Background: RtcB is a new RNA ligase implicated in tRNA splicing and RNA repair.
Results: RtcB has 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-hydroxyl ligase activities.
Conclusion: RtcB seals broken RNAs via a two-step reaction pathway.
Significance: The structure, active site, and chemical mechanism of RtcB are unique.
Keywords: Enzyme mechanisms, Manganese, Mutagenesis site-specific, RNA, RNA splicing, Transfer RNA (tRNA)
Abstract
RtcB enzymes are a newly discovered family of RNA ligases, implicated in tRNA splicing and other RNA repair reactions, that seal broken RNAs with 2′,3′-cyclic phosphate and 5′-OH ends. Parsimony and energetics would suggest a one-step mechanism for RtcB sealing via attack by the O5′ nucleophile on the cyclic phosphate, with expulsion of the ribose O2′ and generation of a 3′,5′-phosphodiester at the splice junction. Yet we find that RtcB violates Occam's razor, insofar as (i) it is adept at ligating 3′-monophosphate and 5′-OH ends; (ii) it has an intrinsic 2′,3′-cyclic phosphodiesterase activity. The 2′,3′-cyclic phosphodiesterase and ligase reactions both require manganese and are abolished by mutation of the RtcB active site. Thus, RtcB executes a unique two-step pathway of strand joining whereby the 2′,3′-cyclic phosphodiester end is hydrolyzed to a 3′-monophosphate, which is then linked to the 5′-OH end to form the splice junction. The energy for the 3′-phosphate ligase activity is provided by GTP, which reacts with RtcB in the presence of manganese to form a covalent RtcB-guanylate adduct. This adduct is sensitive to acid and hydroxylamine but resistant to alkali, consistent with a phosphoramidate bond.
Introduction
RNA 2′,3′-cyclic phosphate and 5′-OH ends figure prominently in RNA metabolism: as the products of RNA cleavage by site-specific endoribonucleases, as substrates for enzymatic RNA end joining during tRNA repair and tRNA splicing, and as substrates for unconventional mRNA splicing during the unfolded protein response (1–6). Broadly speaking, there are two known RNA repair pathways that rectify 2′,3′-cyclic phosphate/5′-OH breaks. The “healing-and-sealing” pathway of RNA repair is a multistep process typically catalyzed by three different enzymes dedicated to 3′ healing, 5′ healing, and ligation, respectively (6, 7). In the healing phase, the 2′,3′-cyclic phosphate end is hydrolyzed to a 3′-OH, and the 5′-OH end is phosphorylated by an NTP-dependent polynucleotide kinase to yield a 5′-monophosphate. The healed 3′-OH and 5′-PO4 termini are then suitable substrates for a “classic” ATP-dependent RNA ligase that restores the 3′, 5′ phosphodiester backbone via three nucleotidyl transfer steps as follows: (i) ligase reacts with ATP to form a ligase-(lysyl-N)-AMP intermediate plus pyrophosphate, (ii) AMP is transferred from ligase-adenylate to the 5′-PO4 RNA end to form an RNA-adenylate intermediate, AppRNA,2 and (iii) ligase catalyzes attack by an RNA 3′-OH on AppRNA to form a 3′, 5′ phosphodiester bond and release AMP (8). The salient principle of classic ligase catalysis is that the high energy of the ATP phosphoanhydride bond is transferred via the enzyme to the RNA 5′ end, thereby activating the 5′ end for phosphodiester synthesis.
RNA end-healing enzymes and classic RNA ligases are found in diverse taxa in all three phylogenetic domains. The healing-and-sealing pathway is responsible for tRNA splicing in fungi and plants (9, 10), for mRNA splicing in the fungal unfolded protein response (4, 5), and for tRNA restriction-repair during bacteriophage infection of Escherichia coli (3). Recent studies highlight a novel bacterial healing-and-sealing pathway that is thought to confer adaptive immunity to tRNA damage inflicted by ribotoxins (11, 12).
A second pathway of RNA repair by “direct ligation” of 2′,3′-cyclic phosphate and 5′-OH ends was discovered nearly 30 years ago via analysis of the mammalian tRNA splicing reaction (13, 14). Specifically, it was shown that the 2′,3′-cyclic phosphate of the cleaved pre-tRNA is incorporated at the splice junction in the mature tRNA. (By contrast, the junction phosphate in yeast tRNA splicing derives from the γ-phosphate of the NTP substrate for the 5′ kinase step.) Direct ligation is thought to be the main pathway of tRNA splicing in animals and archaea (13–15). Earlier this year, three laboratories reported the identification of bacterial, archaeal, and mammalian RtcB proteins as novel RNA ligase enzymes capable of sealing 2′,3′-cyclic phosphate and 5′-OH ends in the absence of ATP (16–18). RtcB homologs are present in metazoa and protozoa but not in fungi and plants. Thus, the phylogenetic distribution of RtcB fortifies its candidacy as a eukaryal tRNA splicing enzyme.
In support of a broad role for RtcB ligases in RNA repair, we've shown that E. coli RtcB is competent and sufficient for tRNA splicing in vivo, by virtue of its ability to complement growth of yeast cells that lack the endogenous healing-and-sealing-type tRNA ligase Trl1 (19). RtcB also protects yeast trl1Δ cells against a fungal ribotoxin that incises the anticodon loop of cellular tRNAs. Moreover, RtcB can replace Trl1 as the catalyst of HAC1 mRNA splicing during the unfolded protein response (19).
RtcB is unique with respect to its tertiary structure and the architecture of its active site (20), thereby suggesting an unprecedented mechanism of end joining. The simplest chemical scheme for the RtcB sealing reaction would entail attack by the O5′ nucleophile on the cyclic phosphate, with expulsion of the ribose O2′ (Fig. 1A). Our findings that RtcB is manganese-dependent distinguishes the RtcB sealing activity from the metal-independent ligation of a 5′-OH strand to 2′,3′-cyclic phosphate strand catalyzed by DNA topoisomerase IB (21). We've speculated that manganese might promote RtcB catalysis by coordinating the O5′ nucleophile to lower its pKa and/or engaging the cyclic phosphate oxygens to stabilize the transition state (18). RtcB has an active site pocket lined by essential histidines and a cysteine (19, 20), suggestive of a metal-binding site.
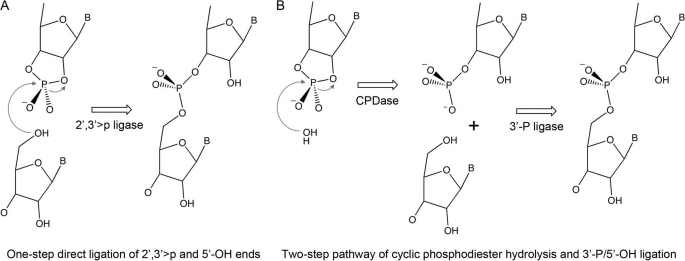
FIGURE 1.
Alternative models for RtcB catalysis. A, one-step direct ligation; B, two-step pathway.
