Background: Expression levels of both IL-17A and transcription factor CREMα are increased in T cells from SLE patients.
Results: In primary human T cells, CREMα binds to the proximal IL17A promoter and induces IL-17A expression by transcriptional activation and epigenetic modifications.
Conclusion: CREMα promotes IL-17A expression.
Significance: Suppression of CREMα expression should mitigate IL-17A-driven inflammatory responses.
Keywords: Chromatin Histone Modification, Chromatin Remodeling, DNA Methylation, Immunology, Interleukin, Promoters, Transcription Factors, CREM, IL-17, SLE
Abstract
IL-17A is a proinflammatory cytokine that is produced by specialized T helper cells and contributes to the development of several autoimmune diseases such as systemic lupus erythematosus (SLE). Transcription factor cAMP-responsive element modulator (CREM)α displays increased expression levels in T cells from SLE patients and has been described to account for aberrant T cell function in SLE pathogenesis. In this report, we provide evidence that CREMα physically binds to a cAMP-responsive element, CRE (−111/−104), within the proximal human IL17A promoter and increases its activity. Chromatin immunoprecipitation assays reveal that activated naïve CD4+ T cells as well as T cells from SLE patients display increased CREMα binding to this site compared with T cells from healthy controls. The histone H3 modification pattern at the CRE site (−111/−104) and neighboring conserved noncoding sequences within the human IL17A gene locus suggests an accessible chromatin structure (H3K27 hypomethylation/H3K18 hyperacetylation) in activated naïve CD4+ T cells and SLE T cells. H3K27 hypomethylation is accompanied by decreased cytosine phosphate guanosine (CpG)-DNA methylation in these regions in SLE T cells. Decreased recruitment of histone deacetylase (HDAC)1 and DNA methyltransferase (DNMT)3a to the CRE site (−111/−104) probably accounts for the observed epigenetic alterations. Reporter studies confirmed that DNA methylation of the IL17A promoter indeed abrogates its inducibility. Our findings demonstrate an extended role for CREMα in the immunopathogenesis of SLE because it contributes to increased expression of IL-17A.
Introduction
IL-17 cytokines are key players in the host defense against bacteria and fungi as well as in the development of several inflammatory and autoimmune diseases (1). Among the six IL-17 isoforms in humans (IL-17A through IL-17F), IL-17A was the first one to be discovered from a CD4+ T cell library and constitutes the prototypic family member (2). IL-17A is produced by T cells, natural killer cells, mast cells, and neutrophils and exerts intense proinflammatory responses, e.g. induction of chemokines, cytokines (e.g. CXCL1, CXCL8, IL-1, and GM-CSF) and recruitment of neutrophils to inflamed tissues (3). Over the last decade, a specialized subset of IL-17-producing T helper cells, denoted Th174 cells, has emerged that plays a pivotal role in the pathogenesis of autoimmune diseases such as rheumatoid arthritis, inflammatory bowel disease, and systemic lupus erythematosus (SLE) (4–6).
IL-17 production and generation of Th17 cells are orchestrated at various levels (7). It has been shown that Th17 differentiation from naïve CD4+ T cells depends on the presence of external signals such as IL-6 and transforming growth factor (TGF)β whereas this T cell lineage requires IL-21 and IL-23 for maintenance (8, 9). At the transcriptional level, IL-17 expression is regulated through the interplay of multiple transcription factors, e.g. retinoic acid-related orphan receptor γt, STAT3, interferon-regulatory factor-4, Runx transcription factor RUNX1, and basic leucine zipper transcription factor activating transcription factor-like (10). Recent studies focused on selective chromatin remodeling events within the murine IL17A locus and displayed that IL-6 and TGFβ promote an increased accessibility of the IL17A locus by histone H3 acetylation of the IL17A promoter region (11). In line with this, Wei et al. identified several conserved noncoding regions within the murine IL17A locus that undergo H3K4 trimethylation in Th17-differentiated cells which is associated with gene activation (12). Nonetheless, epigenetic chromatin or DNA modifications within the human IL17A locus have not been described yet.
In this report, we link the transcription factor cAMP-responsive element modulator (CREM)α to increased IL-17A production in SLE T cells. CREMα belongs to a superfamily of transcription factors that also includes CRE-binding protein (CREB), the inducible cAMP response element repressor, and activating transcription factors. All of these regulators are involved in the cAMP signaling cascade which comprises an evolutionarily conserved pathway that includes stimulus-dependent cAMP production, cAMP binding to the regulatory subunits of protein kinases, nuclear translocation of these kinases and subsequent phosphorylation, and thus, activation of the CREB/CREM/activating transcription factors (13, 14). Upon activation they bind to cAMP-responsive elements (CREs; TGACGTCA) or the CRE 5′-half-site (TGAC) in regulatory gene regions. The multiexonic character of the CREM gene, the usage of various promoters or alternative initiation codons and differential splicing mechanisms result in the presence of multiple CREM isoforms that are expressed in a cell- and development-specific fashion (15–18). CREMα has been demonstrated to repress IL2 transcription in T cells and to contribute to defective IL-2 production in T cells from SLE patients (19). SLE is a chronic autoimmune disease that affects multiple organs and is characterized by severe T cell signaling abnormalities (20). It has been reported that SLE T cells display elevated levels of CREMα due to increased CREM promoter activity that even reflects SLE disease activity (18, 21). Additional immune cell-relevant target genes have been identified that are trans-regulated by CREMα such as transcription factor c-fos, TCR/CD3ζ and antigen-presenting cell molecule CD86 (22–24). Apart from its direct transcriptional effects, CREMα is also involved in epigenetic mechanisms of gene regulation because it may recruit histone deacetylase (HDAC)1 or DNA methyltransferase (DNMT)3a to specific regulatory gene sequences; however, it fails to mobilize histone acetyltransferase activity of p300 (25–27).
IL-17A levels are increased in SLE patients, and IL-17-producing T cells play a central role in the expression of systemic autoimmunity and tissue-specific organ damage in SLE (6, 27, 28). Herein, we show that CREMα binds to a yet unidentified CRE within the proximal IL17A promoter, and T cell activation results in enhanced CREMα recruitment to this site. Furthermore, SLE T cells display increased CREMα binding to this site. We provide evidence that CREMα trans-activates IL17A promoter activity and contributes to several significant epigenetic changes, including histone modifications and cytosine phosphate guanosine (CpG)-DNA methylation within the human IL17A locus. Our data support the importance of CREMα in orchestrating the transcriptional program of SLE T cells and constitute the first report of epigenetic regulation of the human IL17A gene.
