Abstract
Metal-responsive transcription factor 1 (MTF-1) is an essential protein required for mouse embryonic development. We report here the occurrence of sumoylation on MTF-1. Mutational studies demonstrated that sumoylation occurs on the lysine residue at position 627 (Lys627) of mouse MTF-1. Small ubiquitin-like modifier (SUMO)-1 was fused to the C terminus of MTF-1 to mimic the sumoylated form of the protein and it suppressed the transcriptional activity of MTF-1. The nuclear translocation activity, DNA-binding activity, and protein stability of SUMO-fused MTF-1 are similar to that of wild type MTF-1. The level of sumoylation was reduced by metal in a dose- and time-dependent manner. The fact that zinc reduces MTF-1 sumoylation makes the suppressive role of sumoylated MTF-1 in transcription physiologically less significant because the SUMO moiety of MTF-1 is removed when MTF-1 translocates into nucleus. We further identified a SUMO-interacting motif (SIM) on MTF-1. Remarkably, MTF-1 binds sumoylated MTF-1 and/or other cellular factors in a SIM-dependent manner. This interaction was disrupted by treating cells with zinc. Gel permeation chromatography demonstrated that MTF-1 forms SIM-dependent complexes. This cross-interaction transpires in the cytoplasm and markedly reduces upon nuclear translocation. It can therefore be concluded that SUMO conjugation and the SIM on MTF-1 do not play a critical role in suppressing transcriptional activity. Instead, MTF-1 forms complexes with cellular factors through SIM and SUMO moiety in the cytoplasm. The result explores a new understanding for the mode of MTF-1 assembly and regulation in cells.
Keywords: Metals, Post-translational Modification, Protein Self-assembly, Protein-Protein Interactions, Sumoylation, Transcription Factors, Zinc Finger, MTF-1, SUMO-interacting Motif, Cross-interaction
Introduction
Metal-responsive transcription factor 1 (MTF-1)2 is a zinc finger protein that regulates gene expressions in response to various stresses, such as heavy metal exposure (1), oxidative stress (2), and hypoxia (3). MTF-1 genes have been identified in many species from insects to mammals, and encode proteins with high amino acid identities (4–6). The N-terminal region of MTF-1 has six Cys2-His2-type zinc finger motifs that specifically bind to metal responsive elements (MREs) at the promoters of target genes. Three other distinct domains (an acidic, a proline-rich, and a serine/threonine-rich) on MTF-1 are responsible for the transactivation activity of the protein (7, 8).
Metallothioneins (MTs) are the most extensively studied target genes of MTF-1 (7). MTs are low molecular weight, cysteine-rich proteins that play important roles in zinc homeostasis and heavy metal detoxification (9). MTF-1 is responsible for the basal and metal-induced expression of MT genes (10). In addition, MTF-1 regulates the expression of zinc transporter-1 (11), γ-glutamyl-cysteine synthetase heavy chain (12), and placental growth factor (13). It also plays a critical role in embryonic development because disruption of the Mtf-1 gene in mouse leads to oxidative damage-derived impairment of hepatocytic development and fetal lethality (12).
MTF-1 resides mainly in the cytoplasm. Upon metal exposure or stress induction, MTF-1 translocates into the nucleus and binds to MREs (14, 15). Several factors, such as USF-1, USF-2, NF-κB, and HIF-1α, can modulate the transcriptional activity of MTF-1 (16–19). Histone acetyltransferase p300/CBP and transcription factor Sp1 can rapidly form a complex with MTF-1 upon zinc treatment and stimulate gene transcription (20). Alternatively, zinc and cadmium activate MTF-1 by phosphorylating MTF-1 through different kinase cascades. The exact mechanism of the modifications remains unknown (21–23).
Post-translational modification of proteins by small ubiquitin-like modifier (SUMO) has emerged as an important regulatory mechanism of cellular processes (24). Among the three major SUMO isoforms identified in mammalian cells, SUMO-1 is the best characterized isoform. SUMO-1 and ubiquitin have only 18% sequence identity but share similar structural folds (25). Comparable with ubiquitination, SUMO conjugates covalently to substrates via a specific enzymatic cascade, including the heterodimeric SUMO-activating enzyme SAE1/SAE2 (E1), the SUMO-conjugating enzyme Ubc9 (E2), and in some cases, the SUMO E3 ligases (26). Sumoylation of proteins occurs mostly on lysine residues with a ΨKXE (where Ψ is a bulky hydrophobic residue and X is any amino acid) sequence and the modification can be dynamically reversed by the sentrin/SUMO-specific proteases (SENPs) (27). In contrast to the ubiquitination of proteins for degradation, the functional consequences of protein sumoylation are diverse. Sumoylation is involved in the regulation of subcellular protein localization, protein stability, protein-protein interaction, and transcriptional activity of substrate proteins (24).
SUMO-interacting motif (SIM) plays a central role in the process of SUMO conjugation and affects the fate of the conjugated proteins. The best characterized class of SIM has a hydrophobic core ((V/I)-X-(V/I)-(V/I)) flanked by a cluster of negatively charged amino acid residues (28). Recently, more SIM variants are being identified and the preference of these variants for SUMO isoforms to be bound may be different (29). The SIM hydrophobic core can bind to an interaction surface on SUMO via a parallel or antiparallel orientation. The acidic residues adjacent to the core might contribute to the affinity, orientation, or the paralogue specificity of binding (30, 31). The functional roles of SUMO binding to SIM-containing proteins emerged from several recent studies (32, 33). SIM-containing proteins have been implicated in DNA repair, transcriptional activation, nuclear body formation, and protein turnover (28).
Although phosphorylation of MTF-1 has been reported, sumoylation of this protein has never been known. In the present study, we demonstrated that MTF-1 can be modified by SUMO-1. Additionally, MTF-1 has a SIM that cross-interacts with sumoylated MTF-1 and/or other cellular factors. Zinc treatment disrupts this interaction and removes the SUMO from MTF-1. Our results explore a novel mechanism that regulates MTF-1 functions.
EXPERIMENTAL PROCEDURES
Cell Culture
Chinese hamster ovary (CHO) K1 and HEK293 cells were, respectively, cultured as monolayers at 37 °C in McCoy's 5A medium and DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS), 0.22% sodium bicarbonate, 100 units/ml of ampicillin, and 100 mg/ml of streptomycin in 5% CO2, 95% air and 100% humidity. Cell culture reagents were purchased from Invitrogen. Other chemicals in this work were obtained from Sigma, unless otherwise specified. Zinc sulfate and cadmium chloride were purchased from SERVA and Merck, respectively. Anti-His (C-terminal) antibodies were from Invitrogen. His probe (sc-53073), HA probe (sc-805), anti-GFP (sc-8334), and anti-SUMO-1 (sc-5308) antibodies were purchased from Santa Cruz Biotechnology Inc. HA epitope tag antibody (NB600-363) was obtained from Novus Biologicals. Anti-lamin B1 antibodies (ab16048) and MTF-1 antibodies (ab55522) were purchased from Abcam. Anti-tubulin-α antibodies (clone DM1A) were from Thermo Fisher Scientific and anti-FLAG M2 antibodies were purchased from Sigma.
Plasmid Constructions
Mouse MTF-1 (cDNA) was amplified by polymerase chain reaction (PCR) and inserted into a pcDNA4/V5-His plasmid (Invitrogen) that had been restricted with HindIII and AgeI for the MTF-1-His6 fusion protein expression. DNA encoding HA tag with HindIII and EcoRV sites was PCR amplified and inserted into the 5′ end of MTF-1-His6. A stop codon was generated before the His6 tag and the resulting expression plasmid (HA-MTF-1) produced a HA-tagged MTF-1 without C-terminal His6 tagging. Gal4 DNA-binding domain (DBD, residues 1–147) harboring HindIII and AgeI sites was PCR amplified and ligated into the pcDNA4 plasmids (DBDGal4). The MTF-1 transactivation domain (TAD) encompassing residues 313–675 was also amplified, then digested with AgeI and ligated into the 3′ end of DBDGal4 to generate Gal4-TAD-His6. The nuclear localization signal (NLS) of DBDGal4 on this plasmid was then mutated (15RLKK18 to 15ALAA18 and 43KTKR46 to 43ATAA46) to produce a NLS-mutated fusion plasmid (NLSm-TAD-His6). The HA-tagged Gal4-TAD and NLSm-TAD plasmids were produced following the procedures for HA-MTF-1. The GFP-TAD expression plasmid was constructed by inserting the TAD fragment of MTF-1 into the eGFP-C1 plasmid through HindIII and AgeI sites.
