Background: Maturation of urease is assisted by urease accessory proteins UreE, UreF, UreG, and UreH.
Results: Crystal structure of UreF-UreH complex revealed conformational changes of UreF upon complex formation.
Conclusion: Mutagenesis study confirmed that the conformational changes in UreF are essential for recruitment of UreG to the heterotrimeric complex of UreG-UreF-UreH.
Significance: Our results provide a structural basis for understanding urease maturation.
Keywords: Crystal Structure, Enzymes, Metalloenzymes, Metalloproteins, Protein Structure, Urease Accessory Proteins
Abstract
Colonization of Helicobacter pylori in the acidic environment of the human stomach depends on the neutralizing activity of urease. Activation of apo-urease involves carboxylation of lysine 219 and insertion of two nickel ions. In H. pylori, this maturation process involves four urease accessory proteins as follows: UreE, UreF, UreG, and UreH. It is postulated that the apo-urease interacts with UreF, UreG, and UreH to form a pre-activation complex that undergoes GTP-dependent activation of urease. The crystal structure of the UreF-UreH complex reveals conformational changes in two distinct regions of UreF upon complex formation. First, the flexible C-terminal residues of UreF become ordered, forming an extra helix α10 and a loop structure stabilized by hydrogen bonds involving Arg-250. Second, the first turn of helix α2 uncoils to expose a conserved residue, Tyr-48. Substitution of R250A or Y48A in UreF abolishes the formation of the heterotrimeric complex of UreG-UreF-UreH and abolishes urease maturation. Our results suggest that the C-terminal residues and helix α2 of UreF are essential for the recruitment of UreG for the formation of the pre-activation complex. The molecular mass of the UreF-UreH complex determined by static light scattering was 116 ± 2.3 kDa, which is consistent with the quaternary structure of a dimer of heterodimers observed in the crystal structure. Taking advantage of the unique 2-fold symmetry observed in both the crystal structures of H. pylori urease and the UreF-UreH complex, we proposed a topology model of the pre-activation complex for urease maturation.
Introduction
Chronic infection of Helicobacter pylori, a Gram-negative pathogenic bacterium that resides in the human stomach, can lead to stomach ulcer and cancer (1). Survival of H. pylori in the acidic environment of stomach requires the activity of urease that hydrolyzes urea into ammonia, which neutralizes the gastric acid (2, 3).
The structures of urease from various species have been determined (4–7), and the enzyme consists of α, β, and γ subunits. In H. pylori, the ureA gene encodes the β and γ subunits as a fusion protein, and the ureB gene encodes the α subunit (6). The active site of urease contains two nickel ions necessary for catalysis. To become enzymatically active, the apo-urease must undergo post-translational carboxylation of an active site lysine residue followed by insertion of nickel ions (3). In H. pylori, this activation process requires the synergy of four urease accessory proteins as follows: UreE, UreF, UreG, and UreH (8). UreH is the H. pylori ortholog of UreD found in other species (in this study we use the term “UreH(D)” when we refer in general to the homologous UreH or UreD proteins, and we use the term “UreH” when we refer specifically the protein in H. pylori).
How the urease accessory proteins activate urease maturation is only partially understood. UreF was reported to form a complex with UreH(D) (9–12), and the two proteins interact with UreG to form the heterotrimeric complex UreG-UreF-UreH(D) (12, 13). UreG is a SIMIBI (after signal recognition particle, MinD and BioD) class GTPase and is homologous to HypB, a hydrogenase maturation factor responsible for nickel delivery (14). Urease activation was inhibited by addition of the nonhydrolysable GTP analog, suggesting that GTPase activity of UreG is essential for urease activation (15). The apo-urease can form a complex with UreG-UreF-UreH(D) or its components of UreH(D) and UreF-UreH(D) (12, 13, 16, 17). It has been shown that apo-urease can be activated in vitro by adding an excess amount of both carbon dioxide and nickel ion (18). However, addition of nickel ion alone can also partially activate apo-urease in the absence of UreE, UreF, or UreG but not in the absence of UreH(D) (16). The formation of complex with UreF-UreH(D) and UreG-UreF-UreH(D) can increase the rate and the level of in vitro activation of urease (15, 17). Chemical cross-linking experiments suggest that binding of UreF-UreH(D) may induce conformational changes in the urease (19) so that nickel ion and carbon dioxide can access the active site to promote activation of urease (20). The current model proposes that the apo-urease interacts with UreF, UreH(D), and UreG to form a pre-activation complex. UreE, a dimeric nickel-binding protein, then interacts with UreG of the pre-activation complex and triggers the GTP-dependent activation of urease (15, 21, 22).
How UreF, UreH(D), and UreG interact with the apo-urease to assemble the pre-activation complex is poorly understood. Here, we have determined the crystal structure of the H. pylori UreF-UreH complex. By comparison with the crystal structure of a truncated form of H. pylori UreF (23) and by mutagenesis studies, we showed that binding of UreH induces conformational changes in UreF that are essential for the recruitment of UreG and in vivo maturation of urease. Based on the crystal structure of the UreF-UreH complex, a topology model of the pre-activation complex for urease maturation was proposed.
