Background: Microbial secreted α-xylosidases are rare.
Results: An α-xylosidase of Aspergillus niger (AxlA) was purified and expressed in Pichia pastoris.
Conclusion: Native and recombinant AxlA act on pNPαX and isoprimeverose. Together with β-glucosidase, AxlA degrades xyloglucan (XG) heptasaccharide. Together with xyloglucanase, β-galactosidase, and β-glucosidase, it degrades tamarind XG.
Significance: The biochemical function of AxlA is established.
Keywords: Cell Wall, Enzyme Catalysis, Enzyme Purification, Glycoside Hydrolases, Polysaccharide, Biomass, Cellulase, Hemicellulose, Xyloglucan
Abstract
α-Linked xylose is a major component of xyloglucans in the cell walls of higher plants. An α-xylosidase (AxlA) was purified from a commercial enzyme preparation from Aspergillus niger, and the encoding gene was identified. The protein is a member of glycosyl hydrolase family 31. It was active on p-nitrophenyl-α-d-xyloside, isoprimeverose, xyloglucan heptasaccharide (XXXG), and tamarind xyloglucan. When expressed in Pichia pastoris, AxlA had activity comparable to the native enzyme on pNPαX and IP despite apparent hyperglycosylation. The pH optimum of AxlA was between 3.0 and 4.0. AxlA together with β-glucosidase depolymerized xyloglucan heptasaccharide. A combination of AxlA, β-glucosidase, xyloglucanase, and β-galactosidase in the optimal proportions of 51:5:19:25 or 59:5:11:25 could completely depolymerize tamarind XG to free Glc or Xyl, respectively. To the best of our knowledge, this is the first characterization of a secreted microbial α-xylosidase. Secreted α-xylosidases appear to be rare in nature, being absent from other tested commercial enzyme mixtures and from the genomes of most filamentous fungi.
Introduction
Microbial enzymes that catalyze the depolymerization of plant cell wall polysaccharides are important participants in the carbon cycle, recycling fixed carbon into free sugars that ultimately are metabolized to CO2. Cell wall-active microbial enzymes also have many actual or potential biotechnological applications. An emerging major application of microbial enzymes is for deconstruction of lignocellulosic materials such as corn stover for the release of fermentable sugars for ethanol production. The high cost of enzyme mixtures for this purpose is currently a significant barrier to the development of a viable lignocellulosic biofuel industry (1, 2). There is thus a need for more efficient enzyme mixtures that will permit lower, and hence less costly, enzyme loadings.
Effective enzyme mixtures for biomass deconstruction must have all of the catalytic activities necessary to cleave all of the numerous linkages found in plant cell walls. Many microorganisms that live in lignocellulose-rich environments secrete a large number and broad range of cell wall-active enzymes, including cellulases, hemicellulases, pectinases, and proteases. Commercial enzyme mixtures contain 25–150 proteins (3, 4). However, these preparations are not necessarily ideal in their range of catalytic activities or in their relative proportions, especially for diverse pretreatment/feedstock combinations and there is considerable scope for their improvement.
Among the ways in which better enzyme mixtures could be developed is to ensure that they have the necessary enzymes for any particular biomass being processed. Although all higher plant cell walls contain cellulose, different plant species and even different tissues within a plant can have quite different hemicellulose composition and proportions (5). Hemicelluloses that can constitute a major fraction of plant cell walls include xyloglucan, glucuronoarabinoxylan, mannan, galactan, arabinan, mixed-linked glucan, and glucuronoarabinoxylan (6). Collectively, these hemicelluloses contain a number of fermentable monosaccharides, including Glc, Xyl,2 Gal, Ara, Man, Fuc, rhamnose, and uronic acids. Many of these sugars are also found in pectins and wall proteins such as extensins and arabinogalactan proteins.
The major hemicellulose in the primary walls of herbaceous dicotyledons is xyloglucan (XG), comprising a backbone of β1,4-Glc substituted with α1,6-linked Xyl, β-linked Gal, and in some plants, α-linked Fuc (5, 7). Although glucuronoarabinoxylan is a more prominent component than XG of the hemicellulose of cereals (Poaceae family), these species also contain XG. The XG of cereals tends to have a lower degree of Xyl substitution and no Gal or Fuc (7, 8). Considering that a significant fraction of the metabolizable sugars in plant cell walls are in XG, an efficient mixture for biomass deconstruction should have the full range and proper proportions of enzymes needed for its degradation.
The complete deconstruction of XG requires the participation of multiple enzymes. α-Fucosidase is necessary for removal of terminal Fuc in those hemicelluloses that contain it; β-galactosidase removes the penultimate Gal, α-xylosidase (AX) removes the α1,6-linked Xyl residues, and β1,4-glucanases together with β-glucosidase depolymerize the glucan backbone. Some β1,4-glucanases are xyloglucanases, i.e. they can hydrolyze β1,4-glucan linkages in substituted glucans such as XG, whereas other β1,4-glucanases act only on unsubstituted β1,4-glucans such as cellulose (9).
