Background: The Swi5-Sfr1 protein complex is an activator of Rad51 recombinase, which mediates DNA strand exchange in homologous recombination.
Results: Swi5 and Sfr1 form a 1:1 complex, which exhibits an extremely elongated dogleg-shaped structure in solution.
Conclusion: The Swi5-Sfr1 structure is suitable for binding within the helical groove of the Rad51 filament.
Significance: A structural model will advance our understanding of the molecular mechanism of homologous recombination.
Keywords: DNA Repair, Homologous Recombination, Mass Spectrometry (MS), Ultracentrifugation, X-ray Scattering, Yeast, Swi5-Sfr1, Fission Yeast
Abstract
In eukaryotes, DNA strand exchange is the central reaction of homologous recombination, which is promoted by Rad51 recombinases forming a right-handed nucleoprotein filament on single-stranded DNA, also known as a presynaptic filament. Accessory proteins known as recombination mediators are required for the formation of the active presynaptic filament. One such mediator in the fission yeast Schizosaccharomyces pombe is the Swi5-Sfr1 complex, which has been identified as an activator of Rad51 that assists in presynaptic filament formation and stimulates its strand exchange reaction. Here, we determined the 1:1 binding stoichiometry between the two subunits of the Swi5-Sfr1 complex using analytical ultracentrifugation and electrospray ionization mass spectrometry. Small-angle x-ray scattering experiments revealed that the Swi5-Sfr1 complex displays an extremely elongated dogleg-shaped structure in solution, which is consistent with its exceptionally high frictional ratio (f/f0) of 2.0 ± 0.2 obtained by analytical ultracentrifugation. Furthermore, we determined a rough topology of the complex by comparing the small-angle x-ray scattering-based structures of the Swi5-Sfr1 complex and four Swi5-Sfr1-Fab complexes, in which the Fab fragments of monoclonal antibodies were specifically bound to experimentally determined sites of Sfr1. We propose a model for how the Swi5-Sfr1 complex binds to the Rad51 filament, in which the Swi5-Sfr1 complex fits into the groove of the Rad51 filament, leading to an active and stable presynaptic filament.
Introduction
DNA strand exchange is the central reaction of homologous recombination, which is catalyzed by an evolutionally conserved RecA family recombinase. In eukaryotes, two conserved recombinases, Rad51 and Dmc1, are involved in this reaction; Rad51 functions in both mitotic and meiotic recombination and recombination repair, whereas Dmc1 plays a role only in meiotic recombination (1–3). DNA strand exchange is initiated in the single-stranded DNA (ssDNA)2 region and proceeds in essentially three steps: homology search for its homologous intact double-stranded DNA (dsDNA), strand invasion of ssDNA into dsDNA, and the subsequent DNA strand exchange between ssDNA and dsDNA (2, 4). Budding yeast Rad51, as with other recombinases, forms nucleoprotein filaments on ssDNA in an ATP-dependent manner. The resulting structures are referred to as presynaptic filaments, which are the core structures involved in the DNA strand exchange reaction (5). The filament structures of yeast Rad51 and its homologs have been determined by x-ray crystallography and electron microscopy and have shown that the filament structures adopt two conformational states: a compressed inactive state with a helical pitch of ∼80 Å in the absence of nucleotides and an extended active state with a helical pitch of ∼90 Å in the presence of ATP (5–7).
Once ssDNA regions are generated in vivo, they are immediately protected by the binding of an ssDNA-binding protein, replication protein A (RPA) (8–10). Thus, recombinases are unable to directly form presynaptic filaments on RPA-coated ssDNA due to the higher affinity of RPA for ssDNA. To efficiently form presynaptic filaments, accessory proteins are needed to overcome the inhibitory effect of RPA. These proteins are called recombination mediators (9). The first mediator to be identified was Rad52, from both the budding yeast Saccharomyces cerevisiae and humans, which has been shown to assist the Rad51-mediated DNA strand exchange reaction in vitro (11–13). Rad52 physically interacts with Rad51 and RPA and assists Rad51 to displace RPA from ssDNA, leading to the formation of the Rad51 presynaptic filament (14). Although the physical interactions of Rad52 with both RPA and Rad51 are essential for its mediator activity, interestingly, Rad52 itself confers no effect on Rad51 biochemical activities, such as ssDNA- and ATP-binding affinities, which are essential for the DNA strand exchange (15, 16). Therefore, the molecular mechanism of how Rad52 overcomes the RPA inhibition and how it assists Rad51 loading onto ssDNA has not been completely elucidated.
Several other recombination mediators were later identified, including Rad51 paralogs, such as the S. cerevisiae Rad55-Rad57 complex (17) and the human BRCA2 (breast cancer susceptibility gene 2) product(18). However, the mechanism by which these mediators assist recombinase-dependent DNA strand exchange remains to be defined.
