Background: ZO-1 is a scaffolding protein implicated in the assembly of tight junctions.
Results: Structures of core PDZ-SH3-GUK, plus and minus JAM-A peptide, and isolated PDZ are presented.
Conclusion: The SH3 domain is required for JAM-A binding to PDZ3.
Significance: This is the first demonstration for the role of an adjacent domain to the binding of ligands to PDZ domains in the MAGUK family.
Keywords: Cell Polarity, Scaffold Proteins, Structural Biology, Tight Junctions, X-ray Crystallography, JAM-A, MAGUK, PDZ, ZO-1, Crystal Structure
Abstract
Tight junctions are cell-cell contacts that regulate the paracellular flux of solutes and prevent pathogen entry across cell layers. The assembly and permeability of this barrier are dependent on the zonula occludens (ZO) membrane-associated guanylate kinase (MAGUK) proteins ZO-1, -2, and -3. MAGUK proteins are characterized by a core motif of protein-binding domains that include a PDZ domain, a Src homology 3 (SH3) domain, and a region of homology to guanylate kinase (GUK); the structure of this core motif has never been determined for any MAGUK. To better understand how ZO proteins organize the assembly of protein complexes we have crystallized the entire PDZ3-SH3-GUK core motif of ZO-1. We have also crystallized this core motif in complex with the cytoplasmic tail of the ZO-1 PDZ3 ligand, junctional adhesion molecule A (JAM-A) to determine how the activity of different domains is coordinated. Our study shows a new feature for PDZ class II ligand binding that implicates the two highly conserved Phe−2 and Ser−3 residues of JAM. Our x-ray structures and NMR experiments also show for the first time a role for adjacent domains in the binding of ligands to PDZ domains in the MAGUK proteins family.
Introduction
Cells are compartmentalized into distinct structural and functional domains that are linked together through signaling pathways. The organization of individual polypeptides into these functional moieties is often dependent upon so-called “scaffolding molecules,” which spatially and temporally integrate the activity of several molecules by providing a template of modular protein-binding domains (1). The membrane-associated guanylate kinase homologs (MAGUKs)3 are one of the most ubiquitous families of scaffolding molecule associated with plasma membrane complexes (2). They have diverse cellular functions, ranging from membrane trafficking (3) and oncogenic signaling (4) to the assembly of synapses (5) and epithelial and endothelial junctions (6). However, they are all characterized by a core motif of protein-binding domains that include a PDZ domain, an SH3 domain, and a region of homology to guanylate kinase (GUK). These domains are often flanked by additional conserved domains which, in conjunction with the core motif, are important for protein function. Although the structures of individual elements of this core motif have been solved, including the PDZ domain (7) and the SH3/GUK domain pair (8–10), the structure of the entire core motif has not been elucidated.
This is a critical deficiency, because it has become increasingly apparent that the functional activity of MAGUKs may be dependent upon interaction between individual domains within the core motif or between these domains and other regions within the scaffold. Cooperativity within the core motif is perhaps not surprising given the close spatial relationships between the individual domains. The SH3 and GUK domains form a tight intramolecular complex (8–10), and disruption of the intramolecular fold has deleterious effects on ligand binding and cellular function (11–13). However, there have been several observations that suggest that cooperativity among different domains is more than a matter of interdependent protein folding. For example, ligand binding to the GUK domain of the neuronal MAGUK protein PSD93, and the subcellular distribution of this protein, are promoted by ligand engagement of the adjacent PDZ domain (14). Similarly, an intramolecular interaction between the N terminus of SAP97 and the core motif regulates binding of protein ligands to the GUK domain (15) or the SH3 domain (16). These observations suggest that the functional activity of a MAGUK may be regulated by cooperative interactions between different domains and their binding partners.
The tight junction (TJ) is a highly organized cell-cell contact that encircles epithelial cells and creates a barrier between these cells to the movement of ions, macromolecules, and immune cells. The assembly and permeability of this barrier are dependent on the zonula occludens (ZO) MAGUK proteins ZO-1, -2, and -3, although the mechanism by which this occurs is poorly understood (17). Presumably, however, it is based on the ability of ZO proteins to bind to many different components of tight junctions and recruit them into a cross-linked array at the tight junction. ZO proteins, like many MAGUKs, have three PDZ domains at their N terminus. The first of these, PDZ1, binds to the barrier-forming claudin transmembrane proteins. The second, PDZ2, forms a dimerization motif that cross-links ZO proteins into higher order hetero- and homo-oligomers (18). The core PDZ domain, PDZ3, binds to the IgCAM cell adhesion molecule JAM-A, which regulates permeability of TJ to ions and immune cells but is not required for TJ assembly (for review, see Ref. 19). The SH3 domain is required for localization of ZO proteins to the TJ, in a process that is not yet understood (20), whereas the GUK domain binds to the transmembrane protein occludin. Although it is not known how the activity of these different domains is coordinated during TJ assembly, it is known that the PSG core motif is a necessary component of assembly and that elements within the core motif regulate the positioning of TJ proteins within the lateral plasma membrane (17, 20). Thus, the PSG core motif is a critical element of TJ assembly.
ZO proteins also have other cellular roles in epithelial cells. They bind to gap junction connexin proteins via PDZ2 and regulate the size and gating properties of these cell-to-cell channels. They also bind directly to adherens junction proteins, such as ARVCF (PDZ1), AF-6/afadin (SH3) and α-catenin (GUK) and regulate the assembly of actomyosin at the adherens junction in polarized epithelial cells (21–23). However, ZO proteins only localize to the adherens junction early in epithelial biogenesis, prior to the formation of tight junctions, and their localization to gap junctions is spatially distinct from tight junctions. These observations imply that mechanisms exist to regulate the spatial and temporal binding of different ligands (e.g. tight junction versus adherens junction) to ZO proteins within the cell.
