Background: ALDH enzymes metabolize aldehydes in many pathways, including the inactivation of cyclophosphamide.
Results: Covalent inhibitors against ALDH were discovered, and their mechanism of action was determined.
Conclusion: Covalent inhibitors against ALDH potentiate cell killing in cyclophosphamide-resistant cells.
Significance: These inhibitors represent novel research tools and can serve as leads toward therapeutics where increased ALDH activity is associated with disease.
Keywords: Anticancer Drug, Chemical Biology, Crystal Structure, Cysteine-mediated Cross-linking, Enzyme Inhibitors, Mass Spectrometry (MS), Protein Crystallization
Abstract
Human aldehyde dehydrogenases (ALDHs) comprise a family of 17 homologous enzymes that metabolize different biogenic and exogenic aldehydes. To date, there are relatively few general ALDH inhibitors that can be used to probe the contribution of this class of enzymes to particular metabolic pathways. Here, we report the discovery of a general class of ALDH inhibitors with a common mechanism of action. The combined data from kinetic studies, mass spectrometric measurements, and crystallographic analyses demonstrate that these inhibitors undergo an enzyme-mediated β-elimination reaction generating a vinyl ketone intermediate that covalently modifies the active site cysteine residue present in these enzymes. The studies described here can provide the basis for rational approach to design ALDH isoenzyme-specific inhibitors as research tools and perhaps as drugs, to address diseases such as cancer where increased ALDH activity is associated with a cellular phenotype.
Introduction
Members of the aldehyde dehydrogenase (ALDH)2 superfamily catalyze the oxidation of cytotoxic aldehydes utilizing NAD(P)+ as the electron acceptor (1, 2). ALDH family members participate in multiple metabolic pathways, including ethanol oxidation, retinol metabolism, dopamine, and GABA metabolism, as well as branched-chain amino acid metabolism and in the elimination of toxic aldehydes produced from lipid peroxidation (3). Furthermore, recent work has demonstrated that specific ALDH isoenzymes may contribute to the pathology of different diseases. For example, ALDH1A1 is an important biomarker for cancer cells and cancer stem cells (3, 4) and together with ALDH3A1 confers resistance to cyclophosphamide in cancer cells (5). The low activity of ALDH2*2 enzyme present in East Asians is associated with acetaldehyde toxicity when they consume ethanol (6) as well as reduced efficacy of nitroglycerin (7), whereas activation of ALDH2 reduces ischemia reperfusion injury (8–10).
Our groups recently reported the discovery and characterization of a novel activator for ALDH2 and ALDH2*2 (8, 11). The same screening procedure also led to the identification of several inhibitors. Inhibitors of ALDH family members have potential application in alcohol dependence (12), cocaine addiction (13), anxiety (14), and as resensitizing agents for cancers that are resistant to the cyclophosphamide class of compounds (15–17). In this latter regard, siRNA knockdown of ALDH1A1 and/or ALDH3A1 increased sensitivity to cyclophosphamide, with the double knockdown demonstrating the greatest resensitization (5).
Members of the ALDH superfamily possess a highly reactive active site cysteine residue (Cys-302 in ALDH1A1 and ALDH2, Cys-243 in ALDH3A1) that performs a nucleophilic attack on the aldehydic carbonyl group leading to the formation of a covalent thiohemiacetal intermediate. Subsequent hydride transfer to NAD(P)+ generates the reduced cofactor and an acyl enzyme intermediate, which is then hydrolyzed by a water molecule activated by a general base believed to be either Glu-268 or Glu-399 (Glu-209 and Glu-333 in ALDH3A1). In general, the order of substrate addition is sequential with the cofactor preceding the aldehyde into the active site, but this order of addition is relaxed for some aromatic substrates where random substrate binding can occur (18).
In this study, we set out to characterize the mechanism of action of a class of inhibitors identified in our high-throughput screen for modulators of ALDH2 activity. Initial characterization of four related compounds showed similar inhibition properties and time-dependent kinetics. Subsequent crystal structure determination demonstrated a covalent adduct with the active site cysteine of both ALDH2 and ALDH3A1. This observation was confirmed by mass spectrometric analysis of the reaction products. Together, these data support a mechanism of covalent inhibition where the enzymes catalyzes a beta-elimination of the substituted amine, forming a reactive aromatic vinyl ketone compound that subsequently modifies the highly reactive catalytic nucleophile in the active site.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were purchased from Sigma Aldrich unless otherwise stated.
Cloning
The human ALDH3A1 clone was obtained from Open Biosystems and cloned into a pET-28a plasmid with an N-terminal His tag. The single mutants were made by site-directed mutagenesis using a Quikchange II site-directed mutagenesis kit (Agilent, Santa Clara, CA).
Protein Purification
Human ALDH3A1 was expressed and purified as described elsewhere (19). The activity of the enzyme is also tested using a standard assay that measures production of NADPH (see description below). Purification of recombinantly expressed ALDH2 and its C302S, E399Q, and E268Q enzymes has been described elsewhere (20–22).
Inhibitor Screens
The ALDH2 enzyme assay design has been reported previously (19). All tested compounds were dissolved in DMSO and screened at 10 μm concentration prior to addition of the substrate. Assays and computer analysis were carried out by the Stanford University High-Throughput Biosciences Center (Department of Chemical and Systems Biology). 63,500 compounds were screened (8), and some were purchased from ChemBridge for further validation.
Inhibitors
Inhibitors that emerged from the screen were purchased from ChemBridge and were designated as follows: Aldi-1, 3-(1-azepanyl)-1-phenyl-1-propanone hydrochloride; Aldi-2, 3-(dimethylamino)-1-(3-fluoro-4-methoxyphenyl)-1-propanone hydrochloride; Aldi-3, 3-(dimethylamino)-1-(4-ethylphenyl)-1-propanone hydrochloride; and Aldi-4, 1-(4-chlorophenyl)-3-(1-piperidinyl)-1-propanone hydrochloride (named here Aldi-1, -2, -3, and -4 for ALDH inhibitors 1–4).
