Background: NLRP7 is responsible for recurrent hydatidiform moles, but its functional role is unknown.
Results: NLRP7 mutations impair IL-1β and TNF secretion but do not affect IL-1β processing. NLRP7 co-localizes with the microtubule organizing center, and mutations impair cytokine trafficking.
Conclusion: Patients with NLRP7 mutations have abnormal tolerance to aberrant pregnancies.
Significance: We unravel a new mechanism for reproductive wastage.
Keywords: Cytokine, Genetic Diseases, Microtubules, Mutant, Pathogen-associated Molecular Pattern (PAMP)
Abstract
A hydatidiform mole (HM) is a human pregnancy with hyperproliferative placenta and abnormal embryonic development. Mutations in NLRP7, a member of the nucleotide oligomerization domain-like receptor family of proteins with roles in inflammation and apoptosis, are responsible for recurrent HMs. However, little is known about the functional role of NLRP7. Here, we demonstrate that peripheral blood mononuclear cells from patients with NLRP7 mutations and rare variants secrete low levels of IL-1β and TNF in response to LPS. We show that the cells from patients, carrying mutations or rare variants, have variable levels of increased intracellular pro-IL-1β indicating that normal NLRP7 down-regulates pro-IL-1β synthesis in response to LPS. Using transient transfections, we confirm the role of normal NLRP7 in inhibiting pro-IL-1β and demonstrate that this inhibitory function is abolished by protein-truncating mutations after the Pyrin domain. Within peripheral blood mononuclear cells, NLRP7 co-localizes with the Golgi and the microtubule-organizing center and is associated with microtubules. This suggests that NLRP7 mutations may affect cytokine secretion by interfering, directly or indirectly, with their trafficking. We propose that the impaired cytokine trafficking and secretion caused by NLRP7 defects makes the patients tolerant to the growth of these earlier arrested conceptions with no fetal vessels and that the retention of these conceptions until the end of the first trimester contribute to the molar phenotype. Our data will impact our understanding of postmolar choriocarcinomas, the only allograft non-self tumors that are able to invade maternal tissues.
Introduction
Hydatidiform moles (HMs)4 are abnormal human pregnancies with excessive proliferation of trophoblast cells and abnormal embryonic development. The common form of HMs is sporadic, not recurrent, and occurs in 1 in every 250 pregnancies in Western countries (1). Molar pregnancies are usually benign but may degenerate into gestational choriocarcinomas and invade maternal tissues outside the uterus (2). Using linkage analysis and positional cloning on rare familial cases of recurrent moles in which the disease segregates as an autosomal recessive condition, our group identified a maternal gene, NLRP7, responsible for recurrent HMs and associated reproductive wastage (3). To date, 60 NLRP7 mutations have been reported by several groups in patients from different populations and are listed on Infevers (4–10). Patients with recurrent HMs have usually two defective alleles. However, to date, 14 patients with a single defective allele each have been reported, and these patients have better reproductive outcomes, i.e. less recurrent moles, more spontaneous abortions, and more live births than patients with two defective alleles (4, 5, 11, 12). Moreover, we have shown that rare nonsynonymous NLRP7 variants found in the general European population are associated with recurrent reproductive wastage in European patients (4, 5).
NLRP7 is a member of the nucleotide oligomerization domain-like family, a series of cytoplasmic proteins characterized by an N-terminal Pyrin domain, followed by a NACHT domain, and a C-terminal leucine-rich repeat (LRR) region. Although the Pyrin and the LRR domains are involved in protein-protein interactions, NACHT is an NTPase domain found in apoptosis proteins as well as in proteins involved in the transactivation of the MHC class II (13). Recent studies have shown increased NLRP7 expression in testicular (14) and endometrial cancers (15). One in vitro study has shown that overexpressing wild-type NLRP7 inhibits caspase-1-dependent IL-1β processing and secretion (16).
In this study, we demonstrate that patients with NLRP7 mutations and variants secrete significantly lower amounts of IL-1β and TNF than control cells in response to LPS despite their higher intracellular pro-IL-1β synthesis and normal pro-IL-1β processing. Using transient transfections, we demonstrate that overexpression of wild-type NLRP7 inhibits primarily pro-IL-1β synthesis and consequently decreases the amount of intracellular mature IL-1β. We then tested the functional consequences of different mutations and found that only protein-truncating mutations after the N-terminal Pyrin domain significantly increased pro-IL-1β synthesis. Using constructs carrying the different NLRP7 domains, we demonstrate that the three domains are required to confer the full inhibitory activity of the protein with the LRR and the NACHT playing the major role. Within PBMCs, NLRP7 co-localizes with the Golgi apparatus and the microtubule-organizing center (MTOC), which indicates that mutations in this gene may impair cytokine trafficking and secretion. Altogether, our ex vivo and in vitro data demonstrate, first, that NLRP7 mutations and variants impair cytokine secretion potentially by affecting their trafficking and, second, that mutations in NLRP7 impair its capacity to down-regulate pro-IL-1β synthesis in response to LPS.
MATERIALS AND METHODS
Patients and Mutation Analysis
Mutations in patients 4, 723, 428, II.3, and 636, have been described previously (3, 4, 11), and their NLRP7 mutations and nonsynonymous variants are shown in Fig. 1 and supplemental Table 1. Additional new patients were analyzed as described previously, and their reproductive history is shown in supplemental Table 1. We use the term mutations to indicate DNA changes leading to protein truncating mutations or missense mutations that were not found in at least 100 controls of matching ethnicities to those of the patients. We use nonsynonymous variants to indicate changes in amino acids that were also found in controls from the general population. This study was approved by the Institutional Review Board of the McGill University. All patients provided written consent to provide blood samples and participate in the study.