To understand better how MAGUK proteins organize the assembly of protein complexes we have crystallized the entire PSG core motif of the tight junction MAGUK ZO-1. We found that the three domains are arranged in an extended fashion, with domain-domain interactions limited to adjacent domains. Furthermore, to understand better how the activity of different domains is coordinated we have examined how binding of JAM-A to PDZ3 is influenced by the SH3 and GUK domains. Our unexpected finding shows that JAM-A binding to PDZ3 requires residues from the adjacent SH3 domain.
EXPERIMENTAL PROCEDURES
Expression and Purification of ZO-1 Fragments
Recombinant PSG (amino acids 417–803) and PDZ3 (amino acids 417–516) proteins were expressed in Escherichia coli (BL21C41) as a His6-tagged fusion proteins (pET14b vector; Pharmacia). Cells were cultured at 37 °C in 2YT medium, then induced by 0.1 mm Isopropyl β-d-1-thiogalactopyranoside at an A600 nm of 0.6, and left to grow overnight at 22 °C. Cells were then harvested by centrifugation at 5,000 rpm for 30 min. Pellets were resuspended and washed with 100 mm KCl, 50 mm Tris-HCl, pH 7.5. Lysis was accomplished by sonication in a buffer composed of 25 mm Tris-HCl, pH 7.5, 500 mm NaCl, 10% glycerol, 1% Triton X-100, and 1 mm PMSF. After centrifugation at 30,000 rpm for 1 h, the supernatant was loaded onto a 5-ml His-trap nickel-affinity column (GE Healthcare), and the column was washed with 300 ml of a buffer composed of 25 mm Tris-HCl, pH 7.5, and 500 mm NaCl. The bound protein was eluted with an imidazole gradient. Fractions containing the protein were pooled and concentrated. The His6 tag from the PSG construct was cleaved using tobacco etch virus protease, and the cut protein (as well as the uncut PDZ3) was injected onto a S-200 gel filtration column (GE Healthcare) equilibrated with 25 mm Tris-HCl, pH 7.5, 500 mm NaCl, pH 7.5, and 3 mm DTT (for PSG) or 25 mm Tris-HCl, pH 7.5, 200 mm NaCl, pH 7.5 (for PDZ3). The purity of the protein preparations was confirmed using SDS-PAGE and detected through Coomassie staining. PSG and PDZ3 proteins were concentrated to ∼13 mg/ml and ∼6 mg/ml, respectively, and stored at −80 °C until used.
Peptide Synthesis
The 6- and 12-mer peptides (TSSFLV and EGEFKQTSSFLV), named JAM-A_P6 and JAM-A_P12, respectively, and corresponding to the cytoplasmic tail of human JAM-A (sp_Q9Y624) were prepared by Biomatik Corp. (Cambridge, MA). The amino acid composition and purity (>98%) of peptides were verified by mass spectroscopy and HPLC analysis by the manufacturer.
Crystallization
Crystals of the peptide-free PSG were grown at 20 °C using hanging drop vapor diffusion against a reservoir containing 1.2 m ammonium sulfate (AmSO4), 0.1 m sodium acetate, pH 5.0, 60 mm sodium fluoride (NaF). Drops of 2.0 μl were set up at a 1:1 ratio of reservoir to protein solution at a concentration of 7 mg/ml. Prior to data collection, crystals were transferred into a solution containing 2 m AmSO4, 0.1 m sodium acetate, pH 5.0, 60 mm NaF, and 20% glycerol for cryoprotection. Crystals of the PSG+JAM-A complex were grown at 20 °C using hanging drop vapor diffusion against a reservoir containing 9.0% PEG 3350, 0.1 m sodium malonate, pH 7.0. Drops of 2.5 μl were set up at a 1:1.5 ratio of reservoir to protein solution at a concentration of 7.0 mg/ml. Prior to data collection, crystals were soaked 2–5 min in mother liquor and stepwise increasing concentrations of PEG 3350 from 10 to 30% with the addition of 20% glycerol for cryoprotection. Crystals of the PDZ3 domain were grown at 20 °C using sitting drop vapor diffusion against a reservoir containing 25% PEG 1500, 0.1 m MMT (dl-malic acid, MES, and Tris, pH 5.0). Drops of 2.0 μl were set up at a 1:1 ratio of reservoir to protein solution at a concentration of 5 mg/ml. Prior to data collection, crystals were soaked 2–5 min in mother liquor and stepwise increasing concentrations of PEG 1500 from 25 to 40% for cryoprotection.
Data Collection and Processing
X-ray data were obtained under standard cryogenic conditions at the Advanced Photon Source using SERCAT beamline BM-22 for peptide free-PSG and ID-22 for the PSG+JAM-A and PDZ3 proteins. Diffraction data were indexed, scaled, and merged using XDS and XSCALE (24).