The Swi5-Sfr1 complex, another recombinase mediator, was first identified in the fission yeast Schizosaccharomyces pombe and is evolutionally conserved from yeast to humans (19–21). Depletion of Swi5 or Sfr1 in mouse or human cells as well as S. pombe causes defects in recombination repair (22, 23). Swi5 and Sfr1 from S. pombe consist of 85 and 299 amino acids, respectively, and form a very stable complex. This complex stimulates both Rad51- and Dmc1-mediated strand exchange reactions when recombinases are incubated with ssDNA before the addition of RPA (24). However, when recombinases are incubated after the addition of RPA, the strand exchange is inhibited even in the presence of the Swi5-Sfr1 complex because Rad51 cannot form presynaptic filaments on ssDNA already coated with RPA. Rad22 (fission yeast homolog of Rad52) is required for this situation. Note that the addition of Rad22 alone is unable to stimulate Rad51-mediated strand exchange in the absence of Swi5-Sfr1, nor has it any effect on the intrinsic ssDNA-dependent ATPase- and ssDNA-binding activities of Rad51 (25). On the other hand, the Swi5-Sfr1 complex, which is able to bind to Rad51 protomers and the Rad51 presynaptic filament, stimulates directly Rad51-mediated displacement of RPA from ssDNA in the presence of Rad22 (24, 25). In addition, the Swi5-Sfr1 complex stabilizes the presynaptic Rad51 filament on ssDNA in an ATP-dependent manner, likely by decreasing the off-rate of Rad51 from ssDNA (25). These observations indicate that at least two different types of accessory proteins are required for full recombination activity: Rad22, which targets Rad51 to RPA-coated ssDNA, and Swi5-Sfr1, which activates the intrinsic strand exchange ability of the recombinase filament (25). Thus, Swi5-Sfr1 is considered to be the first recombinase activator that acts directly on the presynaptic recombinase filament.
To understand the molecular mechanism of how Swi5-Sfr1 activates the intrinsic activity of Rad51 and stimulates Rad51-mediated strand exchange, we took multiple approaches, including analytical ultracentrifugation (AUC), electrospray ionization mass spectrometry (ESI-MS), and small-angle x-ray scattering (SAXS), in this study. Active and inactive filament structures consisting of the 10 Rad51 monomers were reconstituted in silico based on the known structures of the S. cerevisiae Rad51 monomer (26) and RecA filaments (7). On the basis of the results combined with the structural information, we propose a model in which the Swi5-Sfr1 complex fits into the groove of the Rad51 filament, leading to an active and stable presynaptic filament.
EXPERIMENTAL PROCEDURES
Purification of Proteins
The expression and purification of Swi5-Sfr1 were previously described by Kuwabara et al. (27).
AUC
AUC was performed with an Optima XL-I analytical ultracentrifuge (Beckman Coulter, Brea, CA) with a An-50 Ti rotor (Beckman). For sedimentation velocity AUC experiments, cells with a standard Epon two-channel centerpiece and sapphire windows were used. The sample (400 μl) and reference (420 μl) buffers were loaded into the cells. The rotor temperature was equilibrated at 20 °C in the vacuum chamber for 1–2 h before startup. Absorbance (A280) scans were collected at 10-min intervals during sedimentation at 50 × 103 rpm. The concentrations of the loaded protein solutions in the experiments were 2.7 and 0.6 mg/ml in reference buffer (10 mm Hepes, 200 mm NaCl (pH 7.0), and 2 mm tris(2-carboxyethyl)phosphine). The partial specific volume of the protein, solvent density, and solvent viscosity were calculated from standard tables using the SEDNTERP program (Version 1.09) (28). The resulting scans were analyzed using the continuous distribution (c(s)) analysis module in the SEDFIT program (Version 12.1) (29). Sedimentation coefficient increments of 200 were used in the appropriate range for each sample. The frictional coefficient was allowed to float during fitting. The weight average sedimentation coefficient was obtained by integrating the range of sedimentation coefficients in which peaks were present. The s20,w value, the sedimentation coefficient corrected to 20 °C in pure water, was calculated from the observed sedimentation coefficient value. The c(s) distribution was converted to a c(s20,w) distribution using the SEDFIT program. Sedimentation equilibrium AUC experiments were performed in a six-channel centerpiece with quartz windows. The absorbance wavelength was set at 280 nm, and data were acquired at 20 °C. The concentrations of the loaded protein solutions in the sedimentation equilibrium experiments were 0.4, 0.9, and 1.8 mg/ml in reference buffer. Data were obtained at 8.5 × 103, 12 × 103, and 16 × 103 rpm, respectively. A total equilibration time of 20 h was used for each speed, with a scan taken at 16 and 18 h to ensure that equilibrium had been reached. The optical base line was determined by accelerating at 42 × 103 rpm at the end of data collection. Data analysis was performed by global analysis with Ultraspin software (Medical Research Council Center for Protein Engineering, Cambridge, United Kingdom).
ESI-MS
Prior to ESI-MS, the sample buffer was exchanged for an ammonium acetate buffer by dialysis. A sample was dialyzed against 100 mm ammonium acetate (pH 6.8) with a microdialysis tool (Micro-dialyzer TOR-14K, Nippon Genetics Co., Ltd., Tokyo, Japan) using a dialysis cup with a membrane that had a molecular mass cutoff of 14,000 Da. The sample solution was then diluted with concentrated ammonium acetate, resulting in 10 μm proteins in 2 m ammonium acetate (pH 6.8). An aliquot of 3 μl of the sample solution was deposited in a nano-ESI spray tip (Waters) and placed in the nanoelectrospray ion source. ESI mass spectra were obtained using a Q-Tof 2 mass spectrometer (Waters, Milford, MA) equipped with a nanoelectrospray ion source. To observe noncovalently bound complexes, the pressure in the quadrupole ion guide of the Q-Tof 2 spectrometer was maintained at 8 × 10−3 pascals by throttling down the SpeediValve fitted to the rotary pump for the ion source region. MassLynx Version 4.1 software (Waters) was used for data processing and peak integration.