EXPERIMENTAL PROCEDURES
Construction of Expression Vectors and UreF Mutants
Sequences encoding UreF, UreG, and UreH were amplified from genomic DNA of H. pylori 26695 by PCR. The pRSF-ureH plasmid was created by cloning the coding sequence of UreH into the pRSFDuet vector (Novagen) using NdeI-XhoI restriction sites to create pRSF-ureH. UreG was cloned into pRSF using the same restriction sites to create pRSF-ureG. The coding sequence of UreF was cloned into an in-house pHisSUMO vector using the AgeI-EcoRI restriction sites to create pHisSUMO-ureF. The in-house pHisSUMO vector was constructed by inserting the coding sequence of HisSUMO tag into the pRSETA vector (Invitrogen). The construct was designed such that after removal of the HisSUMO2 tag by SENP1C digestion, UreF retains the native N-terminal residue. To create the expression vector for GST-tagged UreF (pGEX-ureF), the coding sequence of UreF was cloned into the pGEX-6P1 vector (GE Healthcare) using the BamHI-XhoI restriction sites. GST-ureF mutants (pHpA2H-ureF(ΔC20), pHpA2H-ureF(Y48A), and pHpA2H-ureF(R250A)) used in GST pulldown assay were constructed using an overlapping PCR technique. The pHpA2H used in urease activity assay was constructed by cloning a DNA fragment encoding the H. pylori urease operon, ureABIEFGH, into the pRSETA vector using the NdeI-EcoRI restriction sites. Mutations were introduced to the ureF gene in pHpA2H by cloning the DNA sequence containing the mutations using the SphI-BbvCI restriction sites to create the vectors pHpA2H-ureF(ΔC20), pHpA2H-ureF(Y48A), and pHpA2H-ureF(R250A). The pHpAB was constructed by cloning the DNA fragment encoding for the H. pylori urease (ureA and ureB) into the pRSFDuet vector using the NdeI-EcoRI restriction sites.
Protein Expression and Purification
UreF with an N-terminal HisSUMO tag (HisSUMO-UreF) was expressed in Escherichia coli strain BL21(DE3)pLysS (Novagen) in Luria Broth with appropriate antibiotics (100 μg/ml ampicillin and 50 μg/ml chloramphenicol). Bacterial cells were grown at 37 °C and induced with 0.4 mm isopropyl thiogalactopyranoside when the A600 reached 0.5. Cells induced overnight at 25 °C were harvested by centrifugation at 8000 × g at 4 °C for 5 min and were lysed by sonication in buffer A (20 mm Tris/HCl, pH 7.5, 500 mm NaCl, 1 mm 2,2′,2″-phosphanetriyltripropanoic acid, and 40 mm imidazole). After removal of cell debris by centrifugation, the supernatant was loaded onto a 5-ml HisTrap column (GE Healthcare) equilibrated with buffer A. After washing with 10 column volumes of buffer A, HisSUMO-UreF was eluted with 300 mm imidazole in buffer A. The HisSUMO tag on UreF was cleaved by a polyhistidine-tagged small ubiquitin-like modifier protease SENP1C. HisSUMO tag, and the small ubiquitin-like modifier protease was separated from UreF by loading the digestion mixture to a 5-ml HisTrap column equilibrated with buffer A. UreF, collected from the flow-through fraction, was further purified by size exclusion chromatography using HiLoad 26/60 Superdex 75 column (GE Healthcare) in buffer A without imidazole.
BL21(DE3)pLysS (Novagen) co-transformed with pGEX-ureF and pRSF-ureH was used to co-express GST-UreF and UreH following a similar procedure used for expressing HisSUMO-UreF. The GST-UreF-UreH complex was purified by loading the clear cell lysate onto a GST-Trap column (GE Healthcare) equilibrated with buffer B (20 mm Tris, pH 7.5, 200 mm NaCl, and 5 mm DTT). After washing with 10 column volumes of buffer B, the GST-UreF-UreH complex was eluted using 10 mm glutathione in buffer B. GST tag was separated from the complex using PreScission protease and a second pass through the GSTrap column. The UreF-UreH complex was further purified by size exclusion chromatography using HiLoad 26/60 Superdex 200 column (GE Healthcare) in buffer B.
Crystallization and Structure Determination
Both UreF and UreF-UreH complex were concentrated to 10 mg/ml for crystallization. Both native and selenomethionine derivative crystals of UreF were grown in 100 mm BisTris, pH 7.0, and 28% polyethylene glycol monomethyl ether 5000 at 16 °C. Crystals were cryoprotected by soaking in mother liquor containing 15% glycerol before flash freezing them for data collection. X-ray diffraction data for selenomethionine derivative crystals of UreF were collected at the selenium K-absorption edge using beamline I-04 at the Diamond Light Source. Diffraction data of native UreF crystals were collected using an in-house FR-E+ (Rigaku) rotating anode x-ray generator, then integrated, and scaled using MOSFLM and SCALA (24). Crystal structure of UreF was solved using multiwavelength anomalous dispersion phasing method. A total of 17 of 18 selenium sites were located in the asymmetric unit by HYSS (25). Phase calculation was performed using maximum likelihood based method by SOLVE (26) driven by PHENIX.AUTOSOL (27).
Crystals of the UreF-UreH complex were grown in 0.1 m MES, pH 6.0, 16% polyethylene glycol 4000, and 0.15 m ammonium sulfate. Crystals were cryoprotected with 15% glycerol. Diffraction data were collected using in-house FR-E+ (Rigaku) rotating anode x-ray generator, with integration and scaling performed by XDS (28). Solution to the phase problem was found by molecular replacement with PHASER (29) driven by PHENIX.AUTOMR (27), using the native crystal structure of UreF as a search model. The initial model obtained had a translation Z-score of 24.8 and Rfree of 0.549. After density modification with RESOLVE (30), clear electron density of UreH was found (supplemental Fig. S1).
Initial phases for structures of the UreF and UreF-UreH complex were improved using iterative cycles of statistical density modification, noncrystallographic averaging, and automated model building with PHENIX.AUTOBUILD (31). 91 and 88% of UreF and UreF-UreH complex were completed this way with the remaining structure completed by manual model building using COOT (32) and refined using PHENIX.REFINE (27). The final models were checked using MOLPROBITY (33), and diffraction precision index was calculated using SFCHECK (34).
GST Pulldown Assay
GST-UreF or its variants were co-expressed with UreH by co-transforming the plasmid pGEX-ureF and pRSF-ureH into E. coli. UreG was expressed by transforming the plasmid pRSF-ureG into E. coli. To detect interactions between UreF and UreH, bacterial cells co-expressing GST-UreF and UreH were lysed by sonication. To detect interaction between the UreF-UreH complex and UreG, bacterial cells co-expressing GST-UreF and UreH were mixed and co-sonicated with cells expressing UreG. After removal of cell debris by centrifugation, the supernatant was loaded to a GSTrap column (GE Healthcare) equilibrated with buffer B. After extensive washing with 10 column volumes of buffer B, the proteins were eluted with 10 mm glutathione in buffer B and analyzed by 12.5% SDS-PAGE.
