7, and H9 but also to the surface outside the central channel formed by helices H6 and H8 (Fig. 3, A and B). This suggests that the 14-3-3 dimer interacts with pRGS3 not only through the ligand binding groove but also through additional less conserved surfaces outside the central cavity in very good agreement with our SAXS-based structural model (Fig. 3, B and C). In addition, the 14-3-3ζ protein alone shows very similar deuteration kinetics, as in the presence of unphosphorylated RGS3, in agreement with the fact that phosphorylation is required for RGS3 binding to 14-3-3ζ.
FIGURE 3.
HDX-MS reveals binding interactions between 14-3-3ζ and pRGS3. A, HDX kinetics for 14-3-3ζ regions that show slower deuterium exchange kinetics upon the pRGS3 binding (shown in red) mapped on the structural model of the 14-3-3ζ·pRGS3 complex derived from SAXS. Light brown ribbons represent regions that show no significant protection against deuteration in the presence of pRGS3. Deuterium exchange is expressed as percentages relative to the maximum theoretical deuteration level for 14-3-3ζ alone (black squares), 14-3-3ζ in the presence of unphosphorylated RGS3 (red circles), and 14-3-3ζ in the presence of phosphorylated pRGS3 (blue triangles). The time units are in seconds. Only one monomer of the 14-3-3ζ dimer is shown for clarity. B, regions that show slower deuterium exchange upon the pRGS3 binding (shown in red) mapped on a surface representation of the human 14-3-3ζ isoform structure (21). C, surface representation of human 14-3-3ζ shaded according the sequence conservation. Residues that are totally conserved among all human isoforms are colored in yellow. The black ellipses in panels B and C indicate the specificity region believed to be involved in protein-protein interactions and responsible for isoform-selective contacts (44, 55, 56).
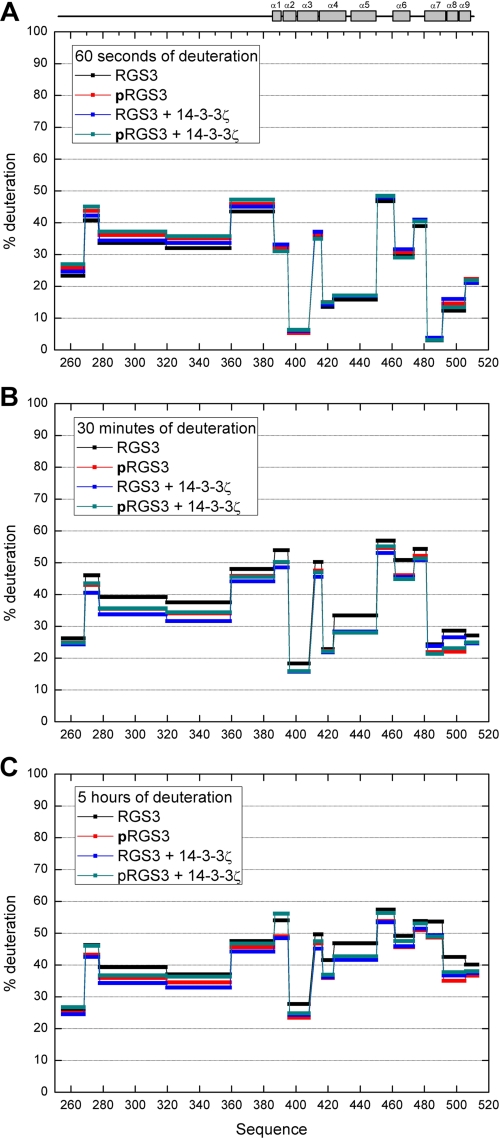
The HDX was also measured for both pRGS3 and RGS3 in the presence and absence of 14-3-3ζ (a map with the coverage of RGS3 upon pepsin digestion is shown in supplemental Fig. S3). However, we were unable to identify any pRGS3 peptides with a significant change in the deuteration kinetics upon the 14-3-3ζ protein binding. Such insensitivity of the backbone amide hydrogens in the pRGS3 binding interface to the formation of a complex with 14-3-3ζ might be a result of the predominantly electrostatic and side chain-mediated interactions between these two proteins. A similar effect was also observed, for example, for the heteromeric complex containing UBC9 and SUMO-1 (45) or in the study involving p47phox (46). In addition, the observed fast deuteration kinetics (Fig. 4) as well as the theoretical prediction of the RGS3 disorder (supplemental Fig. S1) suggest that the N-terminal part of RGS3 is unstructured and, therefore, highly flexible. This might be yet another factor contributing to the absence of HDX difference in the pRGS3 as the very fast deuteration kinetics would be less sensitive to the formation of a complex.