Of the aforementioned enzymes, secreted fungal and bacterial examples of most are known and have been well studied. AX, on the other hand, represents somewhat of an anomaly in our understanding of cell wall active depolymerases, because relatively few microbial AX enzymes have been described. All of the enzymes belong to glycosyl hydrolase family 31 (CAZy database), which also includes enzymes with a number of other activities, especially α-glucosidase (10). Substrates used to assay AX enzymes include p-nitrophenyl-α-xyloside (pNPαX), isoprimeverose (IP), and XG oligo- and polysaccharides. AX enzymes in plants are involved in mobilization of seed storage XG or remodeling of wall XG (11–13). The bacterium Lactobacillus pentosus has an IP utilization operon, which includes an IP transporter and a cytoplasmic AX (14). yicI of Escherichia coli, xylS of Sulfolobus solfataricus, and xyl31A of Cellvibrio japonicus have been demonstrated to encode AX enzymes (15–17). In regard to fungi, cytoplasmic AX enzymes have been characterized from Aspergillus flavus, A. niger, and Penicillium wortmanii, but the encoding genes apparently have not been identified (18–20). A gene (AN7505) encoding an AX from A. nidulans was identified by expression in Pichia pastoris, but it was tested only on pNPαX and was not biochemically characterized further (21).
A salient feature of the microbial AX enzymes studied to date is that there is no strong evidence that any of them are secreted free into the medium, as are most enzymes active on plant cell walls. Some AX enzymes, e.g. those of A. flavus, can be found in the extracellular medium after mycelial lysis has occurred (19). Xyl31A of C. japonicus is partially cytoplasmic and partially anchored to the outer cell wall (15). The cellular location of XylS of S. solfataricus has not been reported, but clustering of its encoding gene with a gene for a putative sugar transporter suggests that it is cytoplasmic, like the AX of L. pentosus (17). Consistent with a cytoplasmic location for most fungal AX enzymes, the majority of the numerous fungal proteins in GenBankTM annotated as belonging to GH31 lack predicted signal peptides. (None of these have been biochemically characterized.)
Secreted enzymes are more likely to survive in harsh and unstable extracellular environments and are therefore generally preferred to cytoplasmic proteins for biotechnological applications. Proteins with quaternary structure, such as the AX enzymes of A. flavus, are also likely to be less robust (20). In this work, we report the identification and characterization of a secreted AX from A. niger.
EXPERIMENTAL PROCEDURES
Fungal Strains, Enzymes, and Substrates
Aspergillus niger strain FGSC A1144 (ATCC 1015) was obtained from the Fungal Genetics Stock Center (Kansas City, MO), Trichoderma reesei (Hypocrea jecorina) strain QM9414 was obtained from the United States Department of Agriculture National Center for Agricultural Utilization Research (Peoria, IL), Fusarium graminearum (Gibberella zeae) strain PH-1 was obtained from Dr. L. P. Hart (Department of Plant Pathology, Michigan State University), and Phanerochaete chrysosporium strain RP-78 was obtained from Dr. D. Cullen (United States Department of Agriculture Forest Products Laboratory, Madison, WI). P. pastoris strain X-33 and plasmid pPicZB were obtained from Invitrogen.
Commercial enzyme preparations (Multifect Pectinase, Multifect Xylanase, Accellerase XY, Accellerase 1000, Accellerase 1500, and Stargen) were obtained from Dupont/Danisco, Inc. (Genencor Division (Rochester, NY)). CTec2 and HTec2 were obtained from Novozymes, Inc. (Franklinton, NC). Isoprimeverose (catalog no. O-IPRM), xyloglucan from tamarind (catalog no. P-XYGLN), and borohydride-reduced XG-derived heptasaccharide (catalog no. O-X3G4R) were purchased from Megazyme Intl. (Wicklow, Ireland). The monosaccharide composition of the XG heptasaccharide and the tamarind XG were reanalyzed by the alditol acetate method (22). For the XG heptasaccharide, total recovery of sugars was 101 ± 2% of the mass and the molar percent composition was 0.2% Ara, 43.2% Xyl, 0.8% Gal, and 55.8% Glc. This is very close to a 4:3 ratio of Xyl:Glc, which is consistent with the manufacturer's stated structure of XXXG, in the nomenclature of Fry et al. (23). Reanalysis of the tamarind XG indicated that it contains 2.3% Ara, 35.1% Xyl, 15.5% Gal, and 47.1% Glc, on a molar basis. This is in good agreement with the manufacturer's stated composition of 4% Ara, 38% Xyl, 16% Gal, and 42% Glc.
Enzyme Assays
p-Nitrophenyl-α-d-glucoside (pNPαG), p-nitrophenyl-α-d-xyloside (pNPαX), p-nitrophenyl-β-d-xyloside, and p-nitrophenyl-β-d-glucoside were purchased from Sigma. Enzyme reactions were performed in 96-well microtiter plates in a total volume of 0.2 ml and absorbances read on a SpectraMax Plus microplate reader (Molecular Devices, Sunnyvale, CA). The influence of pH on AX activity was measured at 37 °C in McIlvaine buffers adjusted to pH values from 2.5 to 7.5 (24). Free Glc and Xyl were measured colorimetrically using enzyme-linked assays in 96-well plates (25). Enzyme kinetics were analyzed by nonlinear curve fitting using GraphPad PrismTM software (La Jolla, CA).