Urease Activation Assay
E. coli was transformed with the wild type (pHpA2H), ureF mutants (pHpA2H-ureF(ΔC20), pHpA2H-ureF(Y48A), and pHpA2H-ureF(R250A)), or the control (pHpAB, pRSETA, and pRSFDuet) plasmids. The bacterial cells were cultured in Luria Broth supplemented with 0.5 mm nickel sulfate, 100 μg/ml ampicillin, and 50 μg/ml chloramphenicol. Cells were induced overnight with 0.4 mm isopropyl thiogalactopyranoside when A600 reached 0.5. 500 μl of overnight cell culture was collected and washed with 50 mm HEPES buffer, pH 7.0. Cells were lysed by sonication, and cell debris was removed by centrifugation. 250 μl of bacterial lysate was then mixed with 250 μl of assay buffer (50 mm HEPES, pH 7.5, and 50 mm urea) and incubated for 30 min at 37 °C. Urease activity was measured by the amount of ammonia released, which was quantified using a phenol/hypochlorite reaction (35). Total protein concentration of bacterial lysate was determined using the protein assay following the manufacturer's protocol (Bio-Rad). Specific activity of the measured sample was calculated as the amount of ammonia released per mg of total protein per min.
Analytical Gel Filtration/Static Light Scattering
After removal of the GST tag from the GST-UreF-UreH complex with PreScission protease (GE Healthcare), the UreF-UreH complex was purified by gel filtration. 100 μl of 3 mg/ml purified UreF or UreF-UreH complex was injected to a Superdex 200 analytical gel filtration column pre-equilibrated with phosphate-buffered saline at a flow rate of 0.5 ml/min. The column was connected downstream to a miniDawn light scattering detector and an Optilab DSP refractometer (Wyatt Technologies). The light scattering data were analyzed using the ASTRA software provided by the manufacturer to obtain the molecular weight of the eluted protein.
RESULTS
UreF Forms a Soluble Complex with UreH
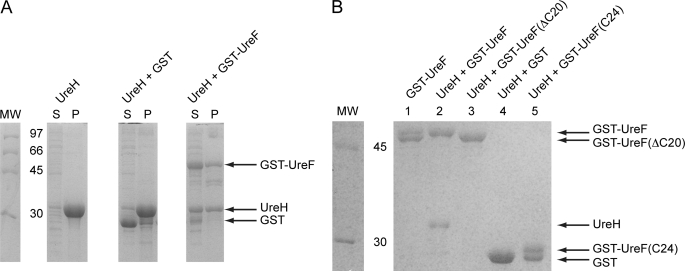
As shown in Fig. 1A, UreH forms insoluble inclusion bodies when expressed alone in E. coli. Based on yeast two-hybrid (10, 36) and tandem affinity purification (9), it has been shown that H. pylori UreF can interact with UreH. We questioned if UreF can solubilize UreH through complex formation. We therefore co-expressed GST or GST-UreF with UreH in E. coli. Bacterial cells were lysed by sonication and separated into soluble and insoluble fractions by centrifugation. We detected a significant portion of UreH in the soluble fraction when it was co-expressed with GST-UreF (Fig. 1A). In contrast, UreH remained largely insoluble when co-expressed with GST.
FIGURE 1.
Conserved C-terminal residues of UreF are essential for the formation of a soluble UreF-UreH complex. A, UreH was expressed alone and co-expressed with GST or GST-UreF. After cell lysis by sonication, the soluble fraction (S) and the pellet (P) were analyzed by SDS-12.5% PAGE with Coomassie Blue staining. UreH was found mainly as inclusion bodies in the pellet when expressed alone or co-expressed with GST. In contrast, a significant portion of UreH was found in the soluble fraction when co-expressed with GST-UreF. B, GST pulldown. UreH was co-expressed with GST, GST-UreF, or its variants. The soluble fraction of the bacterial lysate was loaded to a GSTrap column and eluted with 10 mm glutathione. The protein eluted was analyzed by SDS-12.5% PAGE with Coomassie Blue staining. As a control, bacteria lysate expressing GST-UreF alone was also subjected to similar procedures (lane 1). UreH was co-eluted with GST-UreF (lane 2), and no degradation of UreF was observed. However, UreH was not able to form a soluble complex with GST-UreF(ΔC20) (lane 3), GST (lane 4), and GST-UreF(C24) (lane 5). An additional band with lower molecular weight was observed for GST-UreF (lane 1) and GST-UreF(C24) (lane 5), indicating the presence of degradation.
Next, we attempted to detect the UreF/UreH interaction using a GST pulldown assay. Bacterial cell lysate of E. coli co-expressing GST-UreF and UreH was loaded onto a GSTrap column. After extensive washing, proteins bound to the column were eluted using glutathione. We found that UreH co-eluted with GST-UreF but not with GST (Fig. 1B, lanes 2 and 4). The results suggest that UreF can form a soluble complex with UreH.
C-terminal Residues of UreF Are Essential for UreH Interaction
In our GST pulldowns, we observed a partially degraded band for GST-UreF when the protein was expressed alone (Fig. 1B, lane 1). However, no such degradation was observed when UreF was in complex with UreH (Fig. 1B, lane 2). Mass spectrometry performed on purified UreF sample revealed a major peak at 26209.13 Da that corresponds to the truncated UreF residue 1–233 and a minor peak at 28688.68 Da that corresponds to the full-length UreF. In the previously reported selenomethionine structure of UreF (23), electron density for the residues at the C-terminal end (residue 234–254) was missing. In this study, we have independently determined the crystal structure of native UreF (supplemental Table S1) and observed similar truncation of C-terminal residues in the crystal structures. Taken together, these results suggest that the C-terminal residues were protected from degradation by interacting with UreH. Noteworthy, the C-terminal residues of UreF are highly conserved (supplemental Fig. S2). It is likely that the conserved C-terminal residues play an essential role in the formation of the UreF-UreH complex. To test this hypothesis, we created a UreF variant (UreF(ΔC20)), where the C-terminal residues 235–254 were truncated. GST-UreF(ΔC20) was co-expressed with UreH, and the cell lysate was loaded to a GSTrap column. Our data showed that UreF(ΔC20) failed to form a soluble complex with UreH (Fig. 1B, lane 3), suggesting that the conserved C-terminal residues of UreF is essential for UreF/UreH interaction.