FIGURE 4.
Protection plot showing the deuteration levels of RGS3 and pRGS3 in both the absence and the presence of the 14-3-3ζ protein. A, after 60 s of deuteration. B, after 30 min of deuteration. C, after 5 h of deuteration. Secondary structure elements are indicated at the top.
Validation of the Structural Model by FRET
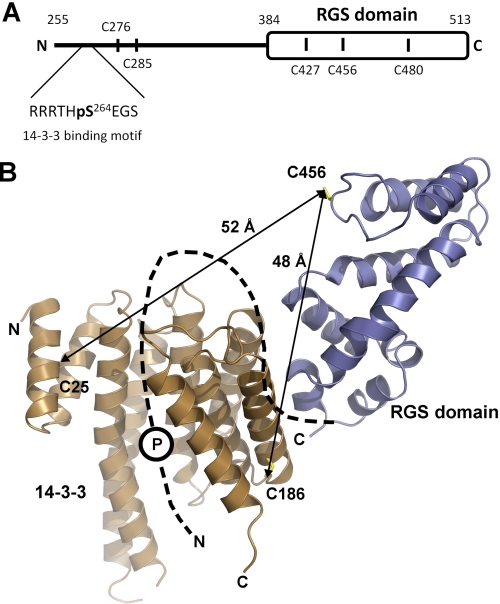
The distances from time-resolved FRET fluorescence experiments were used to further cross-validate the low-resolution structural model of the 14-3-3ζ·pRGS3 complex obtained from SAXS. To prepare well defined pRGS3 and 14-3-3ζ complexes containing only one donor-acceptor pair of fluorophores, we used previously prepared versions of the monomeric form of 14-3-3ζ (mutation S58D) containing a single cysteine residue either at position 25 or at position 189, respectively (26, 28, 30). A mutant of pRGS3 containing a single cysteine residue within the RGS domain at position 456 was used to specifically label the RGS domain (Fig. 5A). The CD spectroscopy was used to check whether the AEDANS-labeled mutant of pRGS3 keeps its native structure, and no significant changes when compared with unlabeled pRGS3 WT were observed (supplemental Fig. S4A). The quantum yield of AEDANS-labeled pRGS3(C456) fluorescence at 22 °C in the presence of unlabeled 14-3-3ζ was determined to be 0.096. The calculated values of the spectral overlap J(λ) between AEDANS and fluorescein, the mean excited state lifetimes (τmean) of AEDANS in the absence and presence of the acceptor (fluorescein), the efficiency of the energy transfer, and the calculated average distances are presented in Table 1. If we consider the size of AEDANS and fluorescein moieties, measured FRET-based distances correspond very well with the structural model of the 14-3-3ζ·pRGS3 complex, thus further corroborating the location of the RGS domain outside the central channel of the 14-3-3ζ dimer (Fig. 5B).
FIGURE 5.
FRET measurements support the SAXS-based structural model of the 14-3-3ζ·pRGS3 complex. A, schematic representation of the primary structure of RGS3 (sequence 255–513). Black vertical bars denote locations of single Cys residues used as sites for AEDANS attachment. B, the SAXS-based structural model of the 14-3-3ζ·pRGS3 complex with the average distances calculated from FRET measurements. Cysteine residues used as sites of AEDANS (Cys-456 of RGS3) and fluorescein (Cys-25 and Cys-189 of 14-3-3ζ) modification are shown as sticks. The missing N-terminal part of RGS3 is schematically shown as a black dashed line. The position of the phosphorylation site Ser-264 is depicted as a circled P. Only one monomer of 14-3-3ζ is shown for clarity.
TABLE 1.