Structure Determination and Refinement
Ternary complex structures were determined via molecular replacement (PHASER) (25) using the structure of human ZO-1 SH3-GUK module (8) (Protein Data Bank code 3LH5) as the search model. The model was used after removing all water molecules, and each domain was positioned separately using rigid body refinement followed by restrained refinement. Iterative refinements were carried out using REFMAC (26), and simulated annealing maps were calculated using CNS (27). Model rebuilding was accomplished using the graphics program COOT 0.7 (28). Final built models comprised the following residues: PSG 417–587, 629–674, and 688–803. Residue Lys629 was modeled as an alanine (residues: 588–628 (U5) and 675–687 were lacking unambiguous electron density); PSG+JAM-A 421–588, 628–684, and 687–802 (residues: 417–420, 589–627 (U5), 685, 686, and 803 were lacking unambiguous electron density); PDZ3 420–512 (residues: 417–419 and 513–516 were lacking unambiguous electron density).
NMR Experiments
NMR experiments were performed on a Bruker 900 MHz spectrometer equipped with a cryogenic triple resonance probe. Experimental conditions were 10 μm protein in 100 mm PO4, pH 7.2, in 90% 1H2O, 10% 2H2O at 25 °C in 3-mm NMR tubes. The WaterLOGSY experiments were performed as described previously (29). The data were collected with a sweep width of 14,370 Hz and a relaxation delay of 2 s. Water was selectively saturated using a 2-ms square-shaped pulse with a mixing time of 2 s. All data were processed by NMRPipe (30) with referencing to the water peak at 4.773 ppm and using a 4-Hz line-broadening window. For determination of the Kd, the data were fit using Kaleidagraph 4.04 to the equation I = Imax − Imax/(1 + L/Kd), where I is the intensity and L is the ligand concentration.
RESULTS AND DISCUSSION
Structure Determination and Overall Fold of the Tandem ZO-1 PDZ3-SH3-GUK Domains
The polypeptide containing the PDZ3-SH3-GUK (residues 417–803) tandem domains of ZO-1 crystallized in the P32 space group and the diffraction pattern exhibited perfect twinning. The PSG structure was solved at 2.85 Å resolution using the molecular replacement method. The asymmetric unit (asu) contained four copies of PSG, but only two had strong electron density for all of the three domains (supplemental Fig. S1). The other two molecules of PSG in the asu did not show traceable density for the terminal GUK domain. Superposition of the complete PSG molecule on the PDZ3-SH3 fragment revealed that there is space for the missing GUK. However, by placing this missing GUK domain on the basis of the modeled PSG, we noticed steric clashes with another GUK from a symmetry-related molecule. This would explain the disorder in the crystal and why we could not trace this domain for two of the four PSG molecules in the asu. The final crystallographic R-factors converged to Rwork 21.1%/Rfree 29.0% (see Table 1 for detailed data collection and refinement statistics).
TABLE 1.
Data collection and refinement statistics
Values in parentheses are for highest resolution shell.
| Parameters | PSG | PSG+JAM-A | PDZ3 |
|---|---|---|---|
| Data collection | |||
| PDB ID | 3TSW | 3TSZ | 3TSV |
| Space group | P32 | P21 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 100.6, 100.6, 182.5 | 50.5, 49.7, 91.8 | 34.5, 45.2, 59.6 |
| α, β, γ (°) | 90, 90, 120 | 90, 101.1, 90 | 90, 90, 90 |
| Resolution (Å) | 87.17-2.85 (3.0-2.83) | 90.1-2.5 (2.57-2.5) | 36.0-1.99 (2.11-1.99) |
| Rmerge (%) | 8.6 (97.6) | 7.3 (55.3) | 5.0 (61.0) |
| I/σI | 17.1 (1.9) | 15.1 (1.6) | 15.1 (2.1) |
| Completeness (%) | 97.6 (86.9) | 96.1 (74.6) | 97.8 (93.3) |
| Redundancy | 5.6 (4.2) | 5.4 (2.4) | 3.2 (3.2) |
| Refinement | |||
| Resolution (Å) | 87.17-2.85 | 90.1-2.5 | 36.0-1.99 |
| No. of reflections | 41,930 | 13,557 | 5,970 |
| Rwork/Rfree (%) | 21.1/29.0 | 23.9/30.9 | 26.8/31.6 |
| No. of atoms | |||
| Protein | 8,532 | 2,730 | 721 |
| Sulfate | 35 | ||
| Peptide | 80 | ||
| Water | 70 | 40 | 37 |
| B-factors (Å2) | |||
| Protein | 73.8 (A) | 50.9 | 36.9 |
| 66.6 (B) | |||
| 76.5 (C) | |||
| 64.7 (D) | |||
| Sulfate | 78.2 | ||
| Peptide | 55.7 | ||
| Water | 44.3 | 50.9 | 38.6 |
| R.m.s.d. | |||
| Bonds length (Å) | 0.012 | 0.007 | 0.009 |
| Bonds angles (°) | 1.565 | 1.058 | 1.146 |
| Ramachandran plot (%) | |||
| Most favored | 87.0 | 87.0 | 87.5 |
| Additionally allowed | 11.0 | 11.4 | 10.0 |
| Generously allowed | 1.3 | 0.6 | 0 |
| Disallowed | 0.6 | 1.0 | 2.5 |
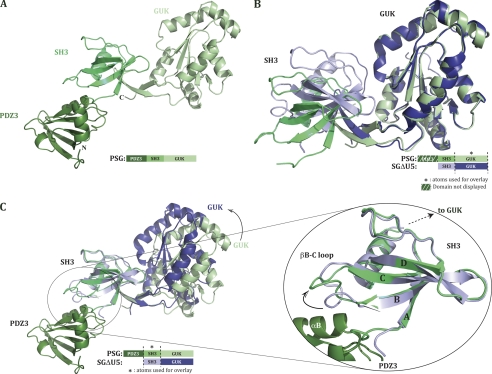
We limit the analysis of the PSG structure to one of the molecules in the asu that had defined density for all three domains (the two molecules with complete density for all three domains are essentially identical, with a root mean square deviation (r.m.s.d.) of 0.71Å over 328 Cα atoms). The overall fold of the tridomain PSG molecule is linear. Interdomain interactions are limited to adjacent domains, such that no interaction between the terminal PDZ3 and GUK domains takes place (Fig. 1A). This extended arrangement of domains was unexpected based on electrostatic considerations. Analysis of the electrostatic potential of the previously solved SH3-GUK domains revealed a high density of positive charge on one of the faces of the GUK domain (supplemental Fig. S2). Intriguingly, the pI of PDZ3 is 4.7. Together, this suggested a possible charge-charge interaction between the PDZ3 domain and the GUK domain. However, our structure of PSG shows that such an interaction does not take place, at least not in the ligand-free state of the molecule. It is still possible that ligand binding to the GUK domain would induce a conformational change that would bring the charge complementary surfaces of the GUK and PDZ3 domains to interact.