To test if the C-terminal residues of UreF alone were enough to account for the formation of the UreF-UreH complex, we fused the residues 231–254 of UreF to the C terminus of GST GST-UreF(C24). Our data showed that GST-UreF(C24) failed to form a soluble complex with UreH (Fig. 1B, lane 5). Interestingly, a partially degraded band was observed for GST-UreF(C24), suggesting that the C-terminal residues were not protected from degradation. These results suggest that the C-terminal residues of UreF alone are not sufficient to form a soluble complex with UreH, and probably other residues of UreF are also involved.
Crystal Structure of UreF-UreH Complex
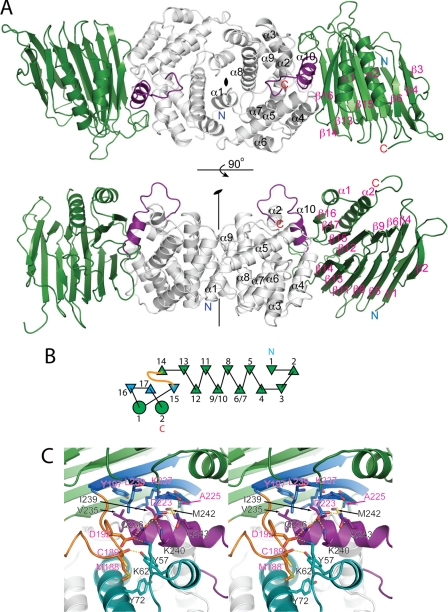
To better characterize the interaction between UreF and UreH, we crystallized the UreF-UreH complex and determined its structure. Solution to the phase problem was found using the structure of UreF as a molecular replacement model. Data collection and refinement statistics are summarized in Table 1. In the structure of the UreF-UreH complex, two copies of UreF and UreH are found in each asymmetric unit (Fig. 2A). The UreF/UreH interaction surface buries ∼2740 Å of solvent-accessible surface area. Around 12 and 11% of the surface area of UreF and UreH, respectively, is buried upon the formation of UreF-UreH complex.
TABLE 1.
Data collection and refinement statistics for structure determination of the UreF-UreH complex
Values in parentheses correspond to the highest resolution shell.
| Protein Data Bank code | 3SF5 |
| Space group | P212121 |
| Unit cell parameters | 70.6 Å, 70.7 Å, 205.5 Å |
| Wavelength | 1.54187 Å |
| Resolution | 50.0 to 2.50 Å (2.65 to 2.50 Å) |
| No. of unique reflections | 36,388 (5529) |
| Redundancy | 7.9 (7.3) |
| Completeness | 99.1% (95.0%) |
| Mosaicity | 0.25° |
| Average I/σ | 23.02 (7.30) |
| Rmerge | 0.057 (0.179) |
| Rwork | 0.173 (0.206) |
| Rfree | 0.217 (0.292) |
| Average B-factor | 31.8 Å2 |
| Root mean square bond lengths | 0.003 Å2 |
| Root mean square angles | 0.692° |
| Ramachandran analysis | Preferred 97.3%; allowed 99.8% |
| Diffraction precision index | 0.2558 Å |
FIGURE 2.
Crystal structure of H. pylori UreF-UreH complex. A, two views of the asymmetric unit differing by a 90° rotation around the horizontal axis. In each asymmetric unit, there are two copies of UreF (white) and UreH (green) related by a noncrystallographic 2-fold symmetry. Secondary structure elements of UreF (black), UreH (pink), N (blue), and C (red) termini are labeled. Residues 234–254 of UreF (purple), absent in the crystal structure of H. pylori UreF, is observed in this complex structure forming helix α10 (residue 236–243) and a structured loop (residue 244–254). B, topology diagram of UreH. Note that β4UreH to β13UreH folds with a β-helix-like topology. The loop region between β14UreH and β15UreH, which docks into the groove formed by α2UreF, α3UreF, and α10UreF, is colored in orange. β15UreH, β16UreH, and β17UreH, which make contact with α10UreF, are colored in blue. C, stereo diagram showing interaction surface between UreF (white) and UreH (green). Loop region formed by residues 179–194 (orange) of UreH docks into a groove formed by helix α2, α3 (cyan), and α11 (purple) of UreF. Helix α10UreF makes extensive contact with β15UreH, β16UreH, and β17UreH. Residues involved in hydrophobic interaction and hydrogen bond formation are labeled and shown in stick representation.
UreH represents a novel fold. A search of proteins with a structure homologous to UreH using DALI yielded SufD (Protein Data Bank code 1VH4), a chaperone protein related to iron-sulfur cluster formation (Z-score 6.4, root mean square deviation of 4.1 Å). However, closer examination showed that similarity lies only in the presence of the β-helix structure. Otherwise, there is little resemblance in terms of the protein fold.
The UreH fold consists of 17 β-strands and 2 α-helices located near the C terminus. The structure is dominated by two mixed strand β-sheets with β-strands 1, 2, 5, 8, 11, 13, and 14 forming β-sheet I and β-strands 3, 4, 6, 7, 9, 10, 12, and 15–17 forming β-sheet II (Fig. 2B). β-Strands 4–13 fold with a β-helix-like topology. The β-strands 6/7 and 9/10 are interrupted by having two proline residues (Pro-111 and Pro-113) between β-strand 9 and 10. Helix α1 and α2 are packed against anti-parallel β-strands 15–17 to form an α/β-like motif located at the C terminus of the β-helix like motif.