Summary of energy transfer measurements
| 14-3-3ζ acceptor site | Label | τmeana,b | Ec | J(λ)d | R0e | R |
|---|---|---|---|---|---|---|
| ns | % | cm3·m−1 | Å | Å | ||
| Cys-25 | Donor only | 13.7 | 13.1 | 1.580 × 10−13 | 38.0 | 52 |
| Donor + acceptor | 11.9 | |||||
| Cys-189 | Donor only | 13.7 | 19.7 | 1.580 × 10−13 | 38.0 | 48 |
| Donor + acceptor | 11.0 |
a Mean lifetimes were calculated as τmean = Σifiτi, where fi is an intensity fraction of the ith lifetime component τi.
b S.D. value is ± 0.1 ns.
c Energy transfer efficiency.
d Spectral overlap integral of the donor fluorescence and acceptor absorption.
e Förster critical distance represents the distance between energy donor and energy acceptor where the FRET efficiency is 50%.
Time-resolved Fluorescence Intensity and Anisotropy Decay Measurements
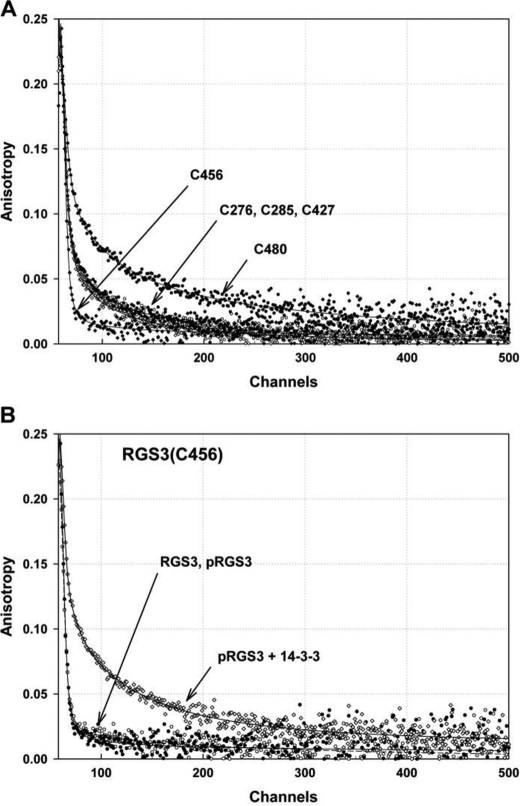
The time-resolved fluorescence intensity and anisotropy decay measurements of AEDANS-labeled pRGS3 were used to investigate the conformational behavior of pRGS3 and its changes upon the 14-3-3ζ binding. The extrinsic fluorophore AEDANS was covalently attached to a single cysteine residue of five different RGS3 mutants to sample various surfaces of the RGS3 molecule (Cys-276, Cys-285, Cys-427, Cys-456, and Cys-480). Residues Cys-276 and Cys-285 are located within the N-terminal half of RGS3 construct, whereas residues Cys-427, Cys-456, and Cys-480 are located in the C-terminal RGS domain (Fig. 5A). Because AEDANS is an environmentally sensitive fluorophore, a variation of its mean excited state lifetime, τmean, sensitively reflects changes in polarity of its microenvironment induced by 14-3-3ζ binding. We have found that all AEDANS-labeled pRGS3 mutants exhibited complex emission decays with multimodal lifetime distributions. The results of the emission decay analysis for all mutants are summarized in Table 2. The 14-3-3ζ protein binding significantly affects the microenvironment around the AEDANS moieties of all pRGS3 mutants with the exception of pRGS3(C480). The largest increase in the mean excited state lifetime was observed for mutant pRGS3(C456), from 9.8 to 12.7 ns, indicating a significant 14-3-3ζ binding-induced decrease in polarity and/or quenching interactions around the AEDANS moiety.
TABLE 2.
Mean excited state lifetimes of AEDANS-labeled single-Cys mutants of pRGS3
| Single-Cys RGS3 mutant | Mean excited state lifetime τmeana |
|
|---|---|---|
| pRGS3 | pRGS3 + 14-3-3ζ | |
| ns | ||
| AEDANS Cys-276 | 13.7 | 12.7 |
| AEDANS Cys-285 | 14.2 | 13.6 |
| AEDANS Cys-427 | 14.3 | 13.8 |
| AEDANS Cys-456 | 9.8 | 12.7 |
| AEDANS Cys-480 | 12.4 | 12.2 |
a The S.D. value is ±0.1 ns.