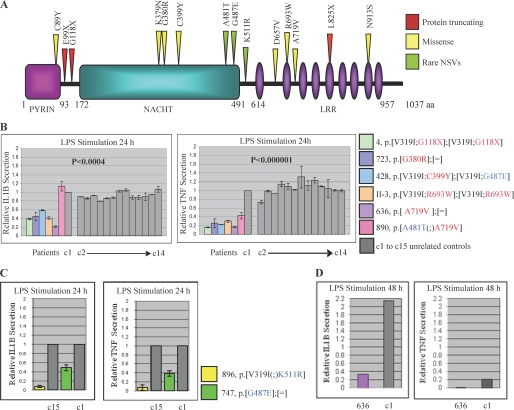
FIGURE 1.
Cytokine secretion by PBMCs from patients with NLRP7 mutations and variants. A, schematic representation of the NLRP7 protein with the various mutations and rare variants analyzed in this study. NLRP7 protein shares three functional domains with several members of the Nod-like receptor (NLR) family, PYD, NACHT, and LRR. (B) PBMCs from patients with NLRP7 mutations have an impaired IL-1β and TNF secretion. Freshly isolated PBMCs (1.5 × 106cells) were stimulated with LPS for 24 h. The cells were centrifuged, and supernatants were collected to measure the amount of IL-1β and TNF released in the media by ELISA. Relative amounts of each cytokine refer to the secreted amounts by patients cells divided by those secreted by control cells. The averages and S.E. were calculated on two to three different ELISA assays on supernatants from the same LPS stimulation. C, PBMCs from two patients with rare NLRP7 variants show also lower IL-1β and TNF secretion than controls. D, IL-1β and TNF secretion by PBMCs from patient 636 stimulated with LPS for 48 h. C1 after 24 h of stimulation was adopted as 1-fold. After 48 h of stimulation, the amount of IL-1β was 2.2 times higher than the level after 24 h and was kept at 2.2 to highlight the accumulation of IL-1β in both control and patient cells. aa, amino acids; NSVs, nonsynonymous variants.
Cytokine Assay
Blood (on K3EDTA) from patients 4, II-3, 428, 636, 723, and 890 with mutations and patients 896, 747, 882, and 993 with rare variants were analyzed in parallel to one sample from a control subject (control 1) within 24 h (h) from withdrawal. Blood from 15 additional controls from unrelated subjects (between 20 and 45 years) from various ethnic groups were later collected and analyzed with control 1. All of the controls did not have family histories of immunological or inflammatory condition and had no family history of recurrent fetal losses. All patients and controls were not pregnant (except for patient 890) or under any type of treatment at the time of blood withdrawal. In addition, a complete white blood cell profile and count was performed on control and patient samples the day of the experiments to exclude any acquired temporary inflammation. PBMCs were isolated using Ficoll-Paque PLUS, and 1.5 × 106 cells were plated in 24-well plates and stimulated with LPS (100 ng/ml) (Sigma, L6529, from Escherichia coli 055:B5) or ultrapure LPS (1 μg/ml (Cedarlane 423(LB) from E. coli 0552:B5).
Cloning and Site-directed Mutagenesis of Human NLRP7 or Plasmid Constructs
Human wtNLRP7 cDNA was cloned into pCR-Blunt-II-TOPO vector (IMAGE ID 40036028, accession no. BC109125; Open Biosystems). The NLRP7 cDNA in the IMAGE clone contained three nonsynonymous variants, Q310R, L311I, and A481T that were corrected by site directed mutagenesis and the corrected vector was considered as our wild-type for this study. The NLRP7 cDNA was FLAG-tagged using the primer 5′-GCGGCTTAAGCCACCATGGACTACAAAGACGACGATGACAAGGGTACCATGACATCGCCCCAG and primer 3′-GCGGCTCGAGTCAGCAAAAAAAGTCACAGCACGGA. FLAG-NLRP7 was inserted into a pcDNA 3.1(+) vector (Invitrogen) using restriction sites AflII and Xho1. Missense mutations and base pair deletions in the NLRP7 gene were generated by site-directed mutagenesis using PfuUltra High-fidelity DNA polymerase AD (Agilent Technologies) the QuikChangeTM site-directed mutagenesis (Stratagene). The integrity of all clones was confirmed by direct sequencing of the whole insert.
Cell Culture and Transfection
One day prior to transfection, HEK293 cells were seeded at a density of 1 × 105 cells per well using 24-well plates in 500 μl of DMEM (Invitrogen) supplemented with 10% of FBS (Invitrogen). The cells were transfected with 400 ng of total plasmid DNA, pre-complexed with the Plus reagent, and diluted in Lipofectamine reagent according to the manufacturer's instructions (Invitrogen). In co-transfection experiments, the pcDNA 3.1(+)-pro-IL-1β vector encoding the human IL-1β protein and the pcDNA 3.1(+)-FLAG-caspase-1 vector encoding the human caspase-1 protein (NM_033292.21) were co-transfected with pcDNA 3.1(+)-FLAG-NLRP7.
Western Blotting
Twenty-four hours post-transfection or -stimulation, cells were lyzed, and proteins were separated on 10% SDS-PAGE, transferred into PVDF membranes, and immunodetected using a monoclonal antibody directed against FLAG (1:2000) (F3165, Sigma), monoclonal antibodies directed against human IL-1β (2022, Cell Signaling Technology) (1:1000), and α-tubulin (2144, Cell Signaling Technology) (1:1000), and β-actin (MAB1501, Chemicon Intl.) (1:1000). Protein bands were revealed using the Hyperfilm ECL Western blotting detection reagents (GE Healthcare) and quantified by NIH ImageJ software.
Nocodazole Treatment
Epstein Barr virus-transformed B lymphocyte cells (1.5 × 106) from one control were plated in 24-well plates, stimulated or not with ultrapure LPS (1 μg/ml) (Cedarlane 423(LB) from E. coli 0552:B5) for 24 h, and treated the next day with 10 μm nocodazole (Sigma, M1404, dissolved in dimethyl sulfoxide), dimethyl sulfoxide alone, or without nocodazole or dimethyl sulfoxide. All treatments were at 37 °C for 30 min in a 5% CO2 humidified atmosphere.