Purification of AX
A column of DEAE-cellulose (Sigma D0909), 3-ml bed volume in a 5-ml syringe, was equilibrated with 25 mm sodium acetate, pH 4.0, and 1 ml of Multifect Pectinase applied and eluted with 25 mm sodium acetate, pH 4.0. Active fractions were combined and loaded onto a cation exchange HPLC column (TSK-Gel SP-5PW, Tosoh Bioscience, Montgomeryville, PA), equilibrated in the same buffer, and eluted with a gradient of 0–0.6 m NaCl in 30 min at a flow rate of 1 ml/min. Fractions containing AX activity were combined, and dry NH4SO4 was added to 1.7 m. This material was applied to a hydrophobic interaction column (TSK-gel Phenyl-5PW, Tosoh BioScience) equilibrated in 25 mm sodium acetate, pH 4.0 + 1.7 m NH4SO4. Proteins were eluted with a 30 min linear gradient to 100% water followed by 20 min of water at a flow rate of 1 ml/min. In some experiments, an additional fractionation step on hydroxyapatite CHT5–1 (10 × 64 mm, Bio-Rad) was included between the cation exchange and hydrophobic interaction steps. Elution conditions were 10 to 500 mm Na2HPO4, pH 7.0, in 30 min at 1 ml/min.
HPLC fractions were analyzed by SDS-PAGE (4–20% acrylamide, Tris-HCl, Bio-Rad). Proteins were visualized with ProtoBlue Safe (National Diagnostics, Atlanta, GA). Proteins were quantitated by the method of Bradford (26) using Bio-Rad protein assay reagent and bovine IgG as standard.
For mass spectrometric proteomics, proteins were excised from SDS-PAGE gels, digested with trypsin, and analyzed at the Michigan State University Proteomics Facility as described (3). For the proteomics analysis of Multifect Pectinase, 100 μg of protein were separated by SDS-PAGE, the gel was divided into four equal portions, and each was processed individually as described (3). The mass spectral data were analyzed using Scaffold software and the A. niger proteome as the query database (version 3.0, Department of Energy Joint Genome Institute, Walnut Creek, CA). Signal peptides were predicted using the SignalP server (version 4.0).
Expression in P. pastoris
A cDNA corresponding to Aspni5|43342 (Department of Energy Joint Genome Institute numbering) was synthesized by GeneArt (Invitrogen) with the addition of restriction sites for PmlI (5′ end) and XbaI (3′ end) and cloned into pPICZB (Invitrogen). P. pastoris was grown and induced as described (4), except with the addition of 1% Casamino acids (Difco Laboratories), which enhanced yield and stability of AxlA. Secretion was driven by the native signal peptide of AxlA. Twenty independent P. pastoris transformants were confirmed by colony PCR, purified by single colony isolation, and grown in 10-ml cultures. The three isolates exhibiting the highest activity on pNPαX were grown in 500-ml cultures and then concentrated and desalted (4). In some cases, AxlA was further purified by cation exchange HPLC as described above.
The expression of β-glucosidase (βG) from T. reesei (Trire2|76672) in P. pastoris was described previously (25). Xyloglucanase (also known as Cel74A; Trire2|49081) and β-galactosidase (Trire2|80240) from T. reesei were expressed by the same method (25). Both of these proteins are present in the secretome of T. reesei (3).
Digestion of XG Heptasaccharide
Each reaction contained 0.5 mg XG-derived heptasaccharide (Megazyme) in a reaction volume of 0.5 ml of sodium acetate (50 mm, pH 5.0). The AxlA and βG were produced in P. pastoris. The final total enzyme concentration was 30 μg/ml, and the reactions were run at 50 °C.
Digestion of Tamarind XG and Optimization with GENPLAT
For digestion of tamarind XG with commercial enzymes, the reaction volume was 0.5 ml, the total protein loading in each assay was 15 μg/g glucan, the reaction time was 24 h, and the reaction temperature was 50 °C.
The mixture optimization experiments with enzymes active on XG used Design ExpertTM software (State-Ease, Inc., Minneapolis, MN) and robotic handling of biomass and enzymes in an integrated platform called GENPLAT (4, 25). A four component quadratic model was used, which involved 15 reactions performed in duplicate. The four components were AX, βG, xyloglucanase, and β-galactosidase. The stock solution of tamarind XG was 2.5 mg/ml in 50 mm citrate buffer, pH 4.8, and the final concentration was 1 mg/ml in a volume of 500 μl. The total protein loading in each reaction was fixed at 15 μg. The reaction plates were incubated at 50 °C for 48 h with end-over-end mixing at 10 rpm, after which 200 μl was transferred to a fresh 96-well plate. Glc and Xyl were measured by enzyme-linked colorimetric assays (25).
RESULTS
Identification and Purification of AX
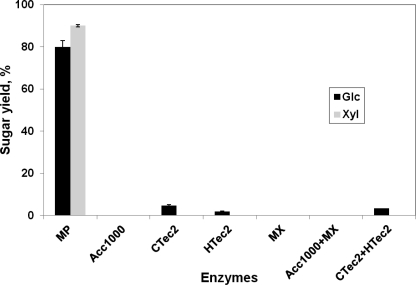
Several fungi grown on a variety of substrates were tested for AX activity. These included Cochliobolus carbonum, F. graminearum, T. reesei, A. niger, and P. chrysosporium. The fungi were grown on ground tamarind seed, solvent-washed corn (Zea mays) stover, solvent-washed pea (Pisum sativum) cell walls, solvent-washed carrot (Daucus carota) cell walls, lactose, or Xyl for 5–14 days in still culture. No activity against pNPαX was seen in any of the resulting culture filtrates. We also examined an assortment of commercial enzyme products, including Accellerase 1000, Accellerase XY, Multifect Xylanase, Multifect Pectinase, Novozyme 188, CTec2, and HTec2. Activity against pNPαX was not seen in any of them except Multifect Pectinase, which had a specific activity of 0.197 μmol/min/mg. Consistent with the presence of AX activity this preparation, and only in this preparation, it could degrade tamarind xyloglucan to free Xyl and Glc (Fig. 1). Among all of the commercial enzyme mixtures tested, Multifect Pectinase was also the only one that showed activity against IP.