As β-sheet II on UreH is longer than β-sheet I, strands 15–17 are exposed and lack the hydrophobic contact of the complementary β-sheet. These strands instead interact with α10 (residue 236–243) of UreF. Val-235UreF, Ile-239UreF, and Met-242UreF on α10UreF make hydrophobic contacts with Tyr-197UreH, Ala-225UreH, Lys-237UreH, and Leu-239UreH located on the exposed β-strands of UreH. In addition, the carboxyl oxygen of Asp-223UreH forms hydrogen bonds with Gln-236UreF, Lys-240UreF, and Gln-243UreF. Gln-243UreF also forms hydrogen bond with the backbone carbonyl oxygen of Gly-224UreF (Fig. 2C).
On the structure of UreF, helices α2UreF, α3UreF, and α10UreF together form a surface groove accepting a loop connecting β14UreH and β15UreH (residues 178–195). Met-188UreH and Cys-189UreH on this loop region make hydrophobic contacts with Tyr-57UreF and Tyr-72UreF in the groove (Fig. 2C). The backbone carbonyl oxygen of Met-188UreH forms a hydrogen bond with the hydroxyl group of Tyr-57UreF (Fig. 2C). Asp-192UreH on this loop region of UreH also forms hydrogen bonds with Lys-62UreF and Gln-236UreF. The inter-molecular interactions between UreF and UreH are summarized in supplemental Table S2.
Conformational Changes of UreF Induced by UreH Binding Are Essential to Formation of UreG-UreF-UreH Complex
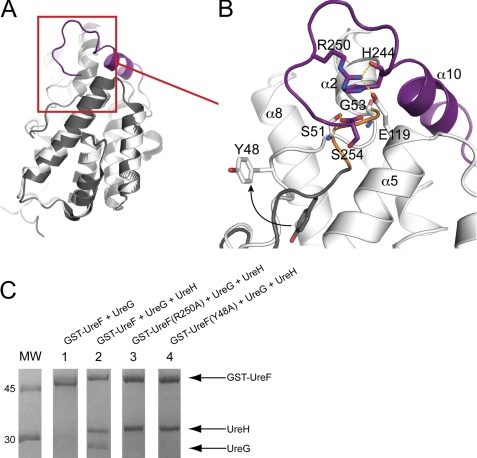
Generally speaking, the structures of UreF before and after complex formation with UreH are largely superimposable, with Cα root mean square deviation of 0.425 Å (Fig. 3A). The most prominent conformational changes involve the ordering of the C-terminal residues of UreF, forming helix α10UreF (residue 236–243), and a structured loop (residue 244–254) (Fig. 3B). These residues are missing in the crystal structure of native UreF but well defined in the crystal structure of the UreF-UreH complex (supplemental Fig. S3). As discussed above, the helix α10UreF is involved in interacting with UreH (Fig. 2C). The structured loop, we refer to as the F-tail loop, is stabilized by a number of interactions (Fig. 3B). Most prominently, the guanidinium group of a highly conserved Arg-250UreF forms hydrogen bonds with Glu-119UreF and backbone carbonyl oxygen of His-244UreF, thus acting like a staple that clips the F-tail loop into position. The formation of the helix α10UreF and the F-tail loop structure justifies the observation that these C-terminal residues of UreF were prone to degradation in the free form but were protected in the UreF-UreH complex (Fig. 1B).
FIGURE 3.
Conformational rearrangements of UreF upon binding with UreH. A, structure of UreF before (gray) and after (white) binding of UreH can be superimposed with a Cα root mean square deviation of 0.425 Å. B, close-up view of regions involved in conformational changes. Upon UreH binding, the C-terminal residues of UreF (purple) become ordered and form helix α10UreF (residues 236–243) and the F-tail loop (residues 244–254). Note that Arg-250 forms hydrogen bonds (dashed lines) with Glu-119 and His-244, stabilizing the F-tail loop. The first turn of helix α2UreF (orange, residue 50–53) uncoils upon complex formation with UreH, allowing the backbone amide groups of Ser-51 and Gly-53 to form hydrogen bonds with the carboxylate group of C-terminal residue Ser-254UreF. After the uncoiling, Tyr-48UreF moves to an exposed position as illustrated by the arrow. C, Y48A and R250A variants of UreF failed to form the heterotrimeric complex of UreG-UreF-UreH. To test if UreF alone can interact with UreG, bacterial cells expressing UreG was mixed with cells expressing GST-UreF and lysed by sonication. Soluble bacterial lysate was loaded onto a GSTrap column and eluted with 10 mm glutathione. The protein eluted was analyzed by SDS-12.5% PAGE with Coomassie Blue staining. No UreG was co-eluted (lane 1) indicating that UreG did not bind to GST-UreF. To test if UreF and its variants can form a heterotrimeric complex with UreH and UreG, cells expressing UreG were mixed with cells co-expressing GST-UreF and UreH instead. Wild-type GST-UreF was co-eluted with UreH and UreG (lane 2), indicating the formation of the heterotrimeric complex. However, GST-UreF(R250A) (lane 3) and GST-UreF(Y48A) (lane 4) were co-eluted only with UreH, suggesting that the variants failed to recruit UreG to form the heterotrimeric complex.
The second conformational changes in UreF involve the uncoiling of the first turn (residues 50–53) of helix α2UreF (Fig. 3B) so that the backbone amide groups of Ser-51 and Gly-53 can form hydrogen bonds with the carboxylate group at the C terminus (Ser-254UreF). As a result, a conserved residue Tyr-48UreF, which is buried in the free form, repositions to an exposed position upon binding of UreH (Fig. 3B).
It has been shown that UreG can interact with UreF-UreH(D) to form a heterotrimeric complex (12, 13, 15, 37). However, there is no evidence demonstrating that UreG can interact with UreF alone. Given that the residues involved in conformational changes in UreF are highly conserved (supplemental Fig. S2), it is likely that the conformational changes induced upon UreH binding are essential for the formation of the UreG-UreF-UreH heterotrimeric complex. To test this hypothesis, we first tested if H. pylori UreF, UreG, and UreH can form a heterotrimeric complex similar to their counterparts in Klebsiella aerogenes (12, 13, 15, 37). Bacterial cells expressing UreG were mixed with cells co-expressing GST-UreF and UreH or with cells expressing GST-UreF alone. Lysate cleared by centrifugation was loaded onto a GSTrap column. After extensive washing, protein was eluted off the column using glutathione. UreG was co-eluted with GST-UreF and UreH (Fig. 3C, lane 2) but not with the GST-UreF control (Fig. 3C, lane 1). This observation shows that UreG can interact with UreF-UreH complex but not with UreF alone.