The one-step transesterification ligation mechanism depicted in Fig. 1A is attractive from an energetics perspective, insofar as the cyclic phosphodiester is a strained energy-rich reactant with a favorable leaving group (the ribose O2′). For this mechanism to be plausible, RtcB would need to exclude water as a nucleophile and have some means to accept a polynucleotide 5′-OH as the only idoneous nucleophile. The mechanism also engenders an explicit prediction that RtcB ought not to ligate a 3′-monophosphate RNA terminus. Here we interrogate and overturn the simple model in Fig. 1A, in favor of a less parsimonious, but mechanistically more interesting, two-step ligation pathway depicted in Fig. 1B, in which RtcB first hydrolyzes the 2′,3′-cyclic phosphodiester end to a 3′-monophosphate and then joins the 3′-monophosphate RNA end to a 5′-OH end to form a 3′→5′ phosphodiester.
EXPERIMENTAL PROCEDURES
Purification of RtcB
pET28b-His10Smt3-RtcB plasmids encoding wild-type RtcB and mutants C78A and H168A were transformed into E. coli BL21-CodonPlus(DE3). Cultures (1 liter) derived from single kanamycin-resistant transformants were grown at 37 °C until the A600 reached 0.6–0.8. The cultures were chilled on ice for 30 min and then adjusted to 0.1 mm isopropyl β-d-thiogalactoside. Incubation was continued at 17 °C for 16 h with constant shaking. Cells were harvested by centrifugation and stored at −80 °C. Preparation of soluble lysates, recovery of the His10Smt3-RtcB proteins by nickel-agarose chromatography, excision of the tags by treatment with the Smt3-specific protease Ulp1, separation of the tag-free RtcB proteins from His10Smt3 by a second round of Ni-agarose chromatography, and final purification of RtcB to by Superdex 200 gel filtration were performed as described previously (18). Protein concentrations were determined using the Bio-Rad dye reagent with BSA as the standard.
RNA Repair Substrates
3′-Phosphorylated RNA oligonucleotides labeled with 32P were prepared by T4 RNA ligase 1 (Rnl1)-mediated addition of [5′-32P]pCp to a 19-mer synthetic oligoribonucleotide. The [5′-32P]pCp donor was generated via enzymatic phosphorylation of unlabeled 3′-CMP by T4 polynucleotide kinase-phosphatase (Pnkp) mutant D167N (kinase active, but devoid of 3′-phosphatase; purified according to Ref. 22) in the presence of [γ-32P]ATP. A kinase reaction mixture (20 μl) containing 50 mm Tris-HCl (pH 8.0), 10 mm MgCl2, 10 mm DTT, 0.1 mm 3′-CMP, 0.1 mm [γ-32P]ATP (130 μCi), 0.1 mg/ml BSA, and 10 μg/ml Pnkp-D167N was incubated at 37 °C for 60 min. The Pnkp was inactivated by heating the mixture at 95 °C for 3 min. The mixture was then adjusted to 40 μl and, in the process, supplemented with 100 μm fresh unlabeled ATP, 50 μm 19-mer oligoribonucleotide 5′-rUGGCUCCGAUAUCACGCUU, and 0.2 mg/ml Rnl1 (purified as described in Ref. 23). The T4 ligase reaction mixture was incubated for 1 h at 37 °C, and the products were resolved by electrophoresis through a 40-cm 20% polyacrylamide gel containing 8 m urea. The 32P-labeled 20-mer oligonucleotide product (5′-HOUGGCUCCGAUAUCACGCUUpCp) was located by autoradiography and excised from the gel. The oligonucleotide was eluted by soaking the gel slice overnight in 0.4 ml of 1 m ammonium acetate, 0.2% SDS, 20 mm EDTA. The eluted oligoribonucleotide was recovered by ethanol precipitation and resuspended in 10 mm Tris-HCl (pH 6.8).
The purified 20-mer was further modified by 5′ phosphorylation and/or cyclization of the 3′-phosphate as follows. Reaction mixtures (100 μl) containing 50 mm Tris-HCl (pH 7.4), 0.1 mm ATP, 10 mm MgCl2, 2 mm DTT, 0.1 mm spermidine, 500 nm 32P-labeled 20-mer RNA, and 10 μg/ml Pnkp-D167N and/or 40 μg/ml E. coli cyclase RtcA (purified according to Ref. 24) were incubated for 30 min at 37 °C. The end-modified reaction products 5′-pUGGCUCCGAUAUCACGCUUpCp, 5′-pUGGCUCCGAUAUCACGCUUpC>p, and 5′-HOUGGCUCCGAUAUCACGCUUpC>p were recovered by phenol/chloroform extraction and ethanol precipitation, then resuspended in 10 mm Tris-HCl (pH 6.8).
A 2′-monophosphate-terminated version of the 20-mer RNA was prepared by reacting 5′-HOUGGCUCCGAUAUCACGCUUpC>p with the C-terminal end-healing domain of plant tRNA ligase, AtRNL-(667–1101) (purified as described in Ref. 6). A reaction mixture (40 μl) containing 50 mm Tris-HCl (pH 7.4), 10 mm MgCl2, 2 mm DTT, 10 μg/ml AtRNL-(667–1104), and 500 nm HOUGGCUCCGAUAUCACGCUUpC>p was incubated at 37 °C for 15 min. The HOUGGCUCCGAUAUCACGCUUpC2′p reaction product was recovered by phenol/chloroform extraction and ethanol precipitation.
Assays of RNA Ligase and Cyclic Phosphodiesterase Activities
Reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 8.0), 2 mm MnCl2, 100 μm GTP, 20 nm RNA repair substrate as specified, and 200 nm RtcB were incubated for 30 min at 37 °C. The reactions were quenched by adding 1 μl of 40 mm EDTA. The products were either analyzed directly by PAGE or digested for 15 min at 37 °C with 1000 units of RNase T1 (Fermentas) prior to PAGE. The samples were mixed with 10 μl of 90% formamide, 0.01% bromphenol blue/xylene cyanol, 50 mm EDTA and then resolved by electrophoresis (at 50 watts of constant power) through a 40-cm 20% polyacrylamide gel containing 8 m urea in 45 mm Tris borate, 1.2 mm EDTA. The 32P-labeled RNAs were visualized by autoradiography of the gel and, where specified, quantified by scanning the gel with a Fuji Film BAS-2500 imager.