EXPERIMENTAL PROCEDURES
Study Subjects and T Cell Culture
All SLE patients included in our studies were female and diagnosed according to the American College of Rheumatology classification criteria (29) and were recruited from the Division of Rheumatology at Beth Israel Deaconess Medical Center, Boston, Massachusetts, after written informed consent under protocol 2006-P-0298. Healthy individuals were chosen as controls. Peripheral venous blood was collected in heparin-lithium tubes, and total human T cells were purified as described before (18). All primary human T cells and human Jurkat T cells were kept in RPMI 1640 medium supplemented with 10% FBS. Naïve CD4+ T cells from healthy controls were purified from total T cell suspension using the Human Naïve CD4+ T Cell Isolation kit II (Miltenyi Biotec). For activation assays, naïve CD4+ T cells were incubated in the absence or presence of plate-bound anti-CD3 and anti-CD28 antibodies (BioXCell; both at 1 μg/ml) for 72 h. All cell culture experiments were performed individually with naïve T cells from single blood donors.
Th Cell Differentiation Assays
Human naïve CD4+ T cells were incubated in the presence of plate-bound anti-CD3 and anti-CD28 antibodies (BioXCell; both at 1 μg/ml) and the following cytokines/antibodies for 3 days: Th1: IL-2 (50 units/ml), IL-12 (10 ng/ml), and anti-IL-4 (5 μg/ml); Th2: IL-2 (50 units/ml), IL-4 (20 ng/ml), anti-IFNγ (10 μg/ml); Th17: IL-21 (25 ng/ml), TGFβ (5 ng/ml), anti-IL-4 (5 μg/ml), anti-IFNγ (10 μg/ml). Subsequently, cells were expanded in a new flask for another 3 days with replenished IL-12 (Th1), IL-4 (Th2), and IL-21/TGFβ (Th17) without any antibodies (30, 31). Differentiation experiments were performed individually with naïve T cells from single blood donors.
Plasmids and Generation of Luciferase Reporter Constructs
An expression plasmid for human CREMα (on a pcDNA3.1/V5-His-TOPO ”backbone”; Invitrogen) was provided by G. N. Europe-Finner (Faculty of Medical Sciences, Newcastle upon Tyne, UK) (32). The DNMT3a expression plasmid has been described before (33). Reporter constructs spanning the proximal 465 and 195 bp of the human IL17A promoter were PCR-amplified and cloned into luciferase vector pGL3-Basic (Promega) using primers with attached restriction sites for MluI and BglII. All plasmid DNA preparations were carried out with DNA purification kits (Qiagen) and sequence-verified (Genewiz, Cambridge, MA). Site-directed mutagenesis at CRE site (−111/−104) within reporter construct IL17Ap(−195)-luc was performed using a DNA oligonucleotide harboring a mutated CRE (5′-gcgacacgccacgtaagctgccacagaagg-3′; MWG Operon) and PfuTurbo® DNA polymerase (Stratagene) according to the manufacturer's instructions.
Luciferase Assays in Primary Human T Cells
Three million primary human T cells were transfected with a total amount of 3 μg of plasmid DNA using the Amaxa Human T cell Nucleofector kit (Lonza) and an Amaxa Nucleofector II device (program U014; Lonza). Effector-reporter transfection experiments were performed at a molar ratio of 3:1. Each reporter experiment included 10 ng of Renilla luciferase construct as an internal control. Five hours after transfection cells were collected and lysed, and luciferase activity was quantified using the Promega Dual Luciferase Assay System (Promega) according to the manufacturer's instructions. Luciferase experiments were repeated at least three times, and values in the bar diagrams are given as mean ± S.D.
mRNA Extraction, Conventional RT-PCR, and Quantitative RT-PCR
Three million primary human T cells were transfected with a total amount of 3 μg of the indicated plasmids using the Amaxa transfection system (Lonza). Total RNA was isolated using the RNeasy Mini kit (Qiagen). Residual genomic DNA contamination was removed by DNase I (Qiagen). RNA was reverse-transcribed into cDNA using the Reverse Transcription system (Promega). Sequences for primers used for regular PCR were as follows: IL-17A forward, 5′-cgaaatccaggatgccc-3′; IL-17A reverse, 5′-gacaccagtatcttctccag-3′; 18 S rRNA forward, 5′-actcaacacgggaaacctca-3′; 18 S rRNA reverse, 5′-aaccagacaaatcgctccac-3′. PCR products were visualized on a 1% agarose gel containing ethidium bromide. Real-time qPCR primer sequences are given in Table 1.
TABLE 1.
Primers for real-time qPCR
| Region | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | Product length | Annealing temperature |
|---|---|---|---|---|
| bp | ºC | |||
| IL-17A | ACCAATCCCAAAAGGTCCTC | CACTTTGCCTCCCAGATCAC | 110 | 60 |
| 18 S rRNA | ACTCAACACGGGAAACCTCA | AACCAGACAAATCGCTCCAC | 110 | 60 |
| IL17A_CNS1 | CTGCCCTTCCCATTTTCCTT | GTTCAGGGGTGACACCATTT | 456 | 55 |
| IL17A_PP | ACATGATATTGACCCATAGC | AGGGCAGAAATTCATGTTCC | 456 | 55 |
| IL17A_PP-CRE | TCATTGGGGGCGGAAATTTTAACCA | GGGCTTTTCTCCTTCTGTGGTCAC | 90 | 55 |
| IL17A_CNS2 | GCGATGCTCTTGCTGATTTG | CCTGGATCTCCATAGTCAGAAC | 352 | 55 |
Electrophoretic Mobility Shift Assays (EMSAs)
Primary T cells (from healthy blood donors) were transfected with either pcDNA3 empty vector or His-tagged CREMα expression plasmids (1 μg of plasmid/1 million cells) using the Amaxa system, and nucleoprotein lysates were prepared 5 h after transfection as reported previously (18). For EMSAs, a double-stranded DNA probe harboring the CRE site (−111/−104) of the human IL17A promoter (5′-cacgtaagtgaccacagaagg-3′) was radiolabeled using the T4-polynucleotide kinase and [γ-32P]ATP. Binding reactions were performed at room temperature for 20 min in HighDensity TBE binding buffer (Invitrogen) containing 5 μg of nuclear protein from the transfected T cells and 1 μg of poly(dI)·poly(dC) in a total volume of 20 μl. Electrophoresis and autoradiography were performed as described previously (18). For supershift assays, 2 μg of polyclonal anti-His6-tagged Ab or unrelated control Ab (both from Abcam) were added to the binding reaction. 50- and 100-fold molar excess of the cold, unlabeled oligonucleotide or a competitor harboring a mutated CRE site (5′-cacgtaagctgccacagaagg-3′) were used for competition assays.