FLAG-tagged SUMO-1, SENP1, HA-SENP2, and Tk-MH100 × 4-Luc plasmids were kind gifts from Dr. H. M. Shih (Academia Sinica, Taiwan). The reporter plasmid containing the 5×MREd-luciferase construct was a generous gift from Dr. G. K. Andrews (Kansas University).
To generate SUMO-fused MTF-1, the human SUMO-1 fragment was amplified by PCR with primers harboring AgeI site. The amplified SUMO-1 fragment was digested with AgeI and ligated into MTF-1-His6 or Gal4-TAD-His6 plasmids to generate MTF-1-SUMO-His6 and Gal4-TAD-SUMO-His6 expression plasmids. Mutations derived from the above constructs and conjugation-defective FLAG-SUMO-1(AA) (Gly96 and Gly97 were changed to Ala) were created by using the QuikChange® site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions.
Transfection and Reporter Gene Assay
Procedures for transfection and reporter gene assays have been described previously (34). For reporter gene assays, expression plasmids encoding a green fluorescence protein (pcDNA3-EGFP, Invitrogen) was included in the DNA mixture for normalization of transfection efficiency. The fluorescence from the GFP of each sample was measured simultaneously. The luciferase activity was normalized to the level of GFP fluorescence. A CMV-Renilla-Luc plasmid (Promega) was used as internal control when the MTF-1-GFP plasmid was transfected in the experiment. Determination of the luciferase and Renilla activities was conducted using a Dual Luciferase® Reporter Assay System (Promega) following the manufacturer's instructions.
Preparation of Whole Cell Extracts
Harvested cells were resuspended in 3 volumes of extraction buffer (20 mm HEPES, pH 7.9, 0.4 m NaCl, 1 mm PMSF, 50 mm NaF, 0.5 mm Na3VO4, 2 μg/ml of aprotinin, 5 μg/ml of leupeptin, 1 μg/ml of pepstatin, and 0.5% Nonidet P-40), and the tube was vigorously rocked at 4 °C for 40 min. N-Ethylmaleimide (20 mm) was added to cell lysates prepared for immunoprecipitation or metal affinity purification and SUMO-1 conjugation detection. Cell debris was removed by centrifugation at 16,000 × g for 10 min. The supernatant (whole cell extracts) was collected and stored at −70 °C. Protein concentration was determined by a dye-binding assay with chemicals purchased from Bio-Rad.
Separation of Cytosolic and Nuclear Fractions
Separation of cytosolic and nuclear fractions was performed as described previously (35) with modifications. Briefly, cells were harvested and cell pellets were resuspended in a hypotonic buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 0.5 mm PMSF, 50 mm NaF, 0.5 mm Na3VO4, 2 μg/ml of aprotinin, 5 μg/ml of leupeptin, and 1 μg/ml of pepstatin). The cells were incubated on ice for 15 min, then Nonidet P-40 was added to a final concentration of 0.5%. Followed by vigorous shaking for 10 s, the homogenate was centrifuged at 16,000 × g for 5 min and the supernatant (cytosolic fraction) was collected. The pellet was suspended in extraction buffer (20 mm HEPES, pH 7.9, 0.4 m NaCl, 1 mm PMSF, 50 mm NaF, 0.5 mm Na3VO4, and protease inhibitors). After shaking vigorously at 4 °C for 40 min, the mixture was centrifuged at 16,000 × g for 20 min. The supernatant was collected as the nuclear fraction.
Chromatin Immunoprecipitation (ChIP) Assay
An EZ ChIPTM Chromatin Immunoprecipitation Kit (Millipore) was used for the assay according to the manufacturer's instructions. Briefly, HEK293 cells (2 × 107) transfected with plasmids encoding HA-MTF-1 and HA-MTF-1-SUMO were treated with or without 100 μm ZnSO4 for 3 h before fixing with 1% formaldehyde. After quenching with 0.125 m glycine, cells were washed and resuspended in lysis buffer, and genomic DNA was sheared to 200–1000 bp in length with an ultrasonic processor (model UP50H, Dr. Hielscher, Germany). Cell debris was removed by centrifugation and 100 μl of the supernatant was diluted with 900 μl of dilution buffer. A 10-μl aliquot from each sample was removed for target DNA quantification (designated as input) and the rest was incubated with 4 μg of anti-HA antibodies overnight at 4 °C. The protein-DNA complexes were pulled down with Magna ChIP protein A magnetic beads (Millipore) at 4 °C for 2 h. After washing with different buffers, the protein-DNA complexes were eluted. The input and the immunoprecipitated samples (with NaCl concentration brought to 0.2 m) were incubated overnight at 65 °C to disrupt the DNA-protein linkages, then treated sequentially with RNase A and proteinase K. DNA in each sample was then extracted by a Wizard® SV Gel and PCR Clean-up System (Promega) before using as templates in PCR. Primers were employed to yield fragments covering the −203 to +28 region of the human MTIIA gene. After 30 cycles of amplification, the products were analyzed on 1.5% agarose gels.
Electrophoresis Mobility Shift Assays (EMSA)
Electrophoresis mobility shift assays were performed as described previously (2). Briefly, 20 μg of whole cell extracts or 5 μg of nuclear extracts were added to 5 μl of 4× binding buffer (48 mm HEPES, pH 7.9, 240 mm KCl, 2 mm DTT, 20 mm MgCl2, and 48% glycerol), 2 μg of dI-dC, and 2 fmol of 32P-labeled probes. The volume of the mixture was brought to 20 μl with H2O and incubated at room temperature for 20 min. After the reaction, 2 μl of EMSA dye (0.25% bromphenol blue, 0.25% xylene cyanol) was added to the sample and the components were separated electrophoretically on 6% native polyacrylamide gels at 4 °C. The running buffer was 1× TGE (25 mm Tris, 0.19 m glycine, 0.5 mm EDTA, pH 8.5). After electrophoresis, the gel was dried and subjected to autoradiography.
Immunoprecipitation
Cell extracts were prepared and added to PBS containing 0.4% Nonidet P-40 and protease inhibitors to make up the 600 μl of working solution. The mixture was treated with ImmunoPure®-immobilized protein A/G-agarose beads (Pierce) for 30 min. The resulting supernatant was collected by centrifugation, then incubated with primary antibodies overnight at 4 °C on a rocking platform. Fresh protein A/G beads were added and the mixture was incubated at 4 °C for 2 h. Thereafter, protein A/G beads were collected and washed three times with PBS containing 0.4% Nonidet P-40. The beads were resuspended in 2× SDS loading buffer (0.125 m Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 0.02% bromphenol blue, 0.2 m DTT) and boiled for 5 min. The resulting supernatants were analyzed by Western blot (36).
Metal Affinity Chromatography
Proteins were isolated with TALONTM resins (Clontech) according to the manufacturer's instructions. Various cell extracts were prepared and diluted in equilibration buffer (50 mm NaH2PO4, pH 7.0, 300 mm NaCl, 10 mm imidazole, 0.4% Nonidet P-40, 10% glycerol, 20 mm N-ethylmaleimide, and protease inhibitors) then incubated with 40 μl of TALON metal affinity resins at 4 °C for 1 h. Thereafter, the TALON resins were collected and washed twice with equilibration buffers containing 20, 30, and 40 mm imidazole sequentially. The resins were then resuspended in SDS loading buffer and boiled for 5 min. The purified samples were analyzed by Western blot.
Column Chromatography
CHO K1 cells were transfected with the designated plasmids and then treated without or with 100 μm ZnSO4 for 6 or 12 h before harvesting. Whole cell extracts were prepared as described above then applied to a prepacked Superose 6 HR 10/30 analytical gel filtration column (GE Healthcare) equilibrated in PBS. The column was developed with PBS at a flow rate of 0.3 ml/min and 0.3-ml fractions were collected. Sixty μl from each fraction were concentrated and analyzed with Western blot. For GFP fluorescence measurements, 200 μl from each fraction were quantified with a Wallac 1420 Multilabel Counter. Gel filtration calibration kits from GE Healthcare were used as protein standards.
Total RNA Extraction and Real-time PCR
Total RNA was extracted with TRIzol reagent (Invitrogen) following the manufacturer's instructions. The extracted RNA (1.5 μg) was reverse transcribed with a RevertAidTM First Strand cDNA Synthesis Kit (Fermentas). The resulting complementary DNA was used for quantitative real-time polymerase chain reaction with the SYBR® Green PCR Master Mix (Applied Biosystems) on an Applied Biosystems 7500 Real-time PCR System. Primer pairs were selected for the amplification of MTIIA: forward, 5′-ATGGATCCCAACTGCTCCTG-3′; reverse, 5′-TCAGGCGCAGCAGCTGCACT-3′ and GAPDH, forward, 5′-GAAGGTGAAGGTCGGAGTC-3′; reverse, 5′-GAAGATGGTGATGGGATTTC-3′.