Antibodies and Fab Preparation
Mice were immunized with the purified Swi5-Sfr1 protein complex. After generating hybridomas, candidate clones were screened by ELISA using plates. Epitopes of the positive clones were mapped by immunoblotting using lysates of Escherichia coli BL21(DE3) cells expressing serial Sfr1 truncation proteins. Clones 7, 19, 49, and 65 recognized amino acids 31–35, 156–160, 1–5, and 76–80 of Sfr1, respectively (see details under “Results”). Cells of these clones (hybridomas) were grown in PFHM-II medium (Invitrogen), and IgGs were purified by passing the supernatants through HiTrap protein A-Sepharose FF (GE Healthcare). The purified IgG was digested with papain, and the resultant Fab fragments were purified by gel filtration chromatography. The purified Fab fragments from clones 7, 19, 49, and 65 were designated as Fab(31–35), Fab(156–160), Fab(1–5), and Fab(76–80), respectively.
Preparation of Swi5-Sfr1-Fab Complexes
A reaction mixture (1.5 ml) containing the purified Swi5-Sfr1 complex (1.2 mg) and Fab fragments (1.415 mg) in gel filtration buffer (5 mm Tris-HCl (pH 7.8), 0.2 m NaCl, 2 mm EDTA, 1 mm DTT, and 17 μg/ml leupeptin) was incubated for 1 h at 4 °C. The sample was concentrated to a volume of 250 μl with an Amicon Ultra concentrator (Millipore), loaded onto a Superdex 200 column (GE Healthcare) equilibrated with gel filtration buffer, and developed using the same buffer. In each sample, 1 ml of peak fraction was collected, followed by the addition of 34 μl of 10 mg/ml leupeptin.
SAXS Measurements
SAXS measurements were carried out at RIKEN structural biology beamline I (BL45XU) at SPring-8 (Hyogo, Japan) (30). Scattering intensities were measured with an R-AXIS IV++ imaging plate detector (Rigaku) at 20 °C with a sample-to-detector distance of 1.5 m, which was then calibrated by the powder diffraction from silver docosanoate. A circular averaging of the scattering intensities was then performed to obtain one-dimensional scattering data (I(q)) as a function of q (q = 4πsinθ/λ, where 2θ is the scattering angle and the x-ray wavelength (λ) = 0.9 Å). To correct interparticle interference effect, I(q) data were collected at two or three concentrations for the Swi5-Sfr1 and Swi5-Sfr1-Fab complexes (i.e. 1.3, 2.1, and 4.7 mg/ml for Swi5-Sfr1; 1.0, 2.7, and 4.3 mg/ml for Swi5-Sfr1-Fab(1–5); 1.8, 2.7, and 3.8 mg/ml for Swi5-Sfr1-Fab(31–35); 1.5 and 2.5 mg/ml for Swi5-Sfr1-Fab(76–80); and 1.5, 3.8, and 5.8 mg/ml for Swi5-Sfr1-Fab(156–160)) in 5 mm Tris-HCl (pH 7.8), 0.2 m NaCl, 2 mm EDTA, 1 mm DTT, and 357 μg/ml leupeptin.
The radius of gyration (Rg) was estimated by fitting I(q) data using Guinier approximation, I(q) = I(0)exp(−q2Rg2/3), where I(0) is the forward scattering at zero scattering angle, in a smaller angle region of qRg < 1.3 by the program PRIMUS (31). The distance distribution function (P(r)) was calculated using the program GNOM (32). The maximum length (Dmax) was estimated from the P(r) function as the distance r, where P(r) = 0 (33). An ab initio low resolution dummy residue model was built with the program GASBOR with no symmetrical restraint (34). The model building was repeated 20 times, and dummy residue models were averaged using the program DAMAVER to obtain general structural information about the proteins (35).
Homology Modeling of Rad51 Filaments
A protomer of Rad51 was modeled with the program MODELLER (36) using the crystal structure of S. cerevisiae Rad51 (Protein Data Bank code 3LDA) (26) as a reference structure. Then, the active filament of Rad51 was constructed by aligning the core domains of Rad51 with those of RecA in the crystal structure of the RecA-ssDNA active filament (Protein Data Bank code 3CMW) (7). The dummy residue model of Swi5-Sfr1 was docked onto the modeled active filament of Rad51 manually. For the construction of the Rad51 inactive filament, the crystal structure of the RecA inactive filament (Protein Data Bank code 1XMS) (37) was used as a reference.
RESULTS AND DISCUSSION
AUC
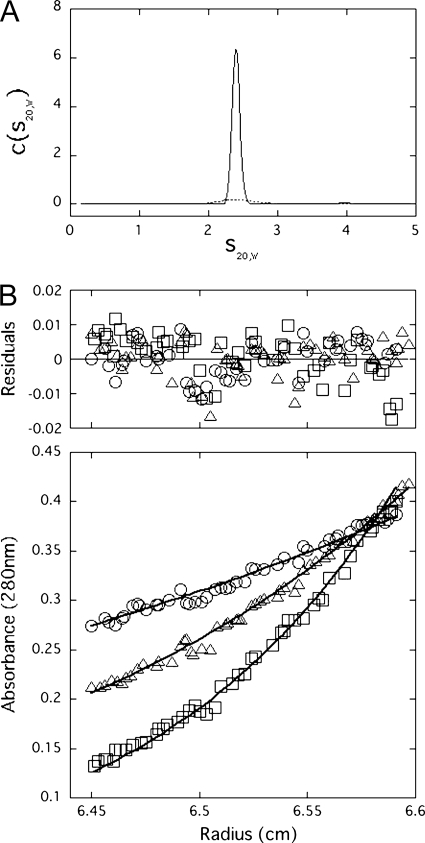
First, we examined the sphericity of Swi5-Sfr1 using sedimentation velocity AUC. Fig. 1A shows the distribution of sedimentation coefficients of Swi5-Sfr1 at concentrations of 2.7 and 0.6 mg/ml. The c(s20,w) distribution of Swi5-Sfr1 showed the presence of a single species in solution with a sedimentation coefficient (s20,w) of 2.4 ± 0.1 S. The c(s20,w) distribution also did not show any significant change at different protein concentrations, suggesting that Swi5-Sfr1 is monodisperse in solution. The frictional ratio (f/f0) was 2.0 ± 0.2, indicating that the Swi5-Sfr1 complex is not spherical. This result prompted us to measure the SAXS of Swi5-Sfr1 to obtain a solution structure of the complex, as described below.