As discussed above, UreH induces conformational changes in UreF in two distinct regions as follows: (i) the formation of the F-tail loop structure; and (ii) the uncoiling of the first turn of helix α2UreF. Next, we created two variants of UreF, GST-UreF(R250A) and GST-UreF(Y48A), to test if the conformational changes observed in UreF are essential for UreG binding. The R250A variant was designed to disrupt two hydrogen bonds that stabilize the F-tail loop (Fig. 3B). In addition, as the uncoiling of the helix α2UreF results in repositioning of Tyr-48UreF to an exposed position near the F-tail loop, the Y48A variant was created to test if this residue is involved in binding UreG. We then tested the ability of these UreF variants to interact with UreH and UreG using the GST pulldown assay as described above. We found that both GST-UreF(R250A) and GST-UreF(Y48A) variants were co-eluted with UreH, suggesting these substitutions did not affect UreF/UreH interaction (Fig. 3C, lanes 3 and 4). In contrast, both variants failed to co-elute with UreG, suggesting that the formation of the F-tail loop structure and Tyr-48UreF are essential for UreG binding.
Formation of UreG-UreF-UreH Complex Is Essential to Urease Maturation
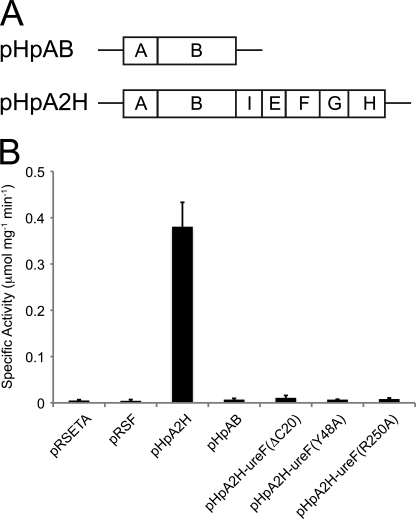
We further explored the functional implications of UreF/UreH and UreG/UreF/UreH interaction by studying the effects of various ureF mutations on urease activation. In vivo urease activation assay was performed by transforming the plasmid pHpA2H containing the entire H. pylori urease operon into E. coli (Fig. 4A). Urease activity was quantified by measuring the amount of ammonia produced. Mutations were introduced to the ureF gene on the plasmid pHpA2H to create pHpA2H-ureF(ΔC20), pHpA2H-ureF(Y48A), and pHpA2H-ureF(R250A). The plasmid pHpAB, which contains only the ureA and ureB genes, was also included as a negative control (Fig. 4A). Urease activity was detected in the case of pHpA2H but not in the case of pHpAB, suggesting that the urease was only activated in the presence of other wild-type urease accessory proteins (Fig. 4B). When the bacteria were transformed with pHpA2H-ureF(ΔC20), pHpA2H-ureF(Y48A), and pHpA2H-ureF(R250A), the urease activity was significantly decreased to the level similar to the negative control of pHpAB. These results suggest that the disruption of the UreG/UreF/UreH interaction abolishes urease maturation in vivo.
FIGURE 4.
Formation of UreG-UreF-UreH complex is essential for urease maturation. A, construction of vectors for urease activation assay. The H. pylori urease operon containing the genes for the urease (ureA and ureB) and urease accessory proteins (ureI, ureE, ureF, ureG, and ureH) were cloned into the pRSETA vector to create pHpA2H. Mutations were introduced to the ureF gene on pHpA2H to create pHpA2H-ureF(ΔC20), pHpA2H-ureF(Y48A), and pHpA2H-ureF(R250A). As a negative control, pHpAB was created by inserting only the urease structural genes (ureA and ureB) into the pRSFDuet vector. B, E. coli was transformed with the empty vector controls (pRSETA or pRSFDuet), pHpAB, pHpA2H, or the ureF mutants of pHpA2H. Urease activity of 0.38 ± 0.05 μmol of NH3/mg/min was detected for pHpA2H. In contrast, urease activity for the empty vector controls, pHpAB, and ureF mutants of pHpA2H-ureF were <0.01 μmol of NH3/mg/min. The urease activity in the bacterial lysate was determined in triplicate.
Oligomerization State of UreF and the UreF-UreH Complex in Solution
We noticed in the crystal structures of both the UreF and UreF-UreH complex that UreF appears as a dimer with essentially the same orientation. This suggests the possibility that the UreF and the UreF-UreH complex may indeed exist as dimers in solution. To determine the oligomeric states of UreF and the UreF-UreH complex in solution, we measured their molecular weights using size exclusion chromatography coupled with static light scattering. UreF was eluted as a single peak with a molecular mass of 43 ± 0.9 kDa (supplemental Fig. S4), which is in-between the molecular mass of monomeric (28.6 kDa) and dimeric (57.2 kDa) UreF. The data suggest that UreF does have a tendency to form dimers in solution, but the dimeric form is in exchange with the monomeric form in solution. In agreement with the oligomerization state observed in the crystal structure, the UreF-UreH complex was eluted as a single peak with a molecular mass of 116 ± 2.3 kDa, which agrees well with the theoretical molecular mass of 116.6 kDa for (UreF/UreH)2 (supplemental Fig. S4). Taken together with the quaternary structure observed in the crystal structure, our observation suggests that the UreF-UreH complex exists as a dimer of heterodimers in solution.