Statistical Analysis
Mean ± S.D. of samples were calculated from the numerical data generated from three independent experiments and significant differences between treatments were determined by Student's t test.
RESULTS
Occurrence and Identification of SUMO-1 Conjugation on MTF-1
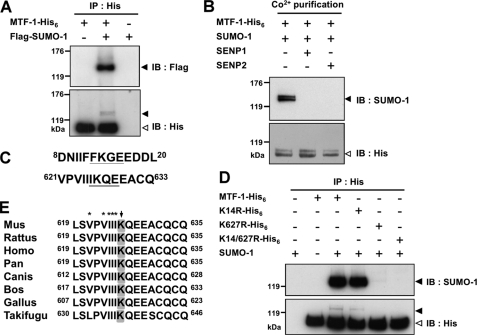
To investigate whether MTF-1 can be sumoylated, plasmids expressing mouse MTF-1 with a His6 tag (MTF-1-His6) and SUMO-1 with a FLAG tag (FLAG-SUMO-1) were co-transfected into CHO K1 cells. The recombinant proteins were immunoprecipitated with anti-His6 antibodies (Fig. 1A). As shown in Fig. 1A, a clear band above 120 kDa was detected by anti-FLAG antibodies. Major and minor MTF-1 bands were observed with antibodies against the His6-tagged antibodies. The electrophoretic mobility of the minor band corresponds to that of the protein identified by anti-FLAG antibodies. We further co-transfected plasmids encoding SENP1 or SENP2 together with the MTF-1-His6 and SUMO-1 encoding plasmids. SENP1 or SENP2 can remove SUMO groups from modified proteins (27). The expressed MTF-1 was purified with metal affinity beads for Western analysis. The results demonstrate that the sumoylated MTF-1 (the band above 120 kDa) is absent in the occurrence of either SENP1 or SENP2 (Fig. 1B). These findings establish that sumoylation of MTF-1 occurs in the cells.
FIGURE 1.
MTF-1 can be sumoylated. CHO K1 cells were transfected with His6-tagged MTF-1 and/or FLAG-tagged SUMO-1 plasmid (A). In addition to MTF-1 and SUMO-1 plasmids, SENP1 or SENP2 plasmids were also co-transfected (B). Cellular MTF-1 was immunoprecipitated with anti-His6 antibodies (A) or pulled down with Co2+-charged metal-affinity beads (B) for Western blot analysis. Proteins were visualized with the designated antibodies. Solid and empty arrows indicate the position of the sumoylated and nonsumoylated MTF-1, respectively. C, mouse MTF-1 primary sequence was analyzed with a computer program and two potential sumoylation sites were predicted. D, the potential sumoylation sites (K14 and K627) on MTF-1 were substituted with arginine singly or in combination (K14R, K627R, and K14R/K627R), then proteins were expressed in the cells. Cell extracts were immunoprecipitated (IP) for Western blotting analysis for sumoylation site identification. E, sequence comparison of potential sumoylation sites on MTF-1 among various species. Arrow indicates the potential sumoylation site. Asterisks indicate the potential region of SIM.
The primary sequence of mouse MTF-1 was analyzed for potential modification site(s) using the consensus sequence for sumoylation (ΨKXE). Two putative sites, one at the N-terminal (Lys14) and the other at the C-terminal (Lys627) region of MTF-1 were identified (Fig. 1C). We then expressed in CHO K1 cells MTF-1 mutants carrying Lys14 and/or Lys627 to arginine substitutions and immunoprecipitated the proteins with anti-His6 antibodies for Western analysis. As shown in Fig. 1D, K14R can still be sumoylated. However, sumoylation of MTF-1 was vastly diminished upon Lys627 substitution. Mutating the consensus glutamate residue to alanine (E629A) also blocked the sumoylation reaction (data not shown).
We further analyzed whether the zinc finger domain of MTF-1 is required for the sumoylation reaction. A new construct was generated with the DNA-binding domain of the Gal4 protein replacing the zinc finger domain and fused with the transactivation domain of MTF-1 (Gal4-TAD-His6). This chimeric protein was expressed and analyzed for sumoylation. Supplemental Fig. S1 shows that the fusion protein is sumoylated and mutations at the sumoylation site (Lys627 and Glu629, designated as Gal4-K627R and Gal4-E629A) abrogate the modification. Sequence alignment among MTF-1 from various species showed that Lys627 is conserved in most of the species (Fig. 1E). However, the corresponding region is absent from zebrafish, tilapia, and Drosophila. Lys14 presents only in mouse MTF-1. This finding indicates that sumoylation of MTF-1 is independent of its DNA-binding motif, but needs the ΨKXE sequence at the C-terminal region of the protein.
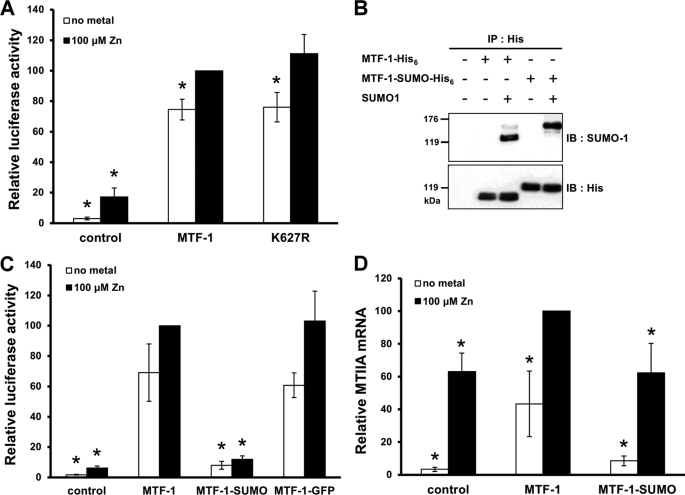
Effect of Sumoylation on the MTF-1 Transcriptional Activity
The biological significance of MTF-1 sumoylation was investigated. Alteration of transcriptional activity was frequently noted for sumoylated transcription factors. We therefore examined initially whether the transcriptional activity of MTF-1 was affected by sumoylation. MTF-1 or its K627R mutant were co-expressed with SUMO-1, and transcriptional activity was evaluated by a reporter gene assay. Fig. 2A shows that mutation at the sumoylation site did not significantly alter the transcriptional activity. The capability of MTF-1 and K627R to activate endogenous target gene expression was investigated. MTF-1 and K627R were co-expressed with SUMO-1 in HEK293 cells, and MTIIA mRNA was analyzed by real-time PCR. No difference was observed in the MTIIA level when MTF-1 or K627R were expressed (supplemental Fig. S2). The Gal4-TAD and its mutants were also used to conduct the reporter gene assay. Distinct from MTF-1, Gal4-TAD activity was not enhanced by the presence of metals (supplemental Fig. S3). However, a noticeable increase in reporter activity was observed when sumoylation of Gal4-TAD was blocked (Gal4-K627R and Gal4-E629A). This result indicates a suppressive role of sumoylated TAD in transcriptional activity.
FIGURE 2.
Effect of sumoylation on MTF-1 transcriptional activity. A, MTF-1 or its K627R mutant were co-transfected with the SUMO-1 and 5×MREd-luciferase reporter plasmids into CHO K1 cells. The cells were treated with or without 100 μm zinc for 6 h, and luciferase activity was measured. B, SUMO-1 and MTF-1-His6 or MTF-1-SUMO-His6 plasmids were co-transfected into CHO K1 cells. The expressed proteins were immunoprecipitated (IP) and then subjected to Western blotting with SUMO-1 antibodies. C, MTF-1-His6, MTF-1-SUMO-His6, or MTF-1-GFP-His6 plasmids were co-transfected with 5×MREd-luciferase reporter plasmids into CHO K1 cells. Relative luciferase activity was determined after adding 100 μm zinc to the transfected cells for 6 h. D, HEK293 cells were transfected with MTF-1-His6 or MTF-1-SUMO-His6 encoding plasmids and treated with 100 μm zinc for 6 h. RNA was extracted and the relative MTIIA mRNA level was determined by quantitative real-time PCR. The MTIIA mRNA level in cells transfected with the MTF-1 plasmid with zinc treatment was designated as 100%. Each value represents a mean ± S.D. of three independent experiments. Asterisks denote significant differences (p < 0.05) from metal-treated cells transfected with MTF-1 (A, C, and D).