FIGURE 1.
A, distribution of sedimentation coefficients (c(s20,w)) for the Swi5-Sfr1 complex by sedimentation velocity AUC. Calculated c(s20,w) is plotted versus s20,w, the sedimentation coefficient corrected to 20 °C in pure water. Experiments were conducted at an initial protein concentration of 2.7 mg/ml (solid line) or 0.6 mg/ml (dashed line). B, sedimentation equilibrium AUC data are shown with the residuals from the best fit to a single-species model. Plots show data obtained at 8.5 × 103 rpm (○), 12 × 103 rpm (△), and 16 × 103 rpm (□).
The sedimentation equilibrium AUC experiments estimated the molecular mass of Swi5-Sfr1 to be 45.0 ± 0.1 kDa using a simple monomeric analysis model (Fig. 1B). This value corresponds to a 1:1 binding stoichiometry of Swi5-Sfr1. However, this value differs from the 2:1 stoichiometry of Swi5-Sfr1 reported previously (24). This discrepancy may be due to the different reducing conditions used in the buffers. We found that a reducing reagent of low efficiency, which was used in the previous study (24), resulted in an association of the Swi5 subunit. When freshly prepared DTT or tris(2-carboxyethyl)phosphine, a more stable reducing agent than DTT, was used in the buffers, the sedimentation equilibrium AUC results indicated that Swi5-Sfr1 exhibits a 1:1 binding stoichiometry, as described above. Therefore, we adopted this 1:1 ratio as the correct binding stoichiometry under physiological conditions. However, considering the relatively small molecular mass of a single Swi5 molecule (9.7 kDa), uncertainty in the binding stoichiometry remained. Therefore, to obtain the conclusive stoichiometry, we decided to conduct ESI-MS experiments, which are capable of accurately measuring the molecular mass of proteins.
ESI-MS of Swi5-Sfr1
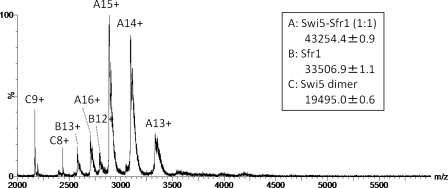
As shown in Fig. 2, multiply charged ions of compound A with a mass of 43,254.4 ± 0.9 Da represented the predominant peaks, whereas minor peaks corresponding to the molecular masses of compounds B (33,506.9 ± 1.1 Da) and C (19,495.0 ± 0.6 Da) were also identified. The theoretical molecular masses of the monomeric Sfr1 peptide with and without the N-terminal Met were calculated to be 33,637.9 and 33,506.7 Da, respectively, and the corresponding 13+ and 12+ charged ions of free Sfr1 are shown in Fig. 2. The major peaks observed in the mass spectrum correspond to 13+∼16+ charged ions of the 1:1 complex of Swi5-Sfr1, whose theoretical mass is 43,254.8 Da. The two peaks labeled C8+ and C9+ suggest that some of the Swi5 population with a theoretical molecular mass of 9748.1 Da exists as a dimer. In the high m/z region of Fig. 2, no significant peak was observed that would suggest a binding stoichiometry other than 1:1. Accordingly, we conclude that Swi5 and Sfr1 preferentially form a 1:1 complex, not a 2:1 complex.
FIGURE 2.
ESI mass spectrum of Swi5-Sfr1. Three species were detected: Swi5-Sfr1 (1:1; A), free Sfr1 (B), and Swi5 dimer (C).
Overall Parameters and Dummy Residue Models of Swi5-Sfr1
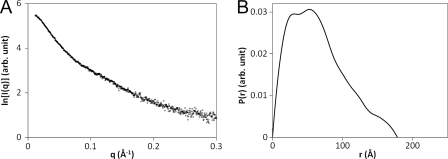
The SAXS profile and overall parameters of Swi5-Sfr1 are shown in Fig. 3A and Table 1, respectively. The radius of gyration (Rg) of Swi5-Sfr1 was 49.0 Å (Table 1), and the P(r) function of Swi5-Sfr1 has a broad peak at 30–60 Å (Fig. 3B). The maximum length (Dmax) of Swi5-Sfr1 was estimated to be 178 Å (Table 1). The dummy residue model of Swi5-Sfr1 constructed by the program GASBOR (34) has a substantially elongated shape, as shown in Fig. 4A. It had been previously reported that limited proteolysis produces a stable C-terminal fragment that lacks 177 amino acids from the N terminus (Sfr1ΔN177) (27). For comparison, Fig. 4B shows the dummy residue model of the Swi5-Sfr1ΔN177 complex. Notably, half of the Swi5-Sfr1 model resembles the Swi5-Sfr1ΔN177 model in size and shape (Fig. 4).
FIGURE 3.
A, SAXS profile of the Swi5-Sfr1 complex. B, pair-distance distribution (P(r)) function of the Swi5-Sfr1 complex.
TABLE 1.