Mass Spectrometry
Complexes for analysis by mass spectrometry were formed by the addition of 20 μm ALDH3A1 or ALDH2 with 40 μm (2-fold molar excess) or 100 μm (5-fold molar excess) of the various inhibitors and incubated overnight at room temperature. Samples (0.5 to 8 μl) were injected into an Agilent 6520 quadrupole time of flight mass spectrometer operating in positive ion electrospray ionization-time of flight mode equipped with an Agilent 1200 series capillary HPLC. The data were analyzed with formula assignments for low molecular weight compounds and deconvolution for proteins using the Mass Hunter/Bioconfirm software suite.
Preparation and Crystallization of ALDH3A1, ALDH2, and Their Complexes with Inhibitors
Crystals of ALDH3A1 at 3 mg/ml in 10 mm HEPES, pH 7.5 were grown from solutions containing 0.2 m potassium acetate and 18% PEG3350 at 25 °C. We prepared the complexes with inhibitors through direct soaking experiments by first equilibrating the crystals in solution containing 2% DMSO overnight, which were then supplemented with 100 μm of the desired inhibitor. The crystals were directly frozen without any further addition of cryoprotectant. Data sets were collected at a wavelength of 0.9869 Å and at 100 K. All diffraction data were indexed, integrated, and scaled using the HKL3000 program suite (23). All refinements were performed using the program packages Refmac5 as implemented in the CCP4 program suite (23), and model inspection and building was accomplished using the program package Coot (24). Molecular replacement was used to solve the structure of the apo form of human ALDH3A1 using the rat ALDH3A1 structure as the search model (Protein Data Bank code 1AD3). For structures complexed with inhibitors, the apo structure was used for direct refinement. Binding of inhibitors was evaluated through inspection of the initial σ-A weighted Fo − Fc electron density maps. The Aldi-1 complexed structure showed Cys-243 in two distinct conformations within the actives sites at 50% occupancy, only one of these conformations was modified covalently.
Crystals of ALDH2 were grown from protein solutions containing 8 mg/ml ALDH2, in 100 mm Na-ACES, pH 6.4–6.6, 100 mm guanidine-HCl, 10 mm MgCl2, and 4 mm dithiothreitol and 16–17% PEG 6000. Introduction of Aldi-3, processing and refinement of the data were performed as outlined above. The structure was solved by using the coordinates of the refined apo enzyme structure of ALDH2 as the refinement model (Protein Data Bank code 1O05) following removal of all solvent molecules. Cys-302 was found in two distinct conformations at 50% occupancy each, only one of those conformations was modified covalently by Aldi-3.
Detailed refinement statistics are provided in Table 2. In the Ramachandran plots, 91.1% (ALDH2-Aldi-3), 93.6% (ALDH3A1) and 92.4% (ALDH3A1-Aldi-1) of all residues are in the most favored regions and there were fewer than 0.5% outliers for all residues.
TABLE 2.
X-ray data collection and refinement statistics for ALDH3A and ALDH2
Data collection and refinement statistics (molecular replacement) are shown below.
| ALDH3A1 (apo)a | ALDH3A1 (Aldi-1)a | ALDH2 (Aldi-3)a | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions | a = 61.41, b = 86.08, and c = 170.40 Å; α = 90.0, β = 90.0, and γ = 90.0° | a = 61.38, b = 85.73, and c = 169.56 Å; α = 90.0, β = 90.0, and γ = 90.0° | a = 140.52, b = 151.05, and c = 177.02 Å; α = 90.0, β = 90.0, and γ = 90.0° |
| Resolution (Å) | 50.0-1.48 (1.51-1.48)b | 50.0-1.51 (1.54-1.51) | 50-2.10 (2.14-2.10) |
| Rsym or Rmerge | 0.058 (0.33) | 0.058 (0.39) | 0.098 (0.53) |
| I/σI | 19.6 (4.1) | 18.6 (2.9) | 18.1 (3.2) |
| Completeness (%) | 97.8 (92.3) | 97.7 (97.8) | 99.7 (99.7) |
| Redundancy | 4.7 (4.0) | 4.4 (3.4) | 5.5 (5.4) |
| Refinement | |||
| Resolution (Å) | 50.0-1.48 | 50.0-1.51 | 50.0-2.10 |
| No. of reflections | 139,344 | 130,505 | 212,188 |
| Rwork/Rfree | 0.16/0.19 | 0.18/0.20 | 0.17/0.23 |
| No. of atoms | |||
| Protein | 7186 | 7224 | 30,543 |
| Ligand/ion | 12 | 32 | 260 |
| Water | 1094 | 1160 | 2597 |
| B-factors | |||
| Protein | 15.1 | 19.1 | 25.5 |
| Ligand/ion | 22.0 | 25.8 | 34.4 |
| Water | 25.4 | 29.7 | 30.9 |
| Root mean square deviations | |||
| Bond lengths (Å) | 0.015 | 0.015 | 0.01 |
| Bond angles | 1.54° | 1.51° | 1.31° |
a One crystal was used for each data collection protocol.
b Values in parentheses are for highest resolution shell.
IC50 Determination Methods for ALDH2, ALDH1A1, and ALDH3A1
IC50 inhibition curves for the inhibitors were measured using the activity of hALDH2, hALDH1A1, and hALDH3A1 as described elsewhere (19). IC50 values were further determined for inhibitors Aldi-1, Aldi-2, and Aldi-4 against propionaldehyde oxidation (ALDH1A1, ALDH2) or benzaldehyde oxidation (ALDH3A1) by measuring NADH production over time at various concentrations ranging from 1 to 100 μm of inhibitors following a 2-min pre-incubation. All reactions were initiated by the addition of substrate aldehyde. The inhibition curves were fit to the four-parameter EC50 equation using SigmaPlot (version 10.0, StatSys). All data represent the average of a minimum of three independent experiments with their S.D.