FIGURE 1.
Crystal structure of ZO-1 PDZ3-SH3-GUK domains. A, structure of PDZ3-SH3-GUK (PSG) represented as a ribbon diagram and in a green color gradient with PDZ3 as the darker domain. The following residues could not be modeled in the structure: 588–628 (U5 region), 675–687, and the side chain of residue Lys629 were poorly ordered and thus modeled as an alanine. B, overlay on GUK domains of PSG and SGΔU5 structures. SGΔU5 is represented in a blue color gradient. C, PSG and SGΔU5 structures overlaid on their SH3 domains. A black arrow indicates the GUK domain movement. The right side shows SH3 domains more specifically and especially the movement of the βB-C loop induced by the presence of helix αB from the PDZ3 domain in the structure.
Interdomain Flexibility
The previous structures of the SG domains of ZO proteins revealed flexibility between the SH3 and GUK domains (that is, the structure of each individual domain is rather rigid, but the relative domain-domain conformation is variable). To reveal any potential influence of the PDZ3 domain on the packing and relative orientation of the SH3 and GUK domains, we superimposed our new PSG structure on the SH3-ΔU5-GUK structure (SGΔU5). The U5 region is a variable surface loop sequence near the C-terminal part of the SH3 domain. It is present in the PSG construct but did not show any traceable electron density. The construct used to solve SGΔU5 lacked the residues of the U5 region. However, our previous observations comparing SG and SGΔU5 indicate that the presence of U5 (without any U5 ligand) does not affect the relative orientation of the SH3 and GUK domains. Hence, the variability in the presence of the U5 region between PSG and SGΔU5 is not expected to influence the interdomain conformation.
When atoms only from the GUK domain are used as the base for overlay of PSG and SGΔU5 (r.m.s.d. on α-carbon positions: 0.65 Å over 150 atoms), no differences are noticed in the overall structure of the GUK domain (Fig. 1B). Yet, the relative position of the SH3 domain to the GUK domain is clearly more open in the PSG structure versus the SGΔU5 structure. Because the PDZ3 domain does not interact with the GUK domain, it is likely that the newly observed SH3 to GUK conformation is due to differences in crystal contacts. However, this observation does validate our earlier interpretation that the relative SH3-GUK conformation is variable.
Just as the GUK domain conformation is identical between that seen in the PSG and SGΔU5 structures, when the two structures are overlaid based only on atoms from their SH3 domains (r.m.s.d. on α-carbon positions: 0.76 Å over 89 atoms), we see a nearly identical SH3 conformation (Fig. 1C). Specifically, the core of the SH3 domain composed of four β-strands and the α-helix remains unchanged (Fig. 1C zoom). Notably, we do notice a difference in the conformation of the loop that connects strands B and C (which we will refer to as βB-C loop). The difference in βB-C loop conformation is due to the presence of the PDZ3 αB-helix. The functional importance of this change in βB-C loop conformation is discussed below.
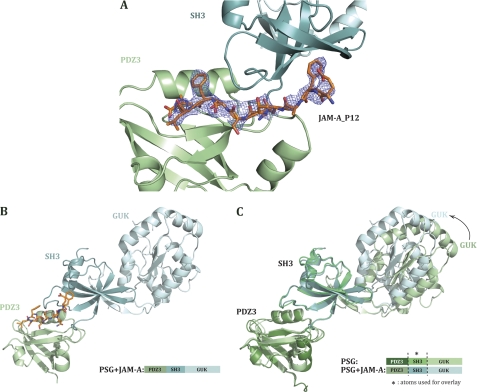
Overall Structure of PSG in Presence of the C-terminal 12-Mer of JAM-A
The scaffolding role of ZO-1 implies that each of the domains binds to various ligands. One reported function of PDZ3 is to bind the C-terminal residues of JAMfamily members (for a recent review of JAM functions see Ref. 19). The impetus for solving the structure of PSG in complex with the C-terminal JAM-A_P12 was to ascertain whether this interaction induces a conformational change in the ZO-1 core motif. The structure of human ZO-1 PDZ3-SH3-GUK in complex with JAM-A_P12 (PSG+JAM-A) was determined to 2.5 Å resolution using the molecular replacement method. The complex crystallized in conditions different from that used for PSG alone, adopted the P21 space group, and contained a single PSG molecule in the asu. We could trace the entire length of PSG, and the electron density for the bound peptide was clear for 8 of the 12 C-terminal residues of JAM-A_P12 (Fig. 2A). The final crystallographic R-factors converged to Rwork 23.9%/Rfree 30.9% (see Table 1 for detailed data collection and refinement statistics).