DISCUSSION
Urease maturation requires the formation of a pre-activation complex involving the binding of UreG, UreF, and UreH(D) to the apo-urease (15). In this study, we have determined the crystal structure of the H. pylori UreF-UreH complex. Upon complex formation, the C-terminal residues of UreF, which were missing in the crystal structure of UreF (23), became ordered and formed an extra helix α10UreF that makes extensive contacts with strand 15–17 of UreH (Fig. 2C). In the absence of UreF, the exposed hydrophobic residues on strand 15–17 of UreH may destabilize the protein or cause aggregation, leading to the formation of inclusion bodies (Fig. 1A). Mutagenesis study showed that these C-terminal residues of UreF play an essential role in the UreF/UreH interaction (Fig. 1). In agreement with our result, Kim et al. (38) also showed that truncation of the C-terminal residues of K. aerogenes UreEF fusion protein (corresponding to Lys-240 to Ser-254 in H. pylori UreF) abolished its ability to pull down urease and other urease accessory proteins and to activate urease in vivo.
Additional conformational changes in UreF induced by UreH are observed as follows: (i) the conserved residues at the C terminus of UreF form the F-tail loop structure, and (ii) the uncoiling of the first turn of helix α2 exposing the Tyr-48 (Fig. 3). Mutagenesis study showed that substituting the conserved residues Arg-250, which stabilizes the conformation of F-tail loop, and Tyr-48 with alanine breaks the interaction between UreG and the UreF-UreH complex and abolishes urease maturation in vivo. In addition, we showed that UreG only interacts with UreF in the presence of UreH (Fig. 3). This result is corroborated by the fact that previous yeast two-hybrid studies detected only UreF-UreH(D) and UreE/UreG interactions and not UreF/UreG interaction (10, 11). Taken together, we conclude that the conformational changes in UreF induced by UreH are required for the recruitment of UreG to form the heterotrimeric UreG-UreF-UreH(D) complex, which is essential for urease maturation (Fig. 5).
FIGURE 5.
Proposed molecular mechanism for the assembly of the UreG-UreF-UreH complex. In the absence of their interaction partners, UreH forms inclusion bodies, and the C-terminal residues of UreF are unstructured. Moreover, UreF alone does not bind to UreG. To bind UreG, UreF has to first form a complex with UreH. The formation of the UreF-UreH complex induces the formation of the F-tail loop structure and the repositioning of Tyr-48. These conformational changes allow the UreF/UreH to bind UreG to form the heterotrimeric complex of UreG-UreF-UreH.
Our results also suggest how UreG interacts with the UreF-UreH complex to form the heterotrimeric complex. Our mutagenesis study suggests that the F-tail loop and Tyr-48UreF are likely to be involved in UreG interaction. Interestingly, these residues of UreF are located around the 2-fold symmetry axis of the UreF/UreH structure and are highly conserved (Fig. 6). We suggest that they form a binding surface for UreG interaction (Fig. 6). Given that UreG belongs to the SIMIBI class of GTP-binding protein that can undergo nucleotide-dependent dimerization (39, 40), it is likely that UreG may also bind as dimer to the UreF-UreH complex.
FIGURE 6.
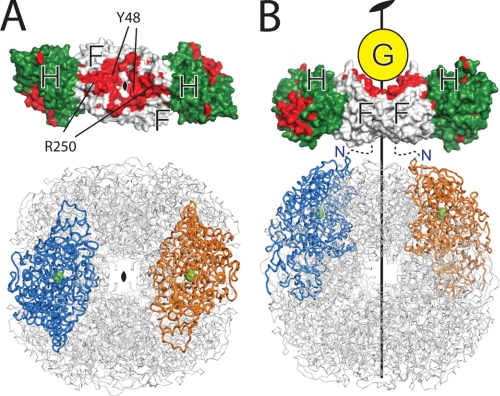
Topology model for the pre-activation complex for urease maturation. A, both the structures of the H. pylori UreF-UreH complex and the urease have a unique 2-fold symmetry. UreF (white) and UreH (green) are shown as a surface representation. Conserved residues (with conservation scores 8–9 determined by the CONSURF server (42)) of UreF and UreH are colored red. The urease is shown in ribbon representation, and the two symmetry-related subunits are colored blue and orange. The nickel ions at the active sites are represented as green spheres. Note that the length of the UreF-UreH complex is similar to the distance between two active sites related by a 2-fold symmetry. B, structures of the H. pylori UreF-UreH complex and the urease are aligned along their unique 2-fold symmetry axes. The C-terminal conserved residues and Tyr-48 are located around the 2-fold symmetry axes, and they constitute the UreG-binding site. We anticipate the urease should bind to the UreF-UreH complex on the opposite site of the UreG-binding site as indicated. The N termini of UreF are indicated.
How the UreG-UreF-UreH(D) complex interacts with urease to form the pre-activation complex for urease maturation is currently not known. Noteworthy, the N terminus of UreF, which is located on the opposite side of the UreG-binding surface (Fig. 6B), was shown to cross-link with the β-subunit of the urease in the K. aerogenes UreF-UreD-urease complex (19). Taken together, these observations suggest that the urease should bind to the UreF-UreH(D) complex on the opposite side of the UreG-binding site (Fig. 6B). We showed that the UreF-UreH complex possibly behaves as a dimer of heterodimers in solution (supplemental Fig. S4). If the oligomeric state of the UreF-UreH complex remains unchanged in the pre-activation complex, the 2-fold symmetry present in the UreF-UreH complex structure implies that the same symmetry should be present on its urease-binding site. Coincidentally, there is only one unique 2-fold symmetry present on the dodecameric structure of H. pylori urease (Fig. 6). We therefore reason that the only plausible interaction topology between the UreF-UreH complex and urease can be found by overlaying the 2-fold symmetry axes of these two structures as shown in Fig. 6B. In support of this idea, we observe that the length of the UreF-UreH complex approximates the distance between the urease active sites (Fig. 6). The conserved residues on UreH, which are likely for interacting with the urease, can be poised to be in contact with the urease active sites (Fig. 6).