Kinetics of Irreversible Inhibition
Enzyme stock solutions each containing ALDH3A1 or ALDH2 and varying concentrations between 0 and 500 μm of Aldi-1 were prepared. At the indicated time points, the remaining enzyme activity was determined. The bi-molecular rate constants for the modification of ALDH2 and ALDH3A1 were determined using the method of Aldridge and Reiner (25).
Caspase-6 Activity Assay
Assays for inhibition of caspase activity were carried out using an established method as described in Berger et al. (26) with minor modifications.
Alcohol Dehydrogenase Activity Assay
Enzymatic activity of alcohol dehydrogenase was monitored by monitoring ADH production at 340 nm using recombinant human ADH1B1 protein. All assays were carried out at 25 °C in 50 mm sodium pyrophosphate buffer, pH 9.0, 2.5 mm NAD+ using 30 μg of ADH and 10 mm of ethanol as a substrate. Aldi-1 or Aldi-2 (10 μm), pyrazole (10 μm, a known ADH inhibitor), or DMSO were added immediately prior to the kinetic assays.
Colorimetric MTT Assay for Cell Survival and Proliferation
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay was used for cell survival and proliferation. A549 cells were seeded at 5000 cells per well in 96-well plates 24 h before the start of the first treatment. Cells were treated twice with ALDH inhibitors in the presence or absence of mafosfamide. For the first treatment, ALDH inhibitors were added for 5 h followed by a second treatment for 19 h. Mafosfamide (125 μm, ASTA Z 7557, Asta Werke, Bielefeld, Germany) was added once to the cells alone or in combination with the ALDH inhibitors. MTT assay was carried out 19 h after the addition of the second dose of inhibitors. After the treatments, 0.01 ml of MTT (Millipore CT01-5, 50 mg/ml in PBS) solution was added to each well, and the cells were incubated for 4 h at 37 °C for the reduction of MTT to occur. Isopropanol with 0.04N HCl (0.1 ml each) was then added and mixed thoroughly for color development. Within an hour, absorbance was measured at 570 nm and at a reference wavelength at 630 nm. For the calculation, absorbances measured at 570 nm were subtracted with those measured at 630 nm (n = 8–16 for each treatment). The relative percentage was calculated in comparison with DMSO-treated controls based on the subtracted absorbances.
RESULTS
ALDH2 Inhibitor Screens
A high-throughput screen of small molecules was performed to find modulators of ALDH2 activity (8). This screen yielded several ALDH2 activators, including Alda-1(8, 11). The screen also identified an interesting class of ALDH inhibitors. The present study focuses on the characterization of four structurally related inhibitors Aldi-1, -2, -3, and -4 with a core structure of 3-amino-1-phenylpropan-1-one. These inhibitors are structurally related to the known inhibitor 4-hydroxyacetophenone, which comprises the affinity ligand used for purification of ALDH2 (27, 28). The IC50 values for inhibition of ALDH2 were between 5–9 μm for the four inhibitors (Table 1), whereas the IC50 values for ALDH3A1 ranged between 1.7 and 12 μm, and the IC50 values for ALDH1A1 ranged between 2.5 and 7 μm (Table 1).
TABLE 1.
Inhibition constants of Aldi inhibitors for human ALDH isoenzymes
| Inhibitor | Average IC50 values (μm) |
||
|---|---|---|---|
| ALDH1A1 | ALDH2 | ALDH3A1 | |
| Aldi-1 | 2.2 (0.3)a | 8.6 (1.2) | 12 (1.0) |
| Aldi-2 | 2.5 (0.1) | 6.4 (1.0) | 1.9 (0.1) |
| Aldi-3 | 7.9 (0.4) | 5.4 (1.3) | 1.7 (0.1) |
| Aldi-4 | 6.8 (0.3) | 8.3 (1.2) | 2.2 (0.3) |
| Ki (m−1 s−1) | |||
| Aldi-1 | NDb | 240 | 30 |
a Values in parentheses are S.D. for three independent assays.
b ND, not determined.
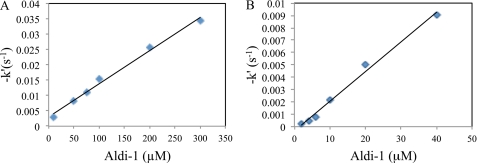
During the initial IC50 experiments, it was noted that the extent of inhibition was variable depending on whether enzyme or substrate was used to initiate the reaction and whether the enzyme and inhibitor were pre-incubated prior to initiating the reaction with substrate. Such order-of-addition and time-dependent effects are often indicative of a covalent means of inhibition. Consequently, we initiated experiments to determine the bi-molecular rate constant for inhibition for the parent compound for this class molecules: Aldi-1. These experiments were consistent with a covalent means of inhibition and bi-molecular rate constants of 30 m−1 s−1 for ALDH3A1 and 240 m−1 s−1 for ALDH2 (Table 1 and Fig. 1).
FIGURE 1.
Time-dependent inhibition of human ALDH3A1 and ALDH2. Shown is a plot of −k′ versus Aldi-1 concentration for ALDH3A1 (A) and for ALDH2 (B). The bimolecular rate constant for inhibition was determined from the slope of these lines using the method of Aldridge and Reiner (25).
ALDH3A1 Crystal Structure
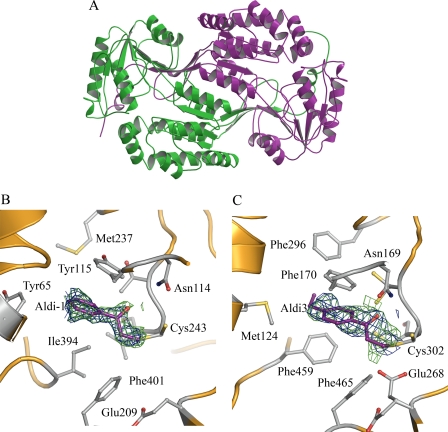
To determine the mechanism by which these inhibitors broadly inhibit ALDH isoenzymes, we examined their binding through the structure determination by x-ray crystallography. The crystal structure of human ALDH3A1 has not been reported previously, although the structure is expected to be highly similar to the rat ALDH3A1 enzyme (1AD3) with which it shares 82% sequence identity. We solved the structure of human ALDH3A1 in its apo enzyme form and in complex with Aldi-1 (Table 2) to 1.5 Å resolution in the P212121 space group. As expected, the enzyme displays a high similarity to the rat ALDH3A1 enzyme (root mean square deviation for all ordered Cα atoms equal to 0.6 Å) and contains all of the structural features that characterize the ALDH family (Fig. 2A) (29). In human ALDH3A1, we detect two distinct conformations for Cys-243, at ∼50% occupancy for each conformation.
