Background: Shedding is a processing mechanism for membrane proteins inducing multifaceted effects.
Results: Proteomic screening identified VIP36 as a shedding target. Overexpression of VIP36 potentiated phagocytosis in a shedding-dependent manner.
Conclusion: VIP36 is a shedding target regulating phagocytosis through its shedding.
Significance: This work revealed new immunological and cell biological functions of shedding and VIP36.
Keywords: ADAM ADAMTS, Lectin, Macrophages, Phagocytosis, Proteomics, Shedding, VIP36
Abstract
Ectodomain shedding is a posttranslational modification mechanism, which liberates extracellular domains of membrane proteins through juxtamembrane processing executed mainly by the ADAM (a disintegrin and metalloprotease) family of metalloproteases. Shedding is a unique and effective mechanism for inducing multifaceted effects through the soluble extracellular domains released and/or the remaining membrane-bound portions; however, the physiological functions of shedding are not yet fully understood. In this study, we performed unbiased proteomic screening for shedding targets in a lipopolysaccharide (LPS)-stimulated macrophage cell line to elucidate a new immunological function of shedding. We identified VIP36 (36-kDa vesicular integral membrane protein), a lectin domain-containing transmembrane protein postulated as a cargo receptor for Golgi-to-endoplasmic reticulum transport, as a new target for shedding and found that the shedding of VIP36 occurs mainly on the cell surface. In addition, we demonstrate that the amount of VIP36 precisely regulates phagocytosis in macrophages and that the shedding of VIP36 is required for this regulation. These results substantially expand our knowledge of the immunological and cell biological functions of both the shedding process and VIP36 itself.
Introduction
Ectodomain shedding, also simply called shedding, is a posttranslational modification mechanism for membrane proteins, which liberates the extracellular domain (ectodomain) of membrane proteins through juxtamembrane processing executed mainly by ADAMs2 (a disintegrin and metalloprotease), a family of membrane-bound metalloproteases (1–3). As a result of processing, shedding converts single membrane proteins into two different portions, a soluble extracellular domain and a membrane-bound remainder, and those proteins play individual roles depending on their cellular localization. For example, heparin-binding EGF-like growth factor, a member of the EGF family of growth factors, is subjected to shedding and thereby converted not only to a soluble growth factor but also to a membrane-bound small fragment that translocates to the inner nuclear membrane and regulates gene expression (4). Shedding also decreases the amounts of cell surface membrane proteins, including receptors and adhesion molecules, and alters the responsiveness of cells to the corresponding ligands. Taken together, shedding is a unique and effective mechanism for inducing multifaceted effects through a simple processing event.
Various membrane proteins have been reported to be shedding targets. Most are membrane proteins whose functions in extracellular space and/or on the cell surface have attracted broad attention. Although shedding is a selective mechanism in which particular membrane proteins are specifically processed depending on physiological conditions, there is to date no known consensus sequence for their cleavage sites. Thus, it is almost impossible to identify new shedding targets by comparing sequence similarity. Taken together, we hypothesize that physiologically meaningful shedding targets remain to be discovered and that identification of new shedding targets will reveal additional physiological functions of the shedding process.
Because unbiased proteomic screening approaches for potential protease substrates have been successful, especially for matrix metalloproteinases(MMPs) (5, 6), a family of metalloproteases distantly related to ADAMs, we herein employed a proteomic method called two-dimensional difference gel electrophoresis (DIGE) to screen for shedding targets on a lipopolysaccharide (LPS)-stimulated macrophage cell line, Raw 264.7. As stimulation of macrophages with LPS derived from Gram-negative bacteria is known to activate shedding of immunologically important cytokines and receptors, including tumor necrosis factor-α (TNF-α) and its receptor (1–3, 7, 8), we expected to identify a new physiological shedding target(s) and a new physiological function(s) of shedding in this system. Indeed, we report here that VIP36 (36-kDa vesicular integral membrane protein) (9) is a shedding target and plays a role in phagocytosis in a shedding-dependent manner.
EXPERIMENTAL PROCEDURES
Antibodies, Plasmids, siRNAs, Chemicals, and Enzymes
Antibodies were purchased from Santa Cruz Biotechnology (anti-M-CSF receptor, sc-692), Roche Applied Science (anti-Myc, 9E10, for Western blotting), Medical and Biological Laboratories (anti-Myc, PL14, for cell surface staining), Sigma (anti-FLAG, F3165; anti-β-actin, A5316), and Abcam (anti-ADAM10, ab39177; anti-ADAM17, ab2051). Anti-VIP36 rabbit polyclonal antibody was raised against a peptide consisting of 11 cytoplasmic amino acids of VIP36 and affinity-purified using CNBr-activated Sepharose 4B (GE Healthcare). Expression plasmids for wild-type human VIP36 (10) and VIP36-like (VIPL) (11) were generous gifts from Dr. Kazuo Yamamoto (University of Tokyo, Japan), and all mutants were constructed using a PCR-based method and subcloned into pcDNA3.1/Zeo(−) (Invitrogen). The amino acid substitutions in point mutants are: aspartic acid 131 to asparagine (D131N, −Lec), asparagine 183 to aspartic acid (N183D, −Gly), asparagine 297 to aspartic acid (N297D), phenylalanine 298 to asparagine (F298N), leucine 299 to methionine (L299M), and serine 301 to leucine (S301L). Primers used and detailed methods are described in the supplemental material. GFP was expressed using a pmaxGFP plasmid (Lonza). All siRNAs were purchased from Ambion (Silencer® select negative control 1 and 2 siRNAs; Silencer® select pre-designed (non-inventoried) siRNAs for ADAM10, ADAM17, and VIP36). LPS, peptide N-glycosidase F, endoglycosidase H, and bafilomycin A1 were purchased from Sigma. Cytochalasin D was purchased from BIOMOL. BB94 was provided by Vernalis.