ChIP Assays
Anti-HDAC1, anti-H3K18ac, and anti-H3K27me3 antibodies, nonspecific normal rabbit and normal mouse IgG were obtained from Upstate (Millipore), and anti-DNMT3a antibody was from Abcam. Polyclonal anti-CREMα antibody detecting human CREMα has been described before (34). ChIP grade protein A/G plus agarose was purchased from Pierce (ThermoScientific). ChIP assay was carried out according to the manufacturer's instructions (Upstate Biotechnology/Millipore). Briefly, 1–2 million cells (either from a total human T cell suspension or from unstimulated or activated naïve human CD4+ T cells, as indicated) were cross-linked with 1% formaldehyde, washed with cold PBS, and lysed in buffer containing protease inhibitors (Roche Applied Science). Cell lysates were sonicated to shear DNA and sedimented, and diluted supernatants were immunoprecipitated with the indicated antibodies. A portion (20%) of the diluted supernatants was kept as “input” (input represents PCR amplification of the total sample). Protein-DNA complexes were eluted in 1% SDS and 0.1 m NaHCO3 and reverse cross-linked at 65 °C. DNA was recovered using the QIAamp DNA Mini kit (Qiagen) and subjected to PCR analysis on an ABI OneStepPlus real-time PCR system. Real-time qPCR primer sequences are given in Table 1. The amount of immunoprecipitated DNA was subtracted by the amplified DNA that was bound by the nonspecific normal IgG and subsequently calculated as relative to the respective input DNA.
IL-17A Secretion Assay
Human naïve CD4+ T cells were activated using anti-CD3/anti-CD28 antibodies for 72 h and PMA (50 ng/ml)/ionomycin (500 ng/ml) for another 5 h. IL-17A secretion assay was performed according to the manufacturer's instructions (Miltenyi Biotec).
Methylated CpG-DNA Immunoprecipitation
The methylated CpG-DNA immunoprecipitation assay was carried out according to the manufacturer's instructions (Zymo Research). Briefly, genomic DNA from T cells obtained from SLE patients and healthy control individuals was purified using the AllPrep RNA/DNA/protein Mini kit (Qiagen), sheared to fragments of ∼200 bp using DNA shearase (Zymo Research). Subsequently, 100 ng of sheared genomic DNA was used for methylated CpG-DNA immunoprecipitation. Methylated DNA was recovered and subjected to PCR analysis on an ABI OneStepPlus real-time PCR system using primers as listed in Table 1. Equal amounts (100 ng) of completely (100%) methylated human DNA and demethylated human DNA (Zymo Research) were included as input and negative control.
Methylation of Reporter Plasmids
To investigate the effects of CpG-DNA methylation on IL17A promoter activity, we methylated both the 195-bp IL17A reporter construct and the empty pGL3 plasmid, using CpG-DNA methylase (Zymo Research) according to the manufacturer's instructions.
Statistical Analysis
A paired two-tailed Student's t test was used for statistical analysis.
RESULTS
Forced CREMα Expression Induces IL17A Gene Expression in Primary Human T Cells
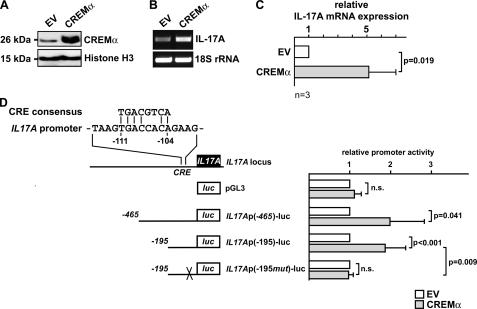
SLE T cells display both increased IL-17A production and elevated cellular CREMα protein levels which have been shown to contribute to decreased IL-2 production in SLE patients. We considered that CREMα may also be involved in IL17A gene regulation. Primary human T cells from healthy individuals display low levels of CREMα; however, to mimic the conditions of increased CREMα levels in SLE T cells, we transiently transfected primary human T cells (from healthy blood donors) with a His6-tagged CREMα expression plasmid and analyzed nuclear CREMα protein expression in these cells 5 h after transfection (Fig. 1A). mRNA obtained from these cells was tested for relative IL-17A expression and proved to express significantly increased IL-17A transcript numbers after forced CREMα expression as assessed by conventional and real-time qRT-PCR (relative increase of 5.13 ± 1.88; p = 0.019; Fig. 1, B and C).
FIGURE 1.
CREMα overexpression induces IL-17A expression by trans-activating the human IL17A promoter. A, primary human T cells were transfected with an expression plasmid encoding human CREMα or pcDNA3 empty vector (EV), respectively. Cells were harvested 5 h after transfection, and nucleoprotein lysates were immunoblotted for CREMα expression. Equal protein load is visualized through histone H3 expression. B and C, pcDNA3 empty vector or CREMα expression plasmid was transfected into primary human T cells, and 5 h after transfection RNA was analyzed for IL-17A and 18 S rRNA expression using regular PCR (B) and real-time qPCR (C). Experiments were performed individually in T cells from four different healthy blood donors. A representative PCR image is shown in B. Bar diagram in C shows the mean relative IL-17A expression (after CREMα overexpression) ± S.D. (error bars) from four experiments. D, alignment of the CRE consensus sequence with the CRE site (−111/−104) of the proximal human IL17A promoter. Schematic below displays the IL17A reporter plasmids used for luciferase assays. IL17Ap(−195mut)-luc indicates a reporter plasmid containing a site-directed mutation at the CRE site (−111/−104). Primary human T cells were transfected with the IL17A reporter plasmids and either pcDNA3 empty vector (white bars) or CREMα expression plasmid (gray bars). Cells were lysed 5 h after transfection, and firefly luciferase activity was measured and normalized by Renilla luciferase activity. For each reporter pcDNA3 EV co-transfection was set to 1, and the relative effect mediated by CREMα was calculated. Each experiment was performed in T cells from at least four different individuals, and values are given as mean ± S.D.
A thorough analysis of the human IL17A promoter yielded a putative CRE site located between 111 and 104 bp upstream of the transcription initiation site, thus denoted CRE site (−111/−104). This site defines an 8-bp sequence that matches 6 nucleotides of the perfect CRE consensus sequence, and the first 4 bp of this site perfectly match the CRE 5′-half-site (TGAC), which has been shown to serve as a minimum binding motif for CREM proteins in other cis-regulatory target sequences (Fig. 1D) (19, 24). To investigate whether CREMα regulates IL-17A expression at the transcriptional level, we performed luciferase assays using reporter constructs spanning the proximal 465 or 195 bp of the human IL17A promoter (Fig. 1D). Indeed, CREMα overexpression resulted in significant up-regulation of IL17A promoter activities in both constructs (-fold increase of 1.96 ± 0.88 and 1.84 ± 0.53, respectively). Next, we mutated the CRE site (−111/−104) within the 195-bp spanning IL17A reporter construct and noted that CREMα did not alter its promoter activity. These results suggest that this site is crucial for CREMα-mediated IL17A transcription in human T cells.