FIGURE 2.
Structure of human ALDH3A1 and ALDH2. A, ribbon representation of the overall structure of dimeric human ALDH3A1 with each monomer colored separately. B, active site of human ALDH3A1 modified with the elimination product of Aldi-1. The electron density map displayed is the original σ-A weighted Fo − Fc map contoured at 3 S.D. (green) and the original 2Fo − Fc map contoured at 1 S.D. (blue) showing the modified Cys-243 in the active site superimposed onto the final refined model of the complex. C, active site of human ALDH2 modified with the elimination product of Aldi-3. The electron density map displayed is the original σ-A weighted Fo − Fc map contoured at 3 S.D. (green) and the original 2Fo − Fc map contoured at 1 S.D. (blue) showing the modified Cys-302 in the active site superimposed onto the final refined model of the complex.
Solving the structure of human ALDH3A1 enabled us to determine how these novel inhibitors exerted their inhibitory effects. The electron density map demonstrated that only a portion of the inhibitor was bound in the crystal structure; due to its proximity to Cys-243, we suspected that a covalent adduct occurs between the inhibitor and ALDH3A1 (Fig. 2B). To test the commonality of this interaction, we also solved the structure of Aldi-3 complexed to ALDH2 to 2.1 Å resolution (Table 2). Similar to that observed in ALDH3A1, the electron density is most consistent with a covalent adduct to Cys-302 (Fig. 2C). However, it was not immediately clear from the electron density which part of the inhibitor was attached to the catalytic cysteines. Nevertheless, the density in both the ALDH3A1 and ALDH2 structures were most consistent with an aromatic ketone and two additional carbons linked to the cysteine residue.
Mass Spectrometry Data
To define the chemical entity bound in the crystal structures, we used mass spectrometry of the unreacted protein and protein incubated in the presence of inhibitor. Quadrupole TOF LC/MS of ALDH3A1, ALDH2, and their inhibitor complexes confirmed that there was an increase in mass of the protein following incubation with the inhibitors. Overall, the ALDH2 protein yielded cleaner MS spectra and a single primary deconvoluted mass of 54,445 (supplemental Fig. S1A). In contrast, ALDH3A1 yielded several peaks in the deconvoluted spectrum, although the mass differences between the primary peaks were identical to the mass differences found for ALDH2. The deconvoluted spectra for ALDH3A1 yielded four peaks with masses of 54,413, 52,445, 52,589, and 52,623 (supplemental Fig. S1E). From the smallest mass peak of ALDH3A1, the next peak is shifted by 32 amu, which most likely represents two oxidations of cysteine residues. The next two peaks are each shifted by ∼178 amu from the first two species. A mass shift of 178 Da is consistent with post-translational modification by α-N-6-phosphogluconoylation of the N-terminal His residue of the His tag (30).
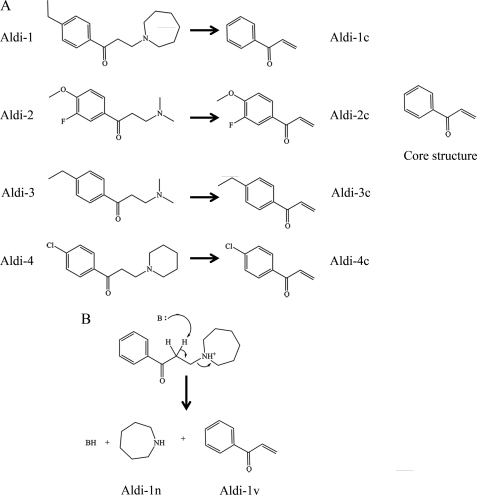
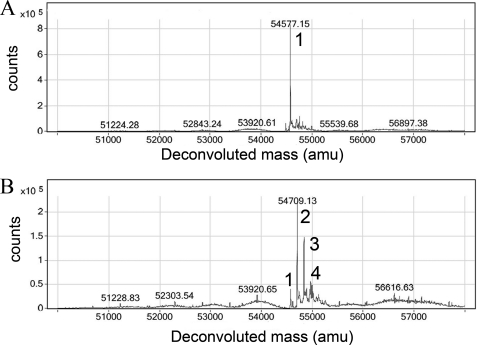
To determine the chemical mechanism of inhibition, our efforts focused on ALDH2, as it was simpler to interpret the mass spectra. Despite the similar phenylpropanone core (Fig. 3), there are numerous differences in structure that would lead to distinct chemical species in the mass spectra. There was an increase in protein mass in all instances when the inhibitors were incubated in the presence of ALDH2 (supplemental Fig. S1, A–D and Table 3). The mass differences between the unmodified and modified proteins were 131.7, 180.0, 159.9, and 166.8 Daltons for incubations with Aldi-1, -2, -3, and -4, respectively (Table 3 and supplemental Fig. S1). These mass differences matched precisely to the resulting vinyl ketone derivatives that would result following β-elimination of the substituted amine group through abstraction of the β-proton by a general base (Fig. 3). The reactive vinyl ketone thus generated reacts covalently with the active site thiol. To confirm this hypothesis and to test the specificity by which the reactive group is generated, we purchased the base vinyl ketone moiety corresponding to the reactive form of Aldi-1 (1-phenyl-2-propen-1-one). Reaction of wild-type ALDH2 with 1-phenyl-2-propen-1-one produced multiple species that differed from the unmodified enzyme by multiples of 132 amu (Table 3 and Fig. 4), matching the molecular weight differential obtained for the Aldi-1. Despite the fact that the Aldi compounds were incubated at 5-fold excess over the concentration of ALDH active sites, only a single cysteine residue was modified based on the combined MS and crystallographic data. In contrast, 1-phenyl-2-propen-1-one only exhibited this level of specific modification at lower concentration ratios (Fig. 4).