FIGURE 1.
Release of free Glc and Xyl from tamarind XG by commercial enzyme mixtures as a percentage of the experimentally determined total Glc and Xyl. MP, Multifect Pectinase; Acc1000, Accellerase 1000; MX, Multifect Xylanase; Acc1000+MX, 50:50 mixture; CTec2+HTec2, 50:50 mixture.
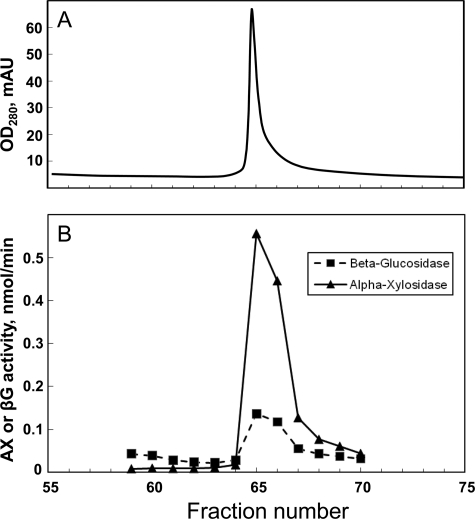
The protein responsible for AX activity was purified by HPLC, the final step of which is shown in Fig. 2. Through three high resolution purification stages, a low level of βG activity was consistently associated with the peak of AX activity (Fig. 2). The peak of AX activity did not contain any α-glucosidase or β-xylosidase activity as measured using pNPαG and p-nitrophenyl-β-d-xyloside, respectively. Later experiments (see below) indicated that the βG activity was probably due to co-purification of a separate enzyme. Their co-elution through multiple purification steps suggests that the two enzymes might form a complex in vivo. Although the secreted proteins of aerobic filamentous fungi are generally considered to be “noncomplexed,” evidence for the formation of complexes between the secreted enzymes of a filamentous fungus has been reported recently (27).
FIGURE 2.
Final purification step of α-xylosidase by hydrophobic interaction chromatography. A, UV trace. B, AX and βG activities against pNPαX and p-nitrophenyl-β-d-glucoside (pNPβG), respectively. mAU, milliabsorbance units.
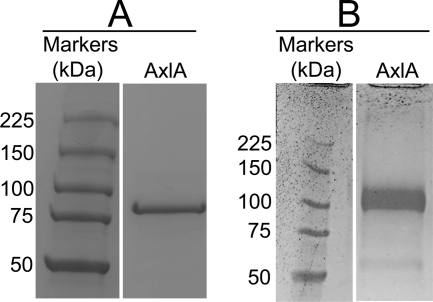
The molecular weight of AX by SDS-PAGE was ∼85 kDa (Fig. 3A). The dominant band was excised and subjected to tryptic digestion and mass spectrometric proteomics based on the whole predicted proteome of A. niger ATCC 1015 as the query database. (Multifect Pectinase is produced by fermentation of A. niger.) Eight unique peptides amounting to 16% coverage of Aspni5|43342 were detected at >95% probability. The only other protein detected, at a lower level, was Aspni5|50997 (two unique peptides, 6% coverage), which is a β-glucosidase in GH family 3. This might account for the residual βG activity co-eluting with AX (Fig. 2), a conclusion that was supported by heterologous expression (see below).
FIGURE 3.
SDS-PAGE of native AxlA (A) and AxlA expressed in Pichia pastoris (B). In each case, the standards and unknowns are from the same gel. The gels were stained with Coomassie Blue.
Unfractionated Multifect Pectinase was also analyzed by mass spectrometric proteomics. At high confidence (95% probability according to Scaffold, and at least two peptides), 132 proteins were identified (supplemental Table S1). More than 90% of the proteins have predicted signal peptides. Both Aspni5|43342 and Aspni5|50997 were detected (supplemental Table S1). Aspni5|56782, not Aspni5|50997, is the most abundant βG in Multifect Pectinase (supplemental Table S1).
Aspni5|43342 is a predicted protein in GH family 31, which includes known α-xylosidases. Alternate designations for this gene and its product are XP_001393647, An09g03300, and CAK40270. On the basis of its weak amino acid similarity to AN7055 of A. nidulans (see below) and its induction by growth on Xyl, Yuan et al. (28) hypothesized that this protein is an α-xylosidase and named it AxlA. Our results provide the first experimental evidence that Aspni5|43342 is, in fact, an α-xylosidase. To minimize confusion in the literature, we use the name AxlA.
By BLASTP against the GenBankTM nonredundant database, AxlA has many orthologs throughout the higher fungi (both ascomycetes and basidiomycetes). Many of these orthologs are annotated as belonging to GH family 31 and as having α-glucosidase or α-xylosidase activity, but with the exception of AN7505 of A. nidulans, there is no supporting biochemical evidence for any of these annotations (21). The top BLASTP hits (all with E-values of 0.0 and percent identities ranging from 52 to 81%) are from several species of Aspergillus, the closely related species Neosartorya fischeri, and two basidiomycetes (Schizophyllum commune XP_003031084 and Serpula lachrymans EGO01163) (supplemental Table S2).
