Background: Listeria monocytogenes is an intracellular pathogen that invades the host cytoplasm.
Results: Cells deficient in TLR2 and NOD2 show defective autophagy of Listeria monocytogenes.
Conclusion: Autophagy of Listeria is dependent on ERK pathway that is perturbed in TLR2- and NOD2-deficient cells.
Significance: Our study establishes the role of innate immune receptors in autophagy of intracellular pathogens.
Keywords: Autophagy, ERK, Macrophages, Nod-like receptors (NLR), Toll-like receptors (TLR), Listeria monocytogenes
Abstract
Listeria monocytogenes is a facultative intracellular pathogen that invades both phagocytic and non-phagocytic cells. Recent studies have shown that L. monocytogenes infection activates the autophagy pathway. However, the innate immune receptors involved and the downstream signaling pathways remain unknown. Here, we show that macrophages deficient in the TLR2 and NOD/RIP2 pathway display defective autophagy induction in response to L. monocytogenes. Inefficient autophagy in Tlr2−/− and Nod2−/− macrophages led to a defect in bacteria colocalization with the autophagosomal marker GFP-LC3. Consequently, macrophages lacking TLR2 and NOD2 were found to be more susceptible to L. monocytogenes infection, as were the Rip2−/− mice. Tlr2−/− and Nod2−/− cells showed perturbed NF-κB and ERK signaling. However, autophagy against L. monocytogenes was dependent selectively on the ERK pathway. In agreement, wild-type cells treated with a pharmacological inhibitor of ERK or ERK-deficient cells displayed inefficient autophagy activation in response to L. monocytogenes. Accordingly, fewer bacteria were targeted to the autophagosomes and, consequently, higher bacterial growth was observed in cells deficient in the ERK signaling pathway. These findings thus demonstrate that TLR2 and NOD proteins, acting via the downstream ERK pathway, are crucial to autophagy activation and provide a mechanistic link between innate immune receptors and induction of autophagy against cytoplasm-invading microbes, such as L. monocytogenes.
Introduction
Listeria monocytogenes is a Gram-positive food-borne pathogen that causes listeriosis, a severe form of gastroenteritis with a possible nervous system infection, particularly in immunocompromised individuals, pregnant women, and neonates (1, 2). L. monocytogenes mediates its own uptake into phagocytic cells and non-phagocytic cells (e.g. enterocytes, hepatocytes, fibroblasts, and endothelial cells) via bacterial invasion factors called internalins (3, 4). Once inside the phagosome, a decrease in pH activates Listeria cytolysin listeriolysin O. Listeriolysin O then blocks phagolysosomal fusion and degrades the vacuolar membrane, leading to the escape of L. monocytogenes into the cytosol (5, 6). 90 min after infection, approximately 80% of the L. monocytogenes are observed in the cytosol (7). Entry into the cytoplasm is also assisted by phosphatidylinositol phospholipase C and phosphatidylcholine phospholipase C two bacterial phospholipases that hydrolyze host lipids to produce diacylglycerol and inositol phosphate, and ceramide, respectively, additionally playing a major role in subverting host cellular responses (8, 9). Three to five hours after infection, L. monocytogenes in the cytosol utilizes its ActA protein to polymerize host actin forming comet-like tail that propels bacterial movement and spread from cell to cell (10).
The innate immune response depends on pathogen recognition receptors for detection of pathogen-associated molecular patterns. These receptors include Toll-like receptors (TLRs),2 RIG-I-like receptors, and Nod-like receptors (NLRs) family of proteins (11). TLRs are transmembrane proteins for sensing extracellular pathogens whereas NLRs sense pathogen-associated molecular patterns in the cytosolic compartment. NLRs consist of more than 20 family members, including Nucleotide Oligomerization domain 1 (NOD1), NOD2, NLRP3, and NLRC4 (12–14). NOD1 is expressed ubiquitously, whereas NOD2 is expressed mainly in the myeloid cells such as macrophages and dendritic cells (DCs) (12). NOD1/NLRC1 and NOD2/NLRC2 recognize peptidoglycan components γ-d-glutamyl-meso-diaminopimelic acid and muramyl dipeptide, respectively (15, 16). L. monocytogenes activates a cytosolic surveillance system that results in the expression of interferon β-regulated genes. Furthermore, host defense against L. monocytogenes is mediated by the secretion of IFN- γ, TNFα, IL-1β, IL-6, IL-12, IL-18, CCL2, MIP2, CXCL1, and the coexpression of costimulatory molecules CD40, CD80, and CD86 on antigen-presenting cells (17, 18).
Autophagy is a highly conserved cellular catabolic process that removes damaged organelles and degrades long-lived proteins during periods of starvation, thereby playing a crucial role during cell survival and death (19–21). Autophagy also has an essential role in the innate defense mechanism, i.e. it eliminates cytoplasm-invading microbes by forming a double-layered membrane that wraps around the cytosolic bacteria so that it can be degraded via fusion with lysosomes (22–26). Autophagy was recently shown to be protective in elimination of bacterial pathogens (27–30). In the context of L. monocytogenes, earlier reports have shown that TLR2 and NOD proteins play a protective role during L. monocytogenes infection. TLR2 is required for macrophage activation (31–33). Similarly, the NOD1-NOD2/RIP2 pathway has been shown to be critical for host defense against L. monocytogenes in vitro and in vivo (34, 35). However, the role of extracellular TLRs and the cytosolic NOD proteins in autophagy of L. monocytogenes remain unknown. Here we show that the innate immune receptors TLR2 and NOD/RIP2 pathways activate autophagy via ERK activation, leading to degradation of L. monocytogenes within autophagosomes.
EXPERIMENTAL PROCEDURES
Reagents
All reagents were obtained from Sigma unless otherwise stated. The following antibodies were used: anti-LC3 from Novus Biologicals, anti-ERK, anti-phospho-pERK, anti-IκB, anti-pIκB (Cell Signaling Technology, Inc.), anti-actin, and anti-tubulin (Sigma). HRP-labeled anti-rabbit and anti-mouse antibodies were obtained from Jackson ImmunoResearch Laboratories, Inc. All fluorescently labeled secondary antibodies were obtained from Molecular Probes (Invitrogen). Rapamycin was obtained from LC Laboratories. NF-κB inhibitor SN50 (catalog no. 481480) and MEK inhibitor PD98059 (catalog no. 513000) were obtained from Calbiochem.
Mice and Macrophage Culture
Nod1−/−, Nod2−/−, Rip2−/−, Tlr2−/−, Tlr4−/− mice backcrossed to the C57BL/6 background for at least 10 generations have been described before (57, 58). All mice were housed in a pathogen-free facility. Bone marrow-derived macrophages were prepared from the femurs of 6- to 10-week-old male mice by using supernatant from L cells as differentiation medium.
Bacterial Strains and Infection of Macrophage/Dendritic Cells and Mice
Wild-type L. monocytogenes and isogenic mutants were grown in brain heart infusion medium at 37 °C overnight to mid-log phase for macrophage infections. Briefly, L. monocytogenes were washed twice with PBS and macrophages were infected for 30 min with a multiplicity of infection of 1:1 unless stated otherwise, and the medium was replaced with fresh medium. After 45 min of infection, gentamicin (10 μg/ml) was added to limit the growth of extracellular bacteria. Where mentioned, CQ (50 μm) was added 30 min after infection. Where mentioned, NF-κB inhibitor SN50 (15 μM) or MEK inhibitor PD98059 (50 μM) were preincubated with macrophages 1 h before infection and maintained during the course of the experiment.