Cell Lines and Transfections
Raw 264.7 was purchased from ATCC and cultured in high glucose DMEM supplemented with 10% fetal bovine serum (FBS), 100 μm 2-mercaptoethanol, and antibiotics. Wild-type and ADAM17−/− MEFs (12) were kindly provided by Dr. Keisuke Horiuchi (Keio University, Tokyo, Japan), and cultured in DMEM supplemented with 10% FBS and antibiotics. Transfections were performed using FuGENE HD (Roche Applied Science) in general, using NucleofectorTM (Lonza) in ADAM10/17 knockdown experiments and using a NEPA21 electroporator (NEPA GENE, Chiba, Japan) in phagocytosis assays.
Sample Preparations for Western Blotting
In general, cell extracts were prepared using extraction buffer solution (20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 2 mm EDTA, 1% Nonidet P-40, 0.1% SDS) supplemented with a protease inhibitor mixture (Sigma). SDS was omitted when extracts were subjected to immunoprecipitation. When conditioned culture media were prepared, cells were cultured in FBS-free medium with or without 20 μm BB94 for 60 min at 37 °C with 5% CO2, and media were centrifuged at 20,400 × g for 10 min to remove cells and debris. Proteins were precipitated by the addition of 0.25 volumes of 100% TCA and briefly washed with −20 °C acetone.
Two-dimensional DIGE Screening for Shedding Targets and Protein Identification
Twenty million Raw 264.7 cells were seeded into a single 15-cm culture dish the day before sample preparation. Cells were washed twice with FBS-free medium and cultured in FBS-free medium with or without 20 μm BB94 for 30 min, and then 100 nm LPS was added and the cells cultured for an additional 30 min at 37 °C with 5% CO2. Conditioned media were collected and centrifuged at 13,130 × g for 30 min to remove cells and debris, and proteins were precipitated by adding 0.561 g/ml ammonium sulfate (80% saturation). Precipitated proteins were solubilized in protein extraction reagent type 4 (Sigma, C0356), purified using a 2-D clean-up kit (GE Healthcare), and solubilized in protein extraction reagent type 4 again. Protein concentrations were determined using protein assay solution (Bio-Rad). Approximately 25 μg of protein were obtained from 2 × 107 cells. Fifty μg each of LPS-treated and LPS+BB94-treated samples were labeled with the interchange of CyDye DIGE Fluor Cy3 and Cy5 (GE Healthcare), combined and two-dimensionally separated using ImmobilineTM DryStrip, pH 4–7, 24 cm (GE Healthcare) and 10% SDS-PAGE gels (20 × 25 cm). Fluorescence images were taken using a Typhoon Trio scanner (GE Healthcare). For mass spectrometric analysis of the spots of interest, gels were stained using a SilverQuestTM silver staining kit (Invitrogen), and the spots were excised and subjected to in-gel digestion using sequencing grade Trypsin (Promega). Digested peptides were subjected to LC-MS/MS (Finnigan LTQ, Thermo Fisher Scientific) analysis.
Cell Surface Staining
Raw 264.7 cells expressing both N-terminally Myc-tagged VIP36 and GFP were cultured with or without 20 μm BB94 for 1 h, briefly washed with phosphate-buffered saline (PBS), and fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. After washing three times with PBS, cells were incubated with 1% bovine serum albumin (BSA) in PBS for 1 h at room temperature and stained with 10 μg/ml anti-Myc mouse monoclonal antibody followed by 6.7 μg/ml Alexa Fluor® 594 goat anti-mouse IgG (Invitrogen) diluted in 1% BSA/PBS. Fluorescence images were taken under a TCS-SP5 confocal fluorescence microscope (Leica).
Phagocytosis Assays
Fifty thousand Raw 264.7 cells per single well of 96-well plates were seeded the day before the assay. Cells were added with 100 μl of complete medium supplemented with 5–10 μg/ml pHrodoTM Escherichia coli (Invitrogen) and 1 μg/ml Hoechst 33342 (Invitrogen), centrifuged at 190 × g for 3 min, and incubated for 30 min at 37 °C with 5% CO2. The medium was then replaced with Dulbecco's PBS with MgCl2 and CaCl2 (Sigma), fluorescence images were taken, and the fluorescence intensities of pHrodoTM-positive granules per single cell were determined using an imaging cytometer, IN Cell Analyzer 2000 (GE Healthcare). In the assays using cells overexpressing VIP36 (see Fig. 5D), GFP was co-expressed with VIP36 mutants, and only GFP-positive cells were analyzed.
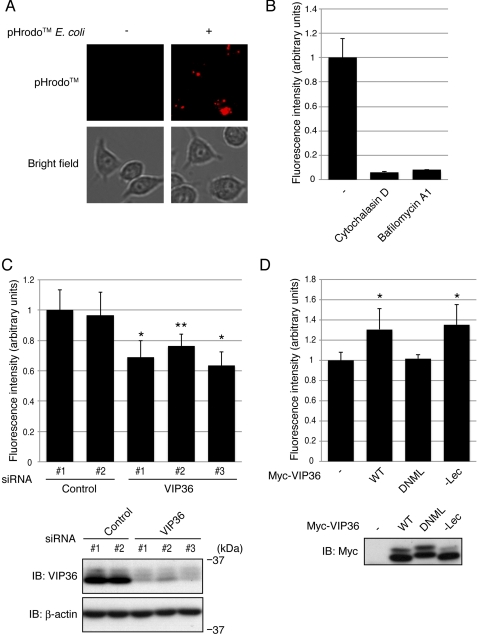
FIGURE 5.
