Background: The properties required for antimicrobial activity of chemokines are unclear.
Results: Native thrombocidin-1 requires a three-dimensional positive patch for activity, but unfolded thrombocidin-1 is active through the N-terminal linear peptide regions.
Conclusion: Native thrombocidin-1 and unfolded thrombocidin-1 exert activity via distinct structural elements.
Significance: Folded and unfolded antimicrobial chemokines can exert activity through different structural elements.
Keywords: Antimicrobial Peptides, Cytokine Action, Peptide Arrays, Peptide Conformation, Protein Denaturation, Protein Domains, Protein Folding, Protein Structure
Abstract
Chemokines (chemotactic cytokines) can have direct antimicrobial activity, which is apparently related to the presence of a distinct positively charged patch on the surface. However, chemokines can retain antimicrobial activity upon linearization despite the loss of their positive patch, thus questioning the importance of this patch for activity. Thrombocidin-1 (TC-1) is a microbicidal protein isolated from human blood platelets. TC-1 only differs from the chemokine NAP-2/CXCL7 by a two-amino acid C-terminal deletion, but this truncation is crucial for antimicrobial activity. We assessed the structure-activity relationship for antimicrobial activity of TC-1. Reduction of the charge of the TC-1-positive patch by replacing lysine 17 with alanine reduced the activity against bacteria and almost abolished activity against the yeast Candida albicans. Conversely, augmentation of the positive patch by increasing charge density or size resulted in a 2–3-fold increased activity against Staphylococcus aureus, Escherichia coli, and Bacillus subtilis but did not substantially affect activity against C. albicans. Reduction of TC-1 resulted in loss of the folded conformation, but this disruption of the positive patch did not affect antimicrobial activity. Using overlapping 15-mer synthetic peptides, we demonstrate peptides corresponding to the N-terminal part of TC-1 to have similar antimicrobial activity as intact TC-1. Although we demonstrate that the positive patch is essential for activity of folded TC-1, unfolded TC-1 retained antimicrobial activity despite the absence of a positive patch. This activity is probably exerted by a linear peptide stretch in the N-terminal part of the molecule. We conclude that intact TC-1 and unfolded TC-1 exert antimicrobial activity via distinct structural elements.
Introduction
Chemokines are important regulators of leukocyte-mediated inflammation and defense and constitute a critical link between innate and adaptive immunity. The general chemokine structure consists of an elongated N-terminal segment, three antiparallel β-strands, and a C-terminal α-helix. Chemokines have a highly constrained conformation that is stabilized by two disulfide bonds (1). Correct folding is essential for specific interactions of chemokines with their receptors (1).
In addition to their chemotactic activity, many chemokines also have direct antimicrobial activity (2–4). Antimicrobial chemokines have a distinct three-dimensional amphipathic structure consisting of a hydrophobic region and a positive patch (4, 5). A similar structure is also characteristic for antimicrobial peptides. An amphipathic structure is supposedly required for microbicidal activity, with hydrophobic domains being essential for membrane interactions and cationic domains providing selective interaction with the negatively charged outer surfaces of microorganisms (6–8).
Strikingly, several antimicrobial chemokines as well as disulfide-containing antimicrobial peptides retain antimicrobial activity when linearized (9–12). Moreover, reduction and unfolding are even required to reveal the full antimicrobial activity of human β-defensin 1 (13). Upon linearization, these proteins can lose their characteristic β-sheet secondary structures (14, 15) that will substantially affect their three-dimensional structure (16). These observations question the necessity of the three-dimensional positive patch for antimicrobial activity and suggest that other structural elements, at least in the linearized proteins, are involved in this antimicrobial activity.
Thrombocidins are microbicidal proteins of human blood platelets (2) derived from platelet chemokines that contribute to innate immunity (17). Thrombocidin-1 (TC-1),2 the most potent thrombocidin, only differs from the chemokine NAP-2/CXCL7 by a C-terminal truncation of an alanine and aspartate residue. This truncation is required to reveal the microbicidal activity of TC-1 (2).
In this study, we assessed the structural elements required for antimicrobial activity of TC-1. To determine the role of the positive patch, we constructed TC-1 substitution variants with an augmented or a decreased positive charge of the patch. To study activity of the unfolded protein, we disrupted the three-dimensional structure of TC-1 by reducing the disulfide bonds, and we confirmed change of structure by NMR.
As unfolding did not reduce antimicrobial activity of TC-1, we studied the activity of short linear peptides covering the entire TC-1 sequence. We identified peptides derived from the TC-1 N-terminal region with antimicrobial activity similar to the activity of full-length TC-1. Thus, our data demonstrate that a three-dimensional positive patch is essential for the antimicrobial activity of folded TC-1, whereas in the unfolded TC-1 a part of the N-terminal region seems responsible for the antimicrobial activity. As reduction of antimicrobial proteins likely occurs in vivo (13), it will be relevant to compare structure-function relationships of other antimicrobial proteins in their native and their unfolded conformation.
EXPERIMENTAL PROCEDURES
Microorganisms
Antimicrobial activity was assessed against the laboratory strains Bacillus subtilis ATCC6633, Staphylococcus aureus 42D, Escherichia coli ML-35p (18), and clinical isolates of Candida albicans and Cryptococcus neoformans (2).
Peptide Synthesis
Overlapping 15-mer peptides covering the entire TC-1 sequence were synthesized and purified as described previously (19). Peptides were named after their first amino acid and the position of this amino acid in TC-1.
Molecular Modeling
The x-ray resolved structure of NAP-1 (Protein Data Bank code 1nap) (20) was used as template for all molecular modeling of TC-1. Ramachandran analyses were determined by Procheck (21). As all substitutions are located at the protein surface, and all TC-1 variants only had marginally reduced Ramachandran scores (supplemental Table S1), the designed substitutions would not significantly alter the fold of the protein. RasWin Molecular Graphics version 2.7.4.2 was used for ribbon representation of TC-1 (22). Swiss model was used for prediction of the three-dimensional structure of TC-1 substitution variants (23). CCP4mg version 1.1.1 (24) and UCSF Chimera version 1.3 (25) were used for electrostatic potential and hydrophobicity plots, respectively.