CREMα Binding to the CRE Site (−111/−104) Is Increased in Activated and SLE T Cells
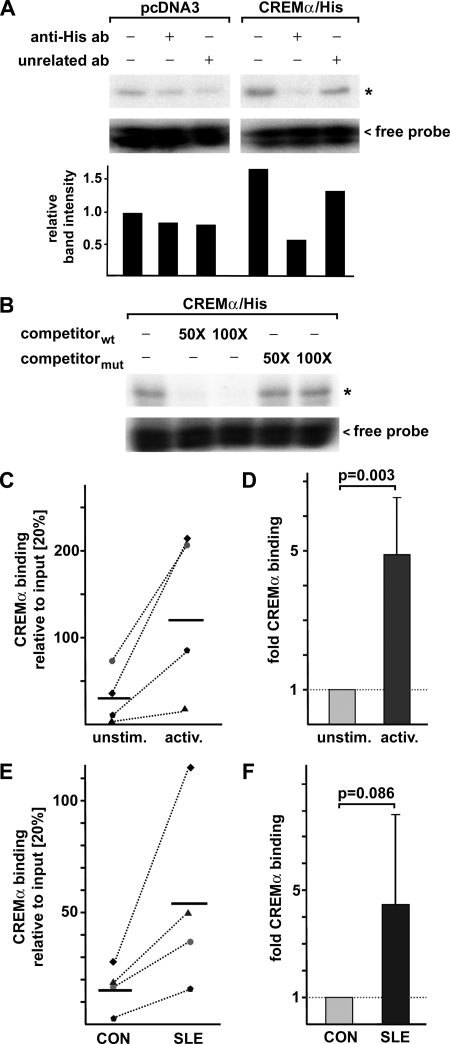
We performed DNA binding studies using a synthetic radiolabeled oligonucleotide harboring the CRE site (−111/−104) of the human IL17A promoter and nuclear protein lysates from primary human T cells that had been transfected either with pcDNA3 empty vector or His6-tagged CREMα for 5 h (Fig. 2, A and B). Binding reactions that included CREMα-transfected nucleoprotein showed a stronger band intensity of a protein-DNA complex compared with the binding reactions with nucleoprotein without CREMα overexpression (∼1.7-fold increase). Specificity of this complex was tested by inclusion of a polyclonal anti-His antibody (detecting the His6-tagged CREMα protein) into the binding reaction which decreased band intensity of this complex whereas an unrelated control antibody barely altered band intensity (Fig. 2A). Competition assays supported our findings, as this complex was absent after addition of the cold, unlabeled probe in a 50- or 100-fold molar excess; however, a cold competitor with a mutated CRE site did not affect complex formation.
FIGURE 2.
CREMα binds to a previously unidentified CRE site within the proximal IL17A promoter. A, primary human T cells were transfected either with pcDNA3 empty vector or CREMα expression plasmid for 5 h. Nucleoprotein lysates were prepared from these cells and used for DNA binding studies using a radiolabeled oligonucleotide harboring the CRE site (−111/−104) of the human IL17A promoter. DNA binding reaction was performed in the absence or presence of polyclonal anti-His6-tagged antibody (as the overexpressed CREMα contains a His6 tag) or an unrelated polyclonal antibody. Band intensities were quantified by densitometry, and relative values are shown (band in first 1 was set to 1.0). B, competition assays were performed with CREMα-containing nucleoprotein lysates, the radiolabeled CRE (−111/−104) probe and the unlabeled wild type (wt) oligonucleotide or a corresponding oligonucleotide harboring a mutated CRE site in 50- or 100-fold molar excess. C, naïve human CD4+ T cells were isolated from four healthy individuals and cultured in the absence or presence of anti-CD3/anti-CD28 antibodies for 72 h. Protein-DNA complexes were cross-linked, and ChIP assays were performed using an anti-CREMα antibody. Immunoprecipitated DNA was analyzed by real-time qPCR amplifying a region that covers the CRE site of the proximal IL17A promoter. Ratios between anti-CREMα immunoprecipitated and input DNA are shown. Dotted lines associate data from corresponding unstimulated and activated cells obtained from the same individual. Horizontal bars represent the mean of the four experiments. D, percentage of anti-CREMα immunoprecipitated DNA in the unstimulated cells from each individual analyzed in C was set to 100%, and the relative change following anti-CD3/anti-CD28 activation was calculated. Values are given as mean ± S.D. (error bars). E, ChIP was performed using total T cells from four matched pairs of SLE patients and healthy controls (CON) and anti-CREMα antibody. Immunoprecipitated DNA was analyzed by real-time qPCR using the same primers as in C. Ratios between anti-CREMα immunoprecipitated and input DNA are shown. Dotted lines associate data from the matched CON/SLE pairs. Horizontal bars represent mean values. F, percentage of anti-CREMα immunoprecipitated DNA in T cells from a control individual was set to 100%, and relative CREMα binding in the corresponding SLE patient was calculated. Values are given as mean ± S.D.
We addressed the in vivo relevance of CREMα binding to CRE site (−111/−104) in the human IL17A promoter by ChIP assays using a CREMα-specific polyclonal antibody. We used naïve CD4+ T cells that were freshly isolated from total human CD4+ T cells and compared them with naïve CD4+ T cells that had been stimulated with anti-CD3 and anti-CD28 antibodies for 72 h (Fig. 2, C and D). We observed significantly increased CREMα binding to this site in activated naïve T cells after individual comparison with CREMα binding in unstimulated cells (-fold increase of 4.9 ± 1.7; p = 0.003; Fig. 2D).
T cells from SLE patients display multiple severe signaling abnormalities resembling a “hyperactivated” phenotype. Thus, we sought to determine CREMα binding to the IL17A promoter in total T cells from a cohort of four SLE patients and healthy control individuals (all female) that were individually matched by age and ethnicity (Fig. 2, E and F). Interestingly, we found a tendency toward higher relative CREMα binding to this site in SLE T cells (after comparison with the matched healthy controls; -fold increase of 4.5 ± 3.4; p = 0.086).
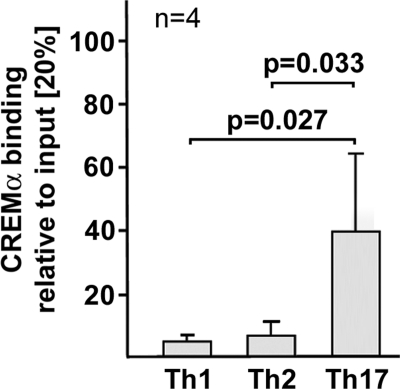
Because our findings suggest a role for CREMα in transcriptional control of the IL17A gene, we analyzed CREMα involvement in T helper (Th) cell differentiation by addition of specific cytokines and antibodies priming toward a Th1 (IL-12 and anti-IL-4), Th2 (IL-4 and anti-IFNγ), and Th17 (TGFβ and IL-21) cell fate decision (29, 30). Compared with Th1 and Th2 conditions, CREMα binding to CRE site (−111/−104) was significantly increased in Th17 differentiated cells (Fig. 3).
FIGURE 3.
CREMα binding to the human IL17A promoter under Th1, Th2, and Th17 differentiation conditions. Naïve human CD4+ T cells were primed toward Th1, Th2, and Th17 lineage decisions by addition of the appropriate cytokines and antibodies for 5 days (as outlined under “Experimental Procedures”). Subsequently, CREMα binding to the IL17A-CRE was quantified by ChIP analysis and qPCR. Ratios between anti-CREMα immunoprecipitated and nonimmunoprecipitated input DNA are shown. Values are given as mean ± S.D. (error bars) from four independent experiments. Cells from each individual were cultured individually.