FIGURE 3.
Mechanism of Aldi-mediated inhibition of ALDH. A, elimination products produced from the parent compounds based on the mass spectrometric analysis. B, proposed mechanism for β-elimination of Aldi-1, which yields two products, Aldi-1v and Aldi-1n.
TABLE 3.
Mass spectrometric results for ALDH2 and ALDH3A1 upon incubation with Aldi compounds
Aldi compounds were incubated in 10 mm Tris-HCl, pH 7.3, for 16 h.
| Sample | Deconvoluted mass | Δm/z |
|---|---|---|
| ALDH2 | 54,445 | |
| ALDH2 + Aldi-1 | 54,577 | 132 |
| ALDH2 + Aldi-2 | 54,626 | 181 |
| ALDH2 + Aldi-3 | 54,607 | 162 |
| ALDH2 + Aldi-4 | 54,612 | 167 |
| ALDH2 + 1-phenyl-2-propen-1-one | 54,577 | 132 |
| 54,709 | 264 | |
| 54,841 | 396 | |
| 54,972 | 527 | |
| ALDH2-E399Q | 54,445 | |
| ALDH2-E399Q + Aldi-3 (2-fold) | 54,605a | 160 |
| ALDH2-E399Q + Aldi-3 (5-fold) | 54,605 | 160 |
| ALDH3A1 | 52,413 | |
| ALDH3A1 + Aldi-3 | 52,571 | 159 |
| ALDH3A1-C243S | 52,395 | |
| ALDH3A1-C243S + Aldi-3 (5-fold) | 52,395 | 0 |
| ALDH3A1-E209Q | 52,411 | |
| ALDH3A1-E209Q + Aldi-3 (2-fold) | 52,572a | 161 |
| ALDH3A1-E209Q + Aldi-3 (5-fold) | 52,572 | 161 |
| ALDH3A1-E333Q | 52,411 | |
| ALDH3A1-E333Q + Aldi-3 (2-fold) | 52,571 | 160 |
| ALDH3A1-E333Q + Aldi-3 (5-fold) | 52,571 | 160 |
a Samples contain ∼10–20% of unmodified protein.
FIGURE 4.
Chemical modification of human ALDH2 by 1-phenyl-2-propen-1-one. A, deconvoluted spectrum of ALDH2 incubated with a 2-fold molar excess of 1-phenyl-2-propen-1-one. B, deconvoluted spectrum of ALDH2 incubated with a 5-fold excess of 1-phenyl-2-propen-1-one. The peaks in each deconvoluted spectra are labeled according to the stoichiometry of modification (1, 2, 3, and 4).
Additional evidence for the elimination mechanism was obtained by identifying the substituted amine product following incubation of ALDH2 with the Aldi-1. We analyzed the ALDH2 after incubation with Aldi-1 incubation, focusing on the lower molecular weight region of the spectrum from 50–1700 m/z to permit detection of small molecules. Following reaction of ALDH2 with Aldi-1, a peak with the appropriate 100.1 mass units was detected (data not shown). Agilent MassHunter Software was used to identify a molecular formula of the small accurate mass ions and a formula of C6H13N was assigned: a mass difference of 0.1 ppm from the expected fragment.
Site-directed mutagenesis was used to confirm both the sites of modification and probe the residues responsible for the specificity of the covalent modification. To determine whether any of the surrounding glutamate residues function as the general base to generate the reactive intermediate, we incubated the ALDH2 and ALDH3A1 mutants with 2- to 5-fold excess of Aldi-1 (Table 3) and repeated the mass spectrometry analysis. We find that none of these mutants completely abolished the ability Aldi-1 to modify the proteins. However, the ALDH2-E399Q and ALDH3-E209Q mutants were not fully modified when the inhibitor was present at two times the protein concentration (supplemental Fig. S1G and Table 3). The ALDH3-E333Q mutant was fully modified by a 2-molar excess of Aldi-1, and all mutants were stoichiometrically modified when a 5-fold molar excess was used, suggesting that these glutamates are not required to generate the reactive vinyl ketone. In contrast, mutation of Cys-243 to Ser in the ALDH3A1 enzyme yields a virtually inactive enzyme, which was not covalently modified by any of these inhibitors as assessed by mass spectrometry (Table 3). Interestingly, mutation of Cys-302 to Ser in ALDH2 actually increased the stoichiometry of modification from one to two and mutation of Glu-268 to Gln in ALDH2 (E268Q) further increases the stoichiometry of modification to three. To further implicate Cys-243 as the activating residue, we attempted to detect and quantitate the levels of the 100.1 amu cleavage product produced upon activation of Aldi-1 by either wild-type or the C243S mutant of ALDH3. We were able to detect the 100.1 amu cleavage products in both reactions, but we were unable to obtain accurate quantitative estimates from these experiments.