Radii of gyration (Rg) and maximum lengths (Dmax) of the Swi5-Sfr1 and Swi5-Sfr1-Fab complexes estimated using SAXS
| Complex | Rg | Dmax |
|---|---|---|
| Å | Å | |
| Swi5-Sfr1 | 49.0 | 178 |
| Swi5-Sfr1-Fab(1–5) | 66.1 | 235 |
| Swi5-Sfr1-Fab(31–35) | 54.4 | 212 |
| Swi5-Sfr1-Fab(76–80) | 52.5 | 197 |
| Swi5-Sfr1-Fab(156–160) | 51.3 | 176 |
FIGURE 4.
Dummy residue models of the Swi5-Sfr1 and Swi5-Sfr1ΔN177 complexes. The models of Swi5-Sfr1 (A) and Swi5-Sfr1ΔN177 (B) complexes are shown in the surface representation. All figures were generated using PyMOL (39).
Swi5-Sfr1 and Fab Fragments of Sfr1-specific Monoclonal Antibodies Form Stable Complexes
The above Swi5-Sfr1 structural data prompted us to generate a rough topology of the N-terminal half of Sfr1 in the Swi5-Sfr1 complex. To this end, we generated monoclonal antibodies by immunizing mice with the Swi5-Sfr1 complex as an antigen. Hybridomas were screened by ELISA, and the positive clones were further analyzed by immunoblotting using purified Swi5-Sfr1 as targets. To determine the recognition sequence of each monoclonal antibody, we made a series of plasmid-based truncation constructs of the Sfr1 protein (40 constructs) in which every five amino acid residues were truncated from the N terminus and expressed in a T7 expression system (supplemental Fig. S1). Using these lysates as target antigens, the recognition sequence of each monoclonal antibody was mapped. Of the positive clones, we selected four clones (7, 19, 49, and 65) that recognized Sfr1 amino acids 31–35, 156–160, 1–5, and 76–80, respectively (supplemental Fig. S1). From these hybridomas, Fab fragments were prepared as described under “Experimental Procedures.” The Fab fragments from clones 7, 19, 49, and 65 were designated as Fab(31–35), Fab(156–160), Fab(1–5), and Fab(76–80), respectively.
To investigate whether the obtained Fab fragments formed stable complexes with Swi5-Sfr1, we performed gel filtration chromatography (supplemental Fig. S2). The Swi5-Sfr1 complex alone eluted at ∼13 ml of elution volume, whereas all four of the samples containing both Swi5-Sfr1 and Fab fragments showed single peaks at ∼12 ml of elution volume, indicating that a stable complex had formed between Swi5-Sfr1 and each Fab fragment. These complexes were then used for subsequent SAXS analysis.
Overall Parameters and Dummy Residue Models of Swi5-Sfr1-Fab Complexes
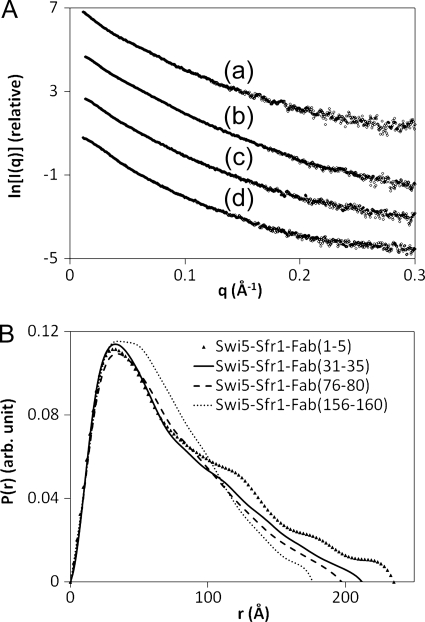
The SAXS profiles for the four Swi5-Sfr1-Fab complexes are shown in Fig. 5A, and the Rg and Dmax of the Swi5-Sfr1-Fab complexes are summarized in Table 1. Among the four complexes, Swi5-Sfr1-Fab(1–5) showed the largest Rg of 66.1 Å, and the Rg decreased in the order of Swi5-Sfr1-Fab(1–5), Swi5-Sfr1-Fab(31–35), Swi5-Sfr1-Fab(76–80), and Swi5-Sfr1-Fab(156–160). The Dmax of the four complexes also exhibited the same order as Rg, and the Dmax of Swi5-Sfr1-Fab(156–160) was comparable with that of Swi5-Sfr1. The P(r) functions for the four complexes also differed significantly from one another (Fig. 5B). These results indicate that the Fab fragments bind to Swi5-Sfr1 from the distal position to the center of Swi5-Sfr1 in the following order: Fab(1–5), Fab(31–35), Fab(76–80), and Fab(156–160).
FIGURE 5.
A, SAXS profiles of the Swi5-Sfr1-Fab complexes containing Fab(1–5) (trace a), Fab(31–35) (trace b), Fab(76–80) (trace c), and Fab(156–160) (trace d). The curves are arbitrarily displaced along the vertical logarithmic axis for clarity. B, pair-distance distribution (P(r)) functions of the Swi5-Sfr1-Fab complexes containing Fab(1–5) (▴), Fab(31–35) (solid line), Fab(76–80) (dashed line), and Fab(156–160) (dotted line). arb., arbitrary.