For in vivo experiments, adult mice (6–10 weeks old) were used. For infection, wild-type and Rip2−/− mice were infected intraperitoneally with 3 × 105 L. monocytogenes. Bacterial loads in the liver and spleen were determined on day 3 after infection by serial dilution of the lysates on brain heart infusion medium.
Western Blotting
At different times after infection, cells were lysed in radioimmune precipitation assay lysis buffer supplemented with complete protease inhibitor mixture (Roche) and PhosSTOP (Roche). Lysates were resolved on SDS-PAGE and transferred to PVDF membranes by electroblotting. Membranes were blocked in 5% nonfat milk and incubated overnight with primary antibody at 4 °C and for 1 h with secondary HRP-tagged antibody at room temperature. The membranes were developed with ECL (Pierce).
Nucleofection, Immunofluorescence, and Transmission Electron Microscopy
Cells were resuspended in nucleofector buffer (Amaxa) at a density of 2 × 106 with 2 μg of GFP-LC3 plasmid. Cells were nucleoporated according to the manufacturer's instructions (Amaxa) and seeded onto glass coverslips. After 4 h, cells were infected with L. monocytogenes at a multiplicity of infection of 1:1. The cells were fixed at the desired time points with 4% paraformaldehyde and processed for immunoflourescence as described previously (59). Cells on coverslips were mounted on slides with ProLong® anti-gold antifade reagent (Invitrogen) and analyzed with a Nikon C1 confocal microscope with a ×40 objective lens using the manufacturer's original software. The images were processed later with the ImageJ program.
For transmission electron microscopy, cells were fixed in a solution of 2.5% glutaraldehyde in 0.1 m cacodylate buffer (pH 7.4). Cells were then embedded and sectioned for transmission electron microscopy by the Cell and Tissue Imaging Core Facility of St. Jude Children's Research Hospital.
Intracellular Growth Curves
Cells were grown on coverslips and infected with L. monocytogenes at a multiplicity of infection of 1:1 as described above. Intracellular growth curves of L. monocytogenes were generated as described previously (60).
Statistics
Quantifications of the number of autophagosomes per cell and LC3 - positive L. monocytogenes were performed by direct visualization on a Nikon C1 confocal microscope. For intracellular growth curves, at least three independent experiments were done in triplicate, and the means ± S.E. are reported. p values were calculated using the two-tailed Student's t test. Densitometry analysis of the immunoblots was performed by the ImageJ program. Bacterial counts of infected mice were analyzed by Mann-Whitney U test.
RESULTS
L. monocytogenes Infection Induces Autophagy in Bone Marrow-derived Macrophages
L. monocytogenes is an intracellular pathogen that escapes the phagosome and replicates in the host cytosol. Recent studies have suggested that cytoplasm-invading bacteria are targets for degradation by autophagy (36). L. monocytogenes was observed to induce autophagy and colocalized with autophagosomes in mouse embryonic fibroblasts and RAW 264.7 macrophages (27, 37). We first evaluated whether L. monocytogenes induces autophagy in bone marrow-derived macrophages by employing autophagy marker, LC3, as a readout (38). LC3 is present in mammalian cells as a cytoplasmic form (LC3-I) or in a membrane-associated form (LC3-II). LC3-I is lipidated by phosphatidylethanolamine (LC3-PE) to the LC3-II form that associates to the autophagosome membrane and migrates on SDS-PAGE with a downward shift in molecular weight.
First, we infected macrophages with L. monocytogenes for 0.5 h at 37 °C. At different times post-infection, protein lysates were collected. As shown in Fig. 1A, control cells showed only basal levels of autophagy. However, there was a time-dependent increase in LC3-II expression after infection with the pathogen. Using this antibody, LC3-I was barely detectable because of the lower sensitivity of the presently available anti-LC3 antibodies for LC3-I (39). Further, this response was not dependent on the bacterial dose because LC3-II expression increased equally well at a higher multiplicity of infection (Fig. 1A, right panel).
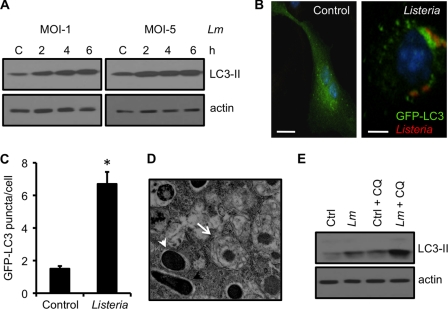
FIGURE 1.
L. monocytogenes infection induces autophagy in bone marrow-derived macrophages. A, Western blot analyses of LC3-II in macrophages either uninfected (C) or infected with L. monocytogenes (Lm) either at a multiplicity of infection of 1:1 or 5:1. Actin was used as a loading control. B and C, confocal microscope images of macrophages nucleofected with the GFP-LC3 plasmid and either uninfected (control) or infected with L. monocytogenes. At 4 h, cells were fixed and analyzed for the number of autophagosomes per cell as shown in C. Scale bars = 10 μm. Values represent mean ± S.E. *, p ≤ 0.05. D, cells infected as described above were fixed in 2.5% gluteraldehyde and processed for transmission electron microscopy. L. monocytogenes, either within an autophagosome (arrow) or free in the cytoplasm (arrowheads) are indicated. E, Western blot analyses of cells infected as described above. At 30 min, a set of control and L. monocytogenes-infected cells were incubated with chloroquine. The lysates were prepared 4 h after infection. Results are representative of three separate experiments.
As the processing of LC3-I to LC3-II increases, there is a corresponding increase in autophagosome formation (40). We next employed GFP-LC3 to observe this phenomenon. Uninfected macrophages transfected with GFP-LC3 showed diffuse cytoplasmic distribution of the protein (Fig. 1B, left panel). However, after infection with L. monocytogenes, an increase in autophagosomes in the form of GFP-LC3 puncta were observed (Fig. 1B, right panel) that was approximately 4 times more than in control cells (Fig. 1C). We also observed autophagy at the ultrastructure level by electron microscopy. As shown in Fig. 1D, autophagosomes showing double-membraned structures were observed after infection with L. monocytogenes.
LC3-II associates with both the outer and the inner membrane of autophagosomes. As autophagosomes mature by fusion with lysosomes, LC3-II present on the inner membrane of this organelle is also degraded (38, 39). An increase in LC3-II levels indicates either enhanced autophagosome formation or decreased turnover of LC3-II (or autophagic flux) caused by delayed fusion with lysosomes. To better interpret the increase in LC3-II levels after L. monocytogenes infection, we treated macrophages with chloroquine (CQ), a lysosomotropic agent that inhibits autophagosome fusion with lysosomes and therefore autophagic degradation. As expected, control cells treated with CQ showed an increase in the expression of the lipidated LC3-II form that showed a similar higher trend when infected macrophages were treated with CQ (Fig. 1E), suggesting the presence of an efficient autophagic flux.