FIGURE 2.
Crystal structure of ZO-1 PDZ3-SH3-GUK+JAM-A domains. A, 2Fo − Fc omit map for JAM-A_P12. SH3 and PDZ3 domains represented in a ribbon diagram. The JAM-A_P12 peptide (orange sticks) conformation is defined in unbiased electron density that was calculated before the peptide was built in (at a contour level of 1 σ). B, structure of PDZ3-SH3-GUK in the presence of human JAM-A C-terminal 12 residues (PSG+JAM-A) represented as a ribbon diagram. PSG+JAM-A is colored in a light blue gradient with SH3 as the darker domain and PDZ3 in pale green. JAM-A peptide is represented in orange sticks. The following residues could not be modeled in the structure: 417–420; 589–627 (U5 region), 685, 686, 803,and the side chains of residues Ser421, Tyr588, Lys629, Arg684, and Arg748 were poorly ordered and thus modeled as alanines. C, overlay on SH3 domains of PSG and PSG+JAM-A structures. A black arrow indicates the GUK domain movement.
Just as apo-PSG, the complex of PSG+JAM-A adopts a linear and extended conformation (Fig. 2B). To identify any conformational changes induced by JAM-A_P12 binding, we overlaid apo-PSG with the peptide-bound PSG (Fig. 2C). The structural overlay based on atoms of either PDZ3 (r.m.s.d. on α-carbon positions: 0.68 Å over 92 atoms) and SH3 (r.m.s.d. on α-carbon positions: 0.89 Å over 168 atoms) showed no conformational changes in the main chain of these domains or in their relative orientation. However, it revealed a change in SH3/GUK orientation to a more closed conformation in PSG+JAM-A compared with PSG (Fig. 2C). In fact, the relative SH3/GUK orientation in PSG+JAM-A is comparable with that seen in the SGΔU5 crystal structure (8). We thus conclude that this change in orientation is more likely due to different crystal packing induced under different crystallization conditions rather than the presence and binding of the JAM-A peptide.
JAM-A_P12 Binding to PSG
PDZ domains can be divided into different classes on the basis of their ligand sequences (for more details, see Ref. 31). ZO-1 PDZ3 domain belongs to class II and recognizes the motif -Φ-X-Φ-COOH (with X defining any amino acid and Φ a hydrophobic amino acid, usually valine, isoleucine, or leucine). In this study and keeping with the nomenclature for residues binding to PDZ motifs, the JAM-A_P12 peptide C-terminal valine residue is designated Val0 and subsequent residues toward the N terminus termed Leu−1, Phe−2, Ser−3, Ser−4, Thr−5, Gln−6, Lys−7, and Phe−8.
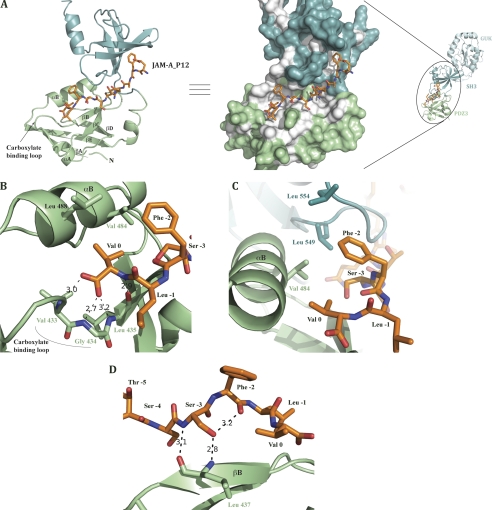
JAM-A_P12 binds a groove on the PDZ3 surface, and the peptide C-terminal residues penetrate a hydrophobic cavity (Fig. 3, A and B). This binding mode is reminiscent of that shown before for peptides binding PDZ class II domains (7), but does deviate in several important aspects. The majority of interactions between the peptide and PDZ3 occur with the last four residues of JAM-A_P12. Previous studies aiming at understanding how PDZ domains recognize such a motif revealed the presence of a loop in the peptide-binding groove termed “carboxylate-binding loop” (for a review, see Fig. 3A and Ref. 31). This well conserved loop usually displays the sequence motif: R/K-X-X-X-G-L-G-F, and the peptide terminal carboxylate is coordinated by a network of hydrogen bonds to main chain amide groups in this loop, as well as an ordered water molecule that is coordinated by the side chain of a conserved Lys/Arg residue at the beginning of the loop (7, 31).
FIGURE 3.
JAM-A peptide binding site. A, left, PSG domains (GUK domain is not displayed) from PSG+JAM-A structure are represented as a ribbon diagram and the JAM-A peptide as sticks. PDZ3 domain secondary structure elements are labeled as well as the carboxylate-binding loop implicated in peptide binding. Right, the Conolly surface of PDZ3 and SH3 domains is shown. Hydrophobic areas are colored in white on the surface. B, JAM-A Val0 is held by hydrogen bonds implicating Val433, Gly434, and Leu435 residues of the carboxylate-binding loop and hydrophobic contacts with Val484 and Leu488. C, JAM-A Phe−2 lies in the hydrophobic pocket and is surrounded by PDZ3 αB Val484 and SH3 leucines 549 and 554. D, JAM-A Ser−3 main chain interacts with PDZ3 βB Leu437. Ser−3 side chain interactions with both PDZ3 βB Leu437 and Phe−3 carbonyl would present Phe−3 to the hydrophobic pocket to strengthen peptide binding.