RESULTS
RtcB Seals Stem-loops with 3′-Monophosphate/5′-OH Breaks
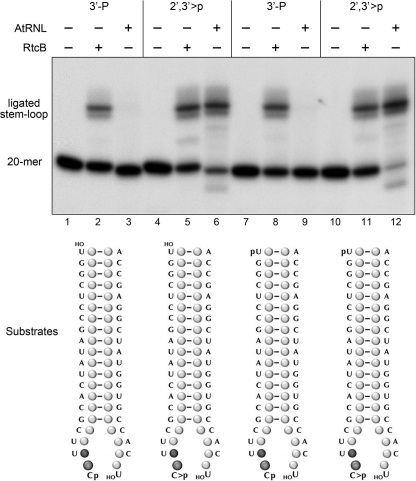
We showed previously that RtcB was adept at ligating broken stem-loop structures with 2′,3′-cyclic phosphate and 5′-OH termini to restore a 3′,5′-phosphodiester at the repair junction (18). Here we queried whether RtcB could seal a 3′-monophosphate end. We used T4 RNA ligase 1 to transfer [5′-32P]pCp to the 3′ end of a 19-mer RNA oligonucleotide to generate a 20-mer strand (HOUGGCUCCGAUAUCACGCUUpCp) with 5′-OH and 3′-monophosphate ends and a single radiolabel between the 3′-terminal and penultimate nucleosides. We used purified E. coli RNA 3′-phosphate cyclase RtcA (24, 25) to convert the 3′-phosphate end of the labeled 20-mer RNA into a 2′,3′-cyclic phosphodiester (HOUGGCUCCGAUAUCACGCUUpC>p). We also converted the 5′-OH termini of these labeled strands into 5′-PO4 ends by reaction with cold ATP and a mutated (phosphatase-dead) version of T4 polynucleotide 5′-kinase/3′-phosphatase (22). The four radiolabeled strands were annealed to a complementary unlabeled 5′-OH terminated 20-mer RNA to form the broken stem-loop substrates depicted in Fig. 2. These substrates were then reacted with either RtcB or plant tRNA ligase (AtRNL) and the products were analyzed by denaturing PAGE. Sealing activity was manifest by the conversion of the labeled 20-mer into a slower-migrating 40-mer stem-loop (Fig. 2). As expected, AtRNL catalyzed efficient sealing of substrates with a 2′,3′-cyclic phosphate end (lanes 6 and 12), but was inert with otherwise identical 3′-monophosphate substrates (lanes 3 and 9). By contrast, RtcB readily ligated breaks with either 3′-monophosphate ends (lanes 2 and 8) or 2′,3′-cyclic phosphate ends (lanes 5 and 11).
FIGURE 2.
RtcB seals 3′-monophosphate/5-OH RNA breaks. Reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 8.0), 0.1 mm ATP, 0.1 mm GTP, 2 mm DTT, 0.1 mm spermidine, 2 mm MnCl2 (for E. coli RtcB; lanes 2, 4, 6, and 8) or 10 mm MgCl2 (for plant tRNA ligase, AtRNL; lanes 3, 6, 9, and 12), 20 nm 32P-labeled broken RNA stem-loop substrates as specified (formed by mixing 20 nm labeled 32P-20-mer with 200 nm unlabeled complementary 20-mer to yield the substrates illustrated at the bottom), and 200 nm RtcB or AtRNL (where indicated by + above the lanes) were incubated for 30 min at 37 °C. The products were resolved by denaturing PAGE and visualized by autoradiography. The radiolabeled RNAs corresponding to the 20-mer substrate strand and the ligated hairpin stem-loop are indicated on the left.
Intramolecular Joining of 3′-Monophosphate and 5′-OH Ends
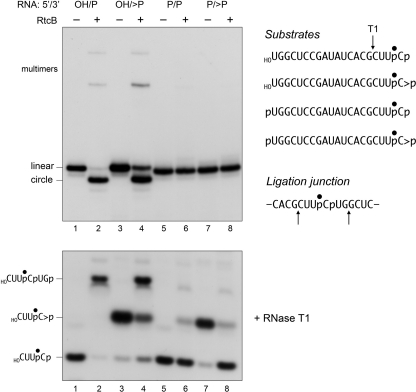
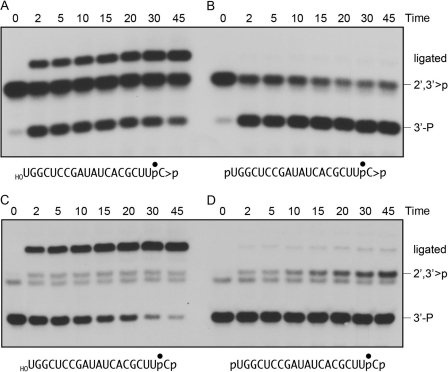
The radiolabeled 20-mer RNAs were reacted with RtcB in the absence of a complementary strand and the products were analyzed by denaturing PAGE (Fig. 3, top panel). The substrates with 5′-OH ends were efficiently converted to a more rapidly migrating circular RNA species as a consequence of intramolecular ligation (lanes 2 and 4). Low levels of more slowly migrating multimers (products of intermolecular ligation) were also generated. There was little difference in the extents of circularization of 5′-OH terminated single strands with 3′-monophosphate versus cyclic phosphate ends. The two substrates with 5′-PO4 ends were not sealed by RtcB (lanes 6 and 8).
FIGURE 3.
RtcB catalyzes intramolecular 3′-phosphate/5′-OH ligation and has a 2′,3′-cyclic phosphodiesterase activity. Reaction mixtures (20 μl) containing 50 mm Tris-HCl (pH 8.0), 0.1 mm GTP, 2 mm MnCl2, 20 nm 32P-labeled 20-mer RNA strand with 5′-OH, 5′-phosphate (5′-P), 3′-phosphate (3′-P), or 2′,3′-cyclic phosphate (>P) termini (depicted at the right, with the radiolabeled phosphate denoted by ●), and 200 nm RtcB (where indicated by + above the lanes) were incubated for 30 min at 37 °C. The reactions were quenched by adding EDTA to 4 mm final concentration. Half of the reaction mixture was analyzed directly by denaturing PAGE; the labeled RNAs were visualized by autoradiography (top panel). The positions of the linear substrates, the ligated circles, and multimeric ligation products are indicated on the left. The other half of the reaction mixture was digested with RNase T1. (The relevant sites of RNase T1 incision of the 20-mer substrates and flanking the ligation junction are indicated by arrows above the RNA sequences at the right.) The T1 digests were then resolved by denaturing PAGE and visualized by autoradiography (bottom panel). The positions of the T1 fragments are indicated on the left.
The input 3′ end structures and the RNA splice junctions were characterized further by digesting the samples with RNase T1 prior to their electrophoresis through a polyacrylamide gel (Fig. 3, bottom panel). RNase T1 incised the 20-mer RNA substrates 3′ of the most distal guanosine to yield 32P-labeled tetranucleotides: either HOCUUpCp or HOCUUpC>p - with distinct electrophoretic mobilities (lanes 1 and 3). (This analysis verified that the efficiency of cyclization of the -UpCp end of the 20-mer during the preparative reaction with RtcA was 95%, i.e. 5% of the RNase T1 product was HOCUUpCp in lane 3.) The key finding was that the reactions with RtcB resulted in depletion of the T1 fragments derived from the substrate strands and the appearance of a new, more slowly migrating T1 fragment (Fig. 3, bottom panel, lanes 2 and 4) that corresponded to a 6-mer oligonucleotide, HOCUUpCpUGp, released by T1 incision at the guanosines flanking the predicted ligation junction (Fig. 3). Note that the T1 junction fragment did not accumulate when RtcB was reacted with non-ligatable substrates with 5′-PO4 ends (lanes 6 and 8).