Because polarized time-resolved emission measurements enable the study of fluorophore movements (47), we used them to examine the effect of 14-3-3ζ binding on segmental dynamics of the AEDANS-labeled pRGS3 mutants. It is likely that the proximity of AEDANS and 14-3-3ζ should restrict AEDANS mobility upon the binding of 14-3-3ζ to pRGS3. The decrease in the fast mobility should depend on both the level of the steric hindrance and the initial rotational freedom of the AEDANS moiety. We found that the phosphorylation of Ser-264 itself has no significant effect on AEDANS fluorescence anisotropy decays. The AEDANS moieties of labeled pRGS3 mutants can be sorted into three groups based on their mobility: (i) very mobile with fast components dominating the anisotropy decays (pRGS3(C456)); (ii) semi-mobile (pRGS3(C276), pRGS3(C285), and pRGS3(C427)); and (iii) rigid with the lowest amplitude of fast decay components (pRGS3(C480)). Fig. 6A shows the differences in the raw AEDANS fluorescence anisotropy decays of all five pRGS3 mutants. As can be seen, the pRGS3(C456) mutant shows the highest mobility, whereas the pRGS3(C480) mutant shows the lowest mobility of the AEDANS moiety.
FIGURE 6.
Fluorescence anisotropy decays of AEDANS-labeled pRGS3 mutants. A, differences in the raw AEDANS fluorescence anisotropy decays of all five pRGS3 mutants. B, changes in the raw AEDANS fluorescence anisotropy decays of the pRGS3(C456) mutant resulting from the interaction with 14-3-3ζ. In both panels, the solid lines represent the best fit.
The mobility of AEDANS moiety of all labeled pRGS3 mutants is significantly reduced upon the 14-3-3ζ protein binding, suggesting either the direct physical contact between the AEDANS moiety and 14-3-3ζ and/or a conformational change of pRGS3 induced by the 14-3-3ζ protein binding. The suppression of the fast components and the related increase in the amplitudes of the slow components are predominantly responsible for the observed changes in the anisotropy decays. The most distinctive change in the mobility was observed for the pRGS3(C456) mutant, which also shows the largest increase in the mean excited state lifetime (Table 2). Fig. 6B demonstrates changes in the raw AEDANS fluorescence anisotropy decays of the pRGS3(C456) mutant resulting from the interaction with 14-3-3ζ. Visual inspection of Fig. 6B reveals that although binding of 14-3-3ζ to the phosphorylated pRGS3(C456) results in a highly significant change in the decay data, the phosphorylation of Ser-264 itself has no effect on AEDANS anisotropy decay.
Residue Cys-456 is located in the loop between helices α5 and α6 (Fig. 5B), which is an important part of the Gα interaction surface of the RGS domain (6). According to our model, this loop is not in direct contact with 14-3-3ζ. We can speculate that observed changes might reflect structural changes in the RGS domain and/or the rearrangement of the segment located between the 14-3-3 binding motif and the RGS domain upon the complex formation.
Circular Dichroism Measurements
CD spectroscopy was used to investigate the possible 14-3-3ζ protein binding-induced changes in the overall structure of pRGS3. However, the far-UV CD spectrum of the 14-3-3ζ·pRGS3 complex shows no significant difference when compared with the sum of the individual CD spectra of pRGS3 and 14-3-3ζ (supplemental Fig. S4B). This suggests that the 14-3-3ζ protein-induced conformational change in the RGS domain of pRGS3, which we observed by fluorescence spectroscopy, affects only a small part of the RGS3 molecule and does not significantly change its overall secondary structure.