Because the sumoylation site on MTF-1 locates at the C-terminal region of the protein, we fused SUMO-1 to the C terminus of MTF-1 (MTF-1-SUMO-His6) to mimic the sumoylated protein for functional studies. Western blot analysis after immunoprecipitating with anti-His6 antibodies reveals that the fusion protein can still be sumoylated despite a SUMO-1 motif had already been constructed at the C terminus of the protein (Fig. 2B).
The constructed gene was delivered into cells for transcriptional activity assays. As shown in Fig. 2C, the MTF-1 activity was drastically reduced after fusing with SUMO-1. This reduction in activity was not due to a steric effect because fusing eGFP to the C terminus of MTF-1 did not alter its transcriptional activity and implying the inhibitory effect is SUMO-1 specific. SUMO-1 was also fused to the C terminus of Gal4-TAD (Gal4-TAD-SUMO-His6). Similarly, sumoylation occurs in this fusion protein (data not shown). Transcriptional activity of the reporter gene was reduced drastically in the occurrence of Gal4-TAD-SUMO-His6 irrespective of the presence or absence of the sumoylation site and metal (supplemental Fig. S4).
We also examined the transcriptional effect of MTF-1-SUMO-His6 on endogenous MT gene expression. The MT gene was not expressed in CHO K1 cells. Therefore, HEK293 cells were used for this study. The MTIIA mRNA in HEK293 were quantified after transfecting the cells with plasmids carrying wild type MTF-1 or MTF-1-SUMO-His6. Fig. 2D shows that MTIIA mRNA increased with metal treatments (a function of endogenous MTF-1). Overexpression of MTF-1 markedly increased the basal and metal-induced MTIIA mRNA levels. However, cells expressing MTF-1-SUMO-His6 reduced the MTIIA mRNA level to that of the control group. The expression level of the transfected genes in Fig. 2, A, C, and D, are shown in supplemental Figs. S5–S7, respectively.
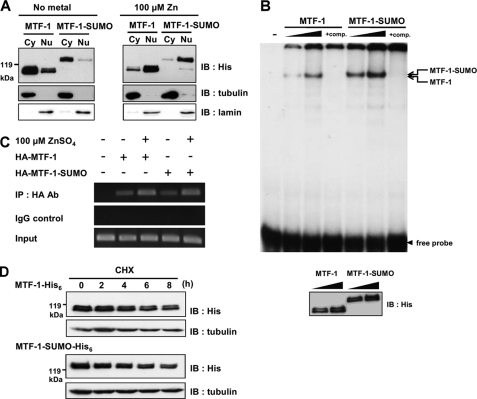
Sumoylation Does Not Alter Nuclear Translocation, MRE-binding Activity, and the Stability of MTF-1
Sumoylation can affect the nuclear translocation activity of a protein (37, 38). MTF-1 has to translocate from the cytoplasm to the nucleus before initiating transcriptional gene regulation. We investigated whether nuclear translocation of MTF-1 was affected by sumoylation leading to a reduction in transcriptional activity. MTF-1-His6 or MTF-1-SUMO-His6 were expressed in cells and the distribution of MTF-1 or MTF-1-SUMO in the cytoplasm and nucleus were examined. MTF-1 was located mainly in the cytoplasm and translocated into the nucleus 3 h after cells were treated with metal. The same result was obtained for MTF-1-SUMO (Fig. 3A). These findings indicate that sumoylation of MTF-1 does not affect the nuclear translocation activity of MTF-1.
FIGURE 3.
Effect of sumoylation on nuclear translocation, MRE-binding activity, and stability of MTF-1. A, MTF-1-His6 or MTF-1-SUMO-His6 were transfected into CHO K1 cells with or without 100 μm zinc treatment for 3 h. Cytosolic (Cy) and nuclear (Nu) extracts were prepared and the expressed proteins were analyzed with Western blot. Tubulin and lamin were used as cytosolic and nuclear markers, respectively. B, MTF-1-His6 or MTF-1-SUMO-His6 were transfected into CHO K1 cells and whole cell extracts were prepared for EMSA with increasing amounts of cell extracts. Unlabeled MREs were used as a competitor to show the specificity of MTF-1/MREs binding. The amount of expressing protein applied to EMSA was analyzed by Western blot with anti-His6 antibodies (lower panel). C, HA-MTF-1 or HA-MTF-1-SUMO were transfected into HEK293 cells with or without 100 μm zinc treatment for 3 h. ChIP assay was conducted to investigate the protein-DNA interaction in vivo and the PCR-amplified fragments of human MTIIA promoter shown. Normal rabbit IgG was used as the control of the assay. D, MTF-1-His6 or MTF-1-SUMO-His6 were transfected into CHO K1 cells. Following administration of 50 μm cycloheximide (CHX) for various time intervals, the quantities of MTF-1 were detected by immunoblotting (IB). Tubulin was employed as loading control.
Because the sumoylated MTF-1 is effective in nuclear translocation, the reduced transcriptional activity may be attributed to the loss of MRE-binding ability. This possibility was examined using electrophoretic mobility shift assay (EMSA) to gauge the interaction between MRE and MTF-1. MTF-1 with and without C-terminal SUMO-1 fusion were expressed and the resulting cell extracts were used for EMSA. As shown in Fig. 3B, the C-terminal SUMO-1 fusion did not alter the MTF-1 ability to bind MRE.
Besides EMSA, the ChIP assay was also conducted to study DNA binding in vivo. MTF-1 and the SUMO-fused proteins were expressed in HEK293 cells and the interaction between MTF-1 and MTIIA promoter was analyzed. ChIP assay shows that both MTF-1 and MTF-1-SUMO bind MTIIA promoters, and the binding increases with the addition of metal (Fig. 3C). These findings demonstrate clearly that sumoylation of MTF-1 does not affect the promoter-binding activity of the proteins.
Protein stability might be altered upon sumoylation (39, 40). MTF-1-His6 and MTF-1-SUMO-His6 constructs were transfected into cells and the expressed proteins were quantified at various time intervals after cycloheximide administration. As shown in Fig. 3D, MTF-1-SUMO-His6 has similar stability as that of the wild type MTF-1-His6.
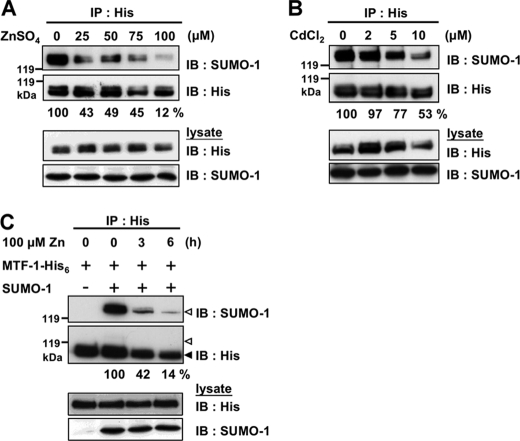
Decline of Sumoylated MTF-1 Level after Metal Exposure
MTF-1 is transcriptionally activated by metal treatment, the level of sumoylated MTF-1 might change upon metal induction. A dose-dependent study was conducted. FLAG-SUMO-1 and MTF-1-His6 genes were co-transfected into cells and MTF-1 was immunoprecipitated after exposing the cells to various concentrations of zinc for 3 h. The results show a dose-dependent reduction of sumoylated MTF-1 with zinc treatment (Fig. 4A). This effect is apparently not zinc specific because addition of cadmium showed the same result (Fig. 4B). A time course study was subsequently performed and cells were treated with 100 μm zinc for 3 or 6 h. Fig. 4C indicates that the decline in sumoylated MTF-1 correlates with the duration of zinc exposure. These findings reveal clearly that metal modulates the level of MTF-1 sumoylation.
FIGURE 4.
Effect of metal exposure on the sumoylation level of MTF-1. SUMO-1 and MTF-1-His6 were co-expressed in CHO K1 cells and MTF-1-His6 was immunoprecipitated (IP) with anti-His6 antibodies after exposing the cells to 25, 50, 75, and 100 μm zinc (A) or 2, 5, and 10 μm cadmium (B) for 3 h. The level of sumoylation on MTF-1 was detected by Western blotting with SUMO-1 antibodies. C, cells transfected with SUMO-1 and MTF-1-His6 were exposed to 100 μm zinc for 3 or 6 h. The level of sumoylation on MTF-1 was detected by Western blotting (IB).