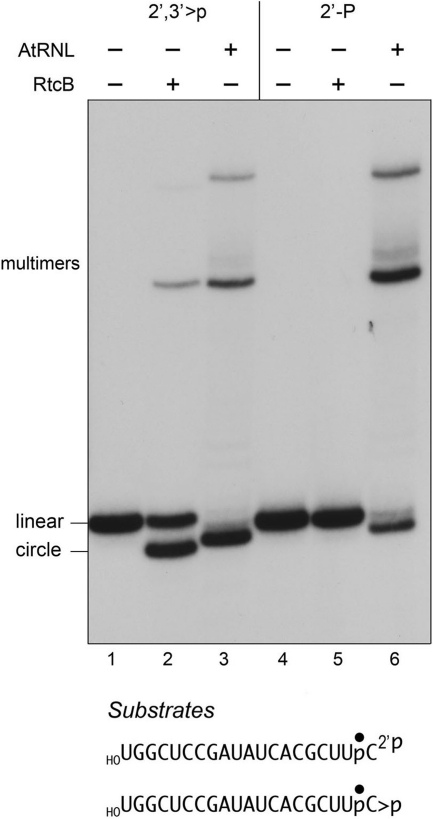
We further queried the specificity of RtcB by asking whether the enzyme could join a 2′-monophosphate end to a 5′-OH. The 2′-monophosphate 20-mer substrate was prepared by treating the radiolabeled HOUGGCUCCGAUAUCACGCUUpC>p strand with the isolated C-terminal end-healing domain of plant tRNA ligase (6), which hydrolyzes the 2′,3′-cyclic phosphodiester to yield a 2′-phosphate, 3′-OH terminus. (By digesting the labeled 20-mer with RNase T1 before and after treatment with the plant ligase healing domain, and then analyzing the the T1 fragments by PAGE, we verified that the initial 2′,3′-cyclic phosphate terminus had been completely converted to a more rapidly migrating phosphomonoester derivative; not shown.) The radiolabeled 2′,3′>p and 2′-phosphate 20-mer strands were then reacted in parallel with RtcB and AtRNL and the products were analyzed by PAGE (Fig. 4). As expected, AtRNL catalyzed efficient sealing of both substrates, yielding a mixture of circular and multimeric ligation products (Fig. 4, lanes 3 and 6). By contrast, RtcB sealed the 2′,3′>p strand to yield a predominant circular ligation product (lane 2) but was unreactive with the 2′-monophosphate substrate (lane 5). The slight differences in electrophoretic mobility of the circular products made by AtRNL versus RtcB reflect the distinct outcomes of the RtcB and AtRNL ligation reactions. Whereas RtcB seals the 2′,3′>p end to yield a splice junction with a conventional 3′→5′ phosphodiester linkage (18), AtRNL sealing at either a 2′,3′>p or 2′-phosphate end yields a splice junction that retains the 2′-phosphate (1). We conclude that RtcB and AtRNL have antipodal RNA end requirements for sealing, indicative of stark differences in the chemistry of end joining.
FIGURE 4.
RtcB cannot seal 2′-monophosphate/5′-OH RNA breaks. RtcB reaction mixtures (20 μl) contained 50 mm Tris-HCl (pH 8.0), 0.1 mm GTP, 2 mm MnCl2, 20 nm 32P-labeled 20-mer RNA strands with 5′-OH and either 2′,3′-cyclic phosphate (>P) or 2′-monophosphate (2′-P) termini (depicted at the bottom, with the radiolabeled phosphate denoted by ●), and 200 nm RtcB. AtRNL reaction mixtures (20 μl) contained 50 mm Tris-HCl (pH 8.0), 0.1 mm ATP, 0.1 mm GTP, 2 mm DTT, 10 mm MgCl2, 20 nm 32P-labeled 20-mer RNA strands, and 200 nm AtRNL. The mixtures were incubated for 30 min at 37 °C, and the reactions were quenched with EDTA. The products were analyzed directly by denaturing PAGE; the labeled RNAs were visualized by autoradiography. The positions of the linear substrates, the ligated circles, and multimeric ligation products are indicated on the left.
RtcB Has 2′,3′-Cyclic Phosphodiesterase Activity
The RNase T1 analysis provided the initially surprising observation that incubation of RtcB with the 5′-phosphorylated 20-mer strand with a cyclic phosphate end, although unproductive with respect to ligation, resulted in a chemical transformation at the 3′ terminus, whereby the original 2′,3′>p was converted to a 3′-monophosphate (Fig. 3, bottom panel, lanes 7 and 8). Thus, in the absence of a polynucleotide 5′-OH, RtcB catalyzes hydrolytic attack on the cyclic phosphodiester. In this experiment, 75% of the input cyclic phosphate was hydrolyzed. We also observed that incubation of RtcB with the 5′-phosphorylated 20-mer strand with a 3′-monophosphate end resulted in a low but appreciable conversion to a T1 fragment that comigrated with the cyclic phosphate species (Fig. 3, bottom panel, lanes 5 and 6).
The time course of the 2′,3′-cyclic phosphodiesterase (CPDase) activity of RtcB is shown in Fig. 5B. Opening of the cyclic end was relatively rapid and efficient (65% of the input 2′,3′>p substrate was hydrolyzed in 2 min), and the reaction attained an end point with 80% of the cyclic ends opened. The inference that the CPDase reaction is reversible was consistent with the kinetic profile of the reaction of RtcB with a 5′-phosphorylated, 3′-monophosphate strand, wherein we saw a slow but steady conversion of the 3′-phosphate end to a species that comigrated with the 2′,3′>p T1 fragment (Fig. 5D). The extent of this conversion was 15% at 45 min. The RtcB reaction mixtures did not include ATP, which is the requisite high energy substrate for RNA 3′-cyclization catalyzed by bacterial RtcA (24, 25). Given that RtcA gene expression is repressed in E. coli under standard growth conditions (25), and the purity of the recombinant RtcB preparation (Fig. S2), we thought it unlikely that the 3′-phosphate cyclization observed for RtcB could be caused by RtcA contamination. To confirm this idea, we purposefully included 0.1 mm ATP in the RtcB reaction mixtures and examined the kinetic profile of cyclization, assuming that ATP would strongly stimulate the generation of cyclic ends if RtcA was the culprit (25). However, we found no effect of ATP on the rate or yield of cyclic phosphate product (not shown). Thus, we surmise that RtcB has an intrinsic CPDase activity that favors the forward hydrolysis reaction under the conditions of our assays.
FIGURE 5.
Time course of ligation and CPDase activities. Reaction mixtures (80 μl) containing 50 mm Tris-HCl (pH 8.0), 0.1 mm GTP, 2 mm MnCl2, 20 nm 32P-labeled 20-mer RNAs as shown (HORNA>p in panel A; pRNA>p in panel B; HORNAp in panel C; pRNAp in panel D), and 200 nm RtcB were incubated at 37 °C. Aliquots (10 μl) were withdrawn at the indicated times (minutes) and quenched immediately with EDTA. The time 0 sample was taken prior to adding RtcB. The quenched mixtures were digested with RNase T1 and then analyzed by denaturing PAGE. The positions of the 3′-P T1 fragment, the 2′,3′>p T1 fragment, and the ligation junction T1 fragment are indicated on the right.
The temporal profile of the RtcB intramolecular ligation reaction with the 5′-OH/2′,3′>p 20-mer substrate was consistent with the intermediacy of a 3′-monophosphate end in the sealing pathway (Fig. 5A). To wit, the 3′-phosphate T1 fragment was more abundant than the ligated junction fragment at the 2-min time point, at which the 3′-phosphate and ligated species comprised 27 and 18% of the total radioactivity, respectively. The 3′-phosphate species declined steadily thereafter (to 11% at 45 min), concomitant with an increase in the abundance of the ligated product (to 52% at 45 min) (Fig. 5A). These results show that RtcB readily executes the hydrolytic CPDase reaction even in the presence of a ligatable 5′-OH RNA end.