Inhibition by Aldi Compounds Is Specific to Aldehyde Dehydrogenase
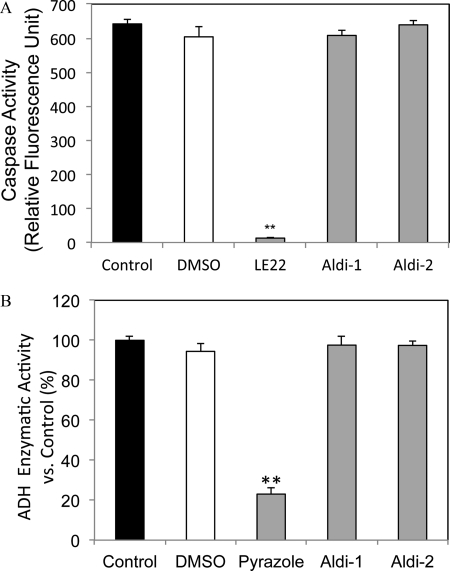
To evaluate whether these covalent inhibitors are specific to aldehyde dehydrogenases, we tested them against caspase-6, a well characterized cysteine protease, alcohol dehydrogenase (human ADH1B1), and protein tyrosine phosphatase 1B, a prototypical tyrosine phosphatase. Each of these enzymes possess active site cysteines that are reactive and susceptible to covalent modification (31–33). In the caspase activity assay, 10 μm Aldi-1 or Aldi-2 showed no inhibitory effect on caspase-6, whereas the caspase-6 specific inhibitor, LE22, completely abolished the activity of this protease (Fig. 5A). Similarly, 10 μm Aldi-1 or Aldi-2 had no effect on the enzymatic activity of human ADH1B1, whereas 10 μm pyrazole significantly inhibited the enzyme at the same concentration (Fig. 5B). Lastly, we also tested whether Aldi-1 could inhibit tyrosine phosphatase protein tyrosine phosphatase 1B. We found no inhibition of protein tyrosine phosphatase 1B at concentrations up to 100 μm and for incubation times up to 60 min (data not shown).
FIGURE 5.
A, caspase-6 activity assay. Caspase activity was measured using a fluorescent caspase-6 substrate probe (Ac-VEID-AFC) in the presence or absence of a caspase inhibitor peptide (LE22, 100 nm) or ALDH inhibitors Aldi-1 or -2 (10 μm). Data are presented as relative fluorescence units at wavelengths of 400 nm excitation/505 mm emission. Black bar, blank control; white bar, DMSO solvent; gray bars, testing compounds; **, p < 0.01 (n = 3). B, inhibition of ADH activity. The activity of ADH1B was measured spectrophotometrically by following the production of NADH at 340 nm (molar ϵ = 6220 liter·mol−1·cm−1) in the presence or absence of the ADH inhibitor pyrazole or Aldi-1 or Aldi-2 (10 μm). Black bar, blank control; white bar, DMSO solvent; gray bars, testing compounds. **, p < 0.01 (n = 3).
Sensitization of Cancer Cell Lines A549 to Mafosfamide by Aldis
ALDH has been identified as an important drug metabolizing enzyme for the detoxification of a commonly used alkylating chemotherapeutic agent, cyclophosphamide (16). More specifically, ALDH1A1 and ALDH3A1 activity was suggested as a predictor of efficacy against cyclophosphamide treatment in breast adenocarcinoma (34). Reduction of ALDH activity increases the sensitivity of the human lung adenocarcinoma cell line A549 to a metabolite of cyclophosphamide, 4-hydroxyperoxycyclophosphamide (5). Inhibition of ALDH may therefore be a potential advantageous strategy in enhancing the efficacy of chemotherapy for a wide range of cancer patients.
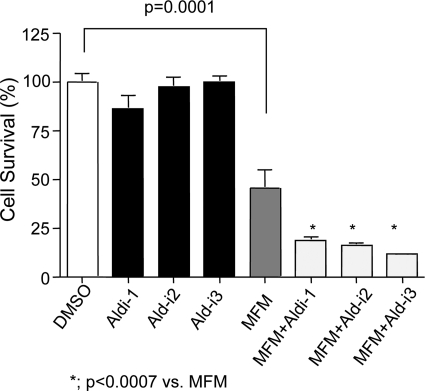
Here, we treated human lung adenocarcinoma A549 cells with mafosfamide, an analog of cyclophosphamide that does not need metabolic activation, in the presence or absence of Aldi-1, -2, or -3 to see whether the inhibitors increase the sensitivity of A549 cells to mafosfamide. In agreement with prior literature, treatment of A549 cells with mafosfamide decreased cell survival (Fig. 6). Treatment with Aldi-1, -2, or -3 alone had no effect in cell survival (Fig. 6). However, combined treatments of mafosfamide with Aldi-1, -2, or -3 significantly decreased cell survival compared with mafosfamide alone.
FIGURE 6.
Sensitization of A549 cancer cells to mafosfamide toxicity with ALDH inhibitors. Human lung adenocarcinoma A549 cells were treated with ALDH inhibitors (Aldi-1 and Aldi-3 at 40 μm and Aldi-2 at 25 μm) in the presence or absence of 125 μm mafosfamide (MFM). Cell viability was determined using MTT assays. DMSO, which was used as a solvent for the inhibitors, was used for the control treatment (*, p < 0.0001 ∼ p < 0.0007; n = 8–16).
Two compounds approved for use in humans possess the same general structural features as the Aldi compounds described here. Oxyfedrine and dyclonine possess aromatic ketones that could form vinyl ketone moieties after β-elimination of their respective amine substituents. Dyclonine is a topical anesthetic found in over-the-counter sore throat medications and oxyfedrine is a vasodilator used most commonly 25 years ago. We tested the ability of dyclonine to inhibit ALDH2 and ALDH3A1 and determined IC50 values of 76 μm for ALDH3A1 and 35 μm for ALDH2. To determine if the mechanism of inhibition involved the same vinyl ketone formation, we subjected ALDH3A1 to mass spectrometry following incubation with dyclonine and find that the mass differential between unmodified and modified protein is 206 amu consistent with a similar mechanism of reactive intermediate formation and covalent modification of the enzyme (data not shown).
DISCUSSION
Aldehyde dehydrogenases are important and ubiquitious detoxication enzymes involved in the metabolism of a variety of aldehyde substrates. ALDH2 has well established roles in the metabolism of acetaldehyde derived from ethanol oxidation as well as lipid peroxidation products and dopamine degradation products (3). ALDH1A1 and ALDH3A1 are perhaps best known for their ability to confer resistance to cyclophosphamide, a commonly used anti-cancer agent (5). Here, we describe the discovery of a novel class of covalent inhibitors that broadly inhibit ALDH isoenzymes through the generation of a reactive vinyl ketone group within their respective active sites. Until the discovery of these compounds, there were no generally applicable inhibitors of ALDH activity.