VIP36 enhances phagocytosis in a shedding-dependent manner. A, Raw 264.7 cells were incubated with pHrodoTM-labeled E. coli for 30 min, and fluorescence images were examined. Top and bottom panels show fluorescence images of pHrodoTM-positive granules and bright field images of Raw 264.7 cells, respectively. B, Raw 264.7 cells were incubated with pHrodoTM-labeled E. coli in the presence of cytochalasin D or bafilomycin A1, and the fluorescence intensities of pHrodoTM-positive granules were determined. C, Raw 264.7 cells transfected with siRNAs targeting VIP36 were incubated with pHrodoTM-labeled E. coli, and the fluorescence intensities of pHrodoTM-positive granules were determined. Triplicate samples were used in each experiment, and the experiments were repeated three times. *, p < 0.0001 and **, p < 0.0005 versus control 1 sample. Bottom panels show the expression levels of endogenous VIP36 and β-actin. D, Raw 264.7 cells expressing wild type (WT), a shedding-resistant mutant (DNML), or a lectin-defective mutant (−Lec) of VIP36 were incubated with pHrodoTM-labeled E. coli, and the fluorescence intensities of pHrodoTM-positive granules were determined. Triplicate samples were used in each experiment, and the experiments were repeated three times. *, p < 0.001 versus control sample. The bottom panel shows the expression levels of Myc-tagged VIP36. Error bars indicate the S.D. p values were calculated by Student's t test.
RESULTS
Proteomic Identification of VIP36 as a Candidate Shedding Target
To establish conditions for the screening of shedding targets in LPS-stimulated Raw 264.7 cells, Raw 264.7 cells were pretreated with or without 20 μm BB94, a hydroxamic acid-derived broad metalloprotease inhibitor, for 30 min and then treated with 100 nm LPS for the indicated times. Cell extracts were then subjected to Western blotting. Levels of cell-associated macrophage colony-stimulating factor (M-CSF) receptor (c-fms), a known shedding target in LPS-stimulated macrophages (13), significantly decreased upon 30 min of LPS treatment in the absence of BB94 (Fig. 1A, closed arrowhead). This decrease was completely suppressed by pretreatment with BB94 (Fig. 1A), confirming that a metalloprotease(s) executes the shedding of M-CSF receptor and that pretreatment with BB94 is sufficient to completely suppress the shedding. We thus used LPS-treated and LPS+BB94-treated Raw 264.7 cells as shedding-positive and shedding-negative samples, respectively, to be compared using a proteomic method for the screening of shedding targets. The effects of LPS-induced events other than shedding were excluded as both samples were treated with LPS.
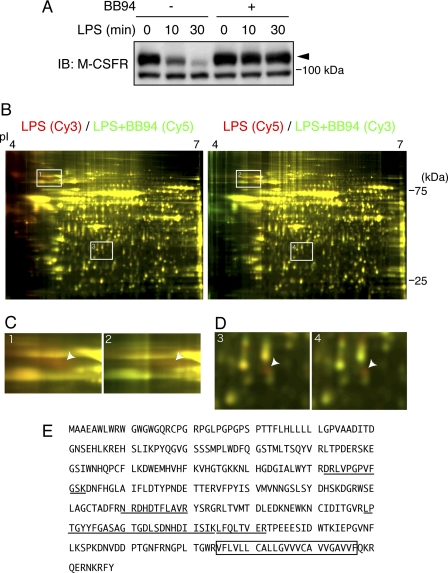
FIGURE 1.
Proteomic identification of VIP36 as a shedding target candidate. A, a mouse macrophage cell line, Raw 264.7, was pretreated with 20 μm BB94 for 30 min and then treated with 100 nm LPS for the indicated times. Cell extracts were subjected to Western blotting (IB) with an antibody against the M-CSF receptor (M-CSFR), a known shedding target. The closed arrowhead indicates the full-length M-CSF receptor. B, conditioned media from LPS-treated (LPS) and LPS+BB94-treated (LPS+BB94) Raw 264.7 cells were collected separately and subjected to two-dimensional DIGE to screen for shedding targets. The proteins were labeled interchangeably with Cy3 and Cy5 as indicated at the top of each image, and the images were pseudo-colored so that shedding targets appear in red. White boxes indicate the positions of red spots enlarged in C and D. C and D, spots that appeared in red in both images were enlarged and are indicated by arrowheads. E, the amino acid sequence of mouse VIP36. The peptides identified by mass spectrometric analysis of the red spots in D are underlined. The transmembrane domain is boxed.
To perform careful comparisons of shedding-positive and -negative samples for the identification of shedding target candidates, we employed two-dimensional DIGE. We labeled proteins in conditioned media from LPS-treated and LPS+BB94-treated Raw 264.7 cells with different fluorescent dyes, Cy3 and Cy5, combined the samples, and separated proteins on single two-dimensional gels. We then searched for proteins present at a higher level in LPS-treated conditioned medium than in LPS+BB94-treated conditioned medium because shedding targets should accumulate in the conditioned medium of shedding-positive samples in the absence of BB94. To avoid any bias derived from a difference in labeling efficiency, we prepared two sets of paired samples: Cy3-labeled LPS-treated and Cy5-labeled LPS+BB94-treated samples and Cy5-labeled LPS-treated and Cy3-labeled LPS+BB94-treated samples. Fig. 1B shows one fluorescence image from each of those samples. The images are pseudo-colored so that the spots of proteins present in higher amounts in LPS-stimulated samples appear red; thus, the red spots in both images are candidates for shedding targets. As shown in Fig. 1B, we found several red spots in both images, two of which are boxed and enlarged in Fig. 1, C and D. Mass spectrometric analysis of the red spot in Fig. 1C identified the M-CSF receptor, a known shedding target (13), demonstrating the feasibility of this screening method. The mass spectrometric analysis of the red spot in Fig. 1D gave us amino acid sequences derived from VIP36 (9), a type I transmembrane protein (Fig. 1E). The shedding of VIP36 has not been reported, but all tryptic peptides identified in mass spectrometric analysis were derived from the extracellular domain of VIP36 (Fig. 1E, underlined). In addition, the molecular mass of the red spot (Fig. 1B, ∼30 kDa) corresponded to that of the extracellular domain of VIP36. These results collectively suggest that VIP36 is subjected to shedding.