The kinetic profile for intramolecular sealing of the 5′-OH/3′-monophosphate 20-mer revealed a simple decay of the 3′-monophosphate substrate concomitant with accumulation of the ligated junction fragment (Fig. 5C). The ligated species comprised 92% of the radiolabeled products at 45 min. Only scant conversion of the 3′-monophosphate to a species comigrating with the 2′,3′>p fragment (to ∼4–5% of the total label) was detectable from 2 to 45 min (Fig. 5C). These data are consistent with a two-step mechanism of RtcB sealing of 2′,3′>p and 5′-OH ends, whereby the CPDase activity of RtcB hydrolyzes the 2′,3′>p to a 3′-monophosphate, after which the ligase activity of RtcB joins the 5′-OH end to the 3′-phosphate end (Fig. 1B).
We queried the ability of RtcB to use an alternative non-water nucleophile for its attack on the 2′,3′-cyclic phosphate terminus, by supplementing the reaction mixtures with increasing concentrations of glycerol (10, 20, and 30% v/v). An apparent glycerololysis reaction was evinced by the appearance of a novel radiolabeled T1 fragment, concomitant with a decline in the abundance of the hydrolysis reaction product HOCUUpCp (supplemental Fig. S1). The yield of this novel species (presumably HOCUUpC3′p-glycerol) was proportional to glycerol concentration and dependent on RtcB. We surmise that glycerol can compete effectively with water as the nucleophile for the RtcB CPDase reaction, i.e. in the presence of 30% glycerol the distribution of radiolabeled CPDase products was 55% hydrolysis versus 45% glycerololysis (supplemental Fig. S1).
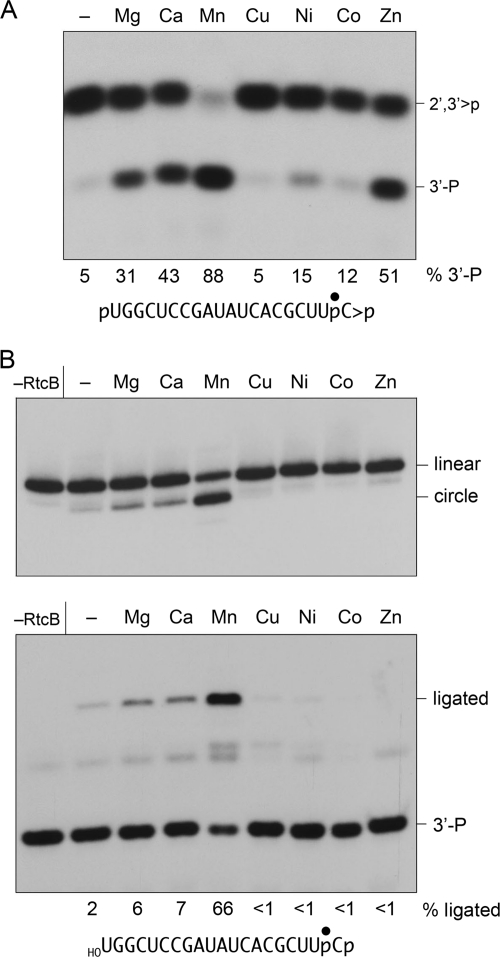
Metal Requirements for the RtcB CPDase and Ligase Activities
We showed previously that sealing of 2′,3′-cyclic phosphate and 5′-OH termini by RtcB depended on manganese (18). Here we examined the metal requirements for the component CPDase and 3′-phosphate/5′-OH ligation reactions. The CPDase reaction per se was metal-dependent; the metal requirement was best satisfied by 2 mm manganese (Fig. 6A). Zinc, calcium, and magnesium were less effective, nickel and cobalt supported feeble activity, and copper was ineffective (Fig. 6A).
FIGURE 6.
Metal specificity of 3′-phosphate ligation and CPDase activities. A, CPDase reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 8.0), 0.1 mm GTP, 20 nm 32P-labeled 20-mer pRNA>p as shown, 200 nm RtcB, and either no divalent cation (lane –) or 2 mm MgCl2, CaCl2, MnCl2, CuCl2, NiCl2, CoCl2, or ZnCl2 as specified were incubated for 30 min at 37 °C. The EDTA-quenched reaction mixtures were digested with RNase T1 and analyzed by PAGE. The percent of total radiolabeled material corresponding to the 3′-P T1 fragment is indicated below each lane. (The 3′-P T1 fragment comprised 5% of the total in a no-enzyme control; not shown.) B, ligase reaction mixtures (20 μl) containing 50 mm Tris-HCl (pH 8.0), 0.1 mm GTP, 20 nm 32P-labeled 20-mer HORNAp as shown, 200 nm RtcB, and either no divalent cation (lane −) or 2 mm MgCl2, CaCl2, MnCl2, CuCl2, NiCl2, CoCl2, or ZnCl2 as specified were incubated for 30 min at 37 °C. Enzyme was omitted from a control reaction mixture (−RtcB). Half of the reaction mixture was analyzed directly by denaturing PAGE (top panel). The other half of the reaction mixture was digested with RNase T1 and then resolved by denaturing PAGE (bottom panel). The extents of ligation are indicated below each lane in the bottom panel.
RtcB was more fastidious in its metal requirement for 3′-phosphate/5-OH ligation, as assayed by the formation of intramolecular circles (Fig. 6B, top panel) and of RNase T1 fragments derived from the substrate and the ligation junction (Fig. 6B, bottom panel). The ligase metal requirement was satisfied by 2 mm manganese, but not by zinc. Copper, nickel, and cobalt were inactive in ligation, whereas magnesium and calcium supported only low levels of sealing activity above that of the no-metal control reaction (Fig. 6B).
Effect of RtcB Mutations on CPDase and Ligase Activities
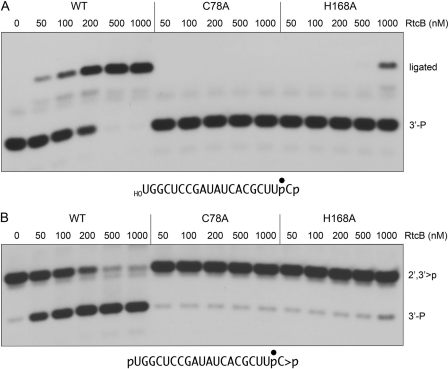
Previously, we used the crystal structure of Pyrococcus horikoshii RtcB (20) to guide a mutational analysis of conserved residues in E. coli RtcB that we thought might comprise an active site. A water in the putative archaeal RtcB active site, coordinated by the equivalents of essential E. coli RtcB side chains Asp-75, Cys-78, and His-168 (19), was regarded as a potential mimic of the enzyme-bound metal cofactor. Here we purified the mutant C78A and H168A RtcB proteins to apparent homogeneity via the same sequence of chromatography steps and gel-filtration used to purify the wild-type RtcB enzyme (supplemental Fig. S2) and then tested the wild type and mutants for their ability to execute the component CPDase and 3′-phosphate ligation steps of the proposed RtcB RNA repair pathway.
An RtcB titration experiment established that the extent of intramolecular sealing of 3′-phosphate and 5′-OH ends was proportional to the concentration of wild-type RtcB from 50 to 500 nm, attaining an end point of 98% end joining at 500–1000 nm (Fig. 7A). By contrast, C78A was inert in 3′-phosphate ligation even at 1000 nm, the highest level of protein tested (Fig. 7A). The H168A mutant displayed 3′-phosphate ligation activity only at 1000 nm input protein (17% of substrate ligated).