Immunofluorescence Staining
Four-well chamber slides (Ultident) were pre-treated with poly-l-lysine (0.1 mg/ml) (Invitrogen) for 2 h at room temperature, and 7.5 × 105 cells were seeded per well and incubated for 15 min at 4 °C. PBMCs were then fixed with 95% alcohol, 5% glacial acetic acid for 15 min at 4 °C, permeabilized with 0.2% Triton X-100 (Sigma) in PBS for 15 min at room temperature, blocked with 3% BSA in PBS for 1 h, and then incubated with each of the primary antibodies: goat anti-NLRP7 (1:100; sc-50642, Santa Cruz Biotechnology), mouse anti-IL-1β (1:100; MAB201, R&D Systems), rabbit anti-giantin (1:100; sc-67168, Santa Cruz Biotechnology), mouse anti-protein disulfide isomerase (1:100; MA3-019, Thermo Scientific), mouse anti-γ-tubulin (1:100) (MA1-850, Thermo Scientific) overnight at 4 °C, followed by incubation with their respective secondary antibody Alexa Fluor-conjugated donkey anti-goat (1:500) (A11055, Invitrogen), donkey anti-mouse (1:500) (A10037, Invitrogen), and donkey anti-rabbit (1:500) (A10042, Invitrogen) for 1 h at room temperature. Finally, the slides were mounted using Vectashield hard-set mounting medium with DAPI (Vector Laboratories). The specificity of the NLRP7 primary antibody was tested after blocking with the NLRP7 peptide (sc-50642 P) according to the manufacturer's instructions. Fluorescence images were captured on an Axioskop 2 plus microscope. Cells were also examined, and images were captured using a laser-scanning confocal microscope (Olympus FluoViewTM FV1000) with an oil immersion objective (UPLSAP, ×100).
Statistical Analyses
Statistical analyses of ELISA measurements were performed using analysis of variance single factor analysis. The frequencies of NLRP7 polar signals were calculated using two-by-two table on the Simple Interactive Statistical Analysis website. p values < 0.05 were considered as statistically significant.
RESULTS
Cells from Patients with NLRP7 Mutations and Variants Secrete Low Levels of IL-1β and TNF
Mutations in NLRP3, another member of the nucleotide oligomerization domain-like receptor proteins, have been shown to be associated with increased IL-1β and TNF secretion from PBMCs of patients with Muckle-Wells and related autoinflammatory disorders (17–19). To investigate the role of NLRP7 in the regulation of cytokine production in an ex vivo cellular model, PBMCs from six patients with previously reported mutations that are not present in the general population (Fig. 1A) and 15 controls (c1 to c15) were stimulated with LPS for 24 h, and the levels of IL-1β and TNF released into the culture supernatants were measured by ELISA. This analysis revealed that the cells of the patients secrete lower amounts of both cytokines than control cells (p values ≤ 0.0004) (Fig. 1B).
Patients with NLRP7 mutations are rare, and most of our patients are referred to our laboratory from international collaborators. Consequently, it is difficult to have access to fresh blood cells from additional patients with mutations. We therefore assessed IL-1β and TNF secretion by PBMCs from nine patients with rare nonsynonymous variants in NLRP7 that were found at higher frequencies in patients than in controls of European ancestry and are therefore associated with recurrent reproductive wastage in the European population (11). Data on four patients (754, 819, 821, and 830) have been reported previously and show statistically lower amounts of secreted IL-1β and TNF. In the current study, we analyzed five additional European patients, 747, 840, 882, 896, and 993, three with variant G487E and two with variant K511R. Again, we found that cells from these patients secrete lower levels of IL-1β and TNF than cells from controls without these variants (Fig. 1C).
PBMCs from patient 636 with one mutation and one control were maintained in the presence of LPS for 48 h and then assayed for IL-1β and TNF secretion. This analysis showed higher concentrations of IL-1β accumulated after 48 h in the supernatants of both patient and control cells, but the ratios of secreted IL-1β by patient and control cells were almost the same (Fig. 1D). For TNF, lower levels were noted after 48 h of stimulation in both patient and control cells and may be due to its degradation in the culture supernatants. Altogether, our data demonstrate that patients with NLRP7 mutations and variants secrete statistically lower levels of IL-1β and TNF than controls and are therefore hyporesponsive to LPS.
Normal IL-1β Processing in ex Vivo-stimulated Patient PBMCs
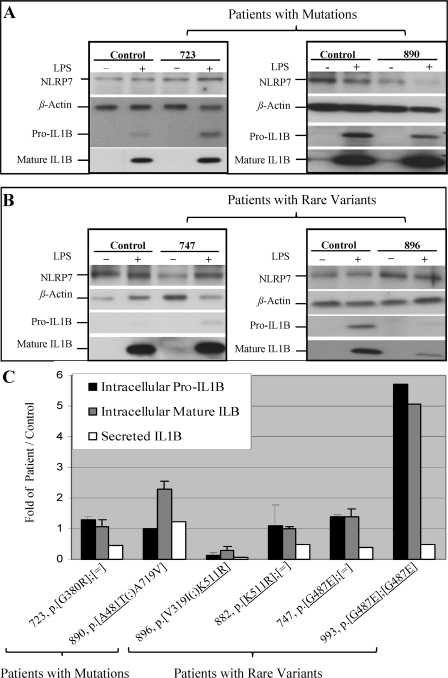
To investigate the causes of the reduced amounts of secreted IL-1β by patients relative to controls cells, we measured the ratios of intracellular pro, mature, and secreted IL-1β in two patients, 723 and 890, with NLRP7 mutations (Fig. 2A) and four patients, 747, 896 (Fig. 2B), 882, and 993, with rare NLRP7 variants versus controls without the mutations or the rare variants. We found variable amounts of synthesized pro-IL-1β in the patients, with four of them having slightly higher amounts of intracellular pro-IL-1β than controls (Fig. 2C). Two patients with rare variants, 896 and 993, had lower and higher amounts of pro-IL-1β than controls, respectively. In five of the six patients, the levels of mature IL-1β mirrored those of pro-IL-1β and none of the patients had defective pro-IL-1β processing. In addition, all the analyzed patients had lower levels of secreted IL-1β than controls with the exception of patient, 890, who was pregnant at the time of blood drawing for LPS stimulation. These data lead us to two conclusions. First, NLRP7 mutations do not affect IL-1β processing but interfere, directly or indirectly, with its secretion in the extracellular milieu. Second, NLRP7 mutations and variants increase the levels of intracellular pro-IL-1β synthesis in response to LPS stimulation, which indicates that normal NLRP7 inhibits pro-IL-1β synthesis and mutations and variants in this gene impair this function. We note that the decrease in cytokine secretion was relatively more important than the increase in pro-IL-1β synthesis and seems to be the major defect of PBMCs from patients carrying NLRP7 mutations and rare variants.