VIP36 Is a Shedding Target
To confirm that VIP36 is truly a shedding target, we first examined the release of the VIP36 extracellular domain into culture medium. Raw 264.7 cells expressing N-terminally Myc-tagged VIP36 were treated with BB94 and/or LPS, and cell extracts (Fig. 2A, top panel) and culture supernatants (Fig. 2A, bottom panel) were subjected to Western blotting using an anti-Myc antibody. Myc-tagged, soluble VIP36 with an apparent molecular mass corresponding to the entire extracellular domain of VIP36 was released into culture medium, and the release of soluble VIP36 was completely suppressed by treatment with BB94 (Fig. 2A, bottom panel, closed arrowhead). These results demonstrate that soluble VIP36 is released as a consequence of shedding executed by a metalloprotease(s) and that VIP36 is truly a shedding target. The release of soluble VIP36 was also observed in bone marrow-derived macrophage (supplemental Fig. S1), confirming the existence of VIP36 shedding in macrophages. We noted that two forms of VIP36 were detected in the cell extract of Raw 264.7, and only the high molecular weight form, which was minor in the absence of BB94, accumulated upon treatment with BB94 (Fig. 2A, top panel, closed arrowhead), indicating that the high molecular weight form of VIP36 is susceptible to shedding.
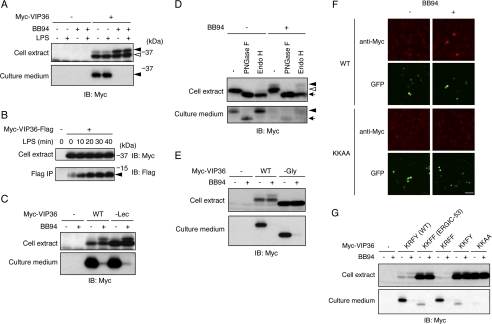
FIGURE 2.
VIP36 is a shedding target. A, N-terminally Myc-tagged VIP36 was expressed in Raw 264.7 cells, and cells were treated with BB94 and/or LPS as indicated. Both cell extracts (top panel) and culture supernatants (bottom panel) were subjected to Western blotting with an anti-Myc antibody. Closed arrowheads indicate the shedding-susceptible form of VIP36 (top panel) and a soluble form of VIP36 (bottom panel), respectively. An open arrowhead indicates the shedding-resistant form of VIP36. B, cytoplasmic FLAG-tagged VIP36 was expressed in Raw 264.7 cells, and cells were treated with LPS for the indicated times. Cell extracts (top panel) and anti-FLAG immunoprecipitates (bottom panel) were subjected to Western blotting (IB) with anti-Myc and anti-FLAG antibodies, respectively. A closed arrowhead indicates the membrane-bound fragment of VIP36. C, a Myc-tagged lectin-defective mutant of VIP36 (−Lec) was expressed in Raw 264.7 cells. Cells were treated with or without BB94, and cell extracts (top panel) and culture supernatants (bottom panel) were subjected to Western blotting with an anti-Myc antibody. D, Myc-tagged VIP36 was expressed in Raw 264.7 cells, and cell extracts (top panel) and culture supernatants (bottom panel) were digested by deglycosylation enzymes as indicated and subjected to Western blotting with an anti-Myc antibody. Closed and open arrowheads in the top panel indicate Endo H-resistant and Endo H-sensitive glycoforms of VIP36, respectively. The arrow in the top panel indicates the deglycosylated form of VIP36. A closed arrowhead and an arrow in the bottom panel indicate the Endo-H-resistant glycoform and deglycosylated form of soluble VIP36, respectively. PNGase F, peptide N-glycosidase F. E, a Myc-tagged glycosylation-defective mutant of VIP36 (−Gly) was expressed in Raw 264.7 cells. Cells were treated with or without BB94, and cell extracts (top panel) and culture supernatants (bottom panel) were subjected to Western blotting with an anti-Myc antibody. F, Raw 264.7 cells expressing Myc-tagged wild-type VIP36 (WT) or C-terminal KKAA mutant of VIP36 (KKAA) were treated with BB94 for 1 h and subjected to cell surface staining using anti-Myc antibody (anti-Myc). Transfected cells were identified as GFP-positive cells (GFP). Bar, 50 μm. G, Myc-tagged C-terminal substitution mutants of VIP36 were expressed in Raw 264.7 cells. Cells were treated with or without BB94, and cell extracts (top panel) and culture supernatants (bottom panel) were subjected to Western blotting with an anti-Myc antibody.
As shown in Fig. 2A, LPS stimulation only slightly increased the amount of soluble VIP36 and only slightly decreased the amount of high molecular weight, cell-associated VIP36. To evaluate the effect of LPS on the shedding of VIP36, we examined another shedding product, the membrane-bound fragment of VIP36. We inserted a FLAG tag immediately after the Lys-347 residue in the cytoplasmic region (see Fig. 4A), based on a previous observation that juxtamembrane insertion of an epitope tag did not change the cellular localization of heparin-binding EGF-like growth factor (4). Raw 264.7 cells expressing a cytoplasmic FLAG-tagged VIP36 were treated with LPS for the indicated times, and cell extracts (Fig. 2B, top panel) as well as anti-FLAG immunoprecipitates (Fig. 2B, bottom panel) were subjected to Western blotting using anti-Myc and anti-FLAG antibodies, respectively. The FLAG-tagged membrane-bound remainder of VIP36 was increased by LPS stimulation in a time-dependent manner (Fig. 2B, closed arrowhead). Taken together, these results demonstrate that VIP36 is constitutively shed to some extent and that LPS further stimulates the shedding of VIP36.
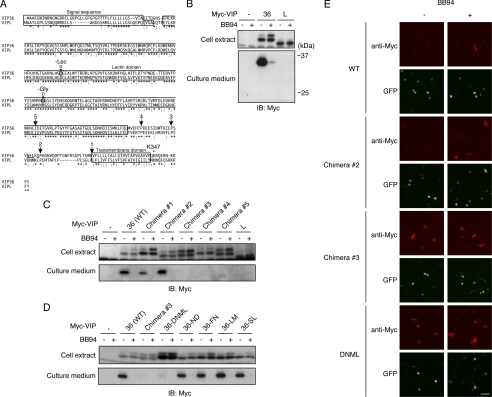
FIGURE 4.