TLR2 Is Required for Autophagy Activation upon L. monocytogenes Infection
Infection with L. monocytogenes led to autophagy induction. Thus, we sought to decipher the molecular sensor for host cells that triggers autophagy after L. monocytogenes infection. We first focused on the role of TLR2 in autophagy activation upon L. monocytogenes infection. Macrophages, either wild-type (WT) or Tlr2−/−, were infected with L. monocytogenes, and LC3 lipidation was evaluated by Western blotting (WB) 2 h and 4 h after infection. Cells deficient in TLR2 showed inefficient autophagy induction after infection with L. monocytogenes (Fig. 2A and B). However, a normal autophagic response was induced with rapamycin, a classical inducer of autophagy (Fig. 2A), thus indicating that autophagy induction only in response to L. monocytogenes is compromised in Tlr2−/− cells. TLR4 has no role in L. monocytogenes recognition. In agreement, Tlr4−/− macrophages showed LC3-II induction comparable with that of WT macrophages (Fig. 2, A and B). Further, autophagic flux in control or infected cells, measured after exposure to CQ, was found to be equally efficient (Fig. 2A).
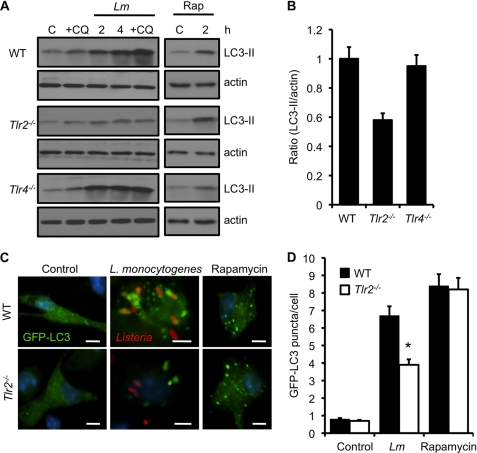
FIGURE 2.
TLR2 is required for autophagy activation upon L. monocytogenes infection. A, Western blot analyses of LC3-II in WT, Tlr2−/−, or Tlr4−/− macrophages that were uninfected (C), infected with L. monocytogenes (Lm) for different periods (in the presence or absence of CQ), or incubated with rapamycin (Rap) (50 μg/ml) for 2 h. Actin was used as a loading control. B, densitometry scanning of the blots showing the ratio of LC3-II to actin in wild-type, Tlr2−/−, or Tlr4−/− macrophages at 4 h. The graph is a representation of three different blots. C, confocal microscope images of WT or TLR2-deficient macrophages transfected with the GFP-LC3 plasmid and uninfected (control), infected with L. monocytogenes for 4 h, or treated with rapamycin (50 μg/ml) for 2 h. Scale bars = 10 μm. D, quantification of the number of GFP-LC3 puncta per cell 4 h after infection as described in C. Results show mean ± S.E. (*, p ≤ 0.05) of experiments done at least three times.
We next used GFP-LC3 to determine the formation of autophagosomes in WT and Tlr2−/− cells. Transfection of WT macrophages showed distinct GFP-LC3 punctate structures after infection with L. monocytogenes (Fig. 2C). Consistent with our WB data, the number of GFP-LC3 puncta were markedly reduced in TLR2-deficient cells infected with L. monocytogenes (Fig. 2, C and D). This was not because of reduced uptake, as it was found to be similar in wild-type and Tlr2−/− macrophages (supplemental Fig. S1A). The induction of autophagosome formation was normal when these cells were treated with rapamycin (Fig. 2, C and D). These data demonstrate that autophagy induction upon L. monocytogenes infection is dependent on TLR2 signaling.
NOD1 and NOD2 Are Required for Autophagy Activation upon L. monocytogenes Infection
Soon after its entry into a phagosome within a host cell, L. monocytogenes escapes into the cytosol, where it is recognized by intracellular receptors NOD1 and NOD2 (35, 41). To resolve whether any of these cytosolic receptors are required for activation of autophagy, we next infected Nod1−/− or Nod2−/− macrophages with L. monocytogenes and evaluated LC3 lipidation by WB at various times post-infection. Cells that were deficient in either NOD1 or NOD2 showed a marked reduction in autophagy induction. However, these cells showed a normal autophagic response to rapamycin (Fig. 3, A and B). Further, autophagic flux in control or infected cells, measured after exposure to CQ, was found to be equally efficient (Fig. 3A).
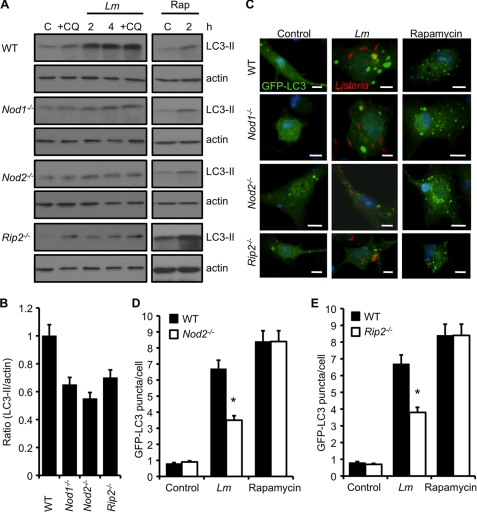
FIGURE 3.
NOD1 and NOD2 are required for autophagy activation upon L. monocytogenes infection. A, Western blot analyses of LC3-II in WT, Nod1−/−, Nod2−/−, or Rip2−/− macrophages that were uninfected (C), infected with L. monocytogenes (Lm) for different periods (in the presence or absence of CQ), or incubated with rapamycin (Rap) (50 μg/ml) for 2 h. Actin was used as a loading control. B, densitometry scanning of the blot showing the ratio of LC3-II to actin in wild-type, Nod1−/−, Nod2−/−, or Rip2−/− macrophages at 4 h. The graph is a representation of three different blots. C, confocal microscope images of WT, NOD1-, NOD2-, or RIP2-deficient macrophages transfected with the GFP-LC3 plasmid. Cells were uninfected, infected with L. monocytogenes for 4 h, or treated with rapamycin (50 μg/ml) for 2 h. Scale bars = 10 μm. D and E, quantification of the number of GFP-LC3 puncta per cell at 4 h post-infection in Nod2−/− macrophages or Rip2−/− macrophages (E) when infected as described in C. Results show mean ± S.E. (*, p ≤ 0.05) of experiments done at least three times.
We next infected GFP-LC3 transfected macrophages with L. monocytogenes. Consistent with our WB data, fewer GFP-LC3 puncta were seen in NOD1- or NOD2- deficient cells infected with L. monocytogenes as compared with WT cells (Fig. 3, C and D). This was not because of reduced uptake, as it was found to be similar in WT and Nod1−/− or Nod2−/− macrophages (supplemental Fig. S1A). The induction of autophagosome formation was normal when Nod1−/− or Nod2−/− cells were treated with rapamycin, indicating that the response is selective for L. monocytogenes infection (Fig. 3, C and D). These results suggest that both NOD1 and NOD2 are essential for induction of autophagy after infection of macrophages with L. monocytogenes.