The Structure of the 14-3-3ζ·pRGS3 Complex Explains the 14-3-3-dependent Inhibition of RGS3 Function
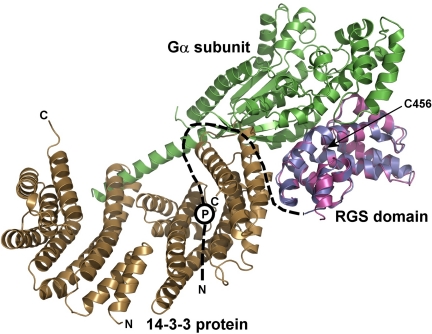
The structural model of the 14-3-3ζ·pRGS3 complex provides a possible explanation for the 14-3-3-dependent inhibition of RGS3 binding to the Gα subunit of G protein. Fig. 7 shows a superimposition of the 14-3-3ζ·pRGS3 complex solution structure with the crystal structure of RGS4 bound to Giα1 (6) using their RGS domains. As can be seen, the 14-3-3 protein binding would sterically occlude the Gα·RGS3 complex formation due to its binding in close proximity to the Gα binding interface of the RGS domain (as demonstrated by steric clashes between the 14-3-3 protein and Gα in Fig. 7). Another factor that can contribute to the inhibition of RGS3 binding to Gα is the 14-3-3 protein-induced conformational change of the RGS domain. Both the time-resolved tryptophan (27) and the time-resolved AEDANS fluorescence measurements (Fig. 6 and Table 2) indicate that the 14-3-3ζ protein binding changes the structure of the RGS domain within its Gα-interacting portion. Residue Cys-456, for which we observed the most significant change in AEDANS fluorescence upon the 14-3-3ζ binding (Fig. 6 and Table 2), is located directly within the Gα interaction surface of the RGS domain (Fig. 7) (6).
FIGURE 7.
The 14-3-3 protein binding sterically occludes the Gα binding interface of the RGS domain of RGS3. The RGS domain of the 14-3-3ζ·pRGS3 complex (shown in blue) is superimposed with the RGS domain of the Giα1·RGS4 complex crystal structure (shown in magenta) (6). The missing N-terminal part of RGS3 is schematically shown as a black dashed line. The approximate position of the phosphorylation site Ser-264 is depicted as a circled P. The Gα subunit is shown in green. As can be seen, the 14-3-3 protein would sterically occlude the Gα·RGS3 complex formation due to its binding in close proximity to the Gα binding interface of the RGS domain (indicated by steric clashes between the 14-3-3ζ protein and Gα). The position of Cys-456 is indicated with an arrow.
DISCUSSION
The goal of this work was to understand in structural terms how the 14-3-3 protein inhibits the interaction between RGS3 and Gα subunit of heterotrimeric G proteins. Because RGS proteins interact with Gα through their RGS domains (5, 6), it is reasonable to speculate that the 14-3-3 protein either sterically occludes the Gα interaction interface of RGS domain and/or changes its structure. The low-resolution solution structure of the 14-3-3ζ·RGS3 complex (Fig. 2D) provides an explanation for the 14-3-3-depedent inhibition of RGS3 function. It shows that the 14-3-3ζ protein, besides binding to the phosphorylated N-terminal 14-3-3 binding motif, interacts with the C-terminal RGS domain in close proximity to the Gα binding interface, and hence, can sterically occlude it. The 14-3-3 protein-induced conformational change in the RGS domain, as suggested by AEDANS fluorescence spectroscopy experiments (Fig. 6 and Table 2), may be an additional factor contributing to the inhibition of RGS3 binding to Gα. A similar mode of 14-3-3-dependent regulation has been suggested for several other proteins: for example, the cell cycle dual specificity phosphatase Cdc25 (48, 49), the class II histone deacetylases (50, 51), or members of the Forkhead box O-class (FOXO) class of transcription factors (52–54). These proteins contain a nuclear localization sequence whose interaction with the nuclear import machinery is inhibited upon the 14-3-3 protein binding. Recently, we have also shown that the forkhead transcription factor FOXO4 is inhibited by a similar mechanism involving the physical occlusion of its DNA binding interface (26).