Construction of Expression Vectors Containing TC-1 Variants
DNA coding for TC-1 was obtained in a two-step PCR protocol. Primers were designed based on the cDNA sequence of platelet basic protein (PBP) (26). First a PBP amplicon was amplified from the human bone marrow cDNA library (Clontech) using Pfu polymerase (Stratagene) and oligonucleotides PBP forward and PBP reverse primers, respectively (supplemental Table S1); primers were from Applied Biosystems. This PBP amplicon served as a template to generate the product coding for TC-1, with primers containing NdeI and BamHI restriction sites to allow ligation of the digested PCR product to NdeI-BamHI-digested pET9a expression vector (Novagen). The ligation products were used to transform E. coli BL21DE3(LysS) cells (Merck) by heat shock. Individual colonies on selective LB agar plates were checked for the presence of an insert by colony PCR using a direct primer recognizing the T7 promoter sequence of the pET9a vector (TAATACGACTCACTATAGGG) and the reverse primer for TC-1 (supplemental Table S2). Both strands of three positive clones were sequenced. Bacteria containing the correct construct were stored in glycerol broth at −80 °C until further use.
Sequences coding for TC-1 variants were generated by PCR using primers containing the desired substitutions. For TC-1 S12K, G13K, and I14K (Fig. 1), primers PKPET S12K, PKPET G13K, and PKPET I14K, respectively, were used as forward primers in combination with reverse primer PKPET-1 (supplemental Table S2). For TC-1 D42K and D49K substitutions, PKPET-1 was used as a forward primer, and PKPET D42K and PKPET D49K as the respective reverse primers. PCR products were cloned into pCR2.1, and the ligation product was used to transform E. coli TOP10 F′ (Invitrogen). Individual colonies on selective LB agar were checked for the presence of a correct TC-1 insert. After digestion with NdeI and BamHI, the TC-1 gene-containing fragments of these clones were purified from agarose gels using QiaExII (Qiagen), ligated to NdeI and BamHI-digested pET9a, and the resulting clones were checked for correct inserts as described above.
FIGURE 1.
Primary amino acid sequence of NAP-2, TC-1, and TC-1 substitution variants. Substituted residues in TC-1 variants are indicated in black.
Expression and Purification of Recombinant Proteins
E. coli BL21DE3(lysS) cells containing pET9a-derived constructs were cultured in 0.5–2 liters of LB medium with chloramphenicol + kanamycin (50 μg/ml each). When cultures had reached an absorbance of 0.3 at 620 nm, isopropyl β-d-thiogalactoside (Roche Applied Science) was added to a final concentration of 0.5 mm to induce expression of the cloned genes. After 5 h of incubation at 37 °C, cells were harvested by centrifugation (5 min, 5000 × g), and pellets were resuspended in lysis buffer (20 mm Tris, pH 7.2, 6 m urea, 2.5% of the original culture volume) and kept at −20 °C overnight. Bacteria were lysed by ultrasonication (5 min, 0 °C); cell debris was removed by centrifugation (1 h, 150,000 × g), and the supernatant was diluted 5-fold in 50 mm HEPES, pH 8.0. Recombinant proteins were purified in a two-step procedure adapted from a method described previously for native thrombocidins (2). As a first step sonicates were applied to a 5-ml SP fast flow column (GE Healthcare) equilibrated in 50 mm HEPES, pH 8.0, at a flow of 2.5 ml/min. Proteins were eluted in 0.5 m NaCl, dialyzed against 0.1% acetic acid, and lyophilized. Proteins were further purified using continuous acid urea-PAGE. Protein concentrations were determined with a BCA protein assay (Pierce) and molecular mass and the presence of intended mutations were verified by MALDI-TOF and Q-TOF analyses.
Biochemical Analysis
Activity of reduced TC-1 was assessed by electrophoretic analysis and gel overlay (see below). Protein (∼1 mg/ml) was pretreated in 100 mm Tris-HCl, pH 8.5, containing 6 m guanidine hydrochloride and 2 mm EDTA, at 50 °C for 30 min. Dithiothreitol (DTT, Sigma) was added (1 mg/ml final concentration), and incubation was allowed to proceed for 4 h under nitrogen at 50 °C. Sulfhydryl groups were alkylated by the addition of iodoacetamide (Sigma; 4 mg/ml final concentration) to prevent reoxidation. Alkylated protein was purified by HPLC using a Hypersil PEP C18 RP column (150 × 4.6 mm; Alltech), lyophilized, and dissolved in 0.01% acetic acid. Efficiency of alkylation was confirmed by mass spectrometry. Protein concentration was determined using a BCA protein assay (Pierce). One-dimensional 1H NMR and far-UV circular dichroism spectra were acquired for folded and reduced TC-1 as described in the supplemental material.
CD Spectroscopy
A J-810 spectropolarimeter (Jasco, Tokyo, Japan) was used to acquire far-UV circular dichroism spectra for TC-1 samples prepared for these and the NMR experiments. Either native TC-1 or reduced TC-1 was dissolved in aqueous solution (90:10 H2O/D2O) or with SDS-d25 micelles (10 mm) at a protein concentration of 0.014 mm and at pH 6.3. Reduced TC-1 was prepared by boiling TC-1 for 15 min followed by incubation with 10 mm DTT-d10 for 1 h at room temperature with gentle shaking. For each sample, 10 CD scans were accumulated and averaged, scanning from 260 to 190 nm at a 200 nm/min scanning rate, 1 nm bandwidth, and 0.5 nm resolution. The mean residue ellipticity, [θ], in degrees cm2 dmol−1 was calculated from [θ] = 100[θ]obs/(lcn), where [θ]obs is the observed ellipticity in millidegrees; l is the path length of the cuvette in centimeters; c is the molar concentration, and n is the number of residues in TC-1.