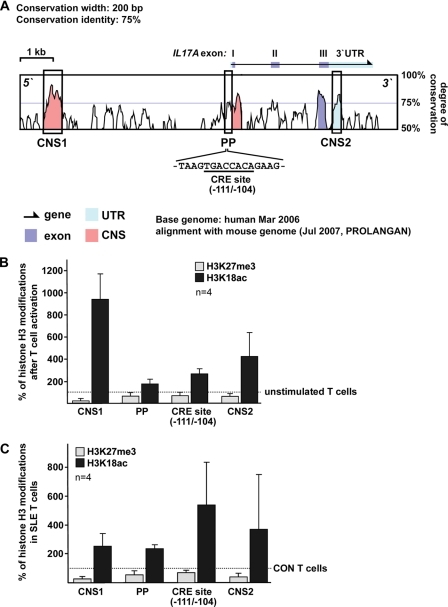
Chromatin Modifications of Conserved Regions of the Human IL17A Gene Locus
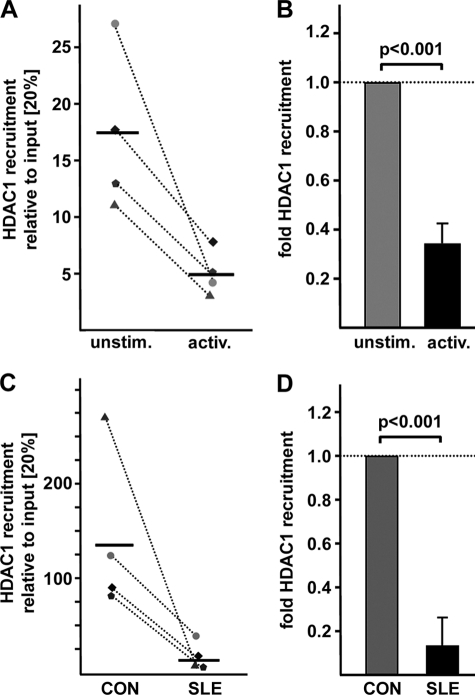
Previous studies have unraveled CREMα involvement in gene regulation at the transcriptional level as well as through remodeling of the chromatin structure by interactions with HDAC1 and p300. To investigate epigenetic patterns across the human IL17A gene, we defined regions of interest, based on bioinformatic approaches. We aligned the mouse and human IL17A genes (VISTA Genome Browser) and searched for conserved noncoding sequences (CNS; Fig. 4A). CNS regions were defined as regions with sequence homology of >75% between the human and mouse genes over a length of at least 200 bp. Based on the degree of sequence conservation we identified three regions of interest, one of which was ∼6 kbp upstream of the IL17A gene, one mapping to the proximal promoter (PP) that contains the aforementioned CRE site (−111/−104), and one within the highly conserved 3′-UTR. We performed ChIP analyses in naïve and activated CD4+ T cells using antibodies against histone H3 modifications reflecting activating “euchromatin” (H3K18ac) or repressive “heterochromatin” (H3K27me3). Throughout the analyzed regions we found decreased H3K27 trimethylation and enriched H3K18 acetylation following T cell activation compared with nonactivated naïve CD4+ T cells (Fig. 4B). Subsequently, we compared these histone modifications in total T cells obtained from SLE patients and matched healthy individuals. Our data argue for a similar pattern in SLE T cells as observed in activated naïve CD4+ T cells (Fig. 4C). SLE T cells display reduced H3K27 trimethylation and increased H3K18 acetylation compared with control T cells (Fig. 4C). Intriguingly, we observed the most prominent differences in H3K18 acetylation when we specifically amplified a region harboring the CRE site (−111/−104) within the proximal promoter. Because CREMα interacts with HDAC1 (25) we analyzed HDAC1 recruitment to this site (Fig. 5). In line with our findings of enriched H3K18 acetylation at the CRE site (−111/−104) in activated naïve CD4+ T cells and total T cells from SLE patients we observed less HDAC1 binding to this site under these conditions (0.31 ± 0.1 after T cell activation; p < 0.001; 0.14 ± 0.13 in SLE CD4+ T cells; p < 0.001). We conclude that CREMα binding to the newly identified CRE site within the human IL17A promoter contributes to significant shifts in the chromatin structure resulting in a gene locus that is more accessible for transcription factors.
FIGURE 4.
Histone H3 modifications at the human IL17A gene in response to T cell activation and in SLE T cells. A, alignment between the human and mouse IL17A gene locus. CNS (pink) were defined as regions with sequence homology of >75% between human and mouse over a length of at least 200 bp. Exons of the human IL17A gene are displayed in blue and conserved UTR regions in turquoise. The sequence of CRE site (−111/−104) within the proximal promoter is shown below. B, histone H3K27 trimethylation (gray bars) and H3K18 acetylation (black bars) was analyzed in unstimulated and activated naïve CD4+ T cells from four different healthy individuals by ChIP assays. The indicated regions of interest within the human IL17A gene were amplified by qPCR, and the proportion of immunoprecipitated DNA was calculated as relative to the input DNA in each sample. Subsequently, the ratio of relative expression was calculated between the activated and the unstimulated naïve T cells (from the same individual). The dotted line represents the methylation or acetylation status in unstimulated cells, for each of which was set to 100%. Changes in the methylation or acetylation status following T cell activation are given in the bar diagram (mean ± S.D. (error bars)). C, histone H3K27 methylation (gray bars) and H3K18 acetylation (black bars) were analyzed in total T cells from four matched control (CON)/SLE pairs by ChIP assays. The indicated regions of interest within the human IL17A gene were amplified by qPCR, and the proportion of immunoprecipitated DNA was calculated relative to the nonimmunoprecipitated input DNA in each sample. Subsequently, the ratio of relative expression was calculated between each SLE patient and the corresponding control individual. The dotted line represents the methylation or acetylation status in control T cells, each of which was set to 100%. Changes in the methylation or acetylation status in the matched SLE patient are given in the bar diagram (mean ± S.D.).
FIGURE 5.
Increased HDAC1 recruitment to the CRE site (−111/−104) after T cell activation and in SLE T cells. A, HDAC1 recruitment to the CRE site (−111/−104) was analyzed in unstimulated and activated naïve CD4+ T cells from four different healthy individuals by ChIP assays using an anti-HDAC1 antibody. Immunoprecipitated DNA was analyzed by real-time qPCR amplifying a region that covers the CRE site of the proximal IL17A promoter. Ratios between anti-CREMα immunoprecipitated and input DNA are shown. Dotted lines associate data from paired unstimulated/activated naïve CD4+ T cell obtained in the same individual. Horizontal bars represent the mean of the four experiments. B, percentage of anti-HDAC1 immunoprecipitated DNA in the unstimulated cells from each individual analyzed in A was set to 100%, and the relative change following anti-CD3/anti-CD28 stimulation was calculated. Values are given as mean ± S.D. (error bars). C, ChIP was performed using total T cells from four individually matched pairs of SLE patients and healthy controls (CON) and anti-HDAC1 antibody. Immunoprecipitated DNA was analyzed by real-time qPCR using the same primers as in A. Ratios between anti-HDAC1 immunoprecipitated and input DNA are shown. Dotted lines associate data from the matched control/SLE pairs. Horizontal bars represent the mean. D, percentage of anti-HDAC1 immunoprecipitated DNA in T cells from the control individual was set to 100%, and relative HDAC1 binding in the corresponding SLE patient was calculated. Values are given as mean ± S.D.