FIGURE 2.
Cells from patients with NLRP7 mutations and rare variants display normal to higher pro-IL-1β synthesis and normal processing. Freshly isolated PBMCs (1.5 × 106 cells) were stimulated with LPS for 24 h. Immunoblots of whole cell lysates show the expression of intracellular pro, mature IL-1β relative to β-actin in patients with NLRP7 mutations (A) and rare variants (B). C, recapitulation of the ratios of pro, mature, and secreted IL-1β by the cells of patients divided by control cells (patient pro-IL-1β/control pro-IL-1β) after signal quantification using ImageJ software. When sufficient amount of cells from patients were available, the averages and S.D. were calculated on two to three different Western blots on cellular lysates from the same LPS stimulation.
NLRP7 Co-localizes with Golgi and MTOC in ex Vivo-stimulated PBMCs with LPS
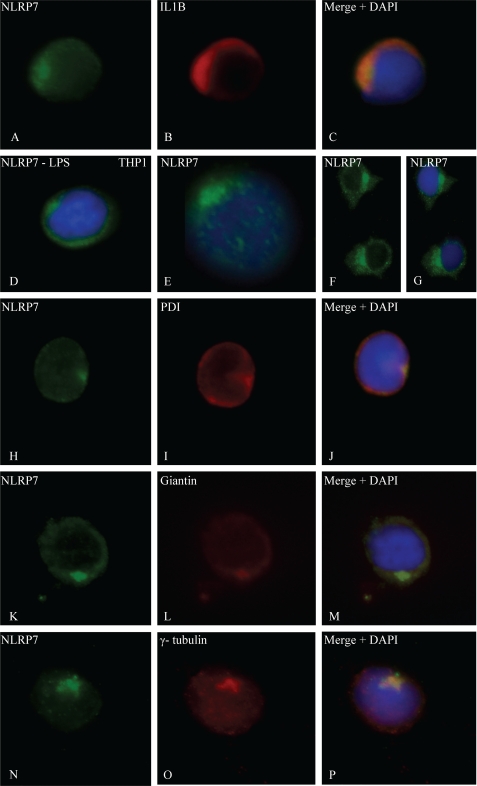
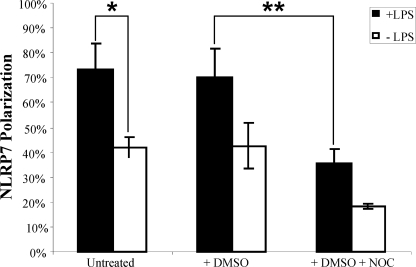
The fact that cells from patients with NLRP7 mutations secrete lower amounts of IL-1β and TNF implies that NLRP7 interferes, directly or indirectly, with the secretion of these cytokines and must be therefore expressed in cells that secrete these cytokines. In peripheral blood mononuclear cells, IL-1β and TNF are mainly secreted by stimulated monocytes. Using immunofluorescence on PBMCs, we confirmed that IL-1β and TNF expression is restricted to LPS-stimulated monocytes and showed NLRP7 expression in monocytes expressing IL-1β (Fig. 3, A–C). NLRP7 expression was also detected in all the other PBMCs subpopulations, and in seven hematopoietic cell lines, Epstein-Barr Virus transformed B-lymphocytes, BJAB, Raji and Ramos (all of B-cell origin), Jurkat (of T-cell origin), THP1 (Fig. 3D and supplemental Fig. S1), and U937 (THP1 and U937 are of monocytic origin). In all LPS-stimulated PBMCs subpopulations, NLRP7 was not distributed homogeneously in the cytoplasm as reported previously in transiently transfected cells overexpressing wild-type NLRP7 cDNA and in human embryonal carcinoma cell lines (14). The strongest NLRP7 signal was detected around the nucleus, restricted to a quarter of the cell circumference suggesting its presence in cellular organelles (Fig. 3, E–G). The polarity of the NLRP7 signal was more prominent after LPS stimulation, being observed in 73% of the cells compared with 42% of unstimulated cells (Fig. 4) (Chi2 = 52.98, p < 0.0001). Using regular immunofluorescence microscopy and antibodies against protein disulfide isomerase, an endoplasmic reticulum resident protein; giantin, a cis-Golgi resident protein; and γ-tubulin, a marker of the MTOC, after LPS stimulation, NLRP7 was found to overlap partially with protein disulfide isomerase (Fig. 3, H–J), but co-localized exactly with giantin (Fig. 3, K–M) and γ-tubulin (Fig. 3, N–P). The co-localization of NLRP7 with giantin and γ-tubulin was also observed after sequential laser confocal microscopy scans (data not shown).
FIGURE 3.
Subcellular localization of NLRP7 and co-localizations. A–C, expression of NLRP7 and IL-1β in a monocyte after LPS stimulation. DNA was counterstained with DAPI (blue). D, NLRP7 is distributed homogeneously in the cytoplasm of unstimulated THP1 cells, whereas it has a predominantly polar signal in stimulated PBMCs (E–G). In E, note the faint microtubule-like network staining emerging from the NLRP7 polar signal. Stimulated PBMCs show overlap between the localization of NLRP7 protein and protein disulfide isomerase (PDI), a marker of the reticulum endoplasmic (H–J). Co-localization of NLRP7 protein in LPS-stimulated THP1 cells with giantin, a marker of the cis-Golgi (K–M). Co-localization in PBMCs of NLRP7 protein with γ-tubulin, a marker of the MTOC (N–P).