Despite the large number of 14-3-3 binding partners, only two crystal structures of 14-3-3 complexes with the protein ligand of a size that exceeds the consensus recognition motifs have been solved: the 14-3-3ζ·serotonin N-acetyltransferase (AANAT) complex (24) and the 14-3-3·C-terminal region of the plant plasma membrane H+-ATPase complex (25). These structures revealed that the primary interactions between 14-3-3 and its protein ligand take place within the ligand binding groove where the segment containing the recognition motif binds, similarly as seen in complexes of 14-3-3 isoforms with short phosphopeptides (21, 22, 44). The interactions between 14-3-3 and its ligand can extend beyond those involving the binding groove. For example, the structure of the 14-3-3ζ·AANAT complex revealed that faces of α-helices H1, H3, H5, H7, and H9 pointing into the central channel, but outside the binding groove, contribute to about 60% of the total binding interface (24). Therefore, the available structural data indicate that the 14-3-3 protein dimer employs the highly conserved surface of the central channel as the main interface allowing ligand binding in a structurally reproducible manner. However, how do we explain that many interactions are quite specific for a certain 14-3-3 isoform or isoforms? Comparative structural analyses of 14-3-3 isoforms suggested regions, including the surface formed by helices H8 and H9 (Fig. 3C), which presumably dictate ligand and dimerization preferences (44, 55, 56). Our results show that the RGS domain of RGS3 interacts with the less conserved regions of helices H6 and H8 outside the central channel of the 14-3-3ζ dimer (Figs. 2D and 3A). The 14-3-3 peptides from helices H6 and H8 show decreased accessibility to the solvent upon the complex formation (Fig. 3A), confirming that they, or their parts, form the binding interface. We believe that this is the first experimental evidence showing that the 14-3-3 protein directly interacts with its binding partner using regions other than that of the central channel. The involvement of such less conserved regions may explain the observed isoform-specific interactions between 14-3-3 and their ligands (reviewed in Ref. 57).
A bioinformatics analysis revealed that >90% of 14-3-3 protein binding partners contain disordered regions and that almost all 14-3-3 binding motifs are inside these unstructured segments (58). The RGS3 protein is not an exception as the theoretical prediction suggests that the N-terminal part of RGS3 containing the 14-3-3 binding motif is disordered (supplemental Fig. S1) and that peptides from this region show fast deuteration kinetics (Fig. 4). It is therefore possible that the 14-3-3ζ binding interface of RGS3 is very plastic and variable when compared with the rigid and well defined binding interface of the 14-3-3 protein dimer. Hence, the solution structure of the 14-3-3ζ·RGS3 complex likely represents the average of many possible configurations simultaneously present in the solution. This could be a relatively common feature of complexes formed by 14-3-3 proteins if we consider the high disorder propensity of 14-3-3 ligands.
Supplementary Material
This work was supported by the Grant Agency of the Academy of Sciences of the Czech Republic (Grant IAA501110801); Grant Agency of Charles University (Grant 28510); Czech Science Foundation (Projects P305/11/0708 and P207/10/1040); Ministry of Education, Youth, and Sports of the Czech Republic Research (Projects MSM0021620857 and MSM0021620835 and Center of Neurosciences LC554); and Academy of Sciences of the Czech Republic (Research Projects AV0Z50110509 and AV0Z50200510).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Throughout this manuscript, pRGS3 denotes RGS3 phosphorylated at Ser-264.
- RGS
- regulator of G protein signaling
- 1,5-IAEDANS
- 5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid
- AEDANS
- 5-(((acetylamino)ethyl)amino) naphthalene-1-sulfonic acid
- SAXS
- small angle x-ray scattering
- HDX
- hydrogen/deuterium exchange
- HDX-MS
- hydrogen/deuterium exchange coupled to mass spectrometry.