CpG-DNA Methylation in Activated T Cells from SLE Patients and Healthy Controls
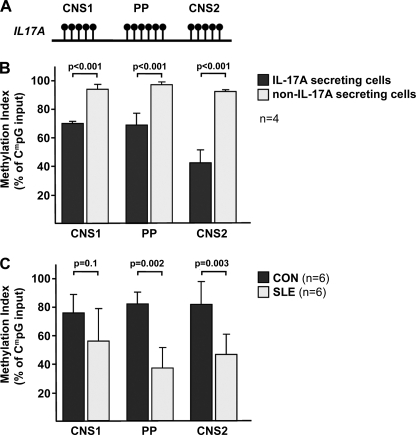
Histone methylation is usually accompanied by concordant CpG-DNA methylation (34). Because the conserved regions and the proximal promoter of the human IL17A gene contain several CpG-rich regions (Fig. 6A), we sought to analyze CpG-DNA methylation patterns across these regions in total T cells from SLE patients and healthy control individuals.
FIGURE 6.
Decreased CpG-DNA methylation in IL-17A-secreting and SLE T cells. A, CpG sites within the CNS and the proximal promoter (PP) of the human IL17A gene are indicated. B, naïve CD4+ T cells from healthy blood donors that had been stimulated with anti-CD3/anti-CD28 antibodies for 72 h followed by stimulation with PMA/ionomycin for another 5 h were subjected to an IL-17A secretion assay. ChIP analyses were performed in both IL-17A-enriched T cells (black bars) and non-IL-17A-secreting T cells (gray bars), using an antibody that specifically detects methylated CpG sequences. Methylated DNA was recovered, and CNS and proximal promoter regions were amplified by real-time qPCR. Completely methylated (input, 100%) and unmethylated human DNA samples (negative control, 0%) were included. Values are given as mean ± S.D. (error bars) from four independent experiments. C, total T cells from six individually matched SLE (gray bars) and healthy control individuals (CON; black bars) were subjected to CpG-DNA immunoprecipitation. The percentage of methylated DNA is given as mean ± S.D.
First, we sorted between IL-17A-secreting and non-IL17A-secreting cells using cytokine capture assays in naïve CD4+ T cells from healthy blood donors that had been sequentially stimulated with anti-CD3/anti-CD28 antibodies for 72 h and with PMA/ionomycin for another 5 h (Fig. 6B). Subsequently, we enriched IL-17A-secreting cells using specific antibodies that yielded a purity of ∼30% within the “IL-17A-secretor” group (as measured by flow cytometry, data not shown). However, cells in the non-IL-17A-secretor group were 100% negative for intracellular IL-17A staining. As expected, CpG-DNA methylation was significantly decreased among the IL-17A-secretors by 58–30%, suggesting an open, active gene locus. Given the technical limitations of this assay which resulted in a decisive contamination of non-IL17A-secreting cells within the IL-17A-secretor group, we assume the real differences between secretors and nonsecretors are actually more pronounced than the observed ones.
Next, we analyzed CpG-DNA methylation of the human IL17A locus in total T cells from a cohort of six female age- and ethnicity-matched SLE and control individuals (Fig. 6C). Total T cells were isolated and activated by subsequent CD3/CD28 and PMA/ionomycin stimulation as described above. Throughout the analyzed regions, SLE T cells displayed less CpG-DNA methylation in response to T cell activation compared with control T cells. Taken together, these findings suggest that IL-17A production in human T cells is tightly controlled by DNA methylation and that T cell activation in SLE patients “opens” the gene locus and increases the accessibility for trans-regulatory factors.
Decreased DNMT3a Recruitment to the CRE Site (−111/−104) within the Human IL17A Promoter
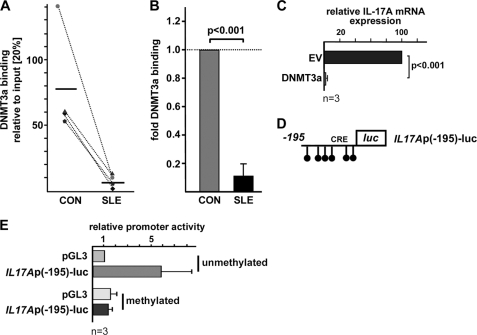
DNMT3a is known to induce de novo DNA methylation and thereby to contribute to transcriptional gene silencing (36). Because CREMα interacts with DNMT3a and appears to recruit it to the human IL2 promoter (27), we tested for the presence of DNMT3a at CRE site (−111/−104) within the human IL17A promoter in four matched female SLE/control T cell pairs. Notably, we detected significantly decreased DNMT3a binding to this CRE site in SLE T cells compared with control T cells (10.6% ± 8.6; p < 0.001; Fig. 7, A and B). Transfection studies confirmed that forced expression of DNMT3a in activated human (Jurkat) T cells decreased IL17A mRNA levels in these cells (Fig. 7C). Because the proximal 195 bp of the human IL17A promoter defines six CpG sequences (Fig. 7D), we next asked whether CpG-DNA methylation of this region affects promoter activity. Methylation of IL17A reporter constructs resulted in distinctly down-regulated promoter activity whereas activity of the empty control reporter was not affected by methylation (Fig. 7E). Our data account for an important role of DNA methylation in the regulation of IL17A gene expression.
FIGURE 7.
DNMT3a recruitment to the CRE site (−111/−104) is decreased in SLE T cells. A, ChIP was performed using total T cells from four matched pairs of SLE patients and healthy controls (CON) and anti-DNMT3a antibody. Immunoprecipitated DNA was analyzed by real-time qPCR using primers detecting a portion of the IL17A promoter harboring the novel CRE site (−111/−104). Ratios between anti-DNMT3a immunoprecipitated and input DNA are shown. Dotted lines associate data from the matched control/SLE pairs. Horizontal bars represent the mean. B, percentage of anti-DNMT3a immunoprecipitated DNA in T cells from the control individual was set to 100%, and relative DNMT3a binding in the corresponding SLE patient was calculated. Values are given as mean ± S.D. (error bars). C, Jurkat T cells were transfected with pcDNA3 empty vector (EV) or an expression plasmid for DNMT3a. Relative IL-17A mRNA expression was analyzed 5 h after transfection. Values are given as mean ± S.D. from three experiments. D, schematic of CpG-DNA methylation sites within the proximal 195 bp of the IL17A reporter construct. E, pGL3-Basic and IL17Ap(−195)-luc reporter plasmids were methylated as outlined under “Experimental Procedures.” Promoter activities of the unmethylated and the methylated reporters were assessed in primary human T cells. Values are given as mean ± S.D. from three experiments.