Among species of Aspergillus, orthologs with strong E-values and percent amino acid identity to AxlA are present in A. flavus, Aspergillus oryzae, Aspergillus terreus, Aspergillus aculeatus, and Aspergillus carbonarius, but not A. fumigatus, A. clavatus, or A. nidulans (Aspergillus Comparative Database (Broad Institute) and DOE Joint Genome Institute) (supplemental Table S2). All of the orthologs in Aspergillus have strongly predicted signal peptides, like AxlA itself. (Reannotation of protein XP_002378848 from A. flavus by reassigning the ATG start codon indicates that it probably also has a signal peptide). AxlA is present in both sequenced strains of A. niger, ATCC 1015 and CBS 513.88, with 100% amino acid identity and 99% nucleotide identity in the coding region (29, 30).
A. nidulans has 10 predicted GH31 genes, five of which have signal peptides. Of these, AN7120 (XP_664724) has the best amino acid identity to AxlA (30%) but no signal peptide. A. niger ATCC 1015 and CBS 513.88 both have seven predicted GH31 genes, the best of which (after AxlA itself) being ANI_1_620014 (also known as Aspni5|55419), with 32% identity.
The AxlA mRNA and protein are induced by growth of A. niger on Xyl compared with maltose (27, 31, 32). AxlA was not included in a genome-wide microarray expresson study comparing A. nidulans, A. oryzae, and A. niger, presumably because it is not common to all three species (33).
After the orthologs in species of Aspergillus, the next best ∼20 hits to AxlA in GenBankTM, with E-values ranging from e-97 to e-23 and percent identities ranging from 22% to 52%, are to a much wider variety of fungi. All of these proteins are hypothetical, and it is not known whether they have AX, α-glucosidase, or other catalytic activities. However, the majority lack predicted signal peptides. This is a strong indication that they are not secreted and are probably functional orthologs of the cytoplasmic AX enzymes of A. flavus, A. niger, and P. wortmanii (18–20). Note that >90% of the proteins in Multifect Pectinase have predicted signal peptides (supplemental Table S1). The encoding genes of the cytoplasmic AX enzymes apparently have not been identified.
T. reesei has only two poor (E-value > e-10 and <25% amino acid identity) BLASTP hits to AxlA (Trire2|121351 and Trir2|69944), and neither of these has a predicted signal peptide. It appears that T. reesei does not have the genetic potential to biosynthesize a secreted AX related to AxlA, which therefore can account for the lack of this enzymatic activity in commercial enzyme mixtures derived from T. reesei (Fig. 1).
AN7505 (XP_680774) of A. nidulans has < 25% amino acid identity to AxlA and lacks a predicted signal peptide. When expressed in P. pastoris fused to a yeast signal peptide, AN7505 was secreted and showed activity against pNPαX but was not further characterized (21). It is therefore not fully established that AN7505 is an AX, and if so, whether its native cellular location is cytoplasmic or extracellular.
Properties of AxlA
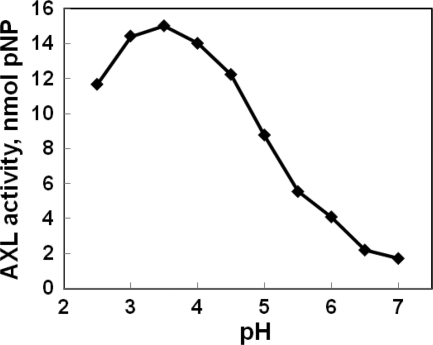
The pH optimum of AX on pNPX was between 3 and 4 (Fig. 4). The temperature optimum was 55 °C. The activity was ∼50% of maximum at 65° (supplemental Fig. S1). The native protein was approximately as active on IP as on the synthetic substrate pNPαX (Table 1).
FIGURE 4.
Dependence of AxlA activity on pH. Assays were performed with 10 mm pNPαX at 50 °C for 30 min.
TABLE 1.
AxlA enzyme kinetics
Nonlinear curve-fitting software (GraphPad Prism) was used to calculate the parameters and confidence intervals.
| Enzyme source | Substrate | Km | Km 95% confidence interval | kcat | kcat 95% confidence interval |
|---|---|---|---|---|---|
| mm | min−1 | ||||
| Native | pNPαX | 3.68 | 1.83–5.53 | 1393 | 1130–1656 |
| Native | IP | 9.78 | 7.73–11.84 | 917 | 834–999 |
| Pichia-expressed | pNPαX | 6.91 | 4.25–9.56 | 1234 | 1047–1420 |
| Pichia-expressed | IP | 4.03 | 3.36–4.71 | 1337 | 1265–1409 |
Heterologous Expression of AxlA
When expressed in P. pastoris, AxlA had an apparent molecular weight of ∼110,000, larger than the native protein. It also ran as a more diffuse band than the native protein (Fig. 3B). Both of these observations suggest that recombinant AxlA is hyperglycosylated when expressed in P. pastoris. (AxlA has 10 predicted N-glycosylation sites.) Nonetheless, the heterologously expressed protein showed kinetic properties similar to the native protein on both pNPαX and IP (Table 1). The protein expressed in P. pastoris had no detectable βG, β-xylosidase, or α-glucosidase activity when assayed with p-nitrophenyl-β-d-glucoside, p-nitrophenyl-β-d-xyloside, or pNPαG, respectively (data not shown). This supports the conclusion that the βG activity seen in the “purified” AX is due to contamination with another protein (Fig. 2).