The ZO-1 PDZ3 domain (true also for the ZO-2 and ZO-3) does not display the well conserved carboxylate-binding loop sequence cited above (sequence is K-G-d-S-Δ-G-d-S, where Δ is a missing residue) or the water molecule. However, keeping with the canonical binding mode, hydrogen bonds are present between the Val0 carboxylate and the amide nitrogens of Val433 and Gly434, both located in the PDZ3 carboxylate-binding loop. The Val0 amide nitrogen also forms a hydrogen bond with the carbonyl of Leu435 (Fig. 3B). The consequence of these Val0 interactions with the carboxylate-binding loop is the presentation of its side chain into a hydrophobic pocket that is bounded by Val484 and Leu488.
In PDZ class II domains, when the peptide residue in position −1 has been shown not to be important for binding (with side chain pointing into solution), residues at the −2 position are invariably important for binding. In the case of JAM-A, the residue at position −2 is a phenylalanine. The structure reveals that Phe−2 is embedded in a continuation of the above mentioned hydrophobic cavity, being surrounded by Val484 from the PDZ3 domain and leucines 549 and 554 from the SH3 domain (Fig. 3C). Notably, Phe−2 does not interact with PDZ3 βB strand as observed in same class PDZ domains (7). Even more interesting, whereas in PDZ class II domains the main chain of the four terminal residues of the bound peptide builds a β-strand interaction with the PDZ domain, JAM-A_P12 interacts in a totally different manner. JAM-A_P12 Ser−3 amide nitrogen interacts with PDZ3 Leu437 carbonyl and Ser−3 side chain hydroxyl interacts with Phe−2 carbonyl backbone (Fig. 3D). Sequence alignment between mammalian JAM proteins indicates a very high conservation of these two residues (supplemental Fig. 3), suggesting a functional importance for these residues. We suggest that the highly conserved Ser−3 is responsible for positioning Phe−2 in the hydrophobic pocket, thus exhibiting a new feature for PDZ class II ligand binding.
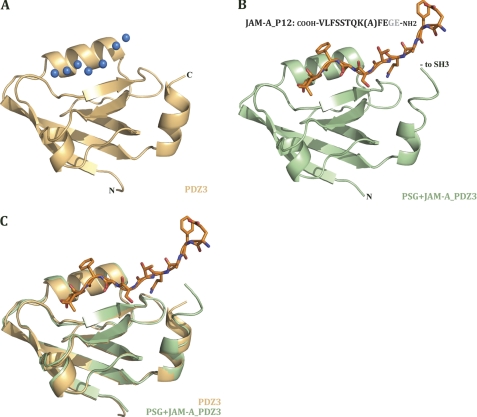
JAM-A_P12 and the Isolated PDZ3 Domain
A guiding goal for this study is to identify relationships between the diverse domains in ZO-1 and how these relationships impact the function of the individual domain. Hence, in addition to solving the structure of the PSG complex with the JAM-A peptide, we also sought to solve the structure of the isolated PDZ3 domain in complex with the peptide. We solved the structure of the ZO-1 PDZ3 domain to 2.0 Å resolution using the model of this domain as seen in our PSG structure as the search model for molecular replacement. See Table 1 for detailed data collection and refinement statistics.
Our crystallization setups of the PDZ3 domain included the JAM-A carboxylic tail, either as the 6- or 12-mer peptides, JAM-A_P6 and JAM-A_P12, respectively. Despite the presence of the peptide, there was no electron density for the peptide at the binding site. Instead, the peptide-binding grove was populated by several water molecules (Fig. 4A). The corresponding view of the PDZ3 domain in complex with JAM-A_P12, as seen in the context of PSG, is shown in Fig. 4B. To understand the reasons for the lack of JAM-A peptide in the crystal structure of the isolated PDZ3 domain, we overlaid the structure of the isolated (and apo) PDZ3 domain on the PDZ3 domain structure as seen in the PSG+JAM-A_P12 structure. This overlay clearly shows no change in the secondary structure of PDZ3 in the presence and absence of ligand (Fig. 4C).
FIGURE 4.
PDZ3 domain with and without JAM-A peptide. A, ribbon diagram of apo-ZO-1 PDZ3 domain. Blue spheres represent water molecules in the ligand free structure that take place in the peptide-binding site. B, ribbon diagram of PDZ3 domain from PSG+JAM-A structure. Peptide is represented as orange sticks, and its sequence is indicated. The lysine side chain is less ordered and was thus modeled as an alanine. The two N-terminal residues of the peptide (gray in the sequence) were not modeled due to disorder and lack of density. C, overlay of isolated apo-PDZ3 domain and PDZ3 from PSG+JAM-A in complex with JAM-A_P12.