DISCUSSION
In this report, we present evidence that the transcription factor CREMα contributes to increased IL-17A production in human T cells and remodeling of the IL17A gene locus. CREMα levels are significantly increased in SLE T cells, most likely through an enhanced transcription of the CREM gene in SLE patients, the levels of which even mirror disease activity (18, 19, 21). Overexpression of CREMα in human T cells results in a robust increase of IL-17A expression which is most likely mediated through a direct transcriptional effect. The proximal IL17A promoter defines a CRE site, and our data indicate in vivo CREMα binding to this site in both activated human naïve CD4+ T cells and total T cells from SLE patients.
Aberrant cytokine expression and an impaired transcriptional network are hallmarks of the pathogenesis of autoimmune diseases such as rheumatoid arthritis, inflammatory bowel disease, and SLE. Of note, SLE is clearly associated with IL-2 deficiency as well as IL-17 overproduction. SLE patients display increased IL-17 levels, and IL-17-producing T cells infiltrate target organs including the kidneys (6). In our efforts to identify common mechanisms that account for altered cytokine expression in SLE T cells, CREMα emerges as a promising candidate. Previous data from our group (19, 25, 27) along with the findings presented herein suggest antithetic CREMα effects on IL2 and IL17A gene regulation at both the transcriptional and epigenetic level. It is of particular interest that in SLE T cells CREMα binding to the promoters of both genes occurs to a comparably increased extent (∼4-fold increase); however, the subsequent implications are diametric. CREMα contributes to decreased IL-2 production in SLE T cells by (i) direct transcriptional repression at a CRE site of the IL2 promoter, (ii) HDAC1 recruitment to the promoter, and (iii) CpG-DNA hypermethylation of the entire IL2 locus as CREMα attracts DNMT3a to the IL2 promoter (27). All these CREMα effects appear to be directly opposed to the mechanisms observed in IL17A gene regulation that eventually result in increased cytokine expression. We propose that CREMα is a novel crucial inducer of IL-17A production by its propensity to activate gene transcription and that CREMα binding is associated with significant histone modifications, including CpG-DNA hypomethylation of the IL17A gene in SLE T cells. It still remains possible that recruitment of CREMα to the IL17A promoter causes histone modifications which facilitate subsequent binding and action of known IL17A transcriptional enhancers, such as interferon-regulatory factor-4, STAT3, and retinoic acid-related orphan receptor γt.
This is the first report to show that the IL17A gene in human T cells from SLE patients is subject to epigenetic remodeling at various levels. Histone modifications such as acetylation, methylation, and phosphorylation rearrange the structure of nucleosomes and thereby regulate the genomic accessibility for transcription factors (37). Acetylation of histone H3 contributes to transcriptional activation whereas trimethylation of H3K27 represses gene transcription. Following T cell activation through CD3/CD28 stimulation, the IL17A gene undergoes histone H3K18 acetylation and histone H3K27 demethylation in naïve CD4+ T cells. We observed the same modifications in SLE T cells compared with T cells from healthy controls. Of note, these effects were reciprocal for the human IL2 gene which appears to be hypermethylated and deacetylated in SLE T cells (27). We propose that HDAC1 is involved in these histone modifications because we found it to be enriched at the IL2 promoter and less recruited to the IL17A promoter in T cells from SLE patients. The role of HDACs in SLE disease expression is still controversial. Mishra et al. (38) reported that HDAC inhibition in lupus-prone mice down-regulated expression of several proinflammatory cytokines and alleviated lupus nephritis; yet, data from our group have claimed that HDAC inhibition may skew T cells toward a “SLE phenotype” (39). Our data, once more, support the concept of HDACs as site- and tissue-specific modulators of gene expression.
Usually, histone and CpG-DNA methylation coincide and are interconnected through several mechanisms (40). CpG-DNA methylation and DNMTs play an important role in the regulation of gene accessibility for transcription factors because they cannot bind to methylated DNA sequences. In general, CpG-DNA methylation is considered to be decreased in patients with SLE and other autoimmune diseases which accounts for an activated gene state and increased expression of several cytokines and molecules that are involved in pathogenesis of SLE and other autoimmune diseases (32, 41–44). CREMα interacts with DNMT3a, which mediates de novo CpG-DNA methylation, and facilitates DNMT3a recruitment to the CRE site of the human IL2 promoter (27) whereas we found that it exerts the opposite effect at the IL17A promoter. Thus, the CREMα-DNMT3a interaction may have site-specific implications which is in line with previous data that displayed that the removal of CpG-DNA methylation is a central event in gene activation during cell differentiation and disease pathogenesis (45). In SLE, several molecules have been linked to CpG-DNA demethylation, including GADD45a, activation-induced deaminase, and methyl-CpG-binding domain 4 (41).
Our data point to a crucial role for CREMα in IL17A gene regulation. Future in vivo analyzes in mice and humans are needed to display its contribution to the development and disease expression of autoimmune conditions. In this context, CREMα may serve as a promising target to correct cytokine and disease expression in patients with SLE and other autoimmune diseases.
Acknowledgment
We thank Melissa Zajdel for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI42269, R01 AI49954, and R01 AI85567 (to G. C. T.). This work was also supported by Deutsche Forschungsgemeinschaft Grant RA1927-1/1 (to T. R.).
- Th cell
- T helper cell
- CNS
- conserved noncoding sequence
- CpG
- cytosine phosphate guanosine
- CRE
- cAMP-responsive element
- CREB
- CRE-binding protein
- CREMα
- CRE modulator α
- DNMT
- DNA methyltransferase
- HDAC
- histone deacetylase
- PMA
- phorbol 12-myristate 13-acetate
- qPCR
- quantitative PCR
- SLE
- systemic lupus erythematosus.