NOD1 and NOD2 signaling pathway involves the downstream adaptor RIP2 kinase. Earlier reports have suggested contradictory results in regard to the role for RIP2 in autophagy induction by stimulation with NOD ligands (42, 43). To investigate whether RIP2 is required for autophagy of L. monocytogenes, Rip2−/− cells were infected with L. monocytogenes. Similar to Nod1−/− cells or Nod2−/− cells, Rip2−/− cells also showed defective LC3-II induction as compared with WT macrophages (Fig. 3, A and B). Further, uptake of L. monocytogenes was found to be similar both in WT and Rip2−/− macrophages (supplemental Fig. S1A). We also examined the formation of GFP-LC3 autophagosomes in Rip2−/− macrophages infected with L. monocytogenes. Fewer autophagosomes were observed in Rip2−/− cells, as indicated by fewer GFP-LC3 puncta (Fig. 3, C and E), suggesting that NOD1/NOD2-dependent autophagy of L. monocytogenes proceeds via RIP2.
Tlr2−/− and Nod2−/− Macrophages Sequester Fewer L. monocytogenes within Autophagosomes
To demonstrate the role of NOD2 in autophagy, we next investigated, at the ultrastructural level, autophagosome formation in WT and NOD2-deficient macrophages infected with L. monocytogenes (Fig. 4A). Four hours post-infection, the Nod2−/− cells contained half the number of autophagosomes per cell than did WT macrophages (Fig. 4B). To confirm this further, we analyzed the colocalization of Listeria with autophagosomes in macrophages that were WT, TLR2-, or NOD2-deficient by using confocal fluorescence microscopy. GFP-LC3 expressing macrophages were infected with L. monocytogenes, and the localization of the pathogen was determined using anti-Listeria antibody (Fig. 4C). After infection, approximately 25% of the pathogen colocalized with GFP-LC3 autophagosomes in WT cells, whereas in TLR2- and NOD2-deficient macrophages, approximately 16 and 15% of the L. monocytogenes were found in GFP-LC3-positive vacuoles, respectively, 4 h after infection (Fig. 4D). Similar results were observed in Rip2−/− cells (supplemental Fig. S2A). L. monocytogenes ActA protein is responsible for actin-based motility in the cytosol, and it has been shown recently that Listeria mutants lacking ActA are more efficiently targeted to autophagosomes (44). Evaluation of autophagy of the Listeria ActA mutant revealed that a remarkably higher percentage (37%) of the mutant Listeria is in autophagosomes in wild-type macrophages (Fig. 4E). However, Tlr2−/− and Nod2−/− macrophages showed only 18 and 16% of the mutant Listeria within autophagosomes, respectively (Fig. 4E). To directly test the role of TLR2 and NOD2 in limiting L. monocytogenes growth, we next compared the growth of the pathogen in WT macrophages with that in Tlr2−/− or Nod2−/− macrophages at various times after infection. A modest but statistically significant increase in colony forming units was observed in Tlr2−/− or Nod2−/− cells (Fig. 4F), suggesting that TLR2 and NOD2 limit the growth of Listeria by autophagy.
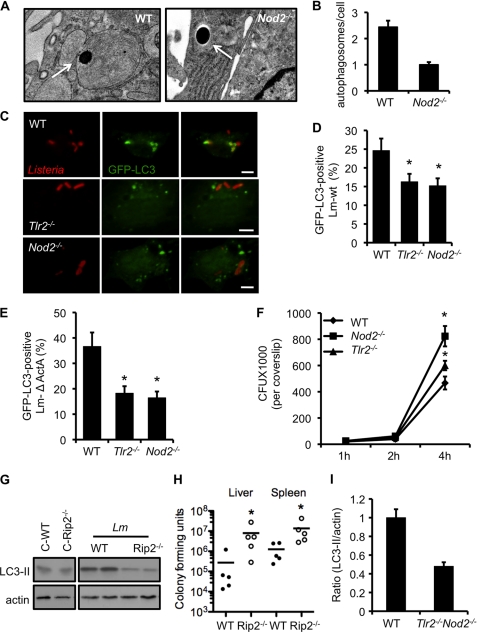
FIGURE 4.
Tlr2−/− and Nod2−/− macrophages sequester fewer L. monocytogenes within autophagosomes. A, electron micrograph of WT and NOD2-deficient macrophages infected with L. monocytogenes. Arrows denote L. monocytogenes either within the autophagosome (left panel) or free in the cytosol (right panel). B, quantification of the number of autophagosomes per cell in macrophages infected as described in A. C, confocal microscope images of GFP-LC3 autophagosomes and Listeria from WT, Tlr2−/−, and Nod2−/− cells 4 h after infection. Scale bars = 10 μm. D, quantification of the GFP-LC3-positive L. monocytogenes wild-type strain in cells infected as described in C. E, quantification of the GFP-LC3-positive Listeria mutant lacking ActA in cells infected as described in C. F, intracellular growth curve of L. monocytogenes in wild-type, Tlr2−/−, and Nod2−/− macrophages. *, p ≤ 0.05. G, wild-type and Rip2−/− mice were infected with 3 × 105 L. monocytogenes. The Western blot analysis shows LC3-II expression in liver homogenates from WT and Rip2−/−-infected mice at day 3 of infection. Actin was used as a loading control. H, wild-type and Rip2−/− mice were infected as described in F. Bacterial loads were determined in liver and spleen at day 3 after infection. Data represent the mean ± S.E. of three independent experiments. *, p ≤ 0.05. I, densitometry scanning of the Western blot analysis showing the ratio of LC3-II to actin in wild-type and Tlr2−/−Nod2−/− macrophages infected with L. monocytogenes at 4 h. The graph is a representation of three different blots.
Next, we asked whether autophagy could restrict the growth of the pathogen in vivo. To perform this, we infected WT and Rip2−/− mice with L. monocytogenes. Liver homogenates, collected 3 days after infection, were assayed for LC3 levels by SDS-PAGE. LC3-II levels in infected WT mice increased as compared with mice that were uninfected (Fig. 4G). However, Rip2−/− mice showed defective autophagy activation, as observed by reduced LC3-II levels in these samples (Fig. 4G). In accordance with reduced autophagy, Rip2−/− mice showed an increased bacterial burden in the liver and spleen (Fig. 4H). In agreement with the role for NOD1/2 adaptor RIP2, TLR2 adaptor MyD88-deficient mice also show increased Listeria growth in vivo (45).
Next, we tested how TLR2 and NOD2 cooperate in activating autophagy in response to L. monocytogenes infection. To test this, we made use of Tlr2−/−Nod2−/− double knockout macrophages. LC3-II levels increased after infection in wild-type cells. However, to our surprise, even the absence of both TLR2 and NOD2 did not abolish LC3-II levels completely (Fig. 4I and supplemental Fig. S2B), indicating the presence of other redundant pathways that are activated to compensate for autophagy induction in the absence of both TLR2 and NOD2 signaling.