FIGURE 4.
Effect of LPS and nocodazole on NLRP7 polarization in Epstein Barr virus-transformed B lymphocyte cells from a control subject. The results represent the percentage of EBV cells with polarized NLRP7 signal after each treatment. The averages and S.D. in these histograms represent cell counts performed by two observers on 50 to 150 cells per condition. The indicated results are representative of data from three different experiments. DMSO, dimethyl sulfoxide; NOC, nocodazole.
To further investigate the potential association of NLRP7 with cytoskeleton proteins, we looked at the NLRP7 signal in Epstein Barr virus-transformed B lymphocytes from a control subject treated with nocodazole, a microtubule depolymerizing agent, for 30 min before fixation and immunofluorescence. We found that nocodazole treatment in the presence or the absence of LPS fragmented the NLRP7 signal and decreased significantly its polarity (Chi2 = 86.34, p < 0.0001) (Fig. 4).
NLRP7 Inhibits Pro-IL-1β Synthesis in Vitro and Nonsense Mutations Impair This Inhibitory Function
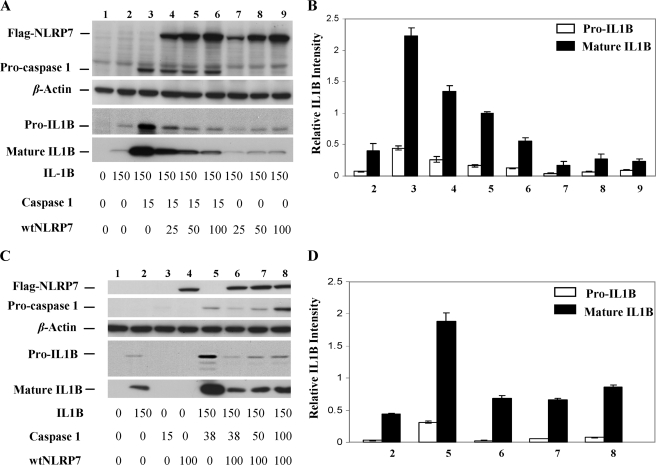
To further characterize the second role of normal NLRP7 in inhibiting pro-IL-1β synthesis, human embryonic kidney (HEK293) cells were co-transfected with vectors expressing pro-IL-1β, procaspase-1, and FLAG-tagged wild-type NLRP7 (FLAG-wtNLRP7) or FLAG-tagged NLRP7 mutants to induce inflammasome formation and IL-1β processing. This analysis demonstrated that in vitro wtNLRP7 also inhibits primarily pro-IL-1β synthesis and decreases the level of mature IL-1β in a dose-dependent manner and only in the presence of caspase-1 (Fig. 5, A and B). We also found that an increased amount of caspase-1 counteracted the inhibitory effect of NLRP7 on pro-IL-1β synthesis (Fig. 5, C and D) and completely abolished it at higher concentrations.
FIGURE 5.
Wild-type NLRP7 inhibits caspase-1-dependent IL-1β production. A, immunoblot of whole cell lysates of HEK293 cells that were transfected simultaneously with expression vectors encoding IL-1β (150 ng), FLAG-caspase-1 (15 ng), and different amounts (25, 50, and 100 ng) of FLAG-wtNLRP7. B, representation of the quantification of signals in A using ImageJ software. This analysis shows that the level of pro-IL-1β inversely correlates with the level of NLRP7 (lanes 3–6). C, immunoblot of whole cell lysates of HEK293 cells that were transfected simultaneously with expression vectors encoding IL-1β (150 ng), different amount of FLAG-caspase-1 (15, 38, 50, and 100 ng) and FLAG-wtNLRP7 (100 ng). D, representation of the quantification of signals in C using ImageJ software. This analysis revealed that a dose-dependent increase of caspase-1 mitigates the inhibitory function of NLRP7 and correlates with an increase of pro-IL-1β synthesis (lanes 6–8). In B and D, the averages and S.D. were calculated on two different Western blots on cellular lysates from different transfection experiments.
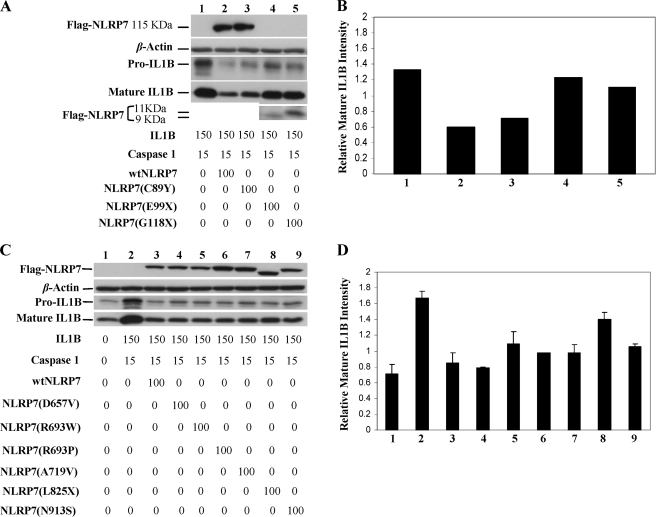
We then tested the effects of fourteen mutations and variants (Fig. 1), of which eight were analyzed in the ex vivo assay. Our data demonstrate that premature stop codons, mainly after the Pyrin domain, compromise significantly the inhibitory function of NLRP7 and lead to a significant increase in the amount of intracellular pro-IL-1β synthesis and consequently the amount of mature IL-1β (p < 0.01) (Fig. 6, A and B). In contrast to nonsense mutations, all the tested missense mutations in the three domains did not alter significantly the inhibitory activity of NLRP7 on pro-IL-1β synthesis in this in vitro system (Fig. 6, C and D) (supplemental Fig. S2).
FIGURE 6.