REFERENCES
- 1. Pappu R., Ramirez-Carrozzi V., Sambandam A. (2011) Immunology 134, 8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yao Z., Painter S. L., Fanslow W. C., Ulrich D., Macduff B. M., Spriggs M. K., Armitage R. J. (1995) J. Immunol. 155, 5483–5486 [PubMed] [Google Scholar]
- 3. Iwakura Y., Nakae S., Saijo S., Ishigame H. (2008) Immunol. Rev. 226, 57–79 [DOI] [PubMed] [Google Scholar]
- 4. Nistala K., Wedderburn L. R. (2009) Rheumatology 48, 602–606 [DOI] [PubMed] [Google Scholar]
- 5. Hundorfean G., Neurath M. F., Mudter J. (March 4, 2011) Inflamm. Bowel Dis. 10.1002/ibd.21677 [DOI] [PubMed] [Google Scholar]
- 6. Crispín J. C., Oukka M., Bayliss G., Cohen R. A., Van Beek C. A., Stillman I. E., Kyttaris V. C., Juang Y. T., Tsokos G. C. (2008) J. Immunol. 181, 8761–8766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen Z., Laurence A., O'Shea J. J. (2007) Semin. Immunol. 19, 400–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bettelli E., Carrier Y., Gao W., Korn T., Strom T. B., Oukka M., Weiner H. L., Kuchroo V. K. (2006) Nature 441, 235–238 [DOI] [PubMed] [Google Scholar]
- 9. Zhou L., Ivanov II, Spolski R., Min R., Shenderov K., Egawa T., Levy D. E., Leonard W. J., Littman D. R. (2007) Nat. Immunol. 8, 967–974 [DOI] [PubMed] [Google Scholar]
- 10. Hirahara K., Ghoreschi K., Laurence A., Yang X. P., Kanno Y., O'Shea J. J. (2010) Cytokine Growth Factor Rev. 21, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Akimzhanov A. M., Yang X. O., Dong C. (2007) J. Biol. Chem. 282, 5969–5972 [DOI] [PubMed] [Google Scholar]
- 12. Wei G., Wei L., Zhu J., Zang C., Hu-Li J., Yao Z., Cui K., Kanno Y., Roh T. Y., Watford W. T., Schones D. E., Peng W., Sun H. W., Paul W. E., O'Shea J. J., Zhao K. (2009) Immunity 30, 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Cesare D., Sassone-Corsi P. (2000) Prog. Nucleic Acid Res. Mol. Biol. 64, 343–369 [DOI] [PubMed] [Google Scholar]
- 14. Sassone-Corsi P. (1995) Annu. Rev. Cell Dev. Biol. 11, 355–377 [DOI] [PubMed] [Google Scholar]
- 15. Delmas V., Laoide B. M., Masquilier D., de Groot R. P., Foulkes N. S., Sassone-Corsi P. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4226–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Foulkes N. S., Duval G., Sassone-Corsi P. (1996) Nature 381, 83–85 [DOI] [PubMed] [Google Scholar]
- 17. Rauen T., Benedyk K., Juang Y. T., Kerkhoff C., Kyttaris V. C., Roth J., Tsokos G. C., Tenbrock K. (2011) J. Biol. Chem. 286, 32366–32372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Juang Y. T., Rauen T., Wang Y., Ichinose K., Benedyk K., Tenbrock K., Tsokos G. C. (2011) J. Biol. Chem. 286, 1795–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Solomou E. E., Juang Y. T., Gourley M. F., Kammer G. M., Tsokos G. C. (2001) J. Immunol. 166, 4216–4222 [DOI] [PubMed] [Google Scholar]
- 20. Tsokos G. C. (2011) N. Engl. J. Med., in press [Google Scholar]
- 21. Kyttaris V. C., Wang Y., Juang Y. T., Weinstein A., Tsokos G. C. (2006) Lupus 15, 840–844 [DOI] [PubMed] [Google Scholar]
- 22. Kyttaris V. C., Juang Y. T., Tenbrock K., Weinstein A., Tsokos G. C. (2004) J. Immunol. 173, 3557–3563 [DOI] [PubMed] [Google Scholar]
- 23. Tenbrock K., Kyttaris V. C., Ahlmann M., Ehrchen J. M., Tolnay M., Melkonyan H., Mawrin C., Roth J., Sorg C., Juang Y. T., Tsokos G. C. (2005) J. Immunol. 175, 5975–5980 [DOI] [PubMed] [Google Scholar]
- 24. Ahlmann M., Varga G., Sturm K., Lippe R., Benedyk K., Viemann D., Scholzen T., Ehrchen J., Müller F. U., Seidl M., Matus M., Tsokos G. C., Roth J., Tenbrock K. (2009) J. Immunol. 182, 4167–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tenbrock K., Juang Y. T., Leukert N., Roth J., Tsokos G. C. (2006) J. Immunol. 177, 6159–6164 [DOI] [PubMed] [Google Scholar]
- 26. Asahara H., Santoso B., Guzman E., Du K., Cole P. A., Davidson I., Montminy M. (2001) Mol. Cell. Biol. 21, 7892–7900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hedrich C. M., Rauen T., Tsokos G. C. (2011) J. Biol. Chem. 286, 43429–43436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nalbandian A., Crispín J. C., Tsokos G. C. (2009) Clin. Exp. Immunol. 157, 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hochberg M. C. (1997) Arthritis Rheum. 40, 1725. [DOI] [PubMed] [Google Scholar]
- 30. Morinobu A., Kanno Y., O'Shea J. J. (2004) J. Biol. Chem. 279, 40640–40646 [DOI] [PubMed] [Google Scholar]
- 31. Yang L., Anderson D. E., Baecher-Allan C., Hastings W. D., Bettelli E., Oukka M., Kuchroo V. K., Hafler D. A. (2008) Nature 454, 350–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bailey J., Tyson-Capper A. J., Gilmore K., Robson S. C., Europe-Finner G. N. (2005) J. Mol. Endocrinol. 34, 1–17 [DOI] [PubMed] [Google Scholar]
- 33. Sunahori K., Juang Y. T., Kyttaris V. C., Tsokos G. C. (2011) J. Immunol. 186, 4508–4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Juang Y. T., Wang Y., Solomou E. E., Li Y., Mawrin C., Tenbrock K., Kyttaris V. C., Tsokos G. C. (2005) J. Clin. Invest. 115, 996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deleted in proof.
- 36. Chédin F. (2011) Prog. Mol. Biol. Transl. Sci. 101, 255–285 [DOI] [PubMed] [Google Scholar]
- 37. Renaudineau Y., Youinou P. (2011) Keio J. Med. 60, 10–16 [DOI] [PubMed] [Google Scholar]
- 38. Mishra N., Reilly C. M., Brown D. R., Ruiz P., Gilkeson G. S. (2003) J. Clin. Invest. 111, 539–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nambiar M. P., Warke V. G., Fisher C. U., Tsokos G. C. (2002) J. Cell. Biochem. 85, 459–469 [DOI] [PubMed] [Google Scholar]
- 40. Brenner C., Fuks F. (2007) Dev. Cell 12, 843–844 [DOI] [PubMed] [Google Scholar]
- 41. Hedrich C. M., Tsokos G. C. (2011) Trends Mol. Med., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Patel D. R., Richardson B. C. (2010) Curr. Opin. Rheumatol. 22, 478–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Y., Zhao M., Yin H., Gao F., Wu X., Luo Y., Zhao S., Zhang X., Su Y., Hu N., Long H., Richardson B., Lu Q. (2010) Arthritis Rheum. 62, 1438–1447 [DOI] [PubMed] [Google Scholar]
- 44. Strickland F. M., Richardson B. C. (2008) Autoimmunity 41, 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bird A. (2003) Nat. Immunol. 4, 208–209 [DOI] [PubMed] [Google Scholar]