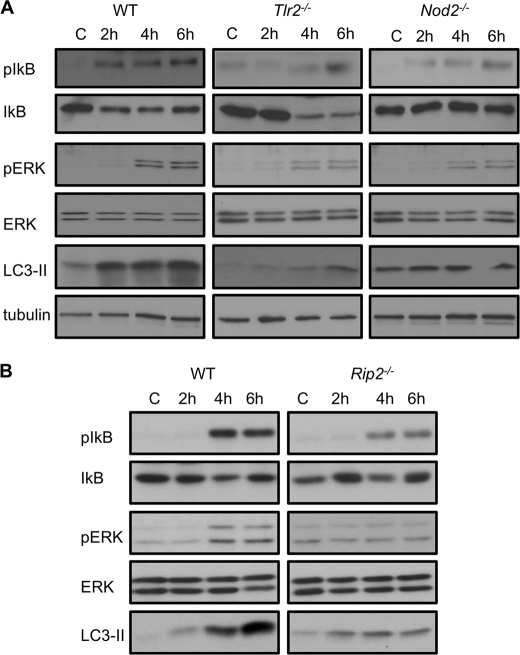
TLR2−/− and Nod2−/− Macrophages Show Defective NF-κB and ERK1/2 Signaling upon Listeria Infection
To determine the signaling mechanisms that regulate autophagy downstream of TLR2 and NOD2 in L. monocytogenes-infected cells, we prepared cell lysates from WT, Tlr2−/−, and Nod2−/− macrophages at various times post-infection and determined the levels of NF-κB and ERK1/2 activation by immunoblotting. At 2 h and later post-infection, we observed increased autophagy induction against L. monocytogenes in wild-type cells that was defective in Tlr2−/− and Nod2−/− cells (Fig. 5A, lower panel). Upon immunoblotting with specific antibodies, we found increased phosphorylation of Iκ-Bα and ERK1/2 in WT cells (Fig. 5A, upper panels). In contrast, these signaling pathways were perturbed in Tlr2−/− and Nod2−/− macrophages infected with L. monocytogenes (Fig. 5A). We also tested these signaling pathways in cells deficient in NOD2 adaptor protein RIP2. As we also demonstrated before (Figs. 3A and 5B, lower panel), Rip2−/− cells were inefficient in LC3-II induction as compared with wild-type cells. Similar to Tlr2−/− and Nod2−/−, cells deficient in RIP2 also displayed perturbed NF-κB and ERK signaling when exposed to L. monocytogenes (Fig. 5B, upper panels).
FIGURE 5.
TLR2−/− and Nod2−/− macrophages show defective NF-κB and ERK1/2 signaling upon Listeria infection. A, Western blots of pIκB, IκB, pERK, ERK, and LC3-II in WT, TLR2-, and NOD2-deficient macrophages that were uninfected (C) or infected with L. monocytogenes for different periods. Tubulin was used as a loading control. B, Western blot analyses of pIκB, IκB, pERK, ERK, and LC3-II in WT and RIP2-deficient macrophages that were uninfected (C) or infected with L. monocytogenes for different periods.
Autophagy of L. monocytogenes Is Dependent on the ERK Signaling Pathway
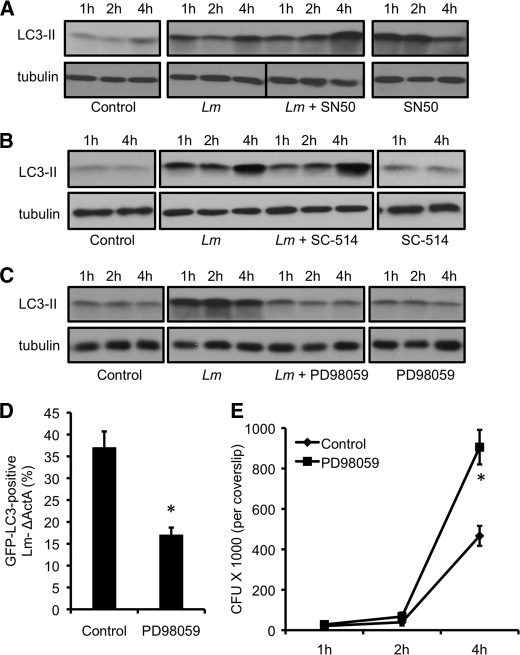
We next sought to test whether the NF-κB and ERK signaling pathways that are defective in Tlr2−/− and Nod2−/− macrophages (Fig. 5A) are directly associated with autophagy of L. monocytogenes. To investigate this, WT macrophages were infected with L. monocytogenes in the presence of inhibitors specific for these signaling pathways. Infection with L. monocytogenes led to an increase in autophagy levels. However, treatment of infected cells with NF-κB inhibitor peptide SN50 did not change LC3-II expression (Fig. 6A). We also confirmed this result by inhibition of NF-κB activation by SC-514, an IKK2 inhibitor, which showed similar results (Fig. 6B), indicating no role for NF-κB in autophagy of Listeria. In contrast, treatment with PD98059, an inhibitor of the MEK signaling pathway abolished any increase in autophagy (Fig. 6C), suggesting the specificity of the MEK/ERK signaling pathway in autophagy of L. monocytogenes infection.
FIGURE 6.
Autophagy of L. monocytogenes is dependent on the ERK signaling pathway. A, Western blot analyses of LC3-II in wild-type macrophages left uninfected (control), infected with L. monocytogenes in the absence or presence of NF-κB inhibitor SN50, or treated with SN50 alone. Tubulin was used as a loading control. B, Western blot analyses of LC3-II in wild-type macrophages left uninfected (control), infected with L. monocytogenes in the absence or presence of NF-κB inhibitor SC-514, or treated with SC-514 alone. Tubulin was used as a loading control. C, Western blots of LC3-II in wild-type macrophages left uninfected (control), infected with L. monocytogenes in the absence or presence of the ERK inhibitor (PD98059), or treated with the inhibitor alone. Tubulin was used as a loading control. D, quantification of GFP-LC3 positive Listeria ActA mutant in wild-type cells in the absence or presence of the ERK inhibitor. *, p ≤ 0.05. E, intracellular growth curve of the L. monocytogenes ActA mutant in wild-type cells in the absence or presence of the ERK inhibitor. *, p ≤ 0.05.
We next employed GFP-LC3 to look for Listeria-targeted autophagosomes in control cells or cells that were exposed to the ERK inhibitor PD98059. In control macrophages, 35% of L. monocytogenes ActA were found within autophagosomes. However, only 20% of Listeria was observed inside GFP-LC3 vacuoles in macrophages treated with PD98059 (Fig. 6D). Consistent with this, more growth of the pathogen was observed in cells treated with the ERK inhibitor (Fig. 6E). We next tested the effect of the ERK inhibitor on Nod2−/− cells that are already defective in autophagy. Exposure of Nod2−/− cells to the ERK inhibitor did not show any change in LC3-II levels (supplemental Fig. S3A). In agreement, no significant effect on colony counts was observed when Nod2−/− cells were exposed to the ERK-inhibitor (supplemental Fig. S3B).