Identification of amino acids required for the shedding of VIP36. A, amino acid sequences of human VIP36 and VIPL are aligned. Conserved and similar amino acids are indicated by asterisks and colons, respectively. Arrows and numbers indicate the positions at which chimeric proteins were constructed. Amino acids substituted in N297D, F298N, L299M, and S301L, and DNML mutants are underlined. Amino acid substitutions in a glycosylation-defective mutant (−Gly) and a lectin-defective mutant (−Lec) are also indicated. A lysine residue where the cytoplasmic FLAG tag is inserted in Fig. 2B is indicated as Lys-347. Signal sequence, lectin domain, and transmembrane domains are boxed. B, Myc-tagged VIP36 or VIPL was expressed in Raw 264.7 cells, and cell extracts (top panel) and culture supernatants (bottom panel) were subjected to Western blotting with an anti-Myc antibody. C and D, Myc-tagged VIP36-VIPL chimeric mutants (C) or substitution mutants of VIP36 (D) were expressed in Raw 264.7 cells, cells were treated with and without BB94 and cell extracts (top panels), and culture supernatants (bottom panels) were subjected to Western blotting with an anti-Myc antibody. ND, N297D; FN, F298N; LM, L299M; SL, S301L. E, Raw 264.7 cells expressing Myc-tagged VIP36 (WT), chimera 2, chimera 3, or DNML mutant were treated with BB94 for 1 h and subjected to cell surface staining using anti-Myc antibody (anti-Myc). Transfected cells were identified as GFP-positive cells (GFP). Bar, 50 μm.
The Lectin Activity of VIP36 Is Dispensable for the Shedding Process
VIP36 is a type I membrane protein and has a lectin domain in its extracellular region (see Fig. 4A) (9). Because VIP36 localizes mainly to the ER and Golgi (14) and the lectin domain of VIP36 has high affinity to an immature type of sugar chain that should be processed in the ER (15, 16), VIP36 is postulated to be a cargo receptor transporting immature glycoproteins from the Golgi to ER in a retrograde manner (17, 18). Thus, we next examined whether the lectin activity of VIP36, critical for the cargo receptor function, is necessary for the shedding. As shown in Fig. 2C, the shedding of a lectin-defective mutant of VIP36 (−Lec, amino acid substitution is indicated in Fig. 4A) (19) was equivalent to that of wild-type VIP36. This result shows that the lectin activity of VIP36 is dispensable for the shedding of VIP36 and that the shedding of VIP36 is independent of its function as a cargo receptor between the ER and Golgi.
VIP36 on the Cell Surface Is Susceptible to Shedding
As mentioned before, Western blotting identifies two forms of cell-associated VIP36, but only the high molecular weight form is susceptible to shedding (Fig. 2A, top panel, closed arrowhead). We wondered what determines the susceptibility of VIP36 to shedding. We first examined whether these two forms are derived from differences in glycosylation by digesting samples with peptide N-glycosidase F, which cleaves all N-linked sugar chains. After peptide N-glycosidase F digestion, both forms of VIP36 collapsed into a single band (Fig. 2D, upper panel, arrow), demonstrating that these two bands represent different glycoforms. We next digested samples with Endo H, which cleaves only immature types of N-linked sugar chains present in the ER and early Golgi. Only the low molecular weight form was sensitive to Endo H (Fig. 2D, upper panel, open arrowhead), indicating that this glycoform is present in the ER or early Golgi. In contrast, the high molecular weight form was resistant to Endo H (Fig. 2D, upper panel, closed arrowhead), indicating that this shedding-susceptible glycoform is likely present in the late Golgi or on the cell surface. Interestingly, N-linked glycosylation was dispensable for the shedding of VIP36 as the shedding of a glycosylation-defective mutant of VIP36 (−Gly, amino acid substitution is indicated in Fig. 4A) was equivalent to that of wild-type VIP36 (Fig. 2E). Taken together, these results indicate that the cellular localization but not the glycosylation of VIP36 determines its susceptibility to shedding.
We next performed cell surface staining to examine the possibility that cell surface VIP36 is susceptible to shedding. Raw 264.7 cells expressing an N-terminally Myc-tagged VIP36 were treated with or without BB94 for 1 h and stained with anti-Myc antibody without cell permeabilization. We detected almost no signal for VIP36 on the cell surface in the absence of BB94 treatment (Fig. 2F, WT, −), but significant VIP36 signals were detected on the cell surface in BB94-treated cells (Fig. 2F, WT, +). The accumulation of cell surface VIP36 signals upon treatment with BB94 is consistent with the shedding-susceptible Endo H-resistant high molecular weight form of VIP36 in Western blotting (Fig. 2, A and D). These results strongly suggest that cell surface VIP36 is susceptible to shedding and that the constant shedding of cell surface VIP36 results in the presence of only low amounts of VIP36 on the cell surface.
To further investigate the relationship between cell surface localization and susceptibility to shedding, we constructed several C-terminal mutants to modify the cellular localization of VIP36. ER-Golgi intermediate compartment (ERGIC)-53 (20), a protein distantly related to VIP36 (21), localizes only between the ER and early Golgi because its C-terminal tetrapeptide, KKFF, contains a dilysine ER retrieval signal, which induces retrograde transport of the protein from the Golgi to ER (20, 22). We substituted the C-terminal tetrapeptide of VIP36, KRFY, with that of ERGIC-53, KKFF, to inhibit the cell surface localization of VIP36 and evaluated the effect of this substitution on shedding. As shown in Fig. 2G, the release of the extracellular domain of the KKFF mutant was severely suppressed, indicating that cell surface localization of VIP36 is required for shedding. We further constructed single amino acid substitution mutants and found that the dilysine ER retrieval signal was critical for the suppression of shedding (Fig. 2G, KRFF and KKFY). Because the two C-terminal aromatic amino acids of ERGIC-53, FF, are known to lessen the ER retrieval signal (22, 23), we further constructed a KKAA mutant to fully activate the dilysine ER retrieval signal and found that shedding of the KKAA mutant was almost completely suppressed (Fig. 2G). Cell surface staining showed that the KKAA mutant did not accumulate on the cell surface in BB94-treated cells (Fig. 2F, KKAA, +), demonstrating that cell surface localization of VIP36 was completely inhibited by this C-terminal mutation. Taken together, these results strongly suggest that the cell surface localization of VIP36 is a critical determinant of its susceptibility to shedding. In addition, we found that all of the ER retrieval signal-containing mutants of VIP36 (KKFF, KKFY, and KKAA) accumulated significantly in cell extracts (Fig. 2G, upper panel), indicating that constant cell surface shedding of VIP36 determines the amount of VIP36 in the cell.