FIGURE 7.
Effect of RtcB active site mutations on 3′-phosphate ligation and CPDase activities. A, ligase reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 8.0), 0.1 mm GTP, 2 mm MnCl2, 20 nm 32P-labeled 20-mer HORNAp as shown and 0, 50, 100, 200, 500, or 1000 nm of wild-type (WT) RtcB or mutants C78A or H168A (see supplemental Fig. S2) were incubated at 37 °C for 30 min. The products were digested with RNase T1 and then analyzed by PAGE. B, CPDase reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 8.0), 0.1 mm GTP, 2 mm MnCl2, 20 nm 32P-labeled 20-mer pRNA>p as shown and 0, 50, 100, 200, 500, or 1000 nm of WT or mutant RtcB proteins were incubated at 37 °C for 30 min. The products were digested with RNase T1 and then resolved by PAGE.
Mutational effects on the CPDase reaction are shown in Fig. 7B. Whereas opening of the cyclic phosphate by wild-type RtcB increased in proportion to enzyme concentration and was nearly complete at 500–1000 nm RtcB, the C78A and H168A mutants were virtually inert at up to 1000 nm input protein. We infer that the CPDase and 3′-phosphate ligase reactions are catalyzed by a common active site.
A Role for GTP in 3′-Phosphate/5′-OH Ligation
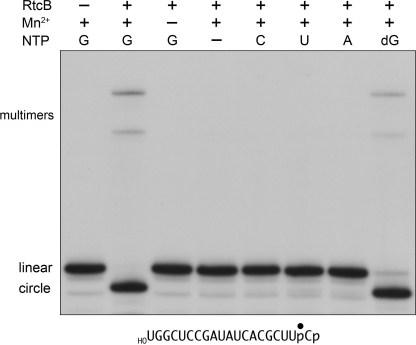
In our initial characterization of RtcB, we noted that 0.1 mm GTP stimulated the sealing of a broken RNA stem-loop with 2′,3′-cyclic phosphate and 5′-OH termini (18). Consequently, we have included GTP in the RtcB reaction mixtures in all experiments presented above. The new findings that RtcB ligates a 3′-phosphate end to a 5′-OH end raised the question of whether GTP might serve as an energy-rich substrate in the ligation reaction, analogous to the way classic RNA ligases and RNA cyclase use ATP for end activation. Thus, we assayed 3′-phosphate/5′-OH sealing by RtcB in the presence and absence of GTP. The extents of product formation are plotted as a function of input RtcB in supplemental Fig. S3, wherein each datum is the average of three separate enzyme titration experiments. We found that RtcB could execute the 3′-phosphate ligation reaction in the absence of GTP; ligation proceeded to near completion at 1000 nm enzyme. However, 3′-phosphate ligase activity was stimulated 3-fold by 0.1 mm GTP (supplemental Fig. S3). By contrast, 0.1 mm GDP had little or no stimulatory effect (supplemental Fig. S3).
If the analogy between RtcB and classic ligases is valid, then one predicts that RtcB would react with GTP to form a covalent enzyme-guanylate intermediate (see below), in which case the observed GTP-independent 3′-phosphate sealing activity would be executed by a population of pre-formed RtcB-GMP (or RtcB·GTP) in the recombinant enzyme preparation. To explore this point further, we modified the purification protocol for recombinant RtcB in an effort to reduce or eliminate any residual nucleotide or metals. To wit: (i) during the second Ni-agarose step, the flow-through fraction containing tag-free RtcB was immediately adjusted to 20 mm EDTA and incubated for 1 h, and (ii) the EDTA-treated RtcB was then purified by gel filtration through a column of Superdex 200 in buffer containing 1 mm EDTA. This new RtcB enzyme preparation was strictly dependent on added manganese and either GTP or dGTP (0.1 mm) for its 3′-phosphate/5′-OH ligation activity (Fig. 8). ATP, CTP, and UTP (0.1 mm) were ineffective in lieu of GTP (Fig. 8).
FIGURE 8.
GTP requirement for 3′-phosphate ligation. Reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 8.0), 2 mm MnCl2 (where indicated by +), 0.1 μm 32P-labeled 20-mer HORNAp (shown at the bottom), 1 μm RtcB (where indicated by +), and either no added NTP (−) or 0.1 mm GTP, ATP, CTP, UTP, or dGTP as specified were incubated at 37 °C for 30 min. The reactions were quenched by adding 10 μl of 90% formamide, 50 mm EDTA. The products were analyzed directly by denaturing PAGE; the labeled RNAs were visualized by autoradiography. The positions of the linear substrates, the ligated circles, and multimeric ligation products are indicated on the left.
RtcB Reacts with GTP to Form a Covalent RtcB-guanylate Adduct
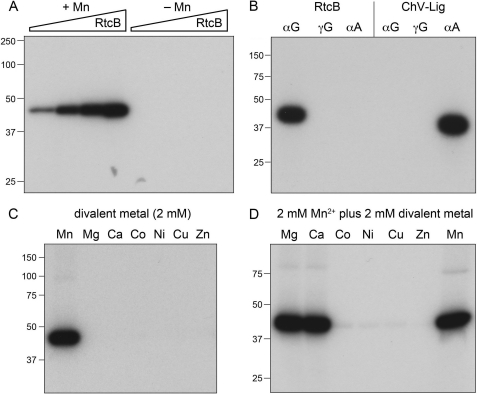
Incubation of RtcB with 10 μm [α-32P]GTP in the presence of 2 mm manganese resulted in the transfer of radiolabel to the RtcB polypeptide to form an SDS-resistant adduct that migrated as a single discrete species of ∼45 kDa during SDS-PAGE (Fig. 9C). The extent of label transfer was proportional to input RtcB concentration (Fig. 9A); ∼10–15% of the added RtcB protomers were 32P-labeled in vitro under these conditions. No label transfer from [α-32P]GTP to RtcB occurred in the absence of manganese (Fig. 9A).
FIGURE 9.
Reaction of RtcB with [α-32P]GTP to form a covalent RtcB-guanylate adduct. A, reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 8.0), 10 μm [α-32P]GTP, either 2 mm MnCl2 (+Mn) or no added divalent cation (−Mn), and increasing concentrations of RtcB (0.5, 1, 2.5, and 5 μm, proceeding from left to right in each titration series) were incubated at 37 °C for 10 min. B, reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 8.0), either 10 μm [α-32P]GTP, 10 μm [γ-32P]GTP, or 10 μm [α-32P]ATP as specified (at equivalent specific radioactivity), and either 5 μm RtcB plus 2 mm MnCl2 or 5 μm Chlorella virus DNA ligase (ChV-Lig) plus 5 mm MgCl2 were incubated at 37 °C for 10 min. C, reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 8.0), 10 μm [α-32P]GTP, 5 μm RtcB, and 2 mm of the indicated divalent cation (as the chloride salt) were incubated at 37 °C for 10 min. D, reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 8.0), 10 μm [α-32P]GTP, 5 μm RtcB, 2 mm MnCl2, and 2 mm of the indicated divalent cation (as the chloride salt) were incubated at 37 °C for 10 min. The reactions were quenched by adding 10 μl of a solution containing 100 mm Tris (pH 6.8), 10 mm EDTA, 200 mm DTT, 4% SDS, 20% glycerol, 0.2% bromphenol blue. The mixtures were then analyzed by SDS-PAGE. Autoradiographs of the dried gels are shown. The positions and sizes of pre-stained marker proteins are indicated on the left.