Nonsense mutations in NLRP7 abolish its inhibitory function. A–C, immunoblots of whole cell lysates of HEK293 cells that were transfected simultaneously with expression vectors encoding IL-1β (150 ng), FLAG-caspase-1 (15 ng), and FLAG-wtNLRP7 (115KDa) or mutant NLRP7 (100 ng) expression vectors. B–D, representations of the quantification of signals in A and C, respectively, using ImageJ software. This analysis showed that missense mutations in the Pyrin and the LRR domains do not alter significantly IL-1β processing. However, nonsense mutations after the Pyrin domain, E99X (9 kDa) and G118X (11 kDa), and in the LRR domain, L825X, compromise significantly the inhibitory effect of NLRP7. In D, the averages and S.D. were calculated on two different Western blots on cellular lysates from different transfection experiments.
We also looked at the NLRP7 subcellular localization in transfected HEK293 cells with FLAG-wtNLRP7 and anti-FLAG antibody. However, in these cells, the level of NLRP7 expression varied between cells and was very high in most of transfected cells, which masked somehow the polarity of the signal (supplemental Fig. S3).
NLRP7 Domains Act Concomitantly to Inhibit Pro-IL-1β Synthesis
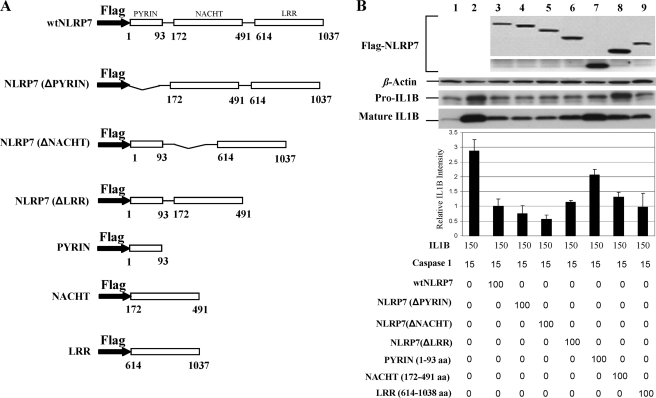
To determine which domain is responsible for inhibiting pro-IL-1β synthesis, we generated constructs containing only the Pyrin, NACHT, or LRR domain of NLRP7 and constructs with a deleted Pyrin, NACHT, or LRR domain. We co-transfected each of these constructs with plasmids expressing pro-IL-1β and pro-caspase-1 and assessed their effects. We show that the Pyrin, NACHT, and LRR domains act concomitantly to reduce the intracellular level of synthesized pro-IL-1β and consequently the intracellular level of mature IL-1β. We found that both the LRR and the NACHT contribute equally to the NLRP7 inhibitory function of pro-IL-1β synthesis (Fig. 7, A–C).
FIGURE 7.
NLRP7 domains work concomitantly to regulate caspase-1 dependent IL-1β production. A, schematic diagram of protein domain structures. B, immunoblot of whole cell lysates of HEK293 cells that cells were transfected simultaneously with expression vectors encoding IL-1β (150 ng), FLAG-caspase-1 (15 ng), FLAG-wtNLRP7 (100 ng), FLAG-tagged NLRP7 domains constructs (100 ng), and FLAG-tagged NLRP7 domain deletion constructs (100 ng). Below are the representation of the quantification of signals using ImageJ software. This analysis revealed that the three NLRP7 domains contribute to the inhibition of IL-1β processing, albeit to different extents. The strongest inhibitory effect is mediated by the NACHT and the LRR domains (lanes 7–9). In B, the averages and S.D. were calculated on two different Western blots on cellular lysates from different transfection experiments. aa, amino acids.
DISCUSSION
Our knowledge of autoinflammatory diseases has expanded significantly in the last decade with the identification of several genes responsible for inflammatory conditions. Among NLRP genes, the most studied one is NLRP3 responsible for three related autoinflammatory diseases. Patients with NLRP3 mutations have constitutive inflammation and their PBMCs secrete higher amounts of IL-1β and TNF than controls upon LPS stimulation (17, 18, 20, 22).
Here, we demonstrate that PBMCs from patients with NLRP7 mutations sense and respond to LPS but secrete lower amounts of IL-1β and TNF than PBMCs from controls. These results are not entirely unexpected because patients with recurrent HMs other than their reproductive problem are in good health and do not manifest inflammatory attacks, fever, or any other clinical sign of inflammation as do patients with NLRP3 mutations. Our cytokine secretion data on patients with NLRP7 mutations and variants are in agreement with previous reports on women with sporadic moles showing their weak cellular mediated immunity in response to phytohemagglutinin and concanavalin A and delayed skin hypersensitivity to dinitrochlorobenzene, purified protein derivates, and recall Candida antigens (23–25).
In addition, we show that the impaired cytokine secretion is not due to an abnormal IL-1β processing as none of the 11 patients with mutations or variants that we have analyzed to date, including those reported previously (11), has an abnormal pro-IL-1β processing. On the contrary, our ex vivo data showed that cells of patients have normal to higher amounts of intracellular pro-IL-1β ranging from 1.1- to 5.6-fold those of controls. This suggest that normal NLRP7, in addition to its role in cytokine secretion, has also another role in down-regulating the intracellular level of pro-IL-1β in response to Toll-like receptor-4 signaling and that mutations and variants in this gene impair this function and consequently lead to higher amounts of pro-IL-1β production. Among the patients analyzed in this study, one with a rare variant in NLRP7, K511R, had a lower level of pro-IL-1β. Similarly, among previously analyzed patients with rare NLRP7 variants, one of five patients (patient 754) displayed lower amount of intracellular pro-IL-1β as compared with control (11). This indicates that patients with rare NLRP7 variants have mutations or variants in other genes affecting pro-IL-1β synthesis and contributing to their disease phenotype.