ADAM10 and ADAM17 Are Dispensable for the Shedding of VIP36
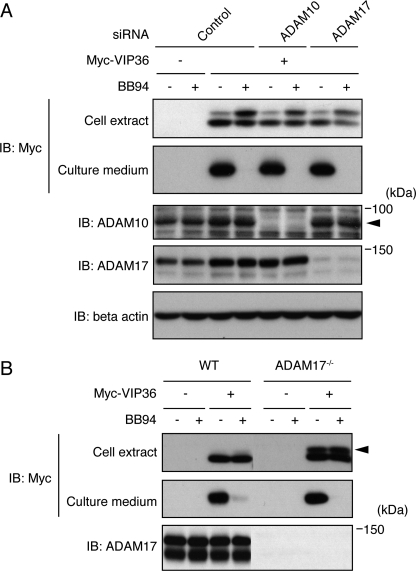
Almost all reported shedding events are performed by members of ADAMs and, among them, ADAM10 and ADAM17 are the two major sheddases executing the majority of reported shedding events (1–3). We therefore next examined whether these two ADAMs participate in the shedding of VIP36. To our surprise, siRNA-mediated knockdown of ADAM10 and ADAM17 failed to suppress the shedding of VIP36 in Raw 264.7 cells (Fig. 3A). Similarly, ADAM17 knock-out MEFs (12) were able to release the ectodomain of VIP36, and this process was completely blocked by BB94 (Fig. 3B). It should be noted, however, that the shedding-susceptible high molecular weight form of VIP36 significantly accumulated in ADAM17 knock-out MEFs (Fig. 3B, closed arrowhead), indicating that ADAM17 participates at least in part in the shedding of VIP36. These results suggest that neither ADAM10 nor ADAM17 is the major sheddase for VIP36.
FIGURE 3.
ADAM10 and ADAM17 are dispensable for the shedding of VIP36. A, Raw 264.7 cells transfected with siRNAs and/or Myc-tagged VIP36 as indicated were treated with BB94, and cell extracts (top and bottom three panels) and culture supernatants (second panel) were subjected to Western blotting (IB) using the antibodies indicated. A closed arrowhead indicates ADAM10. B, both wild-type (WT) and ADAM17−/− MEFs were transfected with Myc-tagged VIP36 and treated with BB94, and cell extracts (top and bottom panels) and culture supernatants (second panel) were subjected to Western blotting (IB) using the antibodies indicated. A closed arrowhead indicates the shedding-susceptible form of VIP36.
Identification of Amino Acids Required for the Shedding of VIP36
VIPL is a homolog of VIP36, which has 68% homology in amino acid sequence with VIP36 (Fig. 4A) (24). We thus wondered whether VIPL is also a shedding target and generated Raw 264.7 cells expressing an N-terminally Myc-tagged VIPL. As shown in Fig. 4B, there was no sign of shedding of VIPL in culture medium, demonstrating that VIPL is not a shedding target. Next we constructed chimeras between VIP36 and VIPL to determine the critical amino acid sequence(s) for the shedding of VIP36. Chimeras were constructed by the recombination of the N-terminal portion of VIP36 and the C-terminal portion of VIPL at the positions indicated in Fig. 4A (arrows and numbers). As shown in Fig. 4C, chimeras 1 and 2 were shed, but chimeras 3, 4, and 5 were not, indicating that the amino acid differences between chimeras 2 and 3 are critical for the shedding of VIP36. As there were only four amino acid differences between chimeras 2 and 3 (Fig. 4A, underlined), we further constructed a substitution mutant in which only these four amino acids were substituted by those of VIPL (DNML, N297D/F298N/L299M/S301L). As shown in Fig. 4D, shedding of VIP36-DNML was almost completely suppressed, indicating that these four amino acids are critical for the shedding of VIP36. Cell surface staining showed that shedding-susceptible mutant, chimera 2, accumulates on the cell surface only in BB94-treated cells, the same as the wild-type VIP36 does (Fig. 4E, WT and chimera 2). On the other hand, two shedding-resistant mutants, chimera 3 and VIP36-DNML, accumulate on the cell surface both in BB94-treated and in non-treated cells (Fig. 4E, chimera 3 and VIP36-DNML), demonstrating that those mutants are properly transported to the cell surface but resistant to shedding in a sequence-dependent manner. Unfortunately, single amino acid substitutions of each of these four positions failed to suppress the shedding of VIP36 (Fig. 4D, N297D, F298N, L299M, and S301L), indicating that multiple amino acids are required for the shedding of VIP36.