REFERENCES
- 1. Lee R. H., Whelan J. P., Lolley R. N., McGinnis J. F. (1988) Exp. Eye Res. 46, 829–840 [DOI] [PubMed] [Google Scholar]
- 2. Willardson B. M., Wilkins J. F., Yoshida T., Bitensky M. W. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1475–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berman D. M., Wilkie T. M., Gilman A. G. (1996) Cell 86, 445–452 [DOI] [PubMed] [Google Scholar]
- 4. Watson N., Linder M. E., Druey K. M., Kehrl J. H., Blumer K. J. (1996) Nature 383, 172–175 [DOI] [PubMed] [Google Scholar]
- 5. Hepler J. R., Berman D. M., Gilman A. G., Kozasa T. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tesmer J. J., Berman D. M., Gilman A. G., Sprang S. R. (1997) Cell 89, 251–261 [DOI] [PubMed] [Google Scholar]
- 7. De Vries L., Gist Farquhar M. (1999) Trends Cell Biol. 9, 138–144 [DOI] [PubMed] [Google Scholar]
- 8. Abramow-Newerly M., Roy A. A., Nunn C., Chidiac P. (2006) Cell. Signal. 18, 579–591 [DOI] [PubMed] [Google Scholar]
- 9. Benzing T., Yaffe M. B., Arnould T., Sellin L., Schermer B., Schilling B., Schreiber R., Kunzelmann K., Leparc G. G., Kim E., Walz G. (2000) J. Biol. Chem. 275, 28167–28172 [DOI] [PubMed] [Google Scholar]
- 10. Benzing T., Köttgen M., Johnson M., Schermer B., Zentgraf H., Walz G., Kim E. (2002) J. Biol. Chem. 277, 32954–32962 [DOI] [PubMed] [Google Scholar]
- 11. Niu J., Scheschonka A., Druey K. M., Davis A., Reed E., Kolenko V., Bodnar R., Voyno-Yasenetskaya T., Du X., Kehrl J., Dulin N. O. (2002) Biochem. J. 365, 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ward R. J., Milligan G. (2005) Mol. Pharmacol. 68, 1821–1830 [DOI] [PubMed] [Google Scholar]
- 13. Abramow-Newerly M., Ming H., Chidiac P. (2006) Cell. Signal. 18, 2209–2222 [DOI] [PubMed] [Google Scholar]
- 14. Fu H., Subramanian R. R., Masters S. C. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 617–647 [DOI] [PubMed] [Google Scholar]
- 15. Bridges D., Moorhead G. B. (2004) Sci. STKE 2004, re10. [DOI] [PubMed] [Google Scholar]
- 16. Mackintosh C. (2004) Biochem. J. 381, 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aitken A. (2006) Semin. Cancer Biol. 16, 162–172 [DOI] [PubMed] [Google Scholar]
- 18. Xiao B., Smerdon S. J., Jones D. H., Dodson G. G., Soneji Y., Aitken A., Gamblin S. J. (1995) Nature 376, 188–191 [DOI] [PubMed] [Google Scholar]
- 19. Liu D., Bienkowska J., Petosa C., Collier R. J., Fu H., Liddington R. (1995) Nature 376, 191–194 [DOI] [PubMed] [Google Scholar]
- 20. Muslin A. J., Tanner J. W., Allen P. M., Shaw A. S. (1996) Cell 84, 889–897 [DOI] [PubMed] [Google Scholar]
- 21. Rittinger K., Budman J., Xu J., Volinia S., Cantley L. C., Smerdon S. J., Gamblin S. J., Yaffe M. B. (1999) Mol. Cell 4, 153–166 [DOI] [PubMed] [Google Scholar]
- 22. Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., Cantley L. C. (1997) Cell 91, 961–971 [DOI] [PubMed] [Google Scholar]
- 23. Petosa C., Masters S. C., Bankston L. A., Pohl J., Wang B., Fu H., Liddington R. C. (1998) J. Biol. Chem. 273, 16305–16310 [DOI] [PubMed] [Google Scholar]
- 24. Obsil T., Ghirlando R., Klein D. C., Ganguly S., Dyda F. (2001) Cell 105, 257–267 [DOI] [PubMed] [Google Scholar]
- 25. Ottmann C., Marco S., Jaspert N., Marcon C., Schauer N., Weyand M., Vandermeeren C., Duby G., Boutry M., Wittinghofer A., Rigaud J. L., Oecking C. (2007) Mol. Cell 25, 427–440 [DOI] [PubMed] [Google Scholar]
- 26. Silhan J., Vacha P., Strnadova P., Vecer J., Herman P., Sulc M., Teisinger J., Obsilova V., Obsil T. (2009) J. Biol. Chem. 284, 19349–19360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rezabkova L., Boura E., Herman P., Vecer J., Bourova L., Sulc M., Svoboda P., Obsilova V., Obsil T. (2010) J. Struct. Biol. 170, 451–461 [DOI] [PubMed] [Google Scholar]