NMR Spectroscopy
One-dimensional 1H NMR spectra were acquired for the samples of native and reduced TC-1 (0.5 mm) in either aqueous solution or with SDS micelles on a Bruker Avance 500 MHz equipped with a Cryo-ProbeTM. 64 scans were acquired at 298 K with a spectral width of 8013 Hz. Water suppression was performed using an excitation sculpting pulse sequence (27).
Antibacterial Activity Testing
Microbicidal Assay
Microbicidal activity was assayed essentially as described by Harwig et al. (28). Overnight cultures in trypticase soy broth (TSB, Difco) were diluted 100-fold in fresh TSB and cultured for 3 h at 37 °C (bacteria) or 30 °C (yeasts). Microorganisms were washed twice with 10 mm phosphate buffer, pH 7.0, + 0.03% (w/v) TSB (incubation buffer), the absorbance at 620 nm was measured, and the microorganisms were diluted to 2 × 106 CFU/ml in incubation buffer, based on absorbance.
To determine the concentration of protein or peptide required to kill at least 99.9% of the inoculum (LC99.9), 25-μl aliquots of 2-fold serially diluted peptide in incubation buffer were prepared in wells of a polypropylene microtiter plate (Costar, Corning), and to each of the wells 25 μl of a microbial suspension containing 2 × 106 CFU/ml was added. To assess the influence of physiological salt concentrations, 0.9% (w/v) NaCl was added to incubations. All tests were performed in duplicate. After 2 h of incubation on a rotary shaker at 150 rpm at 37 °C, duplicate 10-μl aliquots were plated on blood agar plates. The plates were inspected for growth after 24 h.
Radial Diffusion Assay
Microbial suspensions were prepared as described for the liquid bactericidal assay. An inoculum of 107 CFU was mixed with 20 ml of nutrient-poor agarose (0.03% (w/v) TSB in 10 mm sodium phosphate buffer, pH 7.0, with 1% low electroendo-osmosis-agarose (Sigma)) at 45 °C and immediately poured into a 10 × 10-cm culture plate. Wells of 2 mm in diameter were punched in the agarose to which 2.5-μl samples of 250 μm protein solutions were added. Proteins were allowed to diffuse into the agarose for 3 h at 37 °C (bacteria) or 30 °C (yeasts). Subsequently, the agarose was overlaid with 20 ml of double-strength nutrient agar (6% TSB, 1% Bacto-agar (Difco), 45 °C), and plates were incubated overnight at 37 °C (bacteria) or 30 °C (yeasts). To assess the influence of physiological salt concentrations, 0.9% (w/v) NaCl was added to both the nutrient-poor agarose and double strength nutrient agar. The diameter of the zone devoid of microbial growth was measured to calculate the area of growth inhibition.
Gel Overlay Assay
Proteins were separated by AU-PAGE as described by Harwig et al. (29). The gel was washed three times for 8 min in 10 mm phosphate buffer, pH 7.0, and placed on a plate with B. subtilis-inoculated nutrient-poor agarose (see under “Radial Diffusion Assay”), and the plate was incubated for 3 h. After removal of the gel, the agarose was overlaid with double-strength nutrient agar and treated as described for the radial diffusion assay.
RESULTS
Analysis of Positive Patch of TC-1
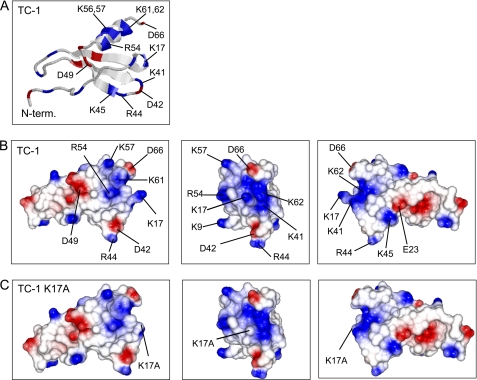
Analysis of TC-1 by electrostatic surface plotting showed that TC-1 has a positive patch composed of the residues Arg-54, Lys-56, Lys-57, Lys-61, and Lys-62 in one face of the C-terminal α-helix; residue Lys-17 in the turn between the N-terminal segment and the first β-strand, and residues Lys-41, Arg-44, and Lys-45 in the turn between the second and third β-strand (Fig. 2, A and B). Within this positive patch, residue Lys-17 appears to fulfill a central role by bridging the positive charges of the loop between the second and third β-strand and those of the C-terminal α-helix (Fig. 2B).
FIGURE 2.
Structural characteristics of TC-1 and TC-1 K17A. A, ribbon model of TC-1 based on the x-ray structure of NAP-2 (20). Electrostatic surface plots of TC-1 (B) and TC-1 K17A (C) were calculated using CCP4mg. Positively and negatively charged surfaces are shown in blue and red, respectively. The left panels correspond to the orientation of TC-1 as shown in A. The middle and right panels show the molecules rotated along their vertical axis by 90° and 180°, respectively.
Disruption of Positive Patch Reduces the Antimicrobial Activity of TC-1
Because Lys-17 occupies a central position in the TC-1 positive patch, substitution of this residue with an uncharged amino acid would disrupt this patch. Electrostatic potential plots show that a substitution of lysine 17 by alanine (K17A) would indeed have a profound effect on the positive patch (Fig. 2C). We therefore constructed a TC-1 K17A variant (Fig. 2B) to assess the importance of the positive patch for antimicrobial activity of TC-1.