VIP36 Enhances Phagocytosis in a Shedding-dependent Manner
Because VIP36 on the cell surface is susceptible to shedding, it is difficult to understand how shedding could regulate the well studied function of VIP36 as a cargo receptor between the ER and Golgi. However, in addition to its function as a cargo receptor, previous studies have shown a relationship between endocytosis and VIP36 (9, 25). Because phagocytosis, which is endocytosis of large solid particles including pathogens, is a major role of macrophages, we examined whether the shedding of VIP36 participates in phagocytosis using pHrodoTM-labeled E. coli (Invitrogen). pHrodoTM is a unique fluorogenic dye that dramatically increases its fluorescence intensity as the pH of its surroundings becomes more acidic, and thus the fluorescence signal of pHrodoTM-labeled E. coli emerges only when they are engulfed and incorporated into an acidic compartment such as the late endosome/lysosome. We incubated Raw 264.7 cells with pHrodoTM-labeled E. coli for 30 min and examined the fluorescence images of the cells. As shown in Fig. 5A, fluorescence signals of pHrodoTM with a granular appearance were observed only inside the cells, and we thus determined the fluorescence intensity of pHrodoTM-positive granules residing in the cell body using an imaging cytometer to quantify the extent of phagocytosis. We found that the fluorescence intensity was strongly reduced by the treatment of inhibitors of phagocytosis including cytochalasin D, which inhibits phagocytosis itself, and bafilomycin A1, which blocks acidification of the endocytic compartment (Fig. 5B), confirming that the fluorescence intensity represents the extent of phagocytosis. We then examined whether endogenous VIP36 participates in phagocytosis using siRNAs targeting VIP36 and found that knockdown of VIP36 using three different siRNAs partially but reproducibly inhibited phagocytosis in accordance with the knockdown level of VIP36 (Fig. 5C), indicating that endogenous VIP36 participates in phagocytosis. We next examined the effect of overexpression of VIP36 on phagocytosis and found that overexpression of wild-type as well as a lectin-defective mutant of VIP36 (WT and −Lec) enhanced phagocytosis (Fig. 5D). In contrast, overexpression of a shedding-resistant mutant of VIP36 (VIP36-DNML) did not enhance phagocytosis (Fig. 5D). These results demonstrate that VIP36 enhances phagocytosis in a shedding-dependent manner. In sum, these results indicate that VIP36 regulates multiple steps of intracellular transport, including retrograde transport of immature glycoproteins from the Golgi to ER and phagocytosis of pathogens from the extracellular space to the endosome.
DISCUSSION
We report herein that VIP36 is subjected to shedding in LPS-stimulated Raw 264.7 cells. Although VIP36 localizes mainly to the ER and Golgi (14) and is postulated to be a cargo receptor for Golgi-to-ER transport (17, 18), we show here that only the Endo H-resistant glycoform of VIP36, which should be present in the late Golgi or on the cell surface, is susceptible to shedding (Fig. 2D) and that VIP36 significantly accumulates on the cell surface upon inhibition of shedding (Fig. 2F). These results clearly demonstrate that cell surface VIP36 is subject to shedding. In addition, C-terminal mutants of VIP36 whose cellular localizations are restricted to between the ER and early Golgi are resistant to shedding and show significant accumulation in cell extracts (Fig. 2G), indicating that a substantial amount of VIP36 translocates to the cell surface and is subjected to shedding. In other words, cell surface shedding of VIP36 hinders the detection of cell surface VIP36 as well as VIP36 functions on the cell surface and/or in the extracellular space. In this study, we have also identified a new function of VIP36 on the cell surface and/or extracellular space: the regulation of phagocytosis.
Knockdown of endogenous VIP36 partially but reproducibly inhibited phagocytosis (Fig. 5C) and overexpression of VIP36 reproducibly enhanced phagocytosis (Fig. 5D) in Raw 264.7 cells, indicating that the amount of VIP36 precisely regulates the rate of phagocytosis. Of note, overexpression of a shedding-defective mutant of VIP36 did not enhance phagocytosis (Fig. 5D, VIP36-DNML), indicating that shedding is indispensable for VIP36 enhancement of phagocytosis. Because VIP36 is subject to shedding on the cell surface, a soluble form of VIP36 should be released into the extracellular space and a membrane-bound portion of VIP36 should be retained on the cell surface, two possible molecular mechanisms that should be considered for the mechanism by which VIP36 enhances phagocytosis. The soluble form of VIP36 may have an opsonizing activity, a binding activity to pathogens to make them edible, as a previous proteomic analysis identified VIP36 in mouse serum (26). Although the lectin activity is dispensable for the enhancement of phagocytosis (Fig. 5D, −Lec), it is possible that a soluble form of VIP36 binds to bacteria in a sugar chain-independent manner. Alternatively, a membrane-bound fragment of VIP36 may be involved in the assembly of phagocytic machinery on the cell surface. It has been reported that the cytoplasmic tail of VIP36 has the ability to induce endocytosis (25) and that antibody-induced clustering of VIP36 causes the accumulation of such clusters in clathrin-coated pits (9), indicating that oligomerization of the cytoplasmic domain of VIP36 stimulates the assembly of endocytotic/phagocytic machinery. It is possible that shedding of VIP36 promotes oligomerization of the membrane-bound fragments of VIP36, including the cytoplasmic domain, upon the removal of the bulky extracellular structure.
We demonstrated that only four amino acids of VIP36 are required for the shedding process. Because these four amino acids exist between the globular ectodomain structure (27) and transmembrane domain of VIP36, the same cleavage site as other shedding targets, it is conceivable that they contain the VIP36 cleavage site. We are now examining the N-terminal amino acid residue of the membrane-bound fragment of VIP36 to determine the exact cleavage site. On the other hand, the identity of the sheddase for VIP36 remains unknown. Knockdown of two major sheddases, ADAM10 and ADAM17, had no effect on the shedding of VIP36 in Raw 264.7 cells. Knock-out of ADAM17 in MEF cells results in accumulation of the shedding-susceptible mature form of VIP36, but shedding of VIP36 was unaffected by the lack of ADAM17. Because the shedding of VIP36 regulates phagocytosis, a role characteristic of macrophages, there is an intriguing possibility that the sheddase of VIP36 is a macrophage-specific metalloprotease(s).
Our results show that the application of proteomic methods is highly effective in the identification of new targets and functions of shedding. In addition, our results demonstrate that screening of shedding targets is possible in many systems using metalloprotease inhibitors. Similar approaches comparing samples prepared under physiological conditions with or without metalloprotease inhibitors will lead to the identification of physiological targets and functions of shedding in future studies.
Supplementary Material
Acknowledgments
We thank Dr. Kazuo Yamamoto for the VIP36 and VIPL expression plasmids; Dr. Keisuke Horiuchi for the wild-type and ADAM17−/− MEFs; and Dr. Shigeki Higashiyama for information about cytoplasmic FLAG tags. We also thank Yuka Setoguchi for technical assistance; AMR Inc. and Medical Proteomics Laboratory of the Institute of Medical Science, University of Tokyo (IMSUT) for LC-MS/MS analysis of shedding targets; and Invitrogen for preparation of anti-VIP36 antibody.