SH3 Domain Is Required for JAM-A_P12 Binding to PDZ3
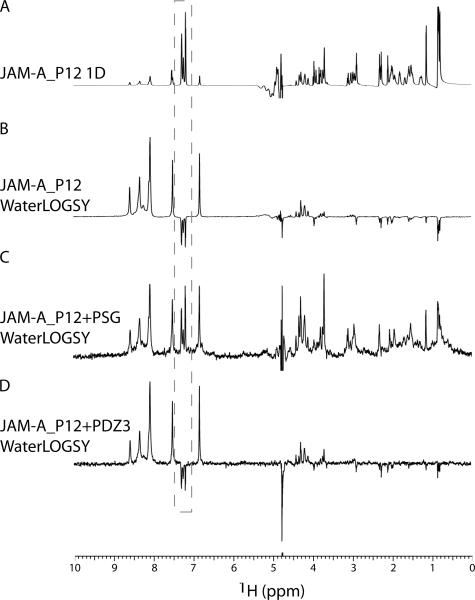
The identical PDZ3 structure in the presence or absence of JAM-A peptide suggested that the reason for lack of peptide binding to the isolated PDZ3 domain is related to features only present in the more complete ZO-1 PSG fragment. As stated above, Phe−2 of JAM-A_P12 was observed to make hydrophobic interactions with leucines 549 and 554 from the SH3 domain. Additionally, Phe−8 binds near the SH3 domain. Both of these phenylalanines are highly conserved in JAM sequences (supplemental Fig. S3). To probe the importance of SH3 residues on peptide binding to PDZ3 and to eliminate the possibility that crystallization conditions or crystal contacts are perturbing the binding of JAM-A peptide to the isolated PDZ3 domain, we conducted NMR experiments that measure the interaction between the peptide and protein in solution. Using the WaterLOGSY technique, which identifies small molecules that bind to proteins, we probed binding of the peptide to the different protein constructs. In Fig. 5A the one-dimensional NMR spectrum of P12 is presented with the side chain resonances of the Phe−2 and Phe−8 boxed in gray. In the absence of protein, nonexchangeable 1H exhibits negative signals in the WaterLOGSY experiment (Fig. 5B). In contrast, in the presence of PSG, the nonexchangeable 1H exhibits positive signals (Fig. 5C), indicative of P12 binding to protein. On the other hand, in the presence of PDZ3, the WaterLOGSY experiment is similar to that of the peptide alone (compare Fig. 5, B and D), which indicates that JAM-A_P12 is not binding to PDZ3 under the examined conditions. We also conducted the same WaterLOGSY experiments using the shorter 6-mer peptide of JAM-A (JAM-A_P6). Again, we detected binding to PSG, but not to the isolated PDZ3 domain (supplemental Fig. S4). To probe for affinity differences between JAM-A_P12 and JAM-A_P6, we performed a WaterLOGSY-based titration (29). We determined the dissociation constant (Kd) for the peptide from PSG. The Kd of JAM-A_P6 is 39 μm, compared with 35 μm for JAM-A_P12 (supplemental Fig. S5). These nearly identical values show that only the terminal 6 residues contribute to the interaction between the peptide and PSG, such that despite sequence conservation of Phe−8, this JAM-A residue does not appear to play a role in the binding to ZO-1.
FIGURE 5.
NMR characterization of JAM-A_P12 binding to PSG. A, one-dimensional NMR spectrum of JAM-A_P12. B, WaterLOGSY of JAM-A_P12. C, WaterLOGSY of JAM-A_P12 in the presence of PSG. D, WaterLOGSY of JAM-A_P12 in the presence of PDZ3. Experimental conditions were 300 μm JAM-A_P12 and ± 10 μm PSG or PDZ3 in 100 mm PO4, pH 7.2, at 25 °C. Resonances corresponding to the side chains of Phe−2 and Phe−8 are boxed in gray.
The weak binding interaction between JAM-A and PDZ3 was not unanticipated and is well within range of binding affinities observed between PDZ3 of MAGUK proteins and their ligands (32, 33). The tight junction is a highly dynamic structure (34, 35), and it had been demonstrated that there is a high turnover of ZO-1 and of its binding partners like claudin and occludin in stable epithelial monolayers (35, 36). This dynamism is likely necessary for the maintenance of the barrier during normal tissue morphogenesis or repair, which would require the remodeling of cell-cell contacts as cells change their spatial relationships to one another. In the gastrointestinal tract, for example, the TJ barrier is constantly maintained despite the fact that the epithelial cells that line the intestine are constantly migrating up the crypt-to-villus axis, and immune cells are moving back and forth across the epithelium. We hypothesize that low affinity, but highly cooperative interactions between ZO proteins and its ligands would promote dynamic regulation of TJ assembly and/or disassembly.
In summary, this is the first demonstration, to our knowledge, for an essential role of adjacent domains to the binding of ligands to PDZ domains in the MAGUK family of proteins. Our crystal structures and NMR data reveal an essential role for the bBC loop in the SH3 domain for JAM-A C terminus binding to the third PDZ domain of ZO-1. Concerning the role of the JAM-A C-terminal residues, we show a critical role for Phe−2 for peptide binding that is facilitated by the conserved Ser−3. Through interactions with the carbonyl group of Phe−2, the hydroxyl group of this serine stabilizes the JAM-A peptide in a binding mode different from the canonical binding mode of type II PDZ domains. In this unique peptide binding conformation, the side chain of Phe−2 is able to interact with residues from the adjacent SH3 domain. The dependence on the SH3 domain for JAM-A binding to ZO-1 suggests a possible communication mechanism between the SH3 domain and the PDZ3 domain. Hence, ligand binding to SH3 may influence the conformation of the βBC loop, and this in turn would influence the affinity of PDZ3 to JAM-A. It would be intriguing to identify those SH3 ligands that can have this modulating effect on JAM-A binding.
Supplementary Material
Acknowledgments
We thank the staff at SERCAT and Benjamin Ramirez of the University of Illinois at Chicago Center for Structural Biology for assistance. We thank Ying Su for technical support.