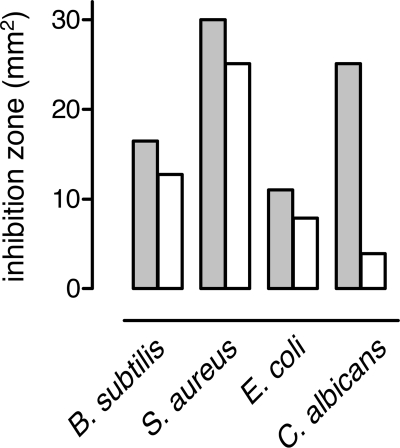
Recombinant TC-1 (rTC-1) had antimicrobial activity against all species tested in radial diffusion assays (Fig. 3). TC-1 K17A almost completely lacked activity against C. albicans and had reduced activity against B. subtilis, S. aureus, and E. coli (Fig. 3). Thus, the cationic residue Lys-17 in the positive patch is important for antimicrobial activity of TC-1, particularly against C. albicans.
FIGURE 3.
Antimicrobial activity of TC-1 and TC-1 K17A. In a radial diffusion assay with the indicated microorganisms, equimolar amounts (2.5 μl of 250 μm preparations) of TC-1 (gray bars) and TC-1 K17A (white bars) were tested. Activity was measured as the area of microbial growth inhibition.
Design of TC-1 Substitution Variants with Augmented Positive Patches
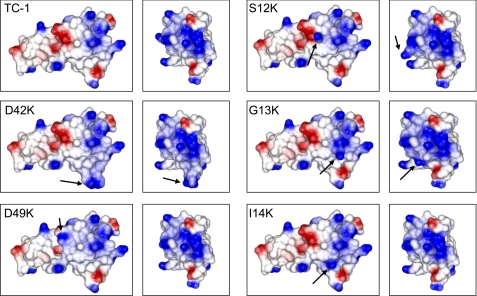
As a complementary approach to assess the importance of the positive patch, we constructed TC-1 substitution variants with augmented positive patches. The negatively charged residue Asp-42 occupies a central position within the positive patch (Fig. 2B), thus reducing the net positive charge of the patch. We constructed a TC-1 D42K substitution variant that has a positive patch with substantially augmented charge density as shown by an electrostatic potential plot of a Swiss Model-generated three-dimensional structure (Fig. 4).
FIGURE 4.
Electrostatic surface plots of thrombocidin variants with enhanced positive patches. Electrostatic surfaces were calculated using CCP4mg, in which positively and negatively charged surfaces are shown in blue and red, respectively. The left panels correspond to the orientation of TC-1 as shown in Fig. 2A. The right panels show the molecules rotated along their vertical axis by 90°.
We also aimed to enlarge the size of the positive patch. The acidic residue Asp-49 is proximal to the positive patch (Fig. 2B), and D49K substitution of TC-1 indeed strongly enlarges the surface area of the positive patch (Fig. 4). TC-1 S12K, G13K, and I14K substitutions result in increased cationicity of the outer surface in the region between the Asp-42 and Asp-49 residues, with Ser-12 located the most distal and Ile-14 the most proximal to the native TC-1 positive patch (Fig. 4).
Antimicrobial Activity of TC-1 Variants with Augmented Positive Patches
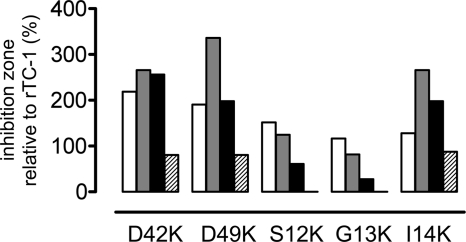
The activity of the TC-1 substitution variants was assessed in a radial diffusion assay, and the area of growth inhibition was compared with that of unmodified TC-1. The TC-1 D42K variant with a larger charge density of the positive patch had a 2.6-fold larger zone of growth inhibition of S. aureus and E. coli compared with TC-1, and 2-fold increased inhibition zone of B. subtilis (Fig. 5). However, the inhibition zone of C. albicans was reduced to 80% that of TC-1. TC-1 D49K, which has a larger positive patch size, produced a 3.4-fold larger zone of growth inhibition of S. aureus than did TC-1 and a 2-fold larger zone of inhibition of B. subtilis and E. coli (Fig. 5). The inhibition zone of C. albicans was reduced to 80% that of TC-1.
FIGURE 5.
Antimicrobial activity of TC-1 variants with augmented positive patches. In a radial diffusion assay equimolar amounts of TC-1 and of the indicated variants were tested, and activity was measured as the area of microbial growth inhibition. Activity of the TC-1 variants against B. subtilis, S. aureus, E. coli, and C. albicans (from left to right) is expressed as inhibition zone size relative to that of TC-1. The areas of growth inhibition of these microorganisms by TC-1 were 16.5, 30.0, 11.0, and 25.1 mm2, respectively.
Of the TC-1 variants with lysine substitutions in the N-terminal segment, only TC-1 I14K had substantially increased antibacterial activity (Fig. 5). The S12K and G13K substitutions exhibited a slight increase in inhibition zones of B. subtilis. Activity of these variants against S. aureus was similar to that of TC-1. With E. coli, inhibition zones of S12K and G13K were 60 and 27% of those of TC-1, respectively. S12K and G13K completely lacked activity against C. albicans (Fig. 5).