Activity of AxlA on XG Heptasaccharide
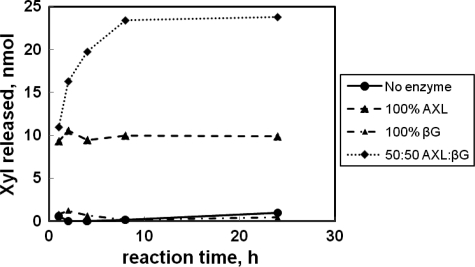
AxlA purified by HPLC (Fig. 2) by itself degraded the heptasaccharide XXXG to free Glc and Xyl (data not shown). However, this preparation contained residual βG activity (Fig. 2). Recombinant AxlA released ∼10 nmol of Xyl, and the quantity did not increase with time (Fig. 5). This proportion of Xyl corresponds to approximately one-third of the total Xyl present in the XG heptasaccharide sample (34 nmol in 12 μl of a reaction volume of 500 μl). This result is consistent with AxlA removing a single Xyl residue from the heptasaccharide to produce GXXG and is further evidence that AxlA does not have intrinsic βG activity. Digestion of the XG heptasaccharide with no enzyme or βG alone released no to little Xyl (Fig. 5). Digestion with a combination of AxlA and βG released 83.4% of the theoretical maximum of Xyl in 10 h (Fig. 5). Thus, βG and AX together are capable of substantially depolymerizing this heptasaccharide repeating unit of native XG.
FIGURE 5.
Digestion of XG heptasaccharide (XXXG) by AxlA, βG, or a 50:50 combination of the two. Concentration of the heptasaccharide was 1 mg/ml. Total protein concentration was 30 μg/ml.
Activity of AxlA on Tamarind XG
Because XG contains β-linked Gal and β-linked Glc in addition to α-linked Xyl, four enzymes were included in the experiment: xyloglucanase, β-glucosidase, and β-galactosidase, all from T. reesei, in addition to AxlA (Table 2). An optimized mixture of the four enzymes was developed using GENPLAT at fixed total protein loading (4, 25). In the first experiment (supplemental Table S3), the lower limit of each enzyme was set to 0%. However, because many combinations failed to yield >5% of Xyl or Glc, a statistically valid model could not be determined. In the second experiment (supplemental Table S4), the lower limit of each enzyme was set to 5%, which gave a statistically valid model for both Glc and Xyl (supplemental Table S5). Complete digestion of tamarind XG was achieved (supplemental Table S4). The optimized proportions of the four enzymes for Glc and Xyl release are shown in Table 2. The corresponding ternary diagrams are shown in supplemental Fig. S2. Of the four enzymes, AxlA was needed in the highest proportions (51% for Glc and 59% for Xyl). The need for a high proportion of AxlA might reflect a lower specific activity, steric hindrance, or the fact that the reactions were run at a suboptimal pH for AxlA (see Fig. 4).
TABLE 2.
Optimal proportions of four hemicellulases for release of Glc and Xyl from tamarind XG
The data on which the model is based are shown in supplemental Table S4 and the statistical validation in supplemental Table S5. The corresponding ternary diagrams are shown in supplemental Fig. S2. Total protein loading was 15 μg/g glucan.
| Product | Optimal enzyme proportions (%) |
Sugar yield | |||
|---|---|---|---|---|---|
| AxlA | Xyloglucanase | βG | β-Galactosidase | ||
| % | |||||
| Glc | 51 | 19 | 5 | 25 | 99 |
| Xyl | 59 | 11 | 5 | 25 | 100 |
DISCUSSION
An α-xylosidase from A. niger was purified and characterized. Evidence that it is secreted include 1) purification from a commercial enzyme preparation composed of the secreted proteins of A. niger, >90% of which have predicted signal peptides, 2) presence of a predicted signal peptide in AxlA itself, and 3) secretion of AxlA from P. pastoris under the control if its native signal peptide.
Previously reported AX enzymes from filamentous fungi are intracellular. Consistent with this, the large majority of proteins (all of which are hypothetical) annotated as being in glycosyl hydrolase family 31 lack predicted signal peptides. Protein AN7505 of A. nidulans is not closely related to AxlA (<25% amino acid identity), is not predicted to be secreted, and has not been characterized beyond showing activity on pNPαX (21). If AN7505 is actually an AX, its native cellular location is probably the cytoplasm, like most other known and presumed fungal AX enzymes. AN7505 needs further analysis to fully understand its catalytic activity, range of substrates, and cellular location.
Despite the abundance of α-linked Xyl in plant cell wall polysaccharides, there has been relatively little previous work on AX enzymes (34). Our analysis suggests that this likely because secreted AX enzymes are rather rare in the microbial world. The available data from both bacteria and fungi suggest that even though most lignocellulolytic microorganisms secrete enzymes that can degrade XG to IP, they transport and degrade IP intracellularly. The rarity of secreted AX enzymes in fungi is illustrated by the example of the commercial enzyme product known as Driselase, which comes from the basidiomycete Irpex lacteus. Although it contains dozens of cell wall-active enzymes, it lacks AX activity (35). This has made it a useful diagnostic tool for studying XG because treatment of plant cell walls with Driselase leads to complete depolymerization of XG only to the level of IP (35).