The mechanism behind the chemical activation of these compounds does not depend on the action of the putative general bases in their active sites (either Glu-268/Glu-209 or Glu-399/Glu-333) because mutation of these residues does not impact their ability to chemically inactivate the enzymes. We cannot conclusively state that cysteine is the sole residue responsible for activation, as we detected some product for the ALDH3-C243A mutant. The available data suggest that either Cys-243 is not essential to the activation of the Aldi inhibitors or the C243S mutant retains sufficient chemical reactivity to activate Aldi-1 but insufficient chemical reactivity to become modified. Prior mutational work on ALDH isoenzymes has found residual enzymatic activity in serine mutants of the active site cysteine (35). We also cannot rule out the possibility that another residue may be involved in activation of the aldi inhibitors, though the only remaining active site residues conserved between ALDH3A1 and ALDH2 are the less reactive Thr-244/186 or Asn-169/114.
That the reactive vinyl ketone intermediates primarily remain localized within the active site and do not escape to a significant extent is supported by the MS data on the basic vinyl ketone compound (1-phenyl-2-propen-1-one), which, when present in solution at greater than equimolar concentrations, results in stoichiometries in excess of those found for the Aldi compounds (Table 3 and Fig. 4). The mutagenesis data on the ALDH3A1 isoenzyme are straightforward to interpret; the presence of Cys-243 is necessary and sufficient to support covalent inhibition by the Aldi compounds, whereas mutation of either Glu-209 or Glu-333 have little effect on chemical inactivation. However, the mutagenesis data on the ALDH2 isoenzyme is more complex, and the simplest explanation is that the combined presence of Glu-268 and Cys-302 direct the modification solely to Cys-302 and preclude modification at either of the adjacent active site cysteine residues (Cys-301 and Cys-303, respectively). However, upon mutation of either Cys-302 or Glu-268, all remaining active site cysteines appear to become reactive, as judged by the increases in stoichiometry of modification from one to two for the C302S mutant (which retains Cys-301 and Cys-303) and from one to three for the E268Q mutant (which retains Cys-301, Cys-302, and Cys-303). That the reaction still remains localized to the active site region even in the E268Q mutant is supported by the presence of an additional four cysteine residues that are surface exposed, yet the stoichiometry does not rise above three under the conditions tested.
To evaluate whether these inhibitors are specific to ALDH, we tested the ability of the Aldi compounds to inhibit other enzymes containing reactive cysteine residues in their active site. The enzymes tested included the cysteine protease, activated caspase-6, the ADH1B1 isoform of alcohol dehydrogenase and protein tyrosine phosphatase 1B (Fig. 6). Each of these enzymes contains a reactive active site cysteine residue and a binding cavity large enough to accommodate these inhibitors, yet none were susceptible to covalent modification. Consequently, we conclude that generation of the reactive intermediate and efficient access to the critical cysteine in the active site of ALDH by this class of inhibitor is highly selective.
Lastly, the expression of either ALDH1A1 or ALDH3A1 individually or in combination is associated with resistance to the cyclophosphamide class of chemotherapeutic agents and manipulation of their expression levels is strongly correlated with changes in the cytotoxic effects of cyclophosphamide (5). These data show that these ALDH inhibitors (Aldi-1, -2, and -3) can indeed increase sensitivity of cancer cells to the treatment of alkylating chemotherapeutic agents of the oxazaphosphorine group such as mafosfamide, 4-hydroperoxycyclophosphamide, and cyclophosphamide, suggesting that these compounds can be used in increasing the efficacy of anti-cancer treatments in other cells lines and may be useful clinically. The lack of general toxicity in this model system is also consistent with their highly selective mode of action. These compounds will serve as powerful research tools to address the contributions of ALDH isoenzymes toward numerous biological pathways and processes. In addition, these inhibitors will serve as starting points for the generation of isoenzyme-selective inhibitors through systematic modification of their aryl substituents to take advantage of the unique active site topologies of the ALDH1A1, ALDH2, and ALDH3A1 isoenzymes.
Supplementary Material
Acknowledgments
We thank Laura E. Edgington and Matthew Bogyo (Department of Pathology, Stanford University) for providing the reagents and technical support for the caspase activity assay. We thank Lanmin Zhai for technical assistance with production of ALDH3A1 and ALDH2 mutants and Dr. Zhong-Yin Zhang and Lily Wu (Department of Biochemistry and Molecular Biology at the Indiana University School of Medicine) for performing the protein tyrosine phosphatase 1B assays. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center, and GM/CA-CAT.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-AA018123 and R21-AA019746 (to T. D. H.), R01-AA018123-S1 (to A. K.-H.), R01-AA11147 (to D. M.-R.) and EY11490 (V. V.). The mass spectrometry work was supported by National Science Foundation MRI Grant 0821661. General Medical/CAncer Collaborative Access Team was supported in whole or in part by NCI Grant Y1-CO-1020 and NIGMS Grant Y1-GM-1104, National Institutes of Health, at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U. S. Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357. Daria Mochly-Rosen is the founder and a shareholder of KAI Pharmaceuticals. However, none of the work described in this study is based on or supported by the company. Che-Hong Chen and Daria Mochly-Rosen are listed as inventors in pending patents filed by Stanford University that are related to this work.
The atomic coordinates and structure factors (codes 3SZA, 3SZB, and 3SZ9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- ALDH
- aldehyde dehydrogenase
- DMSO
- dimethyl sulfoxide
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-(diphenyl tetrazolium bromide)
- amu
- atomic mass unit.