Antibacterial Activity of Reduced TC-1
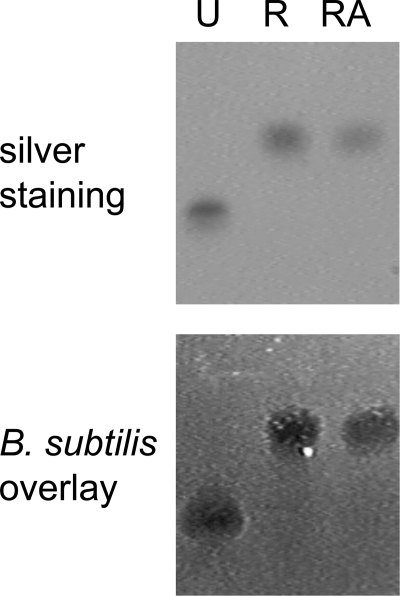
Because several antimicrobial peptides and chemokines retain antimicrobial activity as linearized molecules, we subsequently investigated whether TC-1 would also retain antimicrobial activity upon linearization. Native TC-1 was reduced by DTT treatment or reduced and alkylated. Samples were run on acid urea-PAGE, and the gels were either silver-stained or used for antibacterial activity testing in an overlay assay with B. subtilis. Reduced and reduced/alkylated TC-1 migrated slower in acid urea-PAGE compared with the native protein (Fig. 6, top panel), indicating that the native TC-1 conformation was disrupted. As expected, alkylation, performed to prevent refolding, did not affect protein migration because alkylation does not change the charge of reduced proteins and increases the molecular weight only marginally.
FIGURE 6.
Influence of reduction on antibacterial activity of TC-1. Samples of native TC-1 were left untreated (U), were reduced in DTT (R), or were reduced and alkylated (RA). Samples were analyzed in acid urea gels, which were silver-stained (top panel) or used in an overlay assay with B. subtilis (bottom panel).
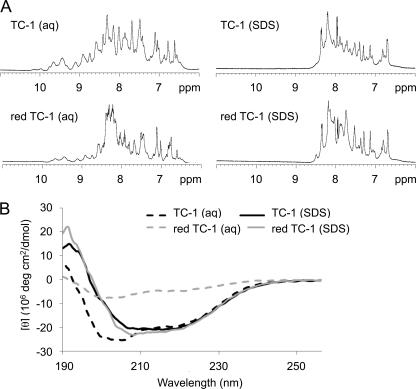
Structural data from NMR spectroscopy and circular dichroism experiments confirmed that reduced TC-1 lost most of its folded structure. The backbone amide region of the one-dimensional NMR spectrum of reduced TC-1 showed a profile very different from that of folded TC-1 with a narrower distribution of peaks (Fig. 7A), indicating loss of folded TC-1 structure upon reduction of the disulfide bonds and disruption of the three-dimensional positive patch. The far-UV CD spectrum of folded TC-1 shows a profile consistent with its expected structure of a three-stranded β-sheet and a C-terminal α-helix (Fig. 7B). The corresponding spectrum for reduced TC-1 showed a much less pronounced profile, indicating major unfolding and supporting the NMR results. Given that many antimicrobial peptides change conformation upon binding to their target membranes (8, 15), the structures of native and reduced TC-1 were also measured in the presence of SDS micelles. The CD and proton NMR spectra for TC-1 indicate that its native β-sheet structure converts into more α-helical elements in the presence of the detergent micelles, with the CD profile showing the prominent double minima at 208 and 222 nm characteristic of α-helical structures (Fig. 7B). A similar membrane-bound conformation was induced for reduced TC-1 (Fig. 7B).
FIGURE 7.
Influence of linearization of TC-1 on three-dimensional structure. A, one-dimensional 1H NMR spectra showing the backbone amide region of the native TC-1 and TC-1 reduced by deuterated DTT-d10 (red TC-1) in aqueous solution and in the presence of SDS micelles. B, corresponding far-UV circular dichroism (CD) spectra of the TC-1 samples.
Interestingly, reduced TC-1 fully retained its antibacterial activity in the overlay assay (Fig. 6, bottom panel). This was confirmed using a quantitative liquid bactericidal assay, in which both folded and reduced TC-1 had an LC99.9 (the lethal concentrations killing 99.9% of an inoculum) of 4 μm for B. subtilis (Table 1). Reduction/alkylation also did not affect activity of TC-1 against S. aureus and only slightly reduced activity against E. coli and against yeasts. The concentration of reduced/alkylated TC-1 required to kill E. coli and C. neoformans was 2-fold higher than of folded TC-1 (Table 1), and the area of growth inhibition of C. albicans in a radial diffusion assay was reduced from 24 mm2 for native to 20 mm2 for unfolded TC-1. When tested at physiological salt concentrations, both native and unfolded TC-1 completely lacked antimicrobial activity (Table 1).
TABLE 1.
Microbicidal activity of TC-1 and TC-derived peptides
Peptides were named after the first residue and its position in TC-1. RA indicates reduced/alkylated.
| Peptide | Sequence | Charge | LC99.9 |
|||
|---|---|---|---|---|---|---|
| B. subtilis | E. coli | S. aureus | C. neoformans | |||
| μm | ||||||
| rTC-1 | AELRCMC… . LAGDES | +4 | 3.8 | 1.9 | 1.9–3.8 | 7.5 |
| RA rTC-1 | AELRCMC… . LAGDES | +4 | 3.8 | 3.8 | 1.9–3.8 | 15 |
| rTC-1a | AELRCMC… . LAGDES | +4 | >60 | >60 | >60 | >60 |
| RA rTC-1a | AELRCMC… . LAGDES | +4 | >60 | >60 | >60 | >60 |
| A1 | AELRCMCIKTTSGIH | +1 | 15 | 30 | >120 | 15 |
| E2 | ELRCMCIKTTSGIHP | +1 | 30 | 60 | >120 | 7.5 |
| L3 | LRCMCIKTTSGIHPK | +3 | 3.8 | 7.5 | 30 | 1.9 |
| R4 | RCMCIKTTSGIHPKN | +3 | 15 | 7.5 | >120 | 3.8 |
| C5 | CMCIKTTSGIHPKNI | +2 | 60 | >120 | >120 | 60 |
| M6 | MCIKTTSGIHPKNIQ | +2 | >120 | >120 | >120 | >120 |
| D51 | DAPRIKKIVQKKLAG | +4 | 30 | 120 | >120 | 30 |
| A52 | APRIKKIVQKKLAGD | +4 | 30 | >120 | 120 | 30 |
| P53 | PRIKKIVQKKLAGDE | +3 | 120 | >120 | >120 | 60–120 |
| R54 | RIKKIVQKKLAGDES | +3 | 60–120 | >120 | >120 | 30 |
a Testing was done with 0.9% NaCl.