The fact that the abnormal low secreted levels of IL-1β mirrored those of TNF suggested that the export pathways of these two cytokines overlap, or at least share common trafficking proteins that are affected, directly or indirectly, by NLRP7 mutations and variants. We therefore looked at NLRP7 subcellular localization in ex vivo-stimulated PBMCs from normal subjects or in THP1 cell line and found that NLRP7 co-localizes with the Golgi apparatus and with the MTOC. This localization was affected by nocodazole treatment, which fragmented the NLRP7 signal and decreased its polarization (Fig. 4). This suggests that mutations and variants in NLRP7 impair cytokine secretion by affecting their trafficking either through the classical secretory pathway via the Golgi apparatus or by disrupting the microtubule network known to play a role in the non-classical pathway of IL-1β secretion. Our data are in line with a previous report showing reduced IL-1β and TNF secretion by normal monocytes stimulated with LPS and treated with histone deacetylase inhibitors, which disrupt microtubules and impair vesicle transportation (26). Also, a defect in factors regulating microtubule and actin filaments organization was suggested by Edwards et al. (27) to underlie postzygotic abnormalities during in vitro culture of embryos from a patient with recurrent moles. This suggestion is in line with the postzygotic aneuploidies and mosaicisms that occur in patients with NLRP7 mutations (4) and the association of another gene from the NLRP family, NLRP5, with abnormal spindle assembly, chromosome misalignment, aneuploidies, and mosaicisms (28, 29).
To confirm the role of NLRP7 in down-regulating pro-IL-1β synthesis, we developed an in vitro assay and assessed first the effect of wtNLRP7. Again, in this in vitro system, we found that wtNLRP7 down-regulates the intracellular level of pro-IL-1β in a dose-dependent manner, which is in agreement with our ex vivo data and previously reported data by Kinoshita et al. (16). However, under our experimental in vitro conditions, wtNLRP7 reduced primarily the amount of pro-IL-1β and subsequently the amount of mature IL-1β. We then tested the effects of constructs containing 13 mutations and rare variants observed in patients, eight of which were analyzed in the ex vivo assay. We found, in vitro, that only the two analyzed nonsense mutations after the Pyrin domain significantly alter the NLRP7 inhibitory function and lead to a higher amounts of pro-and mature IL-1β, whereas all analyzed missense mutations and variants in the three domains did not affect significantly the intracellular level of pro-IL-1β. The lack of impact of missense mutations on pro-IL-1β production, in vitro, may be due to domain redundancy that acts as a compensation mechanism since both the NACHT and the LRR are able to individually mediate an important inhibition of pro-IL-1β synthesis (Fig. 7). Also, it is possible that our in vitro system is not sensitive enough to detect milder defects caused by missense mutations and rare variants. The exact mechanism by which NLRP7 down-regulates pro-IL-1β synthesis in transiently transfected cells with vectors without regulatory promoter regions is not clear, but definitely must involve a number of cellular changes affecting the expression of several endogenous genes that altogether, with the transfected ones, contribute to the down-regulation of pro-IL-1β at the transcriptional and/or translational levels.
We previously have shown the occurrence of all genetic types of moles, diploid biparental (8, 30), diploid androgenetic monopermic (4), and triploid dispermic (21), in patients with NLRP7 mutations. We proposed a two-hit mechanism underlying mole occurrence (4). The first hit is the occurring of various complex abnormalities around the time of fertilization or/and very early in the zygote. The second hit is the retention of these aberrant pregnancies to later gestational stages. Among the analyzed patients in the ex vivo cytokine secretion assay, patient 636 had had triploid dispermic moles (21), patient 428 had had a diploid androgenetic monospermic mole (4), and patient 4 had had diploid biparental moles (30). All genotypic types of moles are basically non-viable conceptions that had arrested earlier than the time of their clinical diagnosis before the differentiation of fetal vessels inside the chorionic villi and the establishment of fetal blood circulation. Despite these abnormalities, these conceptions are retained by the patients until the end of the first trimester of pregnancy. The abnormal retention of such pregnancies is an important feature of this pathology and is by itself an indication of the compromised immune response of the patients. Our data demonstrating the inability of the cells of the patients to secrete cytokines and consequently mount appropriate inflammatory response provide an explanation for their abnormal tolerance of moles, conceptions with no embryos. Our findings will impact our understanding of several forms of reproductive wastage and will also open new avenues of research to better understand the causes of gestational choriocarcinomas, aggressive and malignant tumors that originate generally from the placenta of androgenetic moles. In humans, gestational androgenetic choriocarcinomas are the only tumors of non-self origin that are able to grow for several months and invade several maternal tissues (lung, brain, liver, etc.) without being rejected by the maternal immune system. Our findings about the impaired cytokine secretion by cells from patients with NLRP7 mutations and consequently their delayed rejection of abnormal conceptions will open new avenues of therapeutic research aiming at enhancing the immune response of patients and helping them in rejecting these non-self tumors.
Supplementary Material
Acknowledgments
We thank the patients and controls for cooperation. We also thank Catherine Deveault for technical assistance.
This work was supported by Canadian Institutes of Health Research Grants MOP 86546 and MOP 102469.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1–S3.
- HM
- hydatidiform mole
- MTOC
- microtubule-organizing center
- NACHT
- found in the NAIP, CIITA, HET-E and TP1 family proteins
- LRR
- leucine-rich repeat.
