Background: IL-2 expression is suppressed in SLE T lymphocytes.
Results: CREMα binding to IL2 mediates histone H3K18 deacetylation through HDAC1 and CpG-DNA methylation through DNMT3a.
Conclusion: CREMα mediates epigenetic remodeling of IL2 in SLE T cells.
Significance: Understanding the molecular mechanisms that cause cytokine imbalances in SLE will help to establish target-directed therapeutic approaches.
Keywords: Chromatin Histone Modification, Chromatin Remodeling, DNA Methylation, Immunology, Interleukin, Transcription Factors, CREM, IL-2, SLE, T Cell
Abstract
IL-2 is a key cytokine during proliferation and activation of T lymphocytes and functions as an auto- and paracrine growth factor. Regardless of activating effects on T lymphocytes, the absence of IL-2 has been linked to the development of autoimmune pathology in mice and humans. Systemic lupus erythematosus (SLE) is a multifactorial autoimmune disease and characterized by dysregulation of lymphocyte function, transcription factor and cytokine expression, and antigen presentation. Reduced IL-2 expression is a hallmark of SLE T lymphocytes and results in decreased numbers of regulatory T lymphocytes which play an important role in preventing autoimmunity. Reduced IL-2 expression was linked to overproduction of the transcription regulatory factor cAMP-responsive element modulator (CREM)α in SLE T lymphocytes and subsequent CREMα binding to a CRE site within the IL2 promoter (−180 CRE). In this study, we demonstrate the involvement of CREMα-mediated IL2 silencing in T lymphocytes from SLE patients through a gene-wide histone deacetylase 1-directed deacetylation of histone H3K18 and DNA methyltransferase 3a-directed cytosine phosphate guanosine (CpG)-DNA hypermethylation. For the first time, we provide direct evidence that CREMα mediates silencing of the IL2 gene in SLE T cells though histone deacetylation and CpG-DNA methylation.
Introduction
IL-2 is a pluripotent cytokine that plays a central role during proliferation and activation of T lymphocytes where it functions as an auto- and paracrine growth factor. Regardless of its activating effects on T lymphocytes, the absence of IL-2 has been linked to the development of autoimmune pathology in mice (1) and humans (2).
Systemic lupus erythematosus (SLE)4 is a multifactorial autoimmune disease. It is characterized by multisystem involvement, phases of remission and relapses, and the presence of autoantibodies. Dysregulation of B and T lymphocyte function, transcription factor and cytokine expression, and antigen presentation have been reported (3). Reduced IL-2 production (4, 5) and IL-2 receptor signaling (6, 7) are hallmarks of SLE T lymphocytes (2, 8, 9). Impaired IL-2 expression results in decreased generation of regulatory T lymphocytes which play an important role in preventing autoimmunity. Furthermore, IL-2 regulates activation-induced cell death, which is defective in T lymphocytes from SLE patients (10). Defective cytotoxic CD8+ T cell function in SLE patients has been linked to reduced IL-2 expression and may contribute to higher susceptibility to infections with intracellular pathogens (11).
A dysbalance of CRE-binding protein (CREB) and CRE modulator α (CREMα) contributes to the reduced IL-2 expression in SLE T cells (12). CREMα and CREB share a cis-regulatory element −180 bp upstream the transcriptional start site of IL2 (−180 CRE) (Fig. 1). In resting T lymphocytes, CREB occupies the −180 CRE site; following T cell activation, CREB becomes phosphorylated (pCREB), resulting in increased IL-2 expression (8). SLE patients exhibit increased levels of CREMα compared with healthy controls, and pCREB gets replaced by pCREMα (12), (13), resulting in transcriptional silencing of IL2.
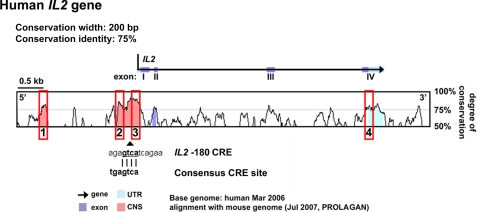
FIGURE 1.