Identification and Characterization of SIM in MTF-1
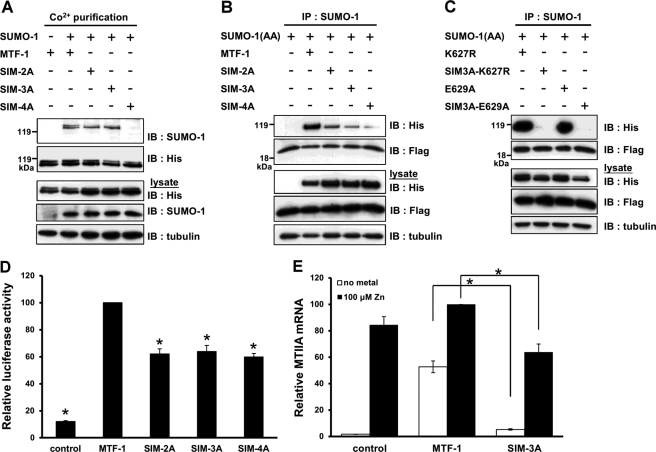
The reduction of MTF-1 sumoylation by zinc masks the biological role of this protein modification. Upon zinc stimulation, MTF-1 is activated and translocated into nucleus. Under this circumstance, the sumoylated MTF-1 is absent from the nucleus and will not exert an inhibitory effect in transcription. The role of MTF-1 sumoylation is thus further investigated. Analysis of mouse MTF-1 primary sequence reveals possible SIM structures (621VPVIII626) located right next to the sumoylation site (Fig. 1E). This structure was identified in all MTF-1 sharing the conserved sumoylation site (Fig. 1E). SIM has a consensus sequence of ((V/I)-X-(V/I)-(V/I)) (28). The hexapeptide, 621VPVIII626, represents two possible SIMs (VPVI and VIII) with two overlapping amino acids (623VI624). Mutations at these two residues will abolish the SIM structure whether one or two SIMs are present.
It has been reported that the presence of SIM can facilitate the sumoylation of proteins (41). However, mutations of 623VI624 or 623VII625 to alanine (SIM-2A or SIM-3A) did not alter the level of MTF-1 sumoylation (Fig. 5A). Changing 623VIII626 to four alanines (SIM-4A) diminished the degree of sumoylation. Because Ile626 overlaps with the conserved sumoylation sequence (626IKQE629, Fig. 1E), mutation at this residue is expected to reduce the sumoylation level.
FIGURE 5.
Identification and functional analysis of SIM. A, MTF-1-His6 or constructs with the SIM mutation were co-transfected with SUMO-1 into CHO K1 cells. The His-tagged proteins were isolated with metal affinity beads and the levels of sumoylation on SIM mutants were analyzed with Western blotting. B, MTF-1-His6 or constructs with the SIM mutation were co-transfected with FLAG-SUMO-1(AA) plasmid. The complexes were isolated by immunoprecipitation (IP) with anti-SUMO-1 antibodies and then subjected to Western blotting (IB) analysis. C, sumoylation-defective MTF-1, K627R, and E629A, and their SIM mutants were co-transfected with FLAG-SUMO-1(AA). Twenty-four h after transfection, the cells were harvested and cell lysates were prepared and subjected to immunoprecipitation with anti-SUMO-1 antibodies. The precipitated products were analyzed with anti-His6 and anti-FLAG antibodies. D, MTF-1 plasmid or constructs with the SIM mutation were co-transfected with 5×MREd-luciferase reporter plasmids into CHO K1 cells. Luciferase activity was determined after exposing the cells with 100 μm zinc for 6 h. E, HEK293 cells were transfected with MTF-1 or SIM-3A plasmids, and treated with or without 100 μm zinc for 6 h before being harvested. Total RNA was extracted and the relative MTIIA mRNA level was determined by real-time PCR. Each value represents a mean ± S.D. of three samples. Asterisks denote significant differences (p < 0.05) with the MTF-1 (D) or between the paired samples (E).
To demonstrate the 621VPVIII626 sequence is a SIM, MTF-1-His6 or the indicated SIM mutants were co-transfected into cells with conjugation-deficient SUMO-1 plasmids (SUMO-1(AA)) and immunoprecipitation was performed with SUMO-1 antibodies after the proteins were expressed. Western blot analysis indicated that the precipitated fraction contained both MTF-1 and SUMO-1 (Fig. 5B). However, the interaction was markedly reduced when SIM was mutated. This result demonstrates the MTF-1 containing the 621VPVIII626 sequence can interact noncovalently with SUMO-1.
Furthermore, SIM-mutated K627R and E629A (SIM-3A-K627R and SIM-3A-E629A) were used to examine the noncovalent interaction of the proteins with SUMO-1(AA). Fig. 5C shows that this interaction can occur on MTF-1 with the sumoylation site abolished, but not with MTF-1 harboring mutated SIM.
With the identification of SIM in MTF-1, the possibility of SIM in maintaining the integral function of MTF-1 was subsequently analyzed. Several SIM-mutated MTF-1 plasmids were expressed in CHO K1 cells and MTF-1 activity was estimated by the luciferase reporter assay. Fig. 5D shows that the transcriptional activity of the mutants was reduced to nearly 60% of that of the wild type MTF-1. We also examined the effect of SIM mutation on endogenous MTIIA gene expression. HEK293 cells were transfected with MTF-1 and SIM-3A, and MTIIA mRNA expression was compared after adding 100 μm zinc for 6 h. MTF-1 induced MTIIA mRNA transcription with and without zinc treatment and this induction was significantly reduced when SIM-3A was expressed in place of the wild type MTF-1 (Fig. 5E). The expression level of the transfected genes was shown in supplemental Figs. S8 and S9. Co-expression of SIM-3A with SUMO-1 also markedly reduced the MTIIA mRNA level in HEK293 cells (supplemental Fig. S10). These results imply that the presence of SIM stimulates MTF-1 activity.
Cross-interaction of MTF-1 through SIM and Disruption of the Interaction by Zinc
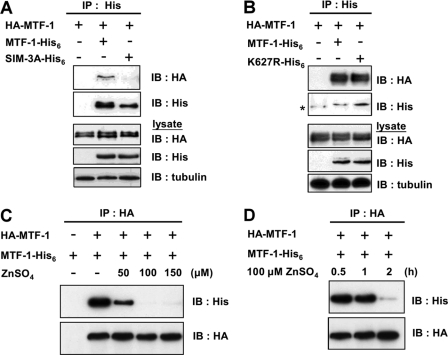
Because MTF-1 contains SIM and can be sumoylated, we investigated whether MTF-1 cross-interacts with its SUMO conjugate. To address this possibility, HA-tagged MTF-1 and His6-tagged MTF-1 were co-expressed in the cells. Following immunoprecipitation by anti-His6 antibodies, the presence of HA-tagged MTF-1 was analyzed. As shown in Fig. 6A, these two proteins interact only when their SIM structure is intact. The presence of a sumoylation site is not required for this interaction because K627R also interacted with MTF-1 (Fig. 6B). Because metal reduces the level of MTF-1 sumoylation, we investigated whether the cross-interaction of MTF-1 was affected by the presence of metal. Fig. 6C shows a dose-dependent decrease in interaction between MTF-1 upon metal treatment. A time course study of the metal effect was also performed. The cross-interaction of MTF-1s reduced dramatically after adding 100 μm zinc for 1–2 h (Fig. 6D). These results demonstrate a cross-interaction of MTF-1s in cells via the SIM, and this interaction is attenuated by zinc.
FIGURE 6.
MTF-1 cross-interacts with each other through SIM and the interaction can be disrupted by zinc treatment. HA-tagged MTF-1 and MTF-1-His6, SIM-3A-His6 (A) or K627R-His6 (B) were co-expressed in CHO K1 cells. Following immunoprecipitation (IP) with anti-His6 antibodies, the presence of HA-tagged MTF-1 was visualized with anti-HA antibodies on Western blot (IB). HA-tagged MTF-1 and His6-tagged MTF-1 were co-expressed in the CHO K1 cells and treated with various concentrations of zinc for 6 h (C) or 100 μm zinc for various time intervals (D). The degree of MTF-1 cross-interaction was analyzed with the anti-His6 antibody after cell extracts were immunoprecipitated with HA antibodies.