TC-1-derived Peptides with Antimicrobial Activity
Our studies with the TC-1 positive patch mutants clearly showed the importance of the three-dimensional patch for the activity of folded TC-1. It was therefore remarkable that unfolding of TC-1, and concomitant disruption of the positive patch, did not markedly affect the antimicrobial activity. Several peptides derived from chemokines and antimicrobial peptides have antimicrobial activity (30–33). To investigate the possibility that linear regions within the protein could be responsible for the antimicrobial activity of linearized TC-1, we screened overlapping 15-mer peptides covering the entire TC-1 sequence. Peptides A1 and D51 indeed had bactericidal activity, causing an ∼3-log decrease in viable counts of E. coli when tested at 60 μm. None of the other peptides tested affected bacterial survival. The active peptides A1 and D51 corresponded to stretches in the N- and C-terminal part of TC-1 respectively. For both regions, 15-mers shifting one residue at a time were synthesized. Peptides A1 to R4 from the N-terminal part of TC-1 were substantially more potent than peptides from the C terminus (Table 1). Peptide L3 was almost as active as rTC-1 (Table 1).
DISCUSSION
Direct antimicrobial activity of chemokines is clearly associated with a distinct positive patch at the surface of the native folded molecule in aqueous solutions (4). These chemokines also have hydrophobic domains, and thus have an amphipathic structure, which is also characteristic for cationic antimicrobial peptides (34). Such an amphipathic structure is deemed essential for antimicrobial activity; a positively charged region is required for selective interaction with negatively charged microbial outer surfaces, whereas a hydrophobic region allows interaction with the apolar part of microbial membranes (35, 36). The mechanism of microbicidal action of these proteins is not fully understood, but it is generally accepted that disruption of microbial membranes (37) or interference with intracellular targets after translocation across the membrane are the most common modes of action (7, 38).
Several antimicrobial peptides and antimicrobial chemokines retain activity as linearized molecules despite the loss of their characteristic β-sheet structures (14, 15). This questions the requirement of the three-dimensional patch of antimicrobial chemokines and shows that other structural elements can also be sufficient for antimicrobial activity of these proteins. Our goal was to investigate the structural elements involved in antimicrobial activity of TC-1.
TC-1 is the most potent antimicrobial protein of human platelets (2). It differs from the chemokine NAP-2/CXCL7 by only a two-amino acid C-terminal truncation. This truncation is required for strong antimicrobial activity (2), but it does not substantially affect chemokine activity (39). In NAP-2, the negatively charged C terminus folds back over the positively charged surface and may thus interfere with antimicrobial activity (40). The C-terminal truncation generating TC-1 reduces the negative charge of the C terminus and thus abolishes interference of the C terminus with the positive patch (40).
The positive patch of TC-1 consists of several residues in the turn between the second and third β-strand, in one face of the C-terminal α-helix, and a single residue (Lys-17) in the turn prior to the first β-strand. Residue Lys-17 occupies a central position between the other two regions and thus links them into a large positive patch. Disruption of the positive patch in TC-1 by substitution of the central cationic residue Lys-17 with an alanine substantially reduced the antimicrobial potency, and vice versa the augmentation of the positive patch enhanced the antimicrobial activity of TC-1. These results clearly demonstrate the importance of this positive patch for the activity of folded TC-1.
The TC-1 K17A substitution had differential effects on the activity against various microorganisms. This substitution nearly abolished antifungal activity but only slightly reduced antibacterial activity. Conversely, augmentation of the positive patch at several positions enhanced antibacterial activity but not antifungal activity. A similar phenomenon has been described for variants of human β-defensins where small modifications in amphipathic structures result in substantially altered antimicrobial specificity (41, 42). It has been proposed that pathogen specificity has driven evolutionary diversification of defensin genes (43). Identification of functional sites involved in pathogen specificity is important to improve our understanding of the mechanism of action of antimicrobial peptides and chemokines and may be utilized to design variants with enhanced activity.
In the presence of SDS micelles, TC-1 loses most of its characteristic β-sheet structure and adopts a conformation with higher α-helical content. Similar conformational changes have been reported for hBD-3 and GCP-2/CXCL6 in membrane-mimicking conditions (14, 15, 44). It has been suggested that these structural modifications are involved in the mode of action of antimicrobial proteins (14, 15). In aqueous conditions, the distinct hydrophilic positive surface patch likely is required for association with the negatively charged outer surface of microorganisms. Upon close contact with microbial membranes, conformational changes likely result in exposure of more apolar residues. The resulting increased hydrophobicity could facilitate subsequent steps in the mode of action of these molecules, i.e. to insert into or transverse the microbial membranes.
Upon reduction, TC-1 loses most of its characteristic β-sheet structure, indicating that the structure of native TC-1 is rigidly held in place by two disulfide bonds. The positive patch of native TC-1 is formed by residues of several secondary structural elements, including residues of the β-sheet. This implies that the patch is disrupted in reduced TC-1. Despite the lack of a positive patch, reduced and unfolded TC-1 had antimicrobial activity comparable with the native protein. This indicates that unfolded TC-1 exerts antimicrobial activity via a distinct mode of action.
Interestingly, linear 15-mer peptides corresponding to the N-terminal part of TC-1 had antimicrobial activity similar to that of intact TC-1. Peptides corresponding to the C-terminal region only had activity at high concentrations. This latter result is in accordance with the previous observation that a peptide corresponding to the entire TC-1 C-terminal α-helical region (residues 50–68) lacks antimicrobial activity (45). Thus, the N-terminal region probably is involved in the antimicrobial activity of reduced and unfolded TC-1.