The hypothesis that secreted AX enzymes are rare among microorganisms is consistent with the preponderance of predicted GH31 proteins without signal peptides in the genomes of sequenced filamentous fungi and with the existence of IP utilization operons in bacteria such as L. pentosus (14). The best BLASTP hits of AxlA to the GenBankTM database are to AX enzymes that have signal peptides, but this is only a small subset of all of the putative fungal GH31 proteins. The orthologs of AxlA with signal peptides are from species of Aspergillus and several basidiomycetes. Species of Aspergillus have many additional predicted GH31 proteins without signal peptides.
AxlA has activity against pNPαX, IP, XG heptasaccharide, and tamarind XG. As a naturally secreted protein, it should be able to tolerate a variety of environmental conditions. It is therefore predicted to be a versatile AX that should find utility in biotechnological applications such as deconstruction of lignocellulosic materials for biofuels. Because herbaceous dicotyledonous plants contain higher amounts of XG than cereals, it might be particularly important for processing biomass from dicot species. AxlA has a pH optimum of <4.0, whereas most cellulase mixtures perform better at pH 4.5–5.0. Therefore, the utility of AX might be improved by finding or engineering an AX with a higher pH optimum.
Supplementary Material
Acknowledgment
We thank Cliff Foster (Great Lakes Bioenergy Research Center, Michigan State University) for polysaccharide analysis.
This work was supported by the U. S. Department of Energy Great Lakes Bioenergy Research Center (Department of Energy Office of Science BER DE-FC02-07ER64494) and by Grant DE-FG02-91ER200021 to the Michigan State University-Plant Research Laboratory from the U. S. Department of Energy, Office of Basic Energy Sciences, Division of Chemical Sciences, Geosciences, and Biosciences.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Figs. S1 and S2.
- Xyl
- xylose
- AX
- α-xylosidase
- βG
- β-glucosidase
- XG
- xyloglucan
- IP
- isoprimeverose
- Ara
- arabinose
- Fuc
- fucose
- pNPαX
- p-nitrophenyl-α-xyloside
- pNPαG
- p-nitrophenyl-α-glucoside
- GH
- glycosyl hydrolase.
REFERENCES
- 1. Banerjee G., Scott-Craig J. S., Walton J. D. (2010) Bioenergy Res. 3, 82–92 [Google Scholar]
- 2. Yang B., Dai Z., Ding S. Y., Wyman C. E. (2011) Biofuels 2, 421–450 [Google Scholar]
- 3. Nagendran S., Hallen-Adams H. E., Paper J. M., Aslam N., Walton J. D. (2009) Fung. Genet. Biol. 46, 427–435 [DOI] [PubMed] [Google Scholar]
- 4. Banerjee G., Car S., Scott-Craig J. S., Borrusch M. S., Bongers M., Walton J. D. (2010) Bioresour. Technol. 101, 9097–9105 [DOI] [PubMed] [Google Scholar]
- 5. Pauly M., Keegstra K. (2008) Plant J. 54, 559–568 [DOI] [PubMed] [Google Scholar]
- 6. Carpita N., McMann M. (2000) in Biochemistry and Molecular Biology of Plants (Buchanan B. B., Gruissem W., Jones R. L. eds) pp. 52–108, American Society of Plant Physiologists, Rockville, MD [Google Scholar]
- 7. Hsieh Y. S., Harris P. J. (2009) Mol. Plant 2, 943–965 [DOI] [PubMed] [Google Scholar]
- 8. Hayashi T. (1989) Annu. Rev. Plant Physiol. Plant Mol. Biol. 40, 139–168 [Google Scholar]
- 9. Grishutin S. G., Gusakov A. V., Markov A. V., Ustinov B. B., Semenova M. V., Sinitsyn A. P. (2004) Biochim. Biophys. Acta 1674, 268–281 [DOI] [PubMed] [Google Scholar]
- 10. Henrissat B., Davies G. J. (1997) Curr. Opin. Struct. Biol. 7, 637–644 [DOI] [PubMed] [Google Scholar]
- 11. Nakai H., Tanizawa S., Ito T., Kamiya K., Kim Y. M., Yamamoto T., Matsubara K., Sakai M., Sato H., Imbe T., Okuyama M., Mori H., Sano Y., Chiba S., Kimura A. (2007) J. Biochem. 142, 491–500 [DOI] [PubMed] [Google Scholar]
- 12. O'Neill R. A., Albersheim P., Darvill A. G. (1989) J. Biol. Chem. 264, 20430–20437 [PubMed] [Google Scholar]
- 13. Sampedro J., Sieiro C., Revilla G., González-Villa T., Zarra I. (2001) Plant Physiol. 126, 910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chaillou S., Lokman B. C., Leer R. J., Posthuma C., Postma P. W., Pouwels P. H. (1998) J. Bacteriol. 180, 2312–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larsbrink J., Izumi A., Ibatullin F. M., Nakhai A., Gilbert H. J., Davies G. J., Brumer H. (2011) Biochem. J. 436, 567–580 [DOI] [PubMed] [Google Scholar]