We found that no label was transferred to RtcB when the enzyme was reacted with 10 μm [γ-32P]GTP (Fig. 9B). Therefore we surmise that the radiolabeled species is most likely a covalent RtcB-[32P]GMP adduct. There was also no detectable label transfer from 10 μm [α-32P]ATP to RtcB (Fig. 9B). As a control for the labeling specificity experiment, we tested the [32P]NTPs for label transfer to Chlorella virus DNA ligase (ChV-Lig), a classic ATP-dependent DNA ligase (26) that reacted with 10 μm [α-32P]ATP to form a covalent ChV-Lig-[32P]AMP intermediate (Fig. 9B). As expected, there was no detectable label transfer to ChV-Lig from either 10 μm [α-32P]GTP or 10 μm [γ-32P]GTP (Fig. 9). (Note that the specific radioactivity of the GTP and ATP substrates used in these experiments was the same, allowing direct comparisons of the extents of label transfer.)
Manganese was the only divalent cation among the seven tested that was effective in supporting the RtcB guanylylation reaction (Fig. 9C). No label transfer to RtcB from [α-32P]GTP was seen when manganese was replaced by 2 mm magnesium, calcium, cobalt, nickel, copper, or zinc (Fig. 9C). We proceeded to conduct metal mixing experiments, in which RtcB guanylylation reactions containing 2 mm manganese were supplemented with 2 mm of another divalent cation. Cobalt, nickel, copper, and zinc virtually abolished RtcB guanylylation in the presence of manganese (Fig. 9D), echoing their inhibitory effects on the overall pathway of manganese-dependent 2′,3′>p/5′-OH RNA sealing (18). By contrast, magnesium and calcium had no effect on RtcB guanylylation in combination with manganese (Fig. 9D), just as they had no negative impact on the overall RNA repair pathway (18). These findings fortify our earlier inferences that the inhibitory “soft” metals (cobalt, nickel, copper, and zinc) out-compete manganese for the His/Cys-rich active site on the enzyme, wherein engaged they are unable to support reaction chemistry. The C78A active mutation that ablated RtcB ligase activity (see above) also abolished the reaction of RtcB with [α-32P]GTP to form the radiolabeled adduct (not shown). Thus, in multiple respects, including metal cofactor specificity, nucleotide specificity, and reliance on Cys-78 in the active site, the properties of the RtcB guanylylation reaction and the RtcB ligation reaction are concordant. Thus, we speculate that the RtcB-GMP complex is a likely intermediate in the 3′-phosphate ligation reaction.
Chemical Sensitivity of the RtcB-guanylate Adduct
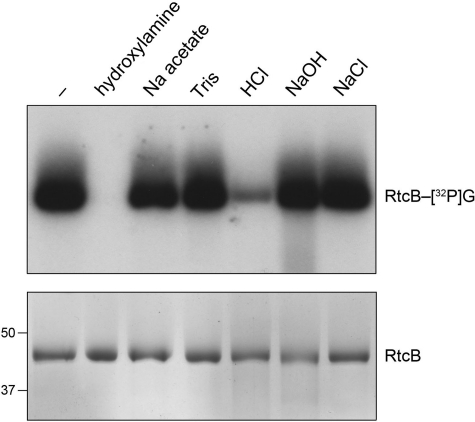
The nature of the linkage between RtcB and the 32P-guanylate was evaluated by chemical sensitivity tests (27), whereby the product of the label-transfer reaction was treated with EDTA and SDS and then exposed to acid, alkali, or hydroxylamine. The protein-guanylate adduct was stable to heating for 5 min at 75 °C in 0.15 m NaOH (Fig. 10). By contrast, 89% of the adduct was hydrolyzed by heating for 5 min at 75 °C in 0.15 m HCl, a treatment to which the RtcB polypeptide was itself stable (Fig. 10). The RtcB-[32P]guanylate adduct was eliminated by exposure for 30 min at 37 °C to 0.6 m hydroxylamine (pH 5.0) but was resistant to treatment in parallel with 0.6 m sodium acetate (pH 5.0) (Fig. 10). The chemical lability of the adduct to acid and hydroxylamine and its resistance to alkali are characteristic of a phosphoramidate bond (27). In the case of RtcB, where the active site is surmised from the crystal structure of the Pyrococcus homolog (20) and the mutational analysis inspired thereby (19), we suspect that the guanylate is attached to one of the essential histidine residues.
FIGURE 10.
Chemical stability of the RtcB-[32P]guanylate adduct. Reaction mixtures (140 μl) containing 50 mm Tris-HCl (pH 8.0), 10 μm [α-32P]GTP, 2 mm MnCl2, and 5 μm RtcB were incubated at 37 °C for 1 min. The mixtures were then adjusted to 0.5% SDS and 5 mm EDTA. Aliquots (20 μl) were withdrawn and either received no further additives (lane −) or were adjusted to 600 mm hydroxylamine (pH 5.0) or 600 mm sodium acetate (pH 5.0), and then incubated at 37 °C for 30 min. Additional aliquots (20 μl) were withdrawn and adjusted to either 150 mm Tris-HCl (pH 7.4), 150 mm HCl, 150 mm NaOH, or 150 mm NaCl; these samples were heated at 75 °C for 5 min; the HCl-treated sample was neutralized by addition of NaOH to 150 mm and the NaOH-treated sample was neutralized by adding HCl to 150 mm. The samples were supplemented with 20 μl of a solution containing 100 mm Tris (pH 6.8), 10 mm EDTA, 200 mm DTT, 4% SDS, 20% glycerol, 0.2% bromphenol blue. One-half of each sample was analyzed by SDS-PAGE, followed by autoradiography of the dried gel to visualize the RtcB-[32P]guanylate adduct (top panel). The other half of each sample was analyzed by SDS-PAGE, and the gel was stained with Coomassie Blue dye to visualize the RtcB polypeptide (bottom panel); the positions of 50- and 37-kDa pre-stained marker proteins are indicated on the left.
DISCUSSION
The present study establishes two facts that weight heavily against the one-step direct ligation mechanism depicted in Fig. 1A: (i) RtcB is adept at joining a 3′-monophosphate to a 5′-OH and (ii) RtcB has a CPDase activity that hydrolyzes a 2′,3′-cyclic phosphate end to a 3′-monophosphate. The available data favor the two-step pathway of RNA sealing depicted in Fig. 1B, in which the CPDase acts upstream of the 3′-phosphate ligase and both activities are intrinsic to RtcB. The CPDase of RtcB is envisioned as a straightforward in-line attack by water (or hydroxyl ion) on the cyclic phosphate apical to the ribose O2′ leaving group. This step is metal-dependent, with manganese being the optimal cofactor. The structure of archaeal RtcB suggests a binding site for one or more metals that is rich in essential histidines and an essential cysteine. The one or more metals could promote CPDase catalysis by coordinating the water/hydroxyl nucleophile, coordinating and promoting expulsion of the ribose 2′ oxyanion leaving group, and/or coordinating the non-bridging phosphate oxygens and stabilizing the transition state.