Similar to TC-1, several chemokines and antimicrobial peptides, including hBD-3, also retain antimicrobial activity after linearization (9–12, 32). For instance, variants of hBD3 lacking disulfides are still antimicrobial, despite the loss of most of the structural elements of the native protein (14, 15). Like TC-1, several short hBD-3 derived synthetic peptides exert antimicrobial activity (46), and the corresponding regions within the protein might thus be involved in the activity of linear hBD-3. In a study on the antimicrobial chemokine CCL28, a variant lacking both cysteines of the CC sequence had similar antimicrobial activity as the native folded protein (9). The authors explain the activity of the linear CCL28 variant by suggesting spontaneous folding into the native conformation (9). As CCL28-derived peptides have antimicrobial activity equivalent to the intact folded protein (9), the activity of linear CCL28 might also be due to peptide regions in the unfolded protein, similar to the case of linearized TC-1.
Human β-defensin 1 (hBD-1) even requires reduction and unfolding to unmask its antimicrobial activity (13). Reduced hBD-1 was shown to be present in several human tissues and to colocalize with thioredoxin, the enzyme system likely involved in hBD-1 reduction (13). The necessity for unfolding to reveal hBD-1 activity indicates that the native protein lacks the characteristics required for antimicrobial activity, whereas the linear molecule does exert such activity. Possibly the reduced protein can spontaneously adopt a conformation with antimicrobial activity or, alternatively, linear peptide stretches within its sequence are responsible for activity of linearized hBD-1.
Because linearization of chemokines and antimicrobial proteins occurs in vivo, linear peptides might also be generated by proteolytic degradation of such linearized proteins. A C-terminal antimicrobial fragment of MIP-3α is actually formed due to proteolysis by cathepsin D (47). Thus, generation of such peptides requires proteolysis. Interestingly, proteolysis is a common and important mechanism in modulation of biological activity of chemokines (48–50). Generation of NAP-2/CXCL7 and connective tissue activating peptide-III (CTAP-III) from their parent molecule, platelet basic protein, requires N-terminal proteolytic processing (51, 52). Additional proteolysis at the C terminus is required to convert NAP-2/CXCL7 and CTAP-III into the microbicidal proteins TC-1 and TC-2, respectively (2). Proteolysis of chemokines is also an important mechanism to dampen inflammatory responses (48). In the case of antimicrobial chemokines, proteolysis would abolish the antimicrobial activity of the intact proteins. However, the generation of antimicrobial peptides from processed chemokines would allow the reduction of chemokine activity while maintaining antimicrobial effects, constituting yet another mechanism of chemokine-mediated fine-tuning of host defenses.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- TC-1
- thrombocidin
- PBP
- platelet basic protein
- TSB
- trypticase soy broth.
REFERENCES
- 1. Fernandez E. J., Lolis E. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 469–499 [DOI] [PubMed] [Google Scholar]
- 2. Krijgsveld J., Zaat S. A., Meeldijk J., van Veelen P. A., Fang G., Poolman B., Brandt E., Ehlert J. E., Kuijpers A. J., Engbers G. H., Feijen J., Dankert J. (2000) J. Biol. Chem. 275, 20374–20381 [DOI] [PubMed] [Google Scholar]
- 3. Cole A. M., Ganz T., Liese A. M., Burdick M. D., Liu L., Strieter R. M. (2001) J. Immunol. 167, 623–627 [DOI] [PubMed] [Google Scholar]
- 4. Yang D., Chen Q., Hoover D. M., Staley P., Tucker K. D., Lubkowski J., Oppenheim J. J. (2003) J. Leukocyte Biol. 74, 448–455 [DOI] [PubMed] [Google Scholar]
- 5. Hoover D. M., Boulegue C., Yang D., Oppenheim J. J., Tucker K., Lu W., Lubkowski J. (2002) J. Biol. Chem. 277, 37647–37654 [DOI] [PubMed] [Google Scholar]
- 6. Zasloff M. (2002) Nature 415, 389–395 [DOI] [PubMed] [Google Scholar]
- 7. Shai Y. (2002) Biopolymers 66, 236–248 [DOI] [PubMed] [Google Scholar]
- 8. Nguyen L. T., Haney E. F., Vogel H. J. (2011) Trends Biotechnol. 29, 464–472 [DOI] [PubMed] [Google Scholar]
- 9. Liu B., Wilson E. (2010) Eur. J. Immunol. 40, 186–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klüver E., Adermann K., Schulz A. (2006) J. Pept. Sci. 12, 243–257 [DOI] [PubMed] [Google Scholar]
- 11. Wu Z., Hoover D. M., Yang D., Boulègue C., Santamaria F., Oppenheim J. J., Lubkowski J., Lu W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8880–8885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mandal M., Jagannadham M. V., Nagaraj R. (2002) Peptides 23, 413–418 [DOI] [PubMed] [Google Scholar]
- 13. Schroeder B. O., Wu Z., Nuding S., Groscurth S., Marcinowski M., Beisner J., Buchner J., Schaller M., Stange E. F., Wehkamp J. (2011) Nature 469, 419–423 [DOI] [PubMed] [Google Scholar]
- 14. Liu S., Zhou L., Li J., Suresh A., Verma C., Foo Y. H., Yap E. P., Tan D. T., Beuerman R. W. (2008) ChemBioChem 9, 964–973 [DOI] [PubMed] [Google Scholar]