Cells Deficient in ERK Signaling Display Defective Autophagy of L. monocytogenes
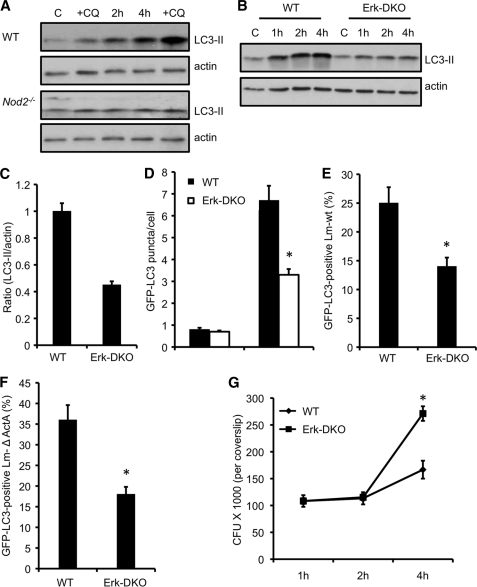
We next confirmed the role for ERK in autophagy of L. monocytogenes by employing Erk1−/−;Erk2fl/fl;CD11c-Cre (Erk-DKO) DCs. As a control, we first tested the role for GM-CSF-differentiated dendritic cells in autophagy activation against L. monocytogenes in WT and Nod2−/− dendritic cells. Similar to our results with macrophages, L. monocytogenes infection activated autophagy in WT DCs. However, Nod2−/− DCs did not show an efficient induction of autophagy (Fig. 7A), suggesting that defective autophagy of Listeria is not restricted solely to Nod2−/− macrophages. Also, treatment with CQ to explore the processing of LC3-II showed an increase in LC3-II lipidation, thereby suggesting the presence of functional autophagolysosomes in these cells. We next exposed WT and Erk-DKO DCs to L. monocytogenes. As shown in Fig. 7B, Erk-DKO DCs displayed defective autophagy activation upon L. monocytogenes infection. Autophagy induction was reduced by almost one-half in knockout cells as compared with WT DCs when exposed to L. monocytogenes (Fig. 7C).
FIGURE 7.
Cells deficient in ERK signaling display defective autophagy of L. monocytogenes. A, Western blot analyses of LC3-II in wild-type and Nod2−/− dendritic cells infected with L. monocytogenes. At 30 min, a set of control and L. monocytogenes-infected cells were incubated with CQ. Results are representative of three separate experiments. B, Western blot analyses of LC3-II in wild-type and Erk1−/−;Erk2fl/fl;CD11c-Cre (ERK-DKO) DCs infected with L. monocytogenes. Results are representative of three separate experiments. C, densitometry scanning of the Western blot analysis showing the ratio of LC3-II to actin in wild-type and ERK-DKO DCs infected with L. monocytogenes 4 h after infection. D, quantification of the number of GFP-LC3 puncta per cell 4 h after infection in wild-type and ERK-DKO DCs. Results show mean ± S.E. (*, p ≤ 0.05) of experiments done at least three times. E, quantification of the GFP-LC3-positive L. monocytogenes wild-type strain in cells infected as described in C. Results show mean ± S.E. (*, p ≤ 0.05) of experiments done at least three times. F, quantification of theGFP-LC3-positive Listeria mutant lacking ActA in cells infected as described in C. Results show mean ± S.E. (*, p ≤ 0.05) of experiments done at least three times. G, intracellular growth curve of L. monocytogenes ΔActA in wild-type and ERK-DKO DCs. *, p ≤ 0.05.
We next employed GFP-LC3 to determine the formation of autophagosomes in WT and Erk-DKO cells. Consistent with our WB data, the number of GFP-LC3 puncta per cell was markedly reduced in Erk-DKO cells (Fig. 7D). This was not because of reduced uptake in knockout cells, as it was similar in both cell types (supplemental Fig. S1B). To confirm further, we enumerated the number of L. monocytogenes localized to GFP-LC3 autophagosomes. In wild-type cells, approximately 25% of L. monocytogenes colocalized with GFP-LC3 autophagosomes, whereas less than 15% of the pathogen localized with the autophagosomes in Erk-DKO cells (Fig. 7E). Because Listeria mutants lacking ActA are more efficiently targeted to autophagosomes (44), we observed approximately 35% of the mutant bacteria in GFP-LC3 vacuoles, whereas 17% of L. monocytogenes lacking ActA were observed in autophagosomes in knockout cells (Fig. 7F). To directly test the role of ERK-dependent autophagy in L. monocytogenes infection, we compared the growth of the Listeria ΔActA mutant in WT and Erk-DKO cells. A significant increase in L. monocytogenes growth was observed in cells lacking the ERK1/2 pathway (Fig. 7G), suggesting the restriction of the bacteria by ERK-dependent autophagy.
DISCUSSION
Autophagy plays a crucial role in innate immune response to intracellular pathogens (24–26, 46). Intracellular pathogens are taken up into phagosomes, wherein many of them are killed by acidification of this compartment after fusion with lysosomes. However, various other intracellular pathogens have evolved strategies to escape phagolysosomal killing. For example, Coxiella survives in an acidic compartment, whereas pathogens like Mycobacteria inhibit phagolysosomal killing and therefore inhabit an endosome/phagosome-like compartment (47). Still others, like L. monocytogenes, evade phagolysosomal killing by escaping into the cytoplasm (48). Although pathogens have evolved mechanisms to evade killing in the phagosome, their mammalian hosts have coevolved alternative mechanisms to limit their growth. Autophagy is one such mechanism that is particularly important in eliminating bacteria that invade cytoplasm. Here we demonstrate that autophagy against L. monocytogenes is mediated by the extracellular TLR2 and intracellular NOD/RIP2 pathways. We showed that autophagy induction through this pathway leads to the sequestration of L. monocytogenes within autophagosomes, leading to decrease in bacterial survival.
Recent studies reported the induction of autophagy by L. monocytogenes (49) and colocalization of L. monocytogenes in autophagosomes in the early phase of infection by mouse embryonic fibroblasts and RAW 264.7 macrophages (27, 36, 37). This process depends on ActA and bacterial phospholipases PI-PLC and PC-PLC. Another recent study described that Listeria-ActA mutants are more efficiently targeted to autophagosomes than wild-type Listeria because ActA helps in recruitment of host actin machinery, thereby escaping autophagy recognition (44). Zhao et al. (50) observed that mice lacking Atg5 in macrophages and granulocytes are more susceptible to L. monocytogenes infection. Similarly, Yano et al. (51) reported an essential role for autophagy in increased survival of Drosophila that depends on the recognition of L. monocytogenes by the peptidoglycan recognition receptor. Collectively, these findings suggest that L. monocytogenes induces autophagy and that Listeria ActA mutants are more efficiently sequestered within autophagosomes during the early phase of infection.
We observed that NOD1 and NOD2 are crucial for autophagic recognition of L. monocytogenes. NOD proteins have been shown very recently to be critical for autophagy induction in response to their specific ligands. Consequently, autophagy was shown to be important in recognition of Shigella and Salmonella in DCs (42, 43). Mutations in NOD2 and an essential autophagy gene, ATG16L1, are associated with Crohn's disease, which results from an excessive inflammatory response to normal gut flora (52–55). In line with this, the induction of autophagy via NOD2 was shown to be dependent on Atg16L1. However, contrasting results were reported in regard to the requirement for RIP2 in autophagy (42, 43). In line with Cooney et al. (43), our study clearly demonstrated a critical role for RIP2 in autophagy of L. monocytogenes in vitro and in vivo.