This research was supported, in whole or in part, by National Institutes of Health Grant DK61397 (to A. S .F. and A. L.). This work was also supported by the National Institutes of Health Division of Intramural Research and the Universities of North Carolina and Illinois.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
The atomic coordinates and structure factors (codes 3TSV, 3TSW, and 3TSZ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MAGUK
- membrane-associated guanylate kinase
- asu
- asymmetric unit
- GUK
- region of homology to guanylate kinase
- JAM-A
- junctional adhesion molecule A
- JAM-A_P6
- 6-mer cytoplasmic tail peptide from JAM-A
- JAM-A_P12
- 12-mer cytoplasmic tail peptide from JAM-A
- PSG
- PDZ3-SH3-GUK core module
- PSG+JAM-A
- PDZ3-SH3-GUK core module in complex with the 12-mer cytoplasmic tail peptide from JAM-A
- r.m.s.d.
- root mean square deviation
- SH3
- Src homology 3
- TJ
- tight junction
- ZO
- zonula occludens.
REFERENCES
- 1. Good M. C., Zalatan J. G., Lim W. A. (2011) Science 332, 680–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Funke L., Dakoji S., Bredt D. S. (2005) Annu. Rev. Biochem. 74, 219–245 [DOI] [PubMed] [Google Scholar]
- 3. Elias G. M., Nicoll R. A. (2007) Trends Cell Biol. 17, 343–352 [DOI] [PubMed] [Google Scholar]
- 4. Javier R. T. (2008) Oncogene 27, 7031–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Montgomery J. M., Zamorano P. L., Garner C. C. (2004) Cell. Mol. Life Sci. 61, 911–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fanning A. S., Anderson J. M. (2009) Ann. N.Y. Acad. Sci. 1165, 113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doyle D. A., Lee A., Lewis J., Kim E., Sheng M., MacKinnon R. (1996) Cell 85, 1067–1076 [DOI] [PubMed] [Google Scholar]
- 8. Lye M. F., Fanning A. S., Su Y., Anderson J. M., Lavie A. (2010) J. Biol. Chem. 285, 13907–13917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McGee A. W., Bredt D. S. (1999) J. Biol. Chem. 274, 17431–17436 [DOI] [PubMed] [Google Scholar]
- 10. Tavares G. A., Panepucci E. H., Brunger A. T. (2001) Mol. Cell 8, 1313–1325 [DOI] [PubMed] [Google Scholar]
- 11. Hoskins R., Hajnal A. F., Harp S. A., Kim S. K. (1996) Development 122, 97–111 [DOI] [PubMed] [Google Scholar]
- 12. Shin H., Hsueh Y. P., Yang F. C., Kim E., Sheng M. (2000) J. Neurosci. 20, 3580–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Woods D. F., Hough C., Peel D., Callaini G., Bryant P. J. (1996) J. Cell Biol. 134, 1469–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brenman J. E., Topinka J. R., Cooper E. C., McGee A. W., Rosen J., Milroy T., Ralston H. J., Bredt D. S. (1998) J. Neurosci. 18, 8805–8813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu H., Reissner C., Kuhlendahl S., Coblentz B., Reuver S., Kindler S., Gundelfinger E. D., Garner C. C. (2000) EMBO J. 19, 5740–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mehta S., Wu H., Garner C. C., Marshall J. (2001) J. Biol. Chem. 276, 16092–16099 [DOI] [PubMed] [Google Scholar]
- 17. Umeda K., Ikenouchi J., Katahira-Tayama S., Furuse K., Sasaki H., Nakayama M., Matsui T., Tsukita S., Furuse M. (2006) Cell 126, 741–754 [DOI] [PubMed] [Google Scholar]
- 18. Fanning A. S., Lye M. F., Anderson J. M., Lavie A. (2007) J. Biol. Chem. 282, 37710–37716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Severson E. A., Parkos C. A. (2009) Ann. N.Y. Acad. Sci. 1165, 10–18 [DOI] [PubMed] [Google Scholar]
- 20. Fanning A. S., Little B. P., Rahner C., Utepbergenov D., Walther Z., Anderson J. M. (2007) Mol. Biol. Cell 18, 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kausalya P. J., Phua D. C., Hunziker W. (2004) Mol. Biol. Cell 15, 5503–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yamamoto T., Harada N., Kano K., Taya S., Canaani E., Matsuura Y., Mizoguchi A., Ide C., Kaibuchi K. (1997) J. Cell Biol. 139, 785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yokoyama S., Tachibana K., Nakanishi H., Yamamoto Y., Irie K., Mandai K., Nagafuchi A., Monden M., Takai Y. (2001) Mol. Biol. Cell 12, 1595–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kabsch W. (2010) Acta Cryst. D66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 27. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 28. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dalvit C., Fogliatto G., Stewart A., Veronesi M., Stockman B. (2001) J. Biomol. NMR 21, 349–359 [DOI] [PubMed] [Google Scholar]
- 30. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 31. Harris B. Z., Lim W. A. (2001) J. Cell Sci. 114, 3219–3231 [DOI] [PubMed] [Google Scholar]
- 32. Saro D., Li T., Rupasinghe C., Paredes A., Caspers N., Spaller M. R. (2007) Biochemistry 46, 6340–6352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen J., Pan L., Wei Z., Zhao Y., Zhang M. (2008) EMBO J. 27, 2113–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sasaki H., Matsui C., Furuse K., Mimori-Kiyosue Y., Furuse M., Tsukita S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3971–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shen L., Weber C. R., Turner J. R. (2008) J. Cell Biol. 181, 683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Raleigh D. R., Boe D. M., Yu D., Weber C. R., Marchiando A. M., Bradford E. M., Wang Y., Wu L., Schneeberger E. E., Shen L., Turner J. R. (2011) J. Cell Biol. 193, 565–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.