- 16. Okuyama M., Mori H., Chiba S., Kimura A. (2004) Protein Expr. Purif. 37, 170–179 [DOI] [PubMed] [Google Scholar]
- 17. Moracci M., Cobucci Ponzano B., Trincone A., Fusco S., De Rosa M., van Der Oost J., Sensen C. W., Charlebois R. L., Rossi M. (2000) J. Biol. Chem. 275, 22082–22089 [DOI] [PubMed] [Google Scholar]
- 18. Matsuo M., Seki T., Mitsuishi Y., Shoun H., Nakahara T. (1996) Biosci. Biotechnol. Biochem. 60, 341–343 [DOI] [PubMed] [Google Scholar]
- 19. Matsushita J., Kato Y., Matsuda K. (1987) Agric. Biol. Chem. 51, 2015–2016 [Google Scholar]
- 20. Yoshikawa K., Yamamoto K., Okada S. (1994) Biosci. Biotechnol. Biochem. 58, 1392–1398 [DOI] [PubMed] [Google Scholar]
- 21. Bauer S., Vasu P., Persson S., Mort A. J., Somerville C. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11417–11422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Foster C. E., Martin T. M., Pauly M. (2010) J. Vis. Exp. doi: 10.3791/1837 [DOI] [Google Scholar]
- 23. Fry S. C., York W. S., Albersheim P., Darvill A., Hayashi T., Joseleau. J. P., Kato Y., Lorences E. P., Maclachlan G. A., McNeil M., Mort A. J., Reid J. S., Seitz H. U., Selvendran R. R., Voragen A. G., White A. R. (1993) Physiol. Plant. 89, 1–3 [Google Scholar]
- 24. McIlvaine T. C. (1921) J. Biol. Chem. 49, 183–186 [Google Scholar]
- 25. Banerjee G., Car S., Scott-Craig J. S., Borrusch M., Aslam N., Walton J. D. (2010) Biotechnol. Bioengineer. 106, 707–720 [DOI] [PubMed] [Google Scholar]
- 26. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 27. Gonzalez-Vogel A., Eyzaguirre J., Oleas G., Callegari E., Navarrete M. (2011) Appl. Microbiol. Biotechnol. 89, 145–155 [DOI] [PubMed] [Google Scholar]
- 28. Yuan X. L., van der Kaaij R. M., van den Hondel C. A., Punt P. J., van der Maarel M. J., Dijkhuizen L., Ram A. F. (2008) Mol. Genet. Genomics 279, 545–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pel H. J., de Winde J. H., Archer D. B., Dyer P. S., Hofmann G., Schaap P. J., Turner G., de Vries R. P., Albang R., Albermann K., Andersen M. R., Bendtsen J. D., Benen J. A., van den, Berg M., Breestraat S., Caddick M. X., Contreras R., Cornell M., Coutinho P. M., Danchin E. G., Debets A. J., Dekker P., van Dijck P. W., van Dijk A., Dijkhuizen L., Driessen A. J., d'Enfert C., Geysens S., Goosen C., Groot G. S., de Groot P. W., Guillemette T., Henrissat B., Herweijer M., van den Hombergh J. P., van den Hondel C. A., van der Heijden R. T., van der Kaaij R. M., Klis F. M., Kools H. J., Kubicek C. P., van Kuyk P. A., Lauber J., Lu X., van der Maarel M. J., Meulenberg R., Menke H., Mortimer M. A., Nielsen J., Oliver S. G., Olsthoorn M., Pal K., van Peij N. N., Ram A. F., Rinas U, Roubos J. A., Sagt C. M., Schmoll M., Sun J., Ussery D., Varga J., Vervecken W., van de Vondervoort P. J., Wedler H., Wösten H. A., Zeng A. P., van Ooyen A. J., Visser J., Stam H. (2007) Nat. Biotechnol. 25, 221–231 [DOI] [PubMed] [Google Scholar]
- 30. Andersen M. R., Salazar M. P., Schaap P. J., van de Vondervoort P. J., Culley D., Thykaer J., Frisvad J. C., Nielsen K. F., Albang R., Albermann K., Berka R. M., Braus G. H., Braus-Stromeyer S. A., Corrochano L. M., Dai Z., van Dijck P. W., Hofmann G., Lasure L. L., Magnuson J. K., Menke H., Meijer M., Meijer S. L., Nielsen J. B., Nielsen M. L., van Ooyen A. J., Pel H. J., Poulsen L., Samson R. A., Stam H., Tsang A., van den Brink J. M., Atkins A., Aerts A., Shapiro H., Pangilinan J., Salamov A., Lou Y., Lindquist E., Lucas S., Grimwood J., Grigoriev I. V., Kubicek C. P., Martinez D., van Peij N. N., Roubos J. A., Nielsen J., Baker S. E. (2011) Genome Res. 21, 885–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jørgensen T. R., Goosen T., Hondel C. A., Ram A. F., Iversen J. J. (2009) BMC Genomics 10, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Oliveira J. M., van Passel M. W., Schaap P. J., de Graaff L. H. (2011) PLoS ONE 6, e20865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andersen M. R., Vongsangnak W., Panagiotou G., Salazar M. P., Lehmann L., Nielsen J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4387–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van den Brink J., de Vries R. P. (2011) Appl. Microbiol. Biotechnol. 91, 1477–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lorences E. P., Fry S. C. (1994) Carbohydr. Res. 263, 285–293 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.