Demonstration of MTF-1 Complex Formation by Column Chromatography
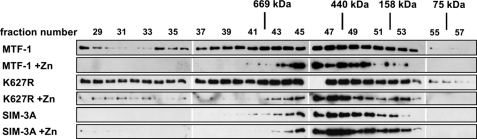
The above results suggest that MTF-1 may form complexes with another MTF-1 and/or other cellular factors. To demonstrate this possibility, cell extracts were separated by molecular sieve column chromatography and the fractions were analyzed with Western blot to determine the size distribution of possible MTF-1 complexes. His6-tagged MTF-1, K627R, and SIM-3A were expressed in the cells with or without the addition of zinc. Cell extracts were prepared for size fractionation and immunoblotting. As shown in Fig. 7, in cells without zinc treatment, MTF-1 and K627R eluted immediately after the void volume (7.5 ml) and spread out in fractions covering a wide range of molecular weights. Noticeably, both proteins did not come out immediately after the void volume and presented only in limited fractions in cells treated with zinc. Furthermore, SIM-3A appeared at similar fractions as those of zinc-treated MTF-1 and K627R even without the addition of zinc (Fig. 7). Interestingly, MTF-1 prepared from zinc-treated cells did not elute off the column with an expected molecular mass (∼80 kDa). We speculate that MTF-1 assumes an elongated structure and/or is associated with unknown cellular macromolecules.
FIGURE 7.
Demonstration of SIM-dependent MTF-1 complex formation with gel permeation chromatography. His6-tagged MTF-1, K627R, or SIM-3A were expressed in CHO K1 cells with or without 100 μm zinc treatment for 6 h. Cell extracts were prepared and separated on an analytical Superose 6 HR 10/30 column. Equal amounts of eluent from each fraction were concentrated and analyzed by immunoblotting using anti-His6 antibodies. The elution positions of the molecular markers were indicated on top of the panel. MTF-1 can be detected after fraction number 26.
To further demonstrate the formation of MTF-1 complexes, other fusion constructs were used. Because SIM and the sumoylation site are located in the TAD of MTF-1, this domain should be sufficient to demonstrate protein cross-interactions. We fused eGFP to TAD (GFP-TAD) or TAD with both SIM (SIM-3A) and K627 mutations (GFP-TAD-SKM). The expressed fusion proteins stay in the cytoplasm because they do not have the NLS. This design can exclude events that occur during zinc-induced translocation. Cells were treated with or without zinc for 6 h and cell extracts were prepared for column chromatography. As shown in supplemental Fig. S11A, the fluorescence of GFP-TAD without zinc treatment appeared broadly in high molecular weight fractions, similar to that of the Western blotting results obtained for MTF-1 in Fig. 7. Interestingly, addition of zinc did not alter the fluorescence profile. However, a single and sharp fluorescence peak was observed for the GFP-TAD-SKM sample at a relatively lower molecular weight region. The presence of GFP-TAD in the eluted fraction was further analyzed by immunoblotting. Supplemental Fig. S11B shows the distribution of GFP-TAD in the eluted fractions was similar to that of MTF-1 (Fig. 7) in the presence and absence of zinc treatment, although GFP-TAD-SKM has a profile similar to MTF-1 after metal treatment. Taking these results together, our study demonstrates clearly that cellular MTF-1 presents in complexes via SIM cross-interactions.
Localize the Occurrence of Cross-interaction
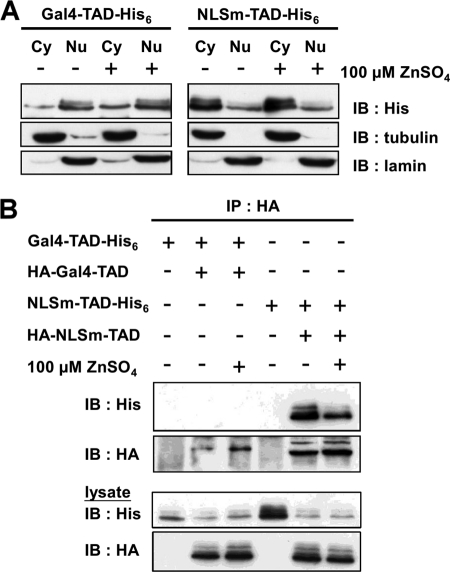
MTF-1 shuttles between the cytoplasm and nucleus. Complex formation apparently occurs in the cytoplasm and addition of zinc directly or indirectly disrupts the cross-interactions. It remains unclear whether this zinc effect occurs in the cytoplasm or during/after the process of nuclear translocation. Zinc treatment dissociates the MTF-1 complexes, whereas it is ineffective in dissociating the GFP-TAD complexes (supplemental Fig. S11). These results imply that cross-interaction occurs in the cytoplasm and dissociates during/after translocation. To address this speculation, HA- and His6-tagged Gal4-TAD and its NLS mutant (NLSm-TAD) were used for the study. Gal4-TAD translocates into nucleus after translation, whereas NLSm-TAD remains in the cytoplasm (Fig. 8A). Different tags of Gal4-TAD or NLSm-TAD were co-transfected into CHO K1 cells and cell extracts were analyzed. Fig. 8B shows that NLSm-TAD was cross-interacted in cells and zinc treatment cannot diminish the interaction. Noticeably, the level of Gal4-TAD cross-interaction was absent with or without zinc treatment. The results suggest that MTF-1 does not present in complex forms after translocating into the nucleus.
FIGURE 8.
Localization of MTF-1 complexes. A, Gal4-TAD or NLSm-TAD were expressed in CHO K1 cells. Cytosolic and nuclear extracts were prepared and cellular distribution of the expressed proteins was analyzed by immunoblotting (IB). Tubulin and lamin were employed as cytosolic (Cy) and nuclear markers (Nu), respectively. B, Gal4-TAD or NLSm-TAD plasmids harboring different tags were co-transfected into CHO K1 cells and cell extracts were prepared. Cross-interaction of Gal4-TAD or NLSm-TAD was analyzed in cells treated with or without 100 μm zinc for 6 h.
DISCUSSION
Biochemical modification of MTF-1 has not been well studied. Phosphorylation is the only modification that has been reported but the biological significance and the site(s) of modification remains unknown (21, 22). Additionally, whether zinc treatment alters the phosphorylation level of MTF-1 remains controversial (22, 42). We report in this study that MTF-1 can be sumoylated and we also identified a functional SIM structure on MTF-1. Because destruction of this SIM did not completely abolish SUMO-1 binding (Fig. 5B), we do not exclude the presence of other SIM(s) in mouse MTF-1. SUMO modification and SIM on transcriptional factors have reportedly reduced the protein activity (43). In contrary, SUMO modification and the SIM on MTF-1 play fewer roles in repressing transcriptional activity. Rather, they cross-interact with cellular factors to form complexes in the cytoplasm. This finding shows a distinct characteristic of SUMO modification and SIM on the assembly and functional regulation of MTF-1.
The proportion of sumoylated MTF-1 is low. This is a common finding and reportedly most substrates have less than 1% of sumoylation at any time point (44). Consequently, detections of sumoylated proteins are often difficult due to the low proportion of conjugation. Co-expression of substrates and enzymes involved in SUMO conjugation is frequently required to verify the occurrence of SUMO modification. For example, sumoylated STAT1 and p53 can only be detected as fusion proteins with Ubc9 (45). In some cases, co-expression of the substrate and SUMO-1 is needed for detecting the modification (46, 47). Despite the low level of sumoylation, the modified proteins can achieve marked biological effects. This phenomenon has been described as “SUMO enigma” (26, 48). Therefore, even though the sumoylated MTF-1 presents in small quantity, its contribution to MTF-1 regulation cannot be ignored.
Mouse MTF-1 is sumoylated at Lys627, which is a conserved residue among MTF-1 of several reported species. This sumoylation site is present in MTF-1 of mammals, avian, and certain species of fish, but not in invertebrates, such as Drosophila or mosquito (4). Interestingly, pufferfish contains the corresponding sumoylation site. However, this site is absent in zebrafish and tilapia (49), which have distinctive carboxyl-terminal sequences. Because the numbers of reported MTF-1 sequences are limited, it is difficult to interpret phylogenetically when the MTF-1 sumoylation site evolved. Noticeably, the presence of the MTF-1 sumoylation site is accompanied with the occurrence of SIM. This coincidence suggests a necessity of both factors for the functional integrity of MTF-1 in a variety of animal species.
Environmental stresses regulate the cellular level of sumoylation. Heat shock causes a global increase in protein sumoylation (50). Hydrogen peroxide treatment results in either an enhancement or inhibition of sumoylation depending on the dosage applied (51). Nitric oxide, another oxidative stress inducer, induces a global desumoylation of cellular proteins (52). Genotoxic agents causing DNA double strand breaks generate varied effects on sumoylation or desumoylation of proteins (53, 54). In this study, metal mediates a reduction in MTF-1 sumoylation. Either SENP1 or SENP2 can remove SUMO-1 from modified proteins and the protein levels of SENP1 and SENP2 are not responsive to metal treatment (data not shown). Sumoylation does not change the MTF-1 stability (Fig. 3D). The reduction in SUMO conjugation on MTF-1 upon metal treatment is probably attributed to enhancing the accessibility of the sumoylated protein to SENP1 or SENP2. Possibly, cytosolic MTF-1 translocates into the nucleus upon metal exposure. The SUMO conjugate on MTF-1 and the unidentified cellular factor(s) can be removed during translocation because SENP1 and SENP2 reside mainly in or near the nuclear pores (27). The cellular factor(s) are retained in the cytoplasm and thus the cross-interaction of MTF-1 does not occur in the nucleus. This speculation may explain why MTF-1 and its K627R mutant have similar activity, because MTF-1 is desumoylated during nuclear translocation and assumes a structure similar to that of the K627R mutant. For GAL4-TAD, it resides in the nucleus after translation. That it can be sumoylated may be due to longer retention in nucleus. The proportion of modified protein exerts a suppressive effect on transcription. Mutation at Lys627 of Gal4-TAD thus enhances the activity.