Autophagy in response to limiting nutrients proceeds through inactivation of mammalian target of rapamycin protein kinase and is dependent on phosphatidylinositol-3 kinase hVps34/Beclin-1 complex (19, 21, 56). However, the upstream signaling components involved that regulate autophagy in response to L. monocytogenes infection have not been identified. Our results show that besides the NOD/RIP2 pathway, TLR2 is also required for autophagy against L. monocytogenes. Consequently, we observed significantly less L. monocytogenes colocalizing with GFP-LC3 autophagosomes in Tlr2−/− macrophages. Further, both NF-κB and ERK signaling pathways are perturbed in Tlr2−/− and Nod2−/− macrophages. However, autophagy against L. monocytogenes was resistant to the NF-κB inhibitor in WT macrophages. In contrast, cells exposed to the pharmacological inhibitor of the MEK/ERK pathway or Erk-DKO cells displayed reduced autophagy activation against L. monocytogenes, implicating MEK/ERK signaling in regulating L. monocytogenes-induced autophagy. Our data showing a critical role for ERK signaling during autophagy of Listeria thus implicates the ERK pathway in initiating antibacterial autophagy for the first time.
In summary, we have elucidated an important link between innate immune receptors and the induction of autophagy against cytoplasm-invading L. monocytogenes. Our data suggest that TLR2 and the NOD/RIP2 axis are essential pathways through which autophagy is initiated to limit the survival of microbes. Consequently, these findings may lead to the development of novel therapeutic interventions that modulate autophagy.
Supplementary Material
Acknowledgments
We thank Anthony Coyle, Ethan Grant, John Bertin (Millennium Pharmaceuticals), Richard Flavell (Yale School of Medicine), and Tak Mak (University of Alberta) for their generous supply of mutant mice and Dan Portnoy (University of California, Berkley) for providing Listeria monocytogenes strains. We thank Mary O'Riordan and Sara Cassidy (University of Michigan, Ann Arbor) for sharing reagents and protocols. We also thank Sharon Frase and Linda Mann (Cell and Tissue Imaging Facility, St. Jude Children's Research Hospital) for technical assistance with electron microscopy.
This work was supported, in whole or in part, by National Institute of Health Grants AR056296 and AI088177.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- TLR
- Toll-like receptor
- NLR
- Nod-like receptor
- DC
- dendritic cell
- CQ
- chloroquine
- WB
- Western blotting.
REFERENCES
- 1. Vázquez-Boland J. A., Kuhn M., Berche P., Chakraborty T., Domínguez-Bernal G., Goebel W., González-Zorn B., Wehland J., Kreft J. (2001) Clin. Microbiol. Rev. 14, 584–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Corr S. C., O'Neill L. A. (2009) Cell Microbiol. 11, 703–709 [DOI] [PubMed] [Google Scholar]
- 3. Ireton K. (2007) Cell Microbiol. 9, 1365–1375 [DOI] [PubMed] [Google Scholar]
- 4. Mengaud J., Ohayon H., Gounon P., Mege R. M., Cossart P. (1996) Cell 84, 923–932 [DOI] [PubMed] [Google Scholar]
- 5. Hamon M., Bierne H., Cossart P. (2006) Nat. Rev. Microbiol. 4, 423–434 [DOI] [PubMed] [Google Scholar]
- 6. Portnoy D. A., Auerbuch V., Glomski I. J. (2002) J. Cell Biol. 158, 409–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Henry R., Shaughnessy L., Loessner M. J., Alberti-Segui C., Higgins D. E., Swanson J. A. (2006) Cell Microbiol. 8, 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gründling A., Gonzalez M. D., Higgins D. E. (2003) J. Bacteriol. 185, 6295–6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Poussin M. A., Goldfine H. (2005) Infect. Immun. 73, 4410–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Monack D. M., Theriot J. A. (2001) Cell Microbiol. 3, 633–647 [DOI] [PubMed] [Google Scholar]
- 11. Creagh E. M., O'Neill L. A. (2006) Trends Immunol. 27, 352–357 [DOI] [PubMed] [Google Scholar]
- 12. Kanneganti T. D., Lamkanfi M., Núñez G. (2007) Immunity 27, 549–559 [DOI] [PubMed] [Google Scholar]
- 13. Kawai T., Akira S. (2009) Int. Immunol. 21, 317–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Anand P. K., Malireddi R. K. S., Kanneganti T. D. (2011) Front. Microbiol. 2, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Girardin S. E., Boneca I. G., Viala J., Chamaillard M., Labigne A., Thomas G., Philpott D. J., Sansonetti P. J. (2003) J. Biol. Chem. 278, 8869–8872 [DOI] [PubMed] [Google Scholar]
- 16. Girardin S. E., Boneca I. G., Carneiro L. A., Antignac A., Jéhanno M., Viala J., Tedin K., Taha M. K., Labigne A., Zähringer U., Coyle A. J., DiStefano P. S., Bertin J., Sansonetti P. J., Philpott D. J. (2003) Science 300, 1584–1587 [DOI] [PubMed] [Google Scholar]
- 17. O'Riordan M., Yi C. H., Gonzales R., Lee K. D., Portnoy D. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13861–13866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McCaffrey R. L., Fawcett P., O'Riordan M., Lee K. D., Havell E. A., Brown P. O., Portnoy D. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11386–11391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Z., Klionsky D. J. (2009) Curr. Top Microbiol. Immunol. 335, 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Glick D., Barth S., Macleod K. F. (2010) J. Pathol. 221, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pattingre S., Espert L., Biard-Piechaczyk M., Codogno P. (2008) Biochimie 90, 313–323 [DOI] [PubMed] [Google Scholar]
- 22. Deretic V. (2005) Trends Immunol. 26, 523–528 [DOI] [PubMed] [Google Scholar]
- 23. Deretic V. (2009) Curr. Opin. Cell Biol. 22, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deretic V., Levine B. (2009) Cell Host Microbe 5, 527–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang J., Brumell J. H. (2009) Curr. Top Microbiol. Immunol. 335, 189–215 [DOI] [PubMed] [Google Scholar]
- 26. Campoy E., Colombo M. I. (2009) Biochim. Biophys. Acta 1793, 1465–1477 [DOI] [PubMed] [Google Scholar]
- 27. Py B. F., Lipinski M. M., Yuan J. (2007) Autophagy 3, 117–125 [DOI] [PubMed] [Google Scholar]
- 28. Nakagawa I., Amano A., Mizushima N., Yamamoto A., Yamaguchi H., Kamimoto T., Nara A., Funao J., Nakata M., Tsuda K., Hamada S., Yoshimori T. (2004) Science 306, 1037–1040 [DOI] [PubMed] [Google Scholar]