The 3′-phosphate ligation step of the RtcB pathway presents a challenge to phosphoryl transfer orthodoxy. It is unappealing on energetic grounds to suppose that the ligation reaction occurs by the direct attack of the terminal O5′ nucleophile on the 3′-monophosphate to yield a 3′,5′-phosphodiester, because such a mechanism calls for the expulsion of one of the non-bridging phosphate oxygens (as hydroxide or, eventually, water) while the bridging phosphomonoester linkage remains intact. Rather, it is more appealing, and less of a departure from classic ligases, to invoke a mechanism entailing RNA 3′ end activation via one or more high energy intermediates. Indeed, we found here that 3′-phosphate ligation by RtcB depends on GTP and that RtcB reacts with GTP in the absence of nucleic acid to from a covalent RtcB-guanylate adduct. These properties are reminiscent of the many classic polynucleotide ligases that act via covalent ligase-adenylate intermediates. Extending the analogy further, we speculate that RtcB-guanylate is the first of two activated ligation reaction intermediates, the second being RNA(3′)pp(5′)G generated by transfer of guanylate from RtcB to the RNA 3′-monophosphate terminus. Guanylylation of the RNA 3′-phosphate would activate it for attack by an RNA 5′-OH end to form the sealed phosphodiester and liberate GMP. This mechanism echoes how RNA cyclase enzymes transfer AMP from a covalent enzyme-adenylate intermediate to an RNA 3′-phosphate to form RNA(3′)pp(5′)A, thereby activating the 3′-phosphate for an attack by the vicinal ribose O2′ to yield a 2′,3′-cyclic phosphate and liberate AMP (24, 28). In this vein, we speculate that the apparent reversal of the CPDase reaction of RtcB (seen when RNA ligation is prevented by a 5′-phosphate) reflects the use of the ribose O2′ as a nucleophile (instead of an RNA 5′-OH end) for attack on RNA(3′)pp(5′)G.
Additional studies will be required to map the site of covalent RtcB guanylylation, validate the intermediacy of RtcB-GMP, and capture the putative (and presumably evanescent) guanylylated RNA intermediate. A complete mechanistic picture will ultimately hinge on obtaining cocrystal structures of RtcB in complexes with manganese, GTP, GMP, and a 3′-phosphate RNA substrate.
Finally, the two-step RtcB pathway in Fig. 1B has important implications for the spectrum of RNA repair reactions that RtcB might perform. Specifically, our findings indicate that the substrates for sealing by RtcB need not be limited to RNAs with 2′,3′>p ends. RNA-damaging ribotoxins and tRNA-splicing endonucleases characteristically generate 2′,3′>p termini as their end-products, and these broken RNAs have been considered the most plausible substrates for splicing or repair by RtcB ligases. However, many other ribonucleases generate 3′-monophosphates, which can now be considered potential RtcB substrates. Indeed, our finding of an intrinsic 3′-phosphate ligase activity in E. coli RtcB provides a plausible explanation for the results of Popow et al. (17), who partially purified a ligase complex containing mammalian RtcB from a HeLa cell extract based on its ability to join a 3′-monophosphate end to a 5′-OH end. These investigators documented by mass spectrometry the presence of human RtcB in the ligase preparation, but they did not detect the presence of the human RNA 3′-phosphate cyclase protein (Rtc1). Based on our findings, we speculate that human RtcB may also exploit a two-step reaction pathway.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM46330.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- AppRNA
- RNA-adenylate intermediate
- Pnkp
- T4 polynucleotide kinase-phosphatase
- AtRNL
- plant tRNA ligase
- CPDase
- 2′,3′-cyclic phosphodiesterase
- ChV-Lig
- Chlorella virus DNA ligase, 2′,3′ > p, 2′,3′ cyclic phosphate.
REFERENCES
- 1. Konarska M., Filipowicz W., Gross H. J. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 1474–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greer C. L., Peebles C. L., Gegenheimer P., Abelson J. (1983) Cell 32, 537–546 [DOI] [PubMed] [Google Scholar]
- 3. Amitsur M., Levitz R., Kaufmann G. (1987) EMBO J. 6, 2499–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sidrauski C., Cox J. S., Walter P. (1996) Cell 87, 405–413 [DOI] [PubMed] [Google Scholar]
- 5. Gonzalez T. N., Sidrauski C., Dörfler S., Walter P. (1999) EMBO J. 18, 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nandakumar J., Schwer B., Schaffrath R., Shuman S. (2008) Mol. Cell 31, 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schwer B., Sawaya R., Ho C. K., Shuman S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2788–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nandakumar J., Shuman S., Lima C. D. (2006) Cell 127, 71–84 [DOI] [PubMed] [Google Scholar]
- 9. Sawaya R., Schwer B., Shuman S. (2003) J. Biol. Chem. 278, 43928–43938 [DOI] [PubMed] [Google Scholar]
- 10. Wang L. K., Schwer B., Englert M., Beier H., Shuman S. (2006) Nucleic Acids Res. 34, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martins A., Shuman S. (2005) RNA 11, 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chan C. M., Zhou C., Huang R. H. (2009) Science 326, 247. [DOI] [PubMed] [Google Scholar]
- 13. Filipowicz W., Shatkin A. J. (1983) Cell 32, 547–557 [DOI] [PubMed] [Google Scholar]
- 14. Laski F. A., Fire A. Z., RajBhandary U. L., Sharp P. A. (1983) J. Biol. Chem. 258, 11974–11980 [PubMed] [Google Scholar]
- 15. Zofallova L., Guo Y., Gupta R. (2000) RNA 6, 1019–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Englert M., Sheppard K., Aslanian A., Yates J. R., 3rd, Söll D. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 1290–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Popow J., Englert M., Weitzer S., Schleiffer A., Mierzwa B., Mechtler K., Trowitzsch S., Will C. L., Lürhmann R., Söll D., Martinez J. (2011) Science 331, 760–764 [DOI] [PubMed] [Google Scholar]
- 18. Tanaka N., Shuman S. (2011) J. Biol. Chem. 286, 7727–7731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tanaka N., Meineke B., Shuman S. (2011) J. Biol. Chem. 286, 30253–30257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Okada C., Maegawa Y., Yao M., Tanaka I. (2006) Proteins 63, 1119–1122 [DOI] [PubMed] [Google Scholar]
- 21. Shuman S. (1998) Mol. Cell 1, 741–748 [DOI] [PubMed] [Google Scholar]
- 22. Wang L. K., Shuman S. (2002) Nucleic Acids Res. 30, 1073–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang L. K., Ho C. K., Pei Y., Shuman S. (2003) J. Biol. Chem. 278, 29454–29462 [DOI] [PubMed] [Google Scholar]
- 24. Tanaka N., Smith P., Shuman S. (2010) Structure 18, 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Genschik P., Drabikowski K., Filipowicz W. (1998) J. Biol. Chem. 273, 25516–25526 [DOI] [PubMed] [Google Scholar]
- 26. Nair P. A., Nandakumar J., Smith P., Odell M., Lima C. D., Shuman S. (2007) Nat. Struct. Mol. Biol. 14, 770–778 [DOI] [PubMed] [Google Scholar]
- 27. Gumport R. I., Lehman I. R. (1971) Proc. Natl. Acad. Sci. U.S.A. 68, 2559–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Filipowicz W., Strugala K., Konarska M., Shatkin A. J. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 1316–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.