REFERENCES
- 1. Vasiliou V., Pappa A. (2000) Pharmacology 61, 192–198 [DOI] [PubMed] [Google Scholar]
- 2. Sládek N. E. (2003) J. Biochem. Mol. Toxicol. 17, 7–23 [DOI] [PubMed] [Google Scholar]
- 3. Vasilou V., Nerbert D. W. (2005) Hum. Genom. 2, 138–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marcato P., Dean C. A., Pan D., Araslanova R., Gillis M., Joshi M., Helyer L., Pan L., Leidal A., Gujar S., Giacomantonio C. A., Lee P. W. (2011) Stem Cells 29, 32–45 [DOI] [PubMed] [Google Scholar]
- 5. Moreb J. S., Mohuczy D., Muhoczy D., Ostmark B., Zucali J. R. (2007) Cancer Chemother. Pharmacol. 59, 127–136 [DOI] [PubMed] [Google Scholar]
- 6. Harada S., Okubo T., Nakamura T., Fujii C., Nomura F., Higuchi S., Tsutsumi M. (1999) Alcohol Clin. Exp. Res. 23, 958–962 [PubMed] [Google Scholar]
- 7. Cheriyan J., Burton T. J., Bradley T. J., Wallace S. M., Mäki-Petäjä K. M., Mackenzie I. S., McEniery C. M., Brown J., Wilkinson I. B. (2009) Br. J. Clin. Pharmacol. 68, 518–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen C. H., Budas G. R., Churchill E. N., Disatnik M. H., Hurley T. D., Mochly-Rosen D. (2008) Science 321, 1493–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Churchill E. N., Disatnik M. H., Mochly-Rosen D. (2009) J. Mol. Cell. Cardiol. 46, 278–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Budas G. R., Disatnik M. H., Chen C. H., Mochly-Rosen D. (2010) J. Mol. Cell. Cardiol. 48, 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perez-Miller S., Younus H., Vanam R., Chen C. H., Mochly-Rosen D., Hurley T. D. (2010) Nat. Struct. Mol. Biol. 17, 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arolfo M. P., Overstreet D. H., Yao L., Fan P., Lawrence A. J., Tao G., Keung W. M., Vallee B. L., Olive M. F., Gass J. T., Rubin E., Anni H., Hodge C. W., Besheer J., Zablocki J., Leung K., Blackburn B. K., Lange L. G., Diamond I. (2009) Alcohol Clin. Exp. Res. 33, 1935–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yao L., Fan P., Arolfo M., Jiang Z., Olive M. F., Zablocki J., Sun H. L., Chu N., Lee J., Kim H. Y., Leung K., Shryock J., Blackburn B., Diamond I. (2010) Nat. Med. 16, 1024–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Overstreet D. H., Knapp D. J., Breese G. R., Diamond I. (2009) Pharmacol. Biochem. Behav. 94, 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hilton J. (1984) Cancer Res. 44, 5156–5160 [PubMed] [Google Scholar]
- 16. Manthey C. L., Landkamer G. J., Sladek N. E. (1990) Cancer Res. 50, 4991–5002 [PubMed] [Google Scholar]
- 17. von Eitzen U., Meier-Tackmann D., Agarwal D. P., Goedde H. W. (1994) Cancer Lett. 76, 45–49 [DOI] [PubMed] [Google Scholar]
- 18. Allali-Hassani A., Weiner H. (2001) Chem. Biol. Interact. 130, 125–133 [DOI] [PubMed] [Google Scholar]
- 19. Parajuli B., Kimble-Hill A. C., Khanna M., Ivanova Y., Meroueh S., Hurley T. D. (2011) Chem. Biol. Interact. 191, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jeng J. J., Weiner H. (1991) Arch. Biochem. Biophys. 289, 214–222 [DOI] [PubMed] [Google Scholar]
- 21. Ni L., Sheikh S., Weiner H. (1997) J. Biol. Chem. 272, 18823–18826 [DOI] [PubMed] [Google Scholar]
- 22. Perez-Miller S. J., Hurley T. D. (2003) Biochemistry 42, 7100–7109 [DOI] [PubMed] [Google Scholar]
- 23. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) Acta Crystallogr. D. Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Aldridge W. N., Reiner E. (1972) Enzyme Inhibitors as Substrates, pp. 1–328, North-Holland Publishing Co., Amsterdam [Google Scholar]
- 26. Berger A. B., Witte M. D., Denault J. B., Sadaghiani A. M., Sexton K. M., Salvesen G. S., Bogyo M. (2006) Mol. Cell. 23, 509–521 [DOI] [PubMed] [Google Scholar]
- 27. Negoro M., Wakabayashi I. (2005) Alcohol Clin. Exp. Res. 29, 304S–308S [DOI] [PubMed] [Google Scholar]
- 28. Ghenbot G., Weiner H. (1992) Protein Expr. Purif. 3, 470–478 [DOI] [PubMed] [Google Scholar]
- 29. Black W., Vasiliou V. (2009) Hum. Genomics 4, 136–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Geoghegan K. F., Dixon H. B., Rosner P. J., Hoth L. R., Lanzetti A. J., Borzilleri K. A., Marr E. S., Pezzullo L. H., Martin L. B., LeMotte P. K., McColl A. S., Kamath A. V., Stroh J. G. (1999) Anal. Biochem. 267, 169–184 [DOI] [PubMed] [Google Scholar]
- 31. Ascenzi P., Salvati L., Bolognesi M., Colasanti M., Polticelli F., Venturini G. (2001) Curr. Protein Pept. Sci. 2, 137–153 [DOI] [PubMed] [Google Scholar]
- 32. Bosron W. F., Yin S. J., Dwulet F. E., Li T. K. (1986) Biochemistry 25, 1876–1881 [DOI] [PubMed] [Google Scholar]
- 33. Kumar S., Zhou B., Liang F., Wang W. Q., Huang Z., Zhang Z. Y. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7943–7948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sreerama L., Sladek N. E. (1997) Clin. Cancer Res. 3, 1901–1914 [PubMed] [Google Scholar]
- 35. Farrés J., Wang T. T., Cunningham S. J., Weiner H. (1995) Biochemistry 34, 2592–2598 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.