Location of Sfr1 in Swi5-Sfr1 Complex
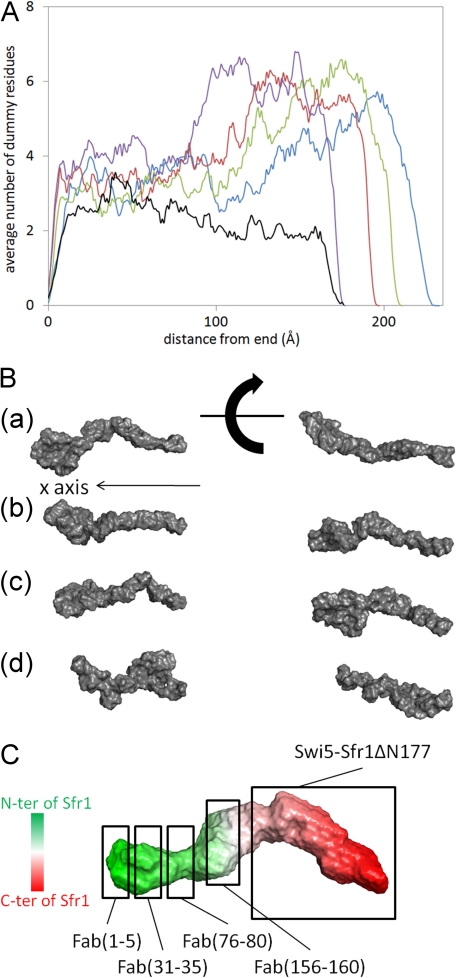
The Rg, Dmax, and P(r) functions of the four Swi5-Sfr1-Fab complexes provided rough topological information about the N-terminal half of Sfr1 in the Swi5-Sfr1 complex. To further assess the topology of Sfr1 in the complex, we constructed dummy residue models of the four Swi5-Sfr1-Fab complexes (Fig. 6B). However, the dummy residue models obtained from independent runs of the program GASBOR (34) for each Swi5-Sfr1-Fab complex could not be used to determine general structural features about the complexes because the models were too unstable to be averaged by the program DAMAVER (35). Instead, we analyzed the binding sites of the Fab fragments as follows. First, the principal axes of the dummy residue models of the Swi5-Sfr1-Fab complexes were calculated. For each model, the longest axis (i.e. corresponding to the smallest principal moment of inertia) was aligned with the x axis, and the number of the dummy residues with x coordinates in the range of iΔx to (i + 1)Δx was counted along the x axis, where i is an integer and Δx was set to 1 Å. The origin was designated as the end located at the smaller half of each model. The numbers of dummy residues along the x axis were averaged for all models. In Fig. 6A, the average numbers of dummy residues along the x axis are plotted for each complex.
FIGURE 6.
Dummy residue models of the Swi5-Sfr1-Fab complexes and the proposed rough topology of Sfr1 in the complex. A, averaged number of dummy residues along the x axis for Swi5-Sfr1-Fab(1–5) (blue), Swi5-Sfr1-Fab(31–35) (green), Swi5-Sfr1-Fab(76–80) (red), Swi5-Sfr1-Fab(156–160) (purple), and Swi5-Sfr1 alone (black). B, representative dummy residue models of the Swi5-Sfr1-Fab complexes containing Fab(1–5) (panel a), Fab(31–35) (panel b), Fab(76–80) (panel c), and Fab(156–160) (panel d). C, proposed location of Sfr1 in the complex. The dummy residue model of the Swi5-Sfr1 complex is shaded gradually from green to red, which corresponds to positions from the N terminus (N-ter) to C terminus (C-ter) of Sfr1. The location of Swi5-Sfr1ΔN177 and the likely binding locations of Fab(1–5), Fab(31–35), Fab(76–80), and Fab(156–160) in the Swi5-Sfr1 complex are boxed.
Swi5-Sfr1-Fab(1–5) has a dummy residue distribution peak at around x = 170–220 Å, indicating that Fab(1–5) is bound to the periphery of Swi5-Sfr1, leading to an increase in the Dmax of Swi5-Sfr1. In Swi5-Sfr1-Fab(31–35) and Swi5-Sfr1-Fab(76–80), broad peaks of the dummy residue distribution appear at around x = 110–210 and 90–200 Å, respectively, which are significantly shorter compared with Swi5-Sfr1-Fab(1–5). The dummy residue distribution of Swi5-Sfr1-Fab(156–160) appears at around x = 85–175 Å and has a broad peak at the shortest x value among the four Fab complexes. Consequently, the binding sites are located from the end to the middle of the Swi5-Sfr1 structure in the following order: 1–5, 31–35, 76–80, and 156–160. This is supported by the fact that the Swi5-Sfr1ΔN177 complex resembles half of the model of the Swi5-Sfr1 complex. These results demonstrate that Sfr1 is positioned within the Swi5-Sfr1 complex according to the topology shown in Fig. 6C.