Sumoylation of MTF-1 may play two roles in the cells. Despite the fact that MTF-1 resides mainly in the cytoplasm and the sumoylated form of MTF-1 is in limited proportion, the possibility that a few sumoylated MTF-1 molecules are present in the nucleus and suppress the target promoters cannot be ruled out. Gal4-TAD moves into the nucleus once synthesized (Fig. 8A). Mutation at the sumoylation site (without the co-expression of SUMO-1 and Ubc9 plasmids) enhances the transcriptional activity of Gal4-TAD (supplemental Fig. S3). These results indicate the suppressive role of sumoylation on the TAD of MTF-1. Because Gal4-TAD can be sumoylated in the nucleus (supplemental Fig. S1), MTF-1 can potentially be modified once retained in the nucleus for a prolonged period. The second role of MTF-1 sumoylation is associated with SIM. Despite presenting in limiting quantity, the SUMO moiety of the modified MTF-1 can bind with the SIM of the adjacent MTF-1 molecule or other cellular factors. This interaction forms complexes of varied sizes. The complexes may retain MTF-1 in the cytoplasm awaiting for stimulatory signals for MTF-1 activation.
The role of SIM on MTF-1 is interesting. Destroying the SIM structure reduced the MTF-1 transcriptional activity (Fig. 5, D and E). This result is somewhat unexpected because studies have shown that SIM can recruit sumoylated proteins and represses transcriptional activity in some transcriptional factors (29, 55). It is not clear whether the SIM is required to bind factors that enhance the transcriptional activity of MTF-1 or destruction of the SIM causes a general effect to reduce the MTF-1 activity. SIM acts as “molecular glue” and interacts noncovalently with SUMO. SUMO and SIM can interact either in a cis or trans manner (56). Because SIM and the consensus sumoylation sequences partially overlap in MTF-1, intermolecular cross-interaction is possible, whereas intramolecular interaction can be ruled out. Noticeably, MTF-1 interacts either with sumoylated MTF-1 or unidentified cellular factor(s) and form complexes in the cytoplasm (Fig. 7). Because this cross-interaction is abolished upon SIM mutation, the SIM on MTF-1 may play dual roles: one for transactivation in nucleus and one for complexes formation in cytoplasm.
Presence of the SUMO conjugation site and SIM on the same protein has been reported in several studies. The promyelocytic leukemia (PML) tumor suppressor is one of the most well studied proteins. It has been demonstrated that the RING domain of the protein is crucial for PML sumoylation (57). The sumoylated PML binds SIM on unmodified PML to form PML-nuclear bodies (NB) and recruits other sumoylated and/or SIM-containing proteins to the PML-NB (58). Daxx is another protein having similar structural characteristics. The SIM in Daxx contributes to its sumoylation, subcellular localization, and repression of sumoylated transcriptional factors. Daxx can also be recruited to PML-NB and interacts with sumoylated PML (55). This binding is reversible and the dissociation of Daxx from PML-NB exerts a trans-repressive effect on other factors. Release of associated proteins from PML-NB can be controlled by a variety of factors and has physiological consequence. For instance, Daxx and Sp100 are released by heat shock and PML is released by cadmium treatment (59).
The SIM on MTF-1 apparently plays different roles. Unlike PML and Daxx, SIM is not crucial for MTF-1 modification because the level of MTF-1 sumoylation does not reduce with SIM mutation (Fig. 5A). Additionally, MTF-1 complexes occur in the cytoplasm but not in the nucleus. Although we did not observe a nucleation of MTF-1 in the cytoplasm under microscopic analysis, complex formation of the proteins is evident (Fig. 7). The formation of the complexes requires the presence of unidentified cellular factors. Sumoylation and/or presence of SIM in the structure can be expected for the factors. Proteins with these characteristics have been reported. Recent studies demonstrate that PML can present in the cytoplasm (60). The cytoplasmic PML interacts with cellular proteins and plays critical roles in cell signaling pathways. For example, cytoplasmic PML physically interacts with Smad2/3 and SARA upon tumor growth factor β (TGF-β) stimulation, and is required for the accumulation of SARA and TGF-β receptor in the early endosome (61). Contradictory to that of TGF-β signaling, MTF-1 associates with other factors before metal exposure and dissociates after zinc stimulation.
In summary, we found in this study that mouse MTF-1 can be sumoylated at a consensus site. The sumoylation of MTF-1 occurs mainly in the cytoplasm. A functional SIM structure is located adjacent to the sumoylation site. Under normal conditions, cytosolic MTF-1(s) (including sumoylated MTF-1) cross-interacts with other cellular factors via the SIM and SUMO moieties to form complexes. These complexes sequestered part of the available cellular MTF-1 and whether they serve particular biological function(s) beside MTF-1 storage remains to be investigated. Upon zinc stimulation, cytosolic MTF-1 and the complexes move toward the nuclear pores for translocation. For the complexes, the SUMO moieties on MTF-1 are removed by SENP or other factors near or on the nuclear pores and the complexes dissociate. MTF-1 is then released from the complexes and translocates into the nucleus. Cross-interaction of MTF-1 in the nuclear does not occur again because of the distinct nuclear environment. The nuclear MTF-1 then recognizes and binds MREs at the promoter of target genes for transcription initiation. Our study explores a novel regulatory mechanism for MTF-1 and represents a distinct functional assembly for a transcriptional factor residing in the cytoplasm before translocation. This mode of regulation may be applied to other cytosolic proteins having a SUMO conjugation site and SIM structure.
Supplementary Material
This work was supported by Grant NSC98-2311-B-007-015-MY3 from the National Science Council, Taiwan, Republic of China.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S11.
- MTF-1
- metal-responsive transcription factor 1
- MRE
- metal responsive element
- MT
- metallothionein
- SUMO
- small ubiquitin-like modifier
- SENP
- sentrin/SUMO-specific protease
- SIM
- SUMO-interacting motif
- DBD
- DNA-binding domain
- TAD
- transactivation domain
- NLS
- nuclear localization signal
- eGFP
- enhanced green fluorescent protein
- PML
- promyelocytic leukemia
- NB
- nuclear bodies.