REFERENCES
- 1. Seckl M. J., Sebire N. J., Berkowitz R. S. (2010) Lancet 376, 717–729 [DOI] [PubMed] [Google Scholar]
- 2. Altieri A., Franceschi S., Ferlay J., Smith J., La Vecchia C. (2003) Lancet Oncol. 4, 670–678 [DOI] [PubMed] [Google Scholar]
- 3. Murdoch S., Djuric U., Mazhar B., Seoud M., Khan R., Kuick R., Bagga R., Kircheisen R., Ao A., Ratti B., Hanash S., Rouleau G. A., Slim R. (2006) Nat. Genet. 38, 300–302 [DOI] [PubMed] [Google Scholar]
- 4. Deveault C., Qian J. H., Chebaro W., Ao A., Gilbert L., Mehio A., Khan R., Tan S. L., Wischmeijer A., Coullin P., Xie X., Slim R. (2009) Hum. Mol. Genet. 18, 888–897 [DOI] [PubMed] [Google Scholar]
- 5. Hayward B. E., De Vos M., Talati N., Abdollahi M. R., Taylor G. R., Meyer E., Williams D., Maher E. R., Setna F., Nazir K., Hussaini S., Jafri H., Rashid Y., Sheridan E., Bonthron D. T. (2009) Hum. Mutat. 30, E629e39 [DOI] [PubMed] [Google Scholar]
- 6. Kou Y. C., Shao L., Peng H. H., Rosetta R., del Gaudio D., Wagner A. F., Al-Hussaini T. K., Van den Veyver I. B. (2008) Mol. Hum. Reprod. 14, 33–40 [DOI] [PubMed] [Google Scholar]
- 7. Puechberty J., Rittore C., Philibert L., Lefort G., Burlet G., Bénos P., Reyftmann L., Sarda P., Touitou I. (2009) Clin. Genet. 75, 298–300 [DOI] [PubMed] [Google Scholar]
- 8. Qian J., Deveault C., Bagga R., Xie X., Slim R. (2007) Hum Mutat 28, 741. [DOI] [PubMed] [Google Scholar]
- 9. Wang C. M., Dixon P. H., Decordova S., Hodges M. D., Sebire N. J., Ozalp S., Fallahian M., Sensi A., Ashrafi F., Repiska V., Zhao J., Xiang Y., Savage P. M., Seckl M. J., Fisher R. A. (2009) J. Med. Genet. 46, 569–575 [DOI] [PubMed] [Google Scholar]
- 10. Milhavet F., Cuisset L., Hoffman H. M., Slim R., El-Shanti H., Aksentijevich I., Lesage S., Waterham H., Wise C., Sarrauste de Menthiere C., Touitou I. (2008) Hum. Mutat. 29, 803–808 [DOI] [PubMed] [Google Scholar]
- 11. Messaed C., Chebaro W., Di Roberto R. B., Rittore C., Cheung A., Arseneau J., Schneider A., Chen M. F., Bernishke K., Surti U., Hoffner L., Sauthier P., Buckett W., Qian J., Lau N. M., Bagga R., Engert J. C., Coullin P., Touitou I., Slim R. (2011) J. Med. Genet. 48, 540–548 [DOI] [PubMed] [Google Scholar]
- 12. Muhlstein J., Golfier F., Rittore C., Hajri T., Philibert L., Abel F., Beneteau C., Touitou I. (2011) Eur. J. Obstet. Gynecol. Reprod. Biol. 157, 197–199 [DOI] [PubMed] [Google Scholar]
- 13. Tschopp J., Martinon F., Burns K. (2003) Nat. Rev. Mol. Cell Biol. 4, 95–104 [DOI] [PubMed] [Google Scholar]
- 14. Okada K., Hirota E., Mizutani Y., Fujioka T., Shuin T., Miki T., Nakamura Y., Katagiri T. (2004) Cancer Sci. 95, 949–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ohno S., Kinoshita T., Ohno Y., Minamoto T., Suzuki N., Inoue M., Suda T. (2008) Anticancer Res. 28, 2493–2497 [PubMed] [Google Scholar]
- 16. Kinoshita T., Wang Y., Hasegawa M., Imamura R., Suda T. (2005) J. Biol. Chem. 280, 21720–21725 [DOI] [PubMed] [Google Scholar]
- 17. Agostini L., Martinon F., Burns K., McDermott M. F., Hawkins P. N., Tschopp J. (2004) Immunity 20, 319–325 [DOI] [PubMed] [Google Scholar]
- 18. Loock J., Lamprecht P., Timmann C., Mrowietz U., Csernok E., Gross W. L. (2010) J. Allergy Clin. Immunol. 125, 500–502 [DOI] [PubMed] [Google Scholar]
- 19. Hoffman H. M., Mueller J. L., Broide D. H., Wanderer A. A., Kolodner R. D. (2001) Nat. Genet. 29, 301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jéru I., Marlin S., Le Borgne G., Cochet E., Normand S., Duquesnoy P., Dastot-Le Moal F., Cuisset L., Hentgen V., Fernandes Alnemri T., Lecron J. C., Dhote R., Grateau G., Alnemri E. S., Amselem S. (2010) Arthritis Rheum. 62, 1176–1185 [DOI] [PubMed] [Google Scholar]
- 21. Slim R., Ao A., Surti U., Zhang L., Hoffner L., Arseneau J., Cheung A., Chebaro W., Wischmeijer A. (2011) Placenta 32, 409–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kastner D. L., Aksentijevich I., Goldbach-Mansky R. (2010) Cell 140, 784–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tomoda Y., Fuma M., Saiki N., Ishizuka N., Akaza T. (1976) Am. J. Obstet. Gynecol. 126, 661–667 [DOI] [PubMed] [Google Scholar]
- 24. Khanna A., Khanna S., Gupta R. M., Gupta S., Khanna A. K. (1985) J. Surg. Oncol. 28, 4–6 [DOI] [PubMed] [Google Scholar]
- 25. Ho P. C., Mak L. W., Lawton J. W., Ma H. K. (1980) J. Reprod. Immunol. 1, 307–319 [DOI] [PubMed] [Google Scholar]
- 26. Carta S., Tassi S., Semino C., Fossati G., Mascagni P., Dinarello C. A., Rubartelli A. (2006) Blood 108, 1618–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Edwards R., Crow J., Dale S., Macnamee M., Hartshorne G., Brinsden P. (1990) Lancet 335, 1030–1031 [DOI] [PubMed] [Google Scholar]
- 28. Zheng P., Dean J. (2009) Proc. Natl. Acad. Sci. 106, 7473–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tong Z. B., Gold L., Pfeifer K. E., Dorward H., Lee E., Bondy C. A., Dean J., Nelson L. M. (2000) Nat. Genet. 26, 267–268 [DOI] [PubMed] [Google Scholar]
- 30. Helwani M. N., Seoud M., Zahed L., Zaatari G., Khalil A., Slim R. (1999) Hum. Genet. 105, 112–115 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.