Model of Swi5-Sfr1 Binding to Rad51 Filament
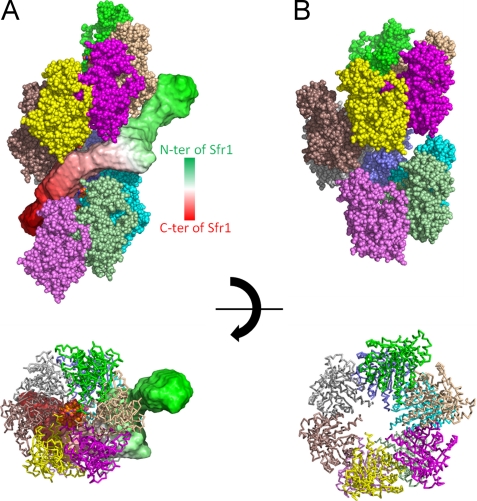
An atomic structure of the S. pombe Rad51 protomer was modeled with the program MODELLER (36) using the crystal structure of S. cerevisiae Rad51 (Protein Data Bank code 3LDA) (26) as a reference structure. Recombinase filaments typically adopt two structures: active and inactive. In the active form, the filament is elongated and has wider grooves, whereas in the inactive form, the filament is a compressed helix with narrower grooves (25). The active Rad51 filament was constructed by aligning the core domains of Rad51 with those of RecA from the crystal structure of the RecA-ssDNA active filament (Protein Data Bank code 3CMW) (26). For the construction of the Rad51 inactive filament, the crystal structure of the RecA inactive filament (Protein Data Bank code 1XMS) (34) was used. The two filament models constructed are consistent with those proposed by electron microscopy (3). Next, the dummy residue model of Swi5-Sfr1 was docked onto the modeled Rad51 filaments manually. Surprisingly, the dummy residue model of the Swi5-Sfr1 complex showed that the dogleg-like shape of the C terminus fits well into the wide groove of the active Rad51 filament (Fig. 7A). The N-terminal domain of the Sfr1 does not enter the groove but plasters itself against the filament. In contrast, the groove of the Rad51 filament in the inactive form is too narrow for the Swi5-Sfr1 complex to enter (Fig. 7B). Thus, because the pitch of the Rad51 filament would be altered as ATP is hydrolyzed, we suggest that the Swi5-Sfr1 complex serves to preserve the ATP-bound active form of the Rad51 filament. Additionally, this model suggests that one Swi5-Sfr1 complex binds to ∼10 Rad51 molecules, which corresponds to the optimum stoichiometry of Rad51 to Swi5-Sfr1 (10:1–20:1) indicated by previous biochemical experiments. When the concentration of the Swi5-Sfr1 complex is increased to more than the optimum concentrations, the activity of strand exchange is significantly reduced (24). At high concentrations of the Swi5-Sfr1 complex, the complex would completely mask the groove of the Rad51 filament, resulting in impairment of the binding of incoming dsDNA. In vivo, the amount of the Swi5-Sfr1 complex in cells is small (20), and there is the possibility of additional factor(s) that regulate(s) the local concentrations of the Swi5-Sfr1 complex in cells.
FIGURE 7.
Model of the Swi5-Sfr1 complex binding to the Rad51 filament. A, active form of the Rad51 filament onto which the dummy residue model of the Swi5-Sfr1 complex is docked. B, inactive form of the Rad51 filament.
Structure of N-terminal Domain of Sfr1
Intrinsically disordered protein prediction (DISOPRED) (38) indicates that the N-terminal half of Sfr1 has high intrinsically disordered protein propensity (supplemental Fig. S3). This prediction is consistent with the following three observations. (a) Limited proteolysis produced a stable C-terminal half-fragment (Sfr1ΔN177) by cleavage of the N-terminal region of Sfr1 (27); (b) Fab-binding sites were mapped from the N-terminal to the central region of Sfr1; and (c) the full-length Swi5-Sfr1 complex could not be crystallized, whereas the crystallization of Swi5 in complex with a 180-amino acid N-terminal deletion mutant of Sfr1 (Sfr1ΔN180) or Sfr1C (Swi5-Sfr1C) was completed successfully (27).
The crystal structure of Swi5-Sfr1C superimposed well onto the dummy residue model of the Swi5-Sfr1ΔN177 complex and the half of the full-length Swi5-Sfr1 model in Fig. 4.3 Although Swi5-Sfr1C possesses the essential ability to stimulate strand exchange, Swi5-Sfr1C requires a concentration 10 times greater than that of the full-length Swi5-Sfr1 complex to achieve the same level of strand exchange activity.4 Additionally, the Sfr1 N-terminal half exhibits DNA-binding activity.4 These observations indicate that the N-terminal half of Sfr1 makes important contributions to the function of the Swi5-Sfr1 complex. Therefore, the solution structure analysis of the full-length Swi5-Sfr1 complex is crucial for the structural investigation of Swi5-Sfr1. If a rigid structure of the Sfr1 N-terminal region is induced upon binding to DNA, Fab fragments that specifically bind to the N-terminal region may inhibit Swi5-Sfr1 functions. Additional experimentation will be required to test this hypothesis.
Conclusions and Perspectives
In this study, we investigated the structure of the full-length Swi5-Sfr1 complex using multiple approaches, including AUC, ESI-MS, and SAXS. We found that Swi5-Sfr1 forms a 1:1 complex and exhibits an extremely elongated dogleg-shaped structure in solution. Taken together, the results lead to a working model for the Swi5-Sfr1 complex, where the Rad51 presynaptic filament is stabilized and activated by the Swi5-Sfr1 complex. Because Swi5 and Sfr1 are widely conserved from yeast to humans, structural insights of the Swi5-Sfr1 complex in S. pombe will be relevant for understanding the structure and function of orthologs in other species. We also showed in this study that SAXS analysis using Fab fragments is a powerful tool for predicting overall protein topology. The methodology developed in this study may be widely applicable for determining the structure of other multisubunit supramolecular complexes in solution.
Supplementary Material
Acknowledgments
We thank Drs. S. Saijo, K. Ito, and T. Hikima for data collection at SPring-8.
This work was supported by grants-in-aids for scientific research on innovative areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT; to S. A., T. S., H. I., M. S., and M. I.) and for scientific research (A (to H. I.) and B (to M. I.)); by the Grand Challenges in Next-Generation Integrated Simulation of Living Matter, part of the Development and Use of the Next-Generation Supercomputer Project of MEXT (to M. I.); by the Japan Society for the Promotion of Science (JSPS); and by research grants for bioscience from the Takeda Science Foundation (to H. I.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
N. Kuwabara and T. Shimizu, unpublished data.
Y. Murayama, Y. Tsutsui, and H. Iwasaki, unpublished data.
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- RPA
- replication protein A
- AUC
- analytical ultracentrifugation
- ESI-MS
- electrospray ionization mass spectrometry
- SAXS
- small-angle x-ray scattering.