REFERENCES
- 1. Lichtlen P., Schaffner W. (2001) Bioessays 23, 1010–1017 [DOI] [PubMed] [Google Scholar]
- 2. Dalton T. P., Li Q., Bittel D., Liang L., Andrews G. K. (1996) J. Biol. Chem. 271, 26233–26241 [DOI] [PubMed] [Google Scholar]
- 3. Murphy B. J., Andrews G. K., Bittel D., Discher D. J., McCue J., Green C. J., Yanovsky M., Giaccia A., Sutherland R. M., Laderoute K. R., Webster K. A. (1999) Cancer Res. 59, 1315–1322 [PubMed] [Google Scholar]
- 4. Zhang B., Egli D., Georgiev O., Schaffner W. (2001) Mol. Cell. Biol. 21, 4505–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Auf der Maur A., Belser T., Elgar G., Georgiev O., Schaffner W. (1999) Biol. Chem. 380, 175–185 [DOI] [PubMed] [Google Scholar]
- 6. Brugnera E., Georgiev O., Radtke F., Heuchel R., Baker E., Sutherland G. R., Schaffner W. (1994) Nucleic Acids Res. 22, 3167–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Radtke F., Heuchel R., Georgiev O., Hergersberg M., Gariglio M., Dembic Z., Schaffner W. (1993) EMBO J. 12, 1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Radtke F., Georgiev O., Müller H. P., Brugnera E., Schaffner W. (1995) Nucleic Acids Res. 23, 2277–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coyle P., Philcox J. C., Carey L. C., Rofe A. M. (2002) Cell. Mol. Life Sci. 59, 627–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heuchel R., Radtke F., Georgiev O., Stark G., Aguet M., Schaffner W. (1994) EMBO J. 13, 2870–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palmiter R. D., Findley S. D. (1995) EMBO J. 14, 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Günes C., Heuchel R., Georgiev O., Müller K. H., Lichtlen P., Blüthmann H., Marino S., Aguzzi A., Schaffner W. (1998) EMBO J. 17, 2846–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Green C. J., Lichtlen P., Huynh N. T., Yanovsky M., Laderoute K. R., Schaffner W., Murphy B. J. (2001) Cancer Res. 61, 2696–2703 [PubMed] [Google Scholar]
- 14. Stitt M. S., Wasserloos K. J., Tang X., Liu X., Pitt B. R., St. Croix C. M. (2006) Vascul. Pharmacol. 44, 149–155 [DOI] [PubMed] [Google Scholar]
- 15. Smirnova I. V., Bittel D. C., Ravindra R., Jiang H., Andrews G. K. (2000) J. Biol. Chem. 275, 9377–9384 [DOI] [PubMed] [Google Scholar]
- 16. Andrews G. K., Lee D. K., Ravindra R., Lichtlen P., Sirito M., Sawadogo M., Schaffner W. (2001) EMBO J. 20, 1114–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cramer M., Nagy I., Murphy B. J., Gassmann M., Hottiger M. O., Georgiev O., Schaffner W. (2005) Biol. Chem. 386, 865–872 [DOI] [PubMed] [Google Scholar]
- 18. Daniels P. J., Andrews G. K. (2003) Nucleic Acids Res. 31, 6710–6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Murphy B. J., Kimura T., Sato B. G., Shi Y., Andrews G. K. (2008) Mol. Cancer Res. 6, 483–490 [DOI] [PubMed] [Google Scholar]
- 20. Li Y., Kimura T., Huyck R. W., Laity J. H., Andrews G. K. (2008) Mol. Cell. Biol. 28, 4275–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. LaRochelle O., Gagné V., Charron J., Soh J. W., Séguin C. (2001) J. Biol. Chem. 276, 41879–41888 [DOI] [PubMed] [Google Scholar]
- 22. Saydam N., Adams T. K., Steiner F., Schaffner W., Freedman J. H. (2002) J. Biol. Chem. 277, 20438–20445 [DOI] [PubMed] [Google Scholar]
- 23. Yu C. W., Chen J. H., Lin L. Y. (1997) FEBS Lett. 420, 69–73 [DOI] [PubMed] [Google Scholar]
- 24. Zhao J. (2007) Cell. Mol. Life Sci. 64, 3017–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bayer P., Arndt A., Metzger S., Mahajan R., Melchior F., Jaenicke R., Becker J. (1998) J. Mol. Biol. 280, 275–286 [DOI] [PubMed] [Google Scholar]
- 26. Wilkinson K. A., Henley J. M. (2010) Biochem. J. 428, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mukhopadhyay D., Dasso M. (2007) Trends Biochem. Sci. 32, 286–295 [DOI] [PubMed] [Google Scholar]
- 28. Kerscher O. (2007) EMBO Rep. 8, 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ouyang J., Shi Y., Valin A., Xuan Y., Gill G. (2009) Mol. Cell 34, 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hecker C. M., Rabiller M., Haglund K., Bayer P., Dikic I. (2006) J. Biol. Chem. 281, 16117–16127 [DOI] [PubMed] [Google Scholar]
- 31. Meulmeester E., Kunze M., Hsiao H. H., Urlaub H., Melchior F. (2008) Mol. Cell 30, 610–619 [DOI] [PubMed] [Google Scholar]
- 32. Merrill J. C., Melhuish T. A., Kagey M. H., Yang S. H., Sharrocks A. D., Wotton D. (2010) PLoS One 5, e8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li Y. J., Stark J. M., Chen D. J., Ann D. K., Chen Y. (2010) Oncogene 29, 3509–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin K. A., Chen J. H., Lee D. F., Lin L. Y. (2005) J. Cell Physiol. 205, 428–436 [DOI] [PubMed] [Google Scholar]
- 35. Chang Y. J., Holtzman M. J., Chen C. C. (2002) J. Biol. Chem. 277, 7118–7126 [DOI] [PubMed] [Google Scholar]
- 36. Yang P. M., Chen H. C., Tsai J. S., Lin L. Y. (2007) Chem. Res. Toxicol. 20, 406–415 [DOI] [PubMed] [Google Scholar]
- 37. Matunis M. J., Coutavas E., Blobel G. (1996) J. Cell Biol. 135, 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yurchenko V., Xue Z., Sadofsky M. J. (2006) Mol. Cell. Biol. 26, 1786–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Minty A., Dumont X., Kaghad M., Caput D. (2000) J. Biol. Chem. 275, 36316–36323 [DOI] [PubMed] [Google Scholar]
- 40. Wei F., Schöler H. R., Atchison M. L. (2007) J. Biol. Chem. 282, 21551–21560 [DOI] [PubMed] [Google Scholar]
- 41. Takahashi H., Hatakeyama S., Saitoh H., Nakayama K. I. (2005) J. Biol. Chem. 280, 5611–5621 [DOI] [PubMed] [Google Scholar]
- 42. Jiang H., Fu K., Andrews G. K. (2004) Biochem. J. 382, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garcia-Dominguez M., Reyes J. C. (2009) Biochim. Biophys. Acta 1789, 451–459 [DOI] [PubMed] [Google Scholar]
- 44. Johnson E. S. (2004) Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 45. Jakobs A., Koehnke J., Himstedt F., Funk M., Korn B., Gaestel M., Niedenthal R. (2007) Nat. Methods 4, 245–250 [DOI] [PubMed] [Google Scholar]
- 46. Suico M. A., Nakamura H., Lu Z., Saitoh H., Shuto T., Nakao M., Kai H. (2006) Biochem. Biophys. Res. Commun. 348, 880–888 [DOI] [PubMed] [Google Scholar]
- 47. Wyatt D., Malik R., Vesecky A. C., Marchese A. (2011) J. Biol. Chem. 286, 3884–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hay R. T. (2005) Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 49. Chen W. Y., John J. A., Lin C. H., Chang C. Y. (2002) Biochem. Biophys. Res. Commun. 291, 798–805 [DOI] [PubMed] [Google Scholar]
- 50. Golebiowski F., Matic I., Tatham M. H., Cole C., Yin Y., Nakamura A., Cox J., Barton G. J., Mann M., Hay R. T. (2009) Sci. Signal. 2, ra24. [DOI] [PubMed] [Google Scholar]
- 51. Bossis G., Melchior F. (2006) Mol. Cell 21, 349–357 [DOI] [PubMed] [Google Scholar]
- 52. Qu J., Liu G. H., Wu K., Han P., Wang P., Li J., Zhang X., Chen C. (2007) PLoS One 2, e1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cheng Z., Ke Y., Ding X., Wang F., Wang H., Wang W., Ahmed K., Liu Z., Xu Y., Aikhionbare F., Yan H., Liu J., Xue Y., Yu J., Powell M., Liang S., Wu Q., Reddy S. E., Hu R., Huang H., Jin C., Yao X. (2008) Oncogene 27, 931–941 [DOI] [PubMed] [Google Scholar]
- 54. Lee Y. K., Thomas S. N., Yang A. J., Ann D. K. (2007) J. Biol. Chem. 282, 1595–1606 [DOI] [PubMed] [Google Scholar]
- 55. Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., Chao T. T., Ho C. C., Chen Y. C., Lin T. P., Fang H. I., Hung C. C., Suen C. S., Hwang M. J., Chang K. S., Maul G. G., Shih H. M. (2006) Mol. Cell 24, 341–354 [DOI] [PubMed] [Google Scholar]
- 56. Matunis M. J., Zhang X. D., Ellis N. A. (2006) Dev. Cell 11, 596–597 [DOI] [PubMed] [Google Scholar]
- 57. Quimby B. B., Yong-Gonzalez V., Anan T., Strunnikov A. V., Dasso M. (2006) Oncogene 25, 2999–3005 [DOI] [PubMed] [Google Scholar]
- 58. Shen T. H., Lin H. K., Scaglioni P. P., Yung T. M., Pandolfi P. P. (2006) Mol. Cell 24, 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nefkens I., Negorev D. G., Ishov A. M., Michaelson J. S., Yeh E. T., Tanguay R. M., Müller W. E., Maul G. G. (2003) J. Cell Sci. 116, 513–524 [DOI] [PubMed] [Google Scholar]
- 60. McNally B. A., Trgovcich J., Maul G. G., Liu Y., Zheng P. (2008) PLoS One 3, e2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lin H. K., Bergmann S., Pandolfi P. P. (2004) Nature 431, 205–211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.