- 15. Chandrababu K. B., Ho B., Yang D. (2009) Biochemistry 48, 6052–6061 [DOI] [PubMed] [Google Scholar]
- 16. Bauer F., Schweimer K., Klüver E., Conejo-Garcia J. R., Forssmann W. G., Rösch P., Adermann K., Sticht H. (2001) Protein Sci. 10, 2470–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dankert J., Krijgsveld J., van Der Werff J., Joldersma W., Zaat S. A. (2001) J. Infect. Dis. 184, 597–605 [DOI] [PubMed] [Google Scholar]
- 18. Lehrer R. I., Barton A., Daher K. A., Harwig S. S., Ganz T., Selsted M. E. (1989) J. Clin. Invest. 84, 553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Koster H. S., Amons R., Benckhuijsen W. E., Feijlbrief M., Schellekens G. A., Drijfhout J. W. (1995) J. Immunol. Methods 187, 179–188 [DOI] [PubMed] [Google Scholar]
- 20. Malkowski M. G., Wu J. Y., Lazar J. B., Johnson P. H., Edwards B. F. (1995) J. Biol. Chem. 270, 7077–7087 [DOI] [PubMed] [Google Scholar]
- 21. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 22. Goodsell D. S. (2005) Curr. Protoc. Bioinformatics 5.6.1–5.4.23 [DOI] [PubMed] [Google Scholar]
- 23. Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 25. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 26. Wenger R. H., Wicki A. N., Walz A., Kieffer N., Clemetson K. J. (1989) Blood 73, 1498–1503 [PubMed] [Google Scholar]
- 27. Callihan D., West J., Kumar S., Schweitzer B. I., Logan T. M. (1996) J. Magn. Reson. B 112, 82–85 [DOI] [PubMed] [Google Scholar]
- 28. Harwig S. S., Ganz T., Lehrer R. I. (1994) Methods Enzymol. 236, 160–172 [DOI] [PubMed] [Google Scholar]
- 29. Harwig S. S., Chen N. P., Park A. S., Lehrer R. I. (1993) Anal. Biochem. 208, 382–386 [DOI] [PubMed] [Google Scholar]
- 30. Björstad A., Fu H., Karlsson A., Dahlgren C., Bylund J. (2005) Antimicrob. Agents Chemother. 49, 3889–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chan D. I., Hunter H. N., Tack B. F., Vogel H. J. (2008) Antimicrob. Agents Chemother. 52, 883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoover D. M., Wu Z., Tucker K., Lu W., Lubkowski J. (2003) Antimicrob. Agents Chemother. 47, 2804–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reynolds N. L., De Cecco M., Taylor K., Stanton C., Kilanowski F., Kalapothakis J., Seo E., Uhrin D., Campopiano D., Govan J., Macmillan D., Barran P., Dorin J. R. (2010) Antimicrob. Agents Chemother. 54, 1922–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pérez-Cañadillas J. M., Zaballos A., Gutiérrez J., Varona R., Roncal F., Albar J. P., Márquez G., Bruix M. (2001) J. Biol. Chem. 276, 28372–28379 [DOI] [PubMed] [Google Scholar]
- 35. Epand R. M., Vogel H. J. (1999) Biochim. Biophys. Acta 1462, 11–28 [DOI] [PubMed] [Google Scholar]
- 36. Matsuzaki K. (1999) Biochim. Biophys. Acta 1462, 1–10 [DOI] [PubMed] [Google Scholar]
- 37. Huang H. W. (2000) Biochemistry 39, 8347–8352 [DOI] [PubMed] [Google Scholar]
- 38. Kobayashi S., Takeshima K., Park C. B., Kim S. C., Matsuzaki K. (2000) Biochemistry 39, 8648–8654 [DOI] [PubMed] [Google Scholar]
- 39. Krijgsveld J. (1999) Thrombocidins, Microbicidal Proteins of Human Blood Platelets, Ph.D. thesis, University of Amsterdam, Amsterdam, The Netherlands [Google Scholar]
- 40. Nguyen L. T., Kwakman P. H., Chan D. I., Liu Z., de Boer L., Zaat S. A., Vogel H. J. (2011) Antimicrob. Agents Chemother. 55, 2074–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pazgier M., Li X., Lu W., Lubkowski J. (2007) Curr. Pharm. Des. 13, 3096–3118 [DOI] [PubMed] [Google Scholar]
- 42. Antcheva N., Boniotto M., Belozersky I., Pacor S., Verga Falzacappa M. V., Crovella S., Tossi A. (2004) Antimicrob. Agents Chemother. 48, 685–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Semple C. A., Rolfe M., Dorin J. R. (2003) Genome Biol. 4, R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Linge H. M., Collin M., Nordenfelt P., Mörgelin M., Malmsten M., Egesten A. (2008) Antimicrob. Agents Chemother. 52, 2599–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nguyen L. T., Chan D. I., Boszhard L., Zaat S. A., Vogel H. J. (2010) Biochim. Biophys. Acta 1798, 1062–1072 [DOI] [PubMed] [Google Scholar]
- 46. Dhople V., Krukemeyer A., Ramamoorthy A. (2006) Biochim. Biophys. Acta 1758, 1499–1512 [DOI] [PubMed] [Google Scholar]
- 47. Hasan L., Mazzucchelli L., Liebi M., Lis M., Hunger R. E., Tester A., Overall C. M., Wolf M. (2006) J. Immunol. 176, 6512–6522 [DOI] [PubMed] [Google Scholar]
- 48. Wolf M., Albrecht S., Märki C. (2008) Int. J. Biochem. Cell Biol. 40, 1185–1198 [DOI] [PubMed] [Google Scholar]
- 49. Berahovich R. D., Miao Z., Wang Y., Premack B., Howard M. C., Schall T. J. (2005) J. Immunol. 174, 7341–7351 [DOI] [PubMed] [Google Scholar]
- 50. Richter R., Bistrian R., Escher S., Forssmann W. G., Vakili J., Henschler R., Spodsberg N., Frimpong-Boateng A., Forssmann U. (2005) J. Immunol. 175, 1599–1608 [DOI] [PubMed] [Google Scholar]
- 51. Brandt E., Van Damme J., Flad H. D. (1991) Cytokine 3, 311–321 [DOI] [PubMed] [Google Scholar]
- 52. Walz A., Baggiolini M. (1990) J. Exp. Med. 171, 449–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.