REFERENCES
- 1. Krogh B. O., Symington L. S. (2004) Annu. Rev. Genet. 38, 233–271 [DOI] [PubMed] [Google Scholar]
- 2. Cox M. M. (2007) Nat. Rev. Mol. Cell Biol. 8, 127–138 [DOI] [PubMed] [Google Scholar]
- 3. Masson J. Y., West S. C. (2001) Trends Biochem. Sci. 26, 131–136 [DOI] [PubMed] [Google Scholar]
- 4. Bianco P. R., Tracy R. B., Kowalczykowski S. C. (1998) Front. Biosci. 3, D570–D603 [DOI] [PubMed] [Google Scholar]
- 5. Galkin V. E., Wu Y., Zhang X. P., Qian X., He Y., Yu X., Heyer W. D., Luo Y., Egelman E. H. (2006) Structure 14, 983–992 [DOI] [PubMed] [Google Scholar]
- 6. VanLoock M. S., Yu X., Yang S., Lai A. L., Low C., Campbell M. J., Egelman E. H. (2003) Structure 11, 187–196 [DOI] [PubMed] [Google Scholar]
- 7. Chen Z., Yang H., Pavletich N. P. (2008) Nature 453, 489–494 [DOI] [PubMed] [Google Scholar]
- 8. Lisby M., Barlow J. H., Burgess R. C., Rothstein R. (2004) Cell 118, 699–713 [DOI] [PubMed] [Google Scholar]
- 9. Sung P., Klein H. (2006) Nat. Rev. Mol. Cell Biol. 7, 739–750 [DOI] [PubMed] [Google Scholar]
- 10. Sung P., Krejci L., Van Komen S., Sehorn M. G. (2003) J. Biol. Chem. 278, 42729–42732 [DOI] [PubMed] [Google Scholar]
- 11. Sung P. (1997) J. Biol. Chem. 272, 28194–28197 [DOI] [PubMed] [Google Scholar]
- 12. New J. H., Sugiyama T., Zaitseva E., Kowalczykowski S. C. (1998) Nature 391, 407–410 [DOI] [PubMed] [Google Scholar]
- 13. Shinohara A., Ogawa T. (1998) Nature 391, 404–407 [DOI] [PubMed] [Google Scholar]
- 14. Sugiyama T., Kowalczykowski S. C. (2002) J. Biol. Chem. 277, 31663–31672 [DOI] [PubMed] [Google Scholar]
- 15. Plate I., Hallwyl S. C., Shi I., Krejci L., Müller C., Albertsen L., Sung P., Mortensen U. H. (2008) J. Biol. Chem. 283, 29077–29085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krejci L., Song B., Bussen W., Rothstein R., Mortensen U. H., Sung P. (2002) J. Biol. Chem. 277, 40132–40141 [DOI] [PubMed] [Google Scholar]
- 17. Symington L. S. (2002) Microbiol. Mol. Biol. Rev. 66, 630–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharan S. K., Morimatsu M., Albrecht U., Lim D. S., Regel E., Dinh C., Sands A., Eichele G., Hasty P., Bradley A. (1997) Nature 386, 804–810 [DOI] [PubMed] [Google Scholar]
- 19. Akamatsu Y., Dziadkowiec D., Ikeguchi M., Shinagawa H., Iwasaki H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15770–15775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Akamatsu Y., Tsutsui Y., Morishita T., Siddique M. S., Kurokawa Y., Ikeguchi M., Yamao F., Arcangioli B., Iwasaki H. (2007) EMBO J. 26, 1352–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ellermeier C., Schmidt H., Smith G. R. (2004) Genetics 168, 1891–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Akamatsu Y., Jasin M. (2010) PLoS Genet. 6, e1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yuan J., Chen J. (2011) J. Biol. Chem. 286, 9888–9893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haruta N., Kurokawa Y., Murayama Y., Akamatsu Y., Unzai S., Tsutsui Y., Iwasaki H. (2006) Nat. Struct. Mol. Biol. 13, 823–830 [DOI] [PubMed] [Google Scholar]
- 25. Kurokawa Y., Murayama Y., Haruta-Takahashi N., Urabe I., Iwasaki H. (2008) PLoS Biol. 6, e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen J., Villanueva N., Rould M. A., Morrical S. W. (2010) Nucleic Acids Res. 38, 4889–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuwabara N., Hashimoto H., Yamada N., Unzai S., Ikeguchi M., Sato M., Murayama Y., Iwasaki H., Shimizu T. (2010) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1124–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laue T. M., Shah B., Ridgeway T. M., Pelletier S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S. E., Rowe A. J., Horton L. C. eds) pp 90–125, Royal Society of Chemistry, Cambridge, United Kingdom [Google Scholar]
- 29. Schuck P., Perugini M. A., Gonzales N. R., Howlett G. J., Schubert D. (2002) Biophys. J. 82, 1096–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fujisawa T., Inoue K., Oka T., Iwamoto H., Uruga T., Kumasaka T., Inoko Y., Yagi N., Yamamoto M., Ueki T. (2000) J. Appl. Crystallogr. 33, 797–800 [Google Scholar]
- 31. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 32. Svergun D. I. (1992) J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 33. Glatter O., Kratky O. (1982) Small-Angle X-Ray Scattering, Academic Press, London: [DOI] [PubMed] [Google Scholar]
- 34. Svergun D. I., Petoukhov M. V., Koch M. H. (2001) Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Volkov V. V., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 860–864 [Google Scholar]
- 36. Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 37. Xing X., Bell C. E. (2004) Biochemistry 43, 16142–16152 [DOI] [PubMed] [Google Scholar]
- 38. Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. (2004) J. Mol. Biol. 337, 635–645 [DOI] [PubMed] [Google Scholar]
- 39. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.