- 28. Silhan J., Obsilova V., Vecer J., Herman P., Sulc M., Teisinger J., Obsil T. (2004) J. Biol. Chem. 279, 49113–49119 [DOI] [PubMed] [Google Scholar]
- 29. Boura E., Silhan J., Herman P., Vecer J., Sulc M., Teisinger J., Obsilova V., Obsil T. (2007) J. Biol. Chem. 282, 8265–8275 [DOI] [PubMed] [Google Scholar]
- 30. Woodcock J. M., Murphy J., Stomski F. C., Berndt M. C., Lopez A. F. (2003) J. Biol. Chem. 278, 36323–36327 [DOI] [PubMed] [Google Scholar]
- 31. Houtman J. C., Brown P. H., Bowden B., Yamaguchi H., Appella E., Samelson L. E., Schuck P. (2007) Protein Sci. 16, 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roessle M. W., Klaering R., Ristau U., Robrahn B., Jahn D., Gehrmann T., Konarev P., Round A., Fiedler S., Hermes C., Svergun D. (2007) J. Appl. Crystallogr. 40, S190–S194 [Google Scholar]
- 34. Guinier A. (1939) Ann. Phys. (Paris) 12, 161–237 [Google Scholar]
- 35. Svergun D. I. (1992) J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 36. Porod G. (1982) in Small-angle X-ray Scattering (Glatter O., Kratky O. eds) pp. 17–51, Academic Press, London [Google Scholar]
- 37. Franke D., Svergun D. I. (2009) J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Volkov V. V., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 860–864 [Google Scholar]
- 39. Strohalm M., Kavan D., Novák P., Volný M., Havlícek V. (2010) Anal. Chem. 82, 4648–4651 [DOI] [PubMed] [Google Scholar]
- 40. Veisova D., Rezabkova L., Stepanek M., Novotna P., Herman P., Vecer J., Obsil T., Obsilova V. (2010) Biochemistry 49, 3853–3861 [DOI] [PubMed] [Google Scholar]
- 41. Obsilova V., Herman P., Vecer J., Sulc M., Teisinger J., Obsil T. (2004) J. Biol. Chem. 279, 4531–4540 [DOI] [PubMed] [Google Scholar]
- 42. Vecer J., Herman P. (2011) J. Fluoresc. 21, 873–881 [DOI] [PubMed] [Google Scholar]
- 43. Melhuish W. H. (1961) J. Phys. Chem. 65, 229–235 [Google Scholar]
- 44. Yang X., Lee W. H., Sobott F., Papagrigoriou E., Robinson C. V., Grossmann J. G., Sundström M., Doyle D. A., Elkins J. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17237–17242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Engen J. R. (2003) Analyst 128, 623–628 [DOI] [PubMed] [Google Scholar]
- 46. Marcoux J., Man P., Petit-Haertlein I., Vivès C., Forest E., Fieschi F. (2010) J. Biol. Chem. 285, 28980–28990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lakowicz J. R. (1999) Principles of Fluorescence Spectroscopy, Second Ed., pp. 321–346, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 48. Davezac N., Baldin V., Gabrielli B., Forrest A., Theis-Febvre N., Yashida M., Ducommun B. (2000) Oncogene 19, 2179–2185 [DOI] [PubMed] [Google Scholar]
- 49. Margolis S. S., Kornbluth S. (2004) Cell Cycle 3, 425–428 [PubMed] [Google Scholar]
- 50. McKinsey T. A., Zhang C. L., Olson E. N. (2001) Mol. Cell. Biol. 21, 6312–6321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang J., Winkler K., Yoshida M., Kornbluth S. (1999) EMBO J. 18, 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 53. Brownawell A. M., Kops G. J., Macara I. G., Burgering B. M. (2001) Mol. Cell. Biol. 21, 3534–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Obsilova V., Vecer J., Herman P., Pabianova A., Sulc M., Teisinger J., Boura E., Obsil T. (2005) Biochemistry 44, 11608–11617 [DOI] [PubMed] [Google Scholar]
- 55. Benzinger A., Popowicz G. M., Joy J. K., Majumdar S., Holak T. A., Hermeking H. (2005) Cell Res. 15, 219–227 [DOI] [PubMed] [Google Scholar]
- 56. Wilker E. W., Grant R. A., Artim S. C., Yaffe M. B. (2005) J. Biol. Chem. 280, 18891–18898 [DOI] [PubMed] [Google Scholar]
- 57. Gardino A. K., Smerdon S. J., Yaffe M. B. (2006) Semin. Cancer Biol. 16, 173–182 [DOI] [PubMed] [Google Scholar]
- 58. Bustos D. M., Iglesias A. A. (2006) Proteins 63, 35–42 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.