UreG has a highly conserved nickel-binding motif shown by mutagenesis to be essential for nickel delivery (41). In the topology of the pre-activation complex we proposed (Fig. 6B), UreG is located at a significant distance away from the urease active site, suggesting that the nickel has to be transferred across a long distance to its final destination. One possibility is that the nickel is transferred from UreG via UreF-UreH(D) to the urease active site (12). Alternatively, chemical cross-linking analysis suggested that the urease β-domain undergoes a hinge-like motion upon binding of the UreF-UreH(D) complex (19). It is possible that the hinge-like motion may facilitate the transfer of nickel from UreG to the β-domain, which then delivers nickel into the active site.
Supplementary Material
Acknowledgments
We thank Carmen Lau for help with cloning, Ka Ming Lee for help with figure preparation, and Chi Kong Lau, Kwok Ho Chan, and Martin Rees for many helpful discussions. We also thank staff members of the Diamond Light Source beamline I-04, in particular, Michael Mrosek, for assistance with data collection and analysis. The equipment used in this study was supported in part by University Grants Council of Hong Kong One-Off Special Equipment Grant SEG CUHK08.
This work was supported by the Collaborative Research Fund of the Research Grants Council of Hong Kong Grant HKU 1/07C.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1 and S2.
The atomic coordinates and structure factors (code 3SF5) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- HisSUMO
- polyhistidine-small ubiquitin-like modifier
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-hydroxymethylpropane-1,3-diol.
REFERENCES
- 1. Warren J. B., Marshall B. (1983) Lancet 1, 1273–1275 [PubMed] [Google Scholar]
- 2. Mobley H. L., Island M. D., Hausinger R. P. (1995) Microbiol. Rev. 59, 451–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carter E. L., Flugga N., Boer J. L., Mulrooney S. B., Hausinger R. P. (2009) Metallomics 1, 207–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jabri E., Carr M. B., Hausinger R. P., Karplus P. A. (1995) Science 268, 998–1004 [PubMed] [Google Scholar]
- 5. Benini S., Rypniewski W. R., Wilson K. S., Miletti S., Ciurli S., Mangani S. (1999) Structure 7, 205–216 [DOI] [PubMed] [Google Scholar]
- 6. Ha N. C., Oh S. T., Sung J. Y., Cha K. A., Lee M. H., Oh B. H. (2001) Nat. Struct. Biol. 8, 505–509 [DOI] [PubMed] [Google Scholar]
- 7. Balasubramanian A., Ponnuraj K. (2010) J. Mol. Biol. 400, 274–283 [DOI] [PubMed] [Google Scholar]
- 8. Maier R. J., Benoit S. L., Seshadri S. (2007) Biometals 20, 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stingl K., Schauer K., Ecobichon C., Labigne A., Lenormand P., Rousselle J. C., Namane A., de Reuse H. (2008) Mol. Cell. Proteomics 7, 2429–2441 [DOI] [PubMed] [Google Scholar]
- 10. Rain J. C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schächter V., Chemama Y., Labigne A., Legrain P. (2001) Nature 409, 211–215 [DOI] [PubMed] [Google Scholar]
- 11. Heimer S. R., Mobley H. L. (2001) J. Bacteriol. 183, 1423–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carter E. L., Hausinger R. P. (2010) J. Bacteriol. 192, 2294–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moncrief M. B., Hausinger R. P. (1997) J. Bacteriol. 179, 4081–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Salomone-Stagni M., Zambelli B., Musiani F., Ciurli S. (2007) Proteins 68, 749–761 [DOI] [PubMed] [Google Scholar]
- 15. Soriano A., Hausinger R. P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11140–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park I. S., Carr M. B., Hausinger R. P. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 3233–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moncrief M. B., Hausinger R. P. (1996) J. Bacteriol. 178, 5417–5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park I. S., Hausinger R. P. (1995) Science 267, 1156–1158 [DOI] [PubMed] [Google Scholar]
- 19. Chang Z., Kuchar J., Hausinger R. P. (2004) J. Biol. Chem. 279, 15305–15313 [DOI] [PubMed] [Google Scholar]
- 20. Quiroz-Valenzuela S., Sukuru S. C., Hausinger R. P., Kuhn L. A., Heller W. T. (2008) Arch. Biochem. Biophys. 480, 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Soriano A., Colpas G. J., Hausinger R. P. (2000) Biochemistry 39, 12435–12440 [DOI] [PubMed] [Google Scholar]
- 22. Bellucci M., Zambelli B., Musiani F., Turano P., Ciurli S. (2009) Biochem. J. 422, 91–100 [DOI] [PubMed] [Google Scholar]
- 23. Lam R., Romanov V., Johns K., Battaile K. P., Wu-Brown J., Guthrie J. L., Hausinger R. P., Pai E. F., Chirgadze N. Y. (2010) Proteins 78, 2839–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 25. Grosse-Kunstleve R. W., Adams P. D. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1966–1973 [DOI] [PubMed] [Google Scholar]
- 26. Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kabsch W. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Terwilliger T. C. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., Adams P. D. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vaguine A. A., Richelle J., Wodak S. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 191–205 [DOI] [PubMed] [Google Scholar]
- 35. Weatherburn M. W. (1967) Anal. Chem. 39, 971–974 [Google Scholar]
- 36. Voland P., Weeks D. L., Marcus E. A., Prinz C., Sachs G., Scott D. (2003) Am. J. Physiol. Gastrointest. Liver Physiol. 284, G96–G106 [DOI] [PubMed] [Google Scholar]
- 37. Park I. S., Hausinger R. P. (1995) J. Bacteriol. 177, 1947–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim J. K., Mulrooney S. B., Hausinger R. P. (2006) J. Bacteriol. 188, 8413–8420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gasper R., Meyer S., Gotthardt K., Sirajuddin M., Wittinghofer A. (2009) Nat. Rev. Mol. Cell Biol. 10, 423–429 [DOI] [PubMed] [Google Scholar]
- 40. Gasper R., Scrima A., Wittinghofer A. (2006) J. Biol. Chem. 281, 27492–27502 [DOI] [PubMed] [Google Scholar]
- 41. Boer J. L., Quiroz-Valenzuela S., Anderson K. L., Hausinger R. P. (2010) Biochemistry 49, 5859–5869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. (2005) Nucleic Acids Res. 33, W299–W302 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.