This work was supported in part by Grant-in-aid for Young Scientists (B) 21790328 (to K. S.) and Grant-in-aid for Scientific Research (S) 19109004 (to Y. O.) from the Japan Society for the Promotion of Science. S. K. is a consultant for Medical and Biological Laboratories, Co. Ltd.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Fig. S1.
- ADAM
- a disintegrin and metalloprotease
- VIP36
- 36-kDa vesicular integral membrane protein
- DIGE
- difference gel electrophoresis
- VIPL
- VIP36-like
- MEF
- mouse embryonic fibroblast
- ER
- endoplasmic reticulum
- ERGIC
- ER-Golgi intermediate compartment
- Endo H
- endoglycosidase H
- Lec
- lectin
- Gly
- glycosylation.
REFERENCES
- 1. Seals D. F., Courtneidge S. A. (2003) Genes Dev. 17, 7–30 [DOI] [PubMed] [Google Scholar]
- 2. Blobel C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 32–43 [DOI] [PubMed] [Google Scholar]
- 3. Huovila A. P., Turner A. J., Pelto-Huikko M., Kärkkäinen I., Ortiz R. M. (2005) Trends Biochem. Sci. 30, 413–422 [DOI] [PubMed] [Google Scholar]
- 4. Hieda M., Isokane M., Koizumi M., Higashi C., Tachibana T., Shudou M., Taguchi T., Hieda Y., Higashiyama S. (2008) J. Cell Biol. 180, 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schilling O., Overall C. M. (2007) Curr. Opin. Chem. Biol. 11, 36–45 [DOI] [PubMed] [Google Scholar]
- 6. Morrison C. J., Butler G. S., Rodríguez D., Overall C. M. (2009) Curr. Opin. Cell Biol. 21, 645–653 [DOI] [PubMed] [Google Scholar]
- 7. Jue D. M., Sherry B., Luedke C., Manogue K. R., Cerami A. (1990) Biochemistry 29, 8371–8377 [DOI] [PubMed] [Google Scholar]
- 8. Leeuwenberg J. F., Dentener M. A., Buurman W. A. (1994) J. Immunol. 152, 5070–5076 [PubMed] [Google Scholar]
- 9. Fiedler K., Parton R. G., Kellner R., Etzold T., Simons K. (1994) EMBO J. 13, 1729–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kawasaki N., Matsuo I., Totani K., Nawa D., Suzuki N., Yamaguchi D., Matsumoto N., Ito Y., Yamamoto K. (2007) J. Biochem. 141, 221–229 [DOI] [PubMed] [Google Scholar]
- 11. Yamaguchi D., Kawasaki N., Matsuo I., Totani K., Tozawa H., Matsumoto N., Ito Y., Yamamoto K. (2007) Glycobiology 17, 1061–1069 [DOI] [PubMed] [Google Scholar]
- 12. Horiuchi K., Kimura T., Miyamoto T., Takaishi H., Okada Y., Toyama Y., Blobel C. P. (2007) J. Immunol. 179, 2686–2689 [DOI] [PubMed] [Google Scholar]
- 13. Rovida E., Paccagnini A., Del Rosso M., Peschon J., Dello Sbarba P. (2001) J. Immunol. 166, 1583–1589 [DOI] [PubMed] [Google Scholar]
- 14. Füllekrug J., Scheiffele P., Simons K. (1999) J. Cell Sci. 112, 2813–2821 [DOI] [PubMed] [Google Scholar]
- 15. Hara-Kuge S., Ohkura T., Seko A., Yamashita K. (1999) Glycobiology 9, 833–839 [DOI] [PubMed] [Google Scholar]
- 16. Kamiya Y., Yamaguchi Y., Takahashi N., Arata Y., Kasai K., Ihara Y., Matsuo I., Ito Y., Yamamoto K., Kato K. (2005) J. Biol. Chem. 280, 37178–37182 [DOI] [PubMed] [Google Scholar]
- 17. Hauri H., Appenzeller C., Kuhn F., Nufer O. (2000) FEBS Lett. 476, 32–37 [DOI] [PubMed] [Google Scholar]
- 18. Schrag J. D., Procopio D. O., Cygler M., Thomas D. Y., Bergeron J. J. (2003) Trends Biochem. Sci. 28, 49–57 [DOI] [PubMed] [Google Scholar]
- 19. Hara-Kuge S., Ohkura T., Ideo H., Shimada O., Atsumi S., Yamashita K. (2002) J. Biol. Chem. 277, 16332–16339 [DOI] [PubMed] [Google Scholar]
- 20. Schindler R., Itin C., Zerial M., Lottspeich F., Hauri H. P. (1993) Eur. J. Cell Biol. 61, 1–9 [PubMed] [Google Scholar]
- 21. Fiedler K., Simons K. (1994) Cell 77, 625–626 [DOI] [PubMed] [Google Scholar]
- 22. Itin C., Schindler R., Hauri H. P. (1995) J. Cell Biol. 131, 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kappeler F., Klopfenstein D. R., Foguet M., Paccaud J. P., Hauri H. P. (1997) J. Biol. Chem. 272, 31801–31808 [DOI] [PubMed] [Google Scholar]
- 24. Nufer O., Mitrovic S., Hauri H. P. (2003) J. Biol. Chem. 278, 15886–15896 [DOI] [PubMed] [Google Scholar]
- 25. Itin C., Kappeler F., Linstedt A. D., Hauri H. P. (1995) EMBO J. 14, 2250–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hood B. L., Zhou M., Chan K. C., Lucas D. A., Kim G. J., Issaq H. J., Veenstra T. D., Conrads T. P. (2005) J. Proteome Res. 4, 1561–1568 [DOI] [PubMed] [Google Scholar]
- 27. Satoh T., Cowieson N. P., Hakamata W., Ideo H., Fukushima K., Kurihara M., Kato R., Yamashita K., Wakatsuki S. (2007) J. Biol. Chem. 282, 28246–28255 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.