- 29. Birmingham C. L., Smith A. C., Bakowski M. A., Yoshimori T., Brumell J. H. (2006) J. Biol. Chem. 281, 11374–11383 [DOI] [PubMed] [Google Scholar]
- 30. Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. (2004) Cell 119, 753–766 [DOI] [PubMed] [Google Scholar]
- 31. Flo T. H., Halaas O., Lien E., Ryan L., Teti G., Golenbock D. T., Sundan A., Espevik T. (2000) J. Immunol. 164, 2064–2069 [DOI] [PubMed] [Google Scholar]
- 32. Machata S., Tchatalbachev S., Mohamed W., Jänsch L., Hain T., Chakraborty T. (2008) J. Immunol. 181, 2028–2035 [DOI] [PubMed] [Google Scholar]
- 33. Seki E., Tsutsui H., Tsuji N. M., Hayashi N., Adachi K., Nakano H., Futatsugi-Yumikura S., Takeuchi O., Hoshino K., Akira S., Fujimoto J., Nakanishi K. (2002) J. Immunol. 169, 3863–3868 [DOI] [PubMed] [Google Scholar]
- 34. Kim Y. G., Park J. H., Shaw M. H., Franchi L., Inohara N., Núñez G. (2008) Immunity 28, 246–257 [DOI] [PubMed] [Google Scholar]
- 35. Park J. H., Kim Y. G., McDonald C., Kanneganti T. D., Hasegawa M., Body-Malapel M., Inohara N., Núñez G. (2007) J. Immunol. 178, 2380–2386 [DOI] [PubMed] [Google Scholar]
- 36. Rich K. A., Burkett C., Webster P. (2003) Cell Microbiol. 5, 455–468 [DOI] [PubMed] [Google Scholar]
- 37. Birmingham C. L., Canadien V., Gouin E., Troy E. B., Yoshimori T., Cossart P., Higgins D. E., Brumell J. H. (2007) Autophagy 3, 442–451 [DOI] [PubMed] [Google Scholar]
- 38. Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Dröge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fésüs L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., González-Estévez C., Gorski S., Gottlieb R. A., Häussinger D., He Y. W., Heidenreich K., Hill J. A., Høyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jäättelä M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovács A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., López-Otín C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Meléndez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Münz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nürnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Tallóczy Z., Tanaka K., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcátegui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mizushima N., Yoshimori T. (2007) Autophagy 3, 542–545 [DOI] [PubMed] [Google Scholar]
- 40. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mosa A., Trumstedt C., Eriksson E., Soehnlein O., Heuts F., Janik K., Klos A., Dittrich-Breiholz O., Kracht M., Hidmark A., Wigzell H., Rottenberg M. E. (2009) Infect. Immun. 77, 2908–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Travassos L. H., Carneiro L. A., Ramjeet M., Hussey S., Kim Y. G., Magalhães J. G., Yuan L., Soares F., Chea E., Le Bourhis L., Boneca I. G., Allaoui A., Jones N. L., Núñez G., Girardin S. E., Philpott D. J. (2010) Nat. Immunol. 11, 55–62 [DOI] [PubMed] [Google Scholar]
- 43. Cooney R., Baker J., Brain O., Danis B., Pichulik T., Allan P., Ferguson D. J., Campbell B. J., Jewell D., Simmons A. (2010) Nat. Med. 16, 90–97 [DOI] [PubMed] [Google Scholar]
- 44. Yoshikawa Y., Ogawa M., Hain T., Yoshida M., Fukumatsu M., Kim M., Mimuro H., Nakagawa I., Yanagawa T., Ishii T., Kakizuka A., Sztul E., Chakraborty T., Sasakawa C. (2009) Nat. Cell Biol. 11, 1233–1240 [DOI] [PubMed] [Google Scholar]
- 45. Torres D., Barrier M., Bihl F., Quesniaux V. J., Maillet I., Akira S., Ryffel B., Erard F. (2004) Infect. Immun. 72, 2131–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Amano A., Nakagawa I., Yoshimori T. (2006) J. Biochem. 140, 161–166 [DOI] [PubMed] [Google Scholar]
- 47. Colombo M. I., Gutierrez M. G., Romano P. S. (2006) Autophagy 2, 162–164 [DOI] [PubMed] [Google Scholar]
- 48. Garcia-del Portillo F., Finlay B. B. (1995) Trends Microbiol. 3, 373–380 [DOI] [PubMed] [Google Scholar]
- 49. Meyer-Morse N., Robbins J. R., Rae C. S., Mochegova S. N., Swanson M. S., Zhao Z., Virgin H. W., Portnoy D. (2010) PLoS ONE 5, e8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao Z., Fux B., Goodwin M., Dunay I. R., Strong D., Miller B. C., Cadwell K., Delgado M. A., Ponpuak M., Green K. G., Schmidt R. E., Mizushima N., Deretic V., Sibley L. D., Virgin H. W. (2008) Cell Host Microbe 4, 458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yano T., Mita S., Ohmori H., Oshima Y., Fujimoto Y., Ueda R., Takada H., Goldman W. E., Fukase K., Silverman N., Yoshimori T., Kurata S. (2008) Nat. Immunol. 9, 908–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kobayashi K. S., Chamaillard M., Ogura Y., Henegariu O., Inohara N., Nuñez G., Flavell R. A. (2005) Science 307, 731–734 [DOI] [PubMed] [Google Scholar]
- 53. Hugot J. P., Chamaillard M., Zouali H., Lesage S., Cézard J. P., Belaiche J., Almer S., Tysk C., O'Morain C. A., Gassull M., Binder V., Finkel Y., Cortot A., Modigliani R., Laurent-Puig P., Gower-Rousseau C., Macry J., Colombel J. F., Sahbatou M., Thomas G. (2001) Nature 411, 599–603 [DOI] [PubMed] [Google Scholar]
- 54. Rioux J. D., Xavier R. J., Taylor K. D., Silverberg M. S., Goyette P., Huett A., Green T., Kuballa P., Barmada M. M., Datta L. W., Shugart Y. Y., Griffiths A. M., Targan S. R., Ippoliti A. F., Bernard E. J., Mei L., Nicolae D. L., Regueiro M., Schumm L. P., Steinhart A. H., Rotter J. I., Duerr R. H., Cho J. H., Daly M. J., Brant S. R. (2007) Nat. Genet. 39, 596–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hampe J., Franke A., Rosenstiel P., Till A., Teuber M., Huse K., Albrecht M., Mayr G., De La Vega F. M., Briggs J., Günther S., Prescott N. J., Onnie C. M., Häsler R., Sipos B., Fölsch U. R., Lengauer T., Platzer M., Mathew C. G., Krawczak M., Schreiber S. (2007) Nat. Genet. 39, 207–211 [DOI] [PubMed] [Google Scholar]
- 56. He C., Klionsky D. J. (2009) Annu. Rev. Genet. 43, 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lamkanfi M., Malireddi R. K., Kanneganti T. D. (2009) J. Biol. Chem. 284, 20574–20581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kanneganti T. D., Lamkanfi M., Kim Y. G., Chen G., Park J. H., Franchi L., Vandenabeele P., Núñez G. (2007) Immunity 26, 433–443 [DOI] [PubMed] [Google Scholar]
- 59. Anand P. K., Anand E., Bleck C. K., Anes E., Griffiths G. (2010) PLoS ONE 5, e10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Portnoy D. A., Jacks P. S., Hinrichs D. J. (1988) J. Exp. Med. 167, 1459–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.