Alignment of the human and mouse IL2 genes. Pink peaks denote CNS sites, purple peaks are exons with sequence identity of >75% over at least 200 bp. Red squares denote conserved noncoding sequences that were determined regions of interest for further analysis of histone modifications and DNA methylation. The previously reported IL2 −180 CRE site is indicated under the alignment.
Disease- and tissue-specific cytokine expression depends largely on a network of transcription factors. To promote physiologic functions, transcription factors bind to corresponding DNA sequences depending on chromatin structure and gene accessibility. Genes can be silenced by cytosine phosphate guanosine (CpG-)DNA methylation through DNA methyltransferases (DNMTs). DNMT1 is responsible for remethylation of hemimethylated CpGs during cell division, whereas DNMT3a and b induce de novo methylation. In SLE, a generally hypomethylated state of T and B lymphocytes has been reported. Still, DNA methylation seems to occur in a region- and tissue-specific manner (3, 14–16). The nucleosome is the basic subunit of chromatin. Modifications of the N-terminal histone tails, such as histone acetylation, methylation, and phosphorylation, support changes in nucleosome arrangement and chromatin structure. Generally, histone acetylation is associated with transcriptional activation while histone trimethylation is repressing transcriptional activity. DNA methylation and histone modifications follow similar patterns and are interconnected by various mechanisms (3).
In the present study, we show that activation of naïve CD4+ T lymphocytes from healthy individuals (through CD3/CD28 signaling) mediates histone H3K18 hyperacetylation and H3K27 hypomethylation that result in an increased IL-2 expression. In contrast, T lymphocytes from SLE patients exhibit reduced H3K18 acetylation and increased H3K27 methylation. Furthermore, histone deacetylase 1 (HDAC1) recruitment to the IL2 promoter co-localized with CREMα binding. We recorded increased CpG-DNA methylation across the IL2 gene in SLE T lymphocytes compared with controls. DNMT3a co-localized with CREMα at the −180 CRE site, and CREMα and DNMT3a physically associated when co-expressed in the same cell. This suggests direct recruitment of DNMT3a to IL2 −180 CRE through CREMα. Overexpression of DNMT3a in human T lymphocytes mediated IL2 gene-wide CpG-DNA methylation which resulted in transcriptional silencing. Thus, we provide a novel mechanism causing transcriptional repression of the IL2 gene in SLE T lymphocytes by CREMα whereby it renders the regulatory region inaccessible to transcription.
EXPERIMENTAL PROCEDURES
Study Subjects and T Cell Culture
All SLE patients included in our studies were diagnosed according to the American College of Rheumatology classification criteria (17) and recruited from the Division of Rheumatology at Beth Israel Deaconess Medical Center, Boston, MA and gave written informed consent under protocol 2006-P-0298 (Table 1). Healthy individuals were chosen as controls. Peripheral venous blood was collected in heparin-lithium tubes, and total human T cells were purified as described before (18). Jurkat and primary human T lymphocytes were kept in RPMI 1640 medium supplemented with 10% FBS.
TABLE 1.
Epidemiologic information and disease activity of included SLE patients
F, female; SLEDAI, systemic lupus erythematosus disease activity index (24) (inactive, 0; mild activity, 1–5; moderate activity, 6–10; high activity, 11–19; very high activity, >20); MMF, mycophenolate mofetil; HCQ, hydroxychloroquine; GC, glucocorticoids; Aza, azathioprine; CmpG-IP, methyl-cytosine-phosphate-guanine immunoprecipitation.
| Patient | Gender | Age | SLEDAI | Treatment | Application |
|---|---|---|---|---|---|
| SLE1 | F | 30 | 4 | MMF, HCQ, GC | CmpG-IP |
| SLE2 | F | 37 | 10 | HCQ, GC | CmpG-IP |
| SLE3 | F | 36 | 10 | MMF, GC | CmpG-IP |
| SLE4 | F | 39 | 8 | CmpG-IP | |
| SLE5 | F | 38 | 36 | GC | CmpG-IP |
| SLE6 | F | 54 | 14 | HCQ | CmpG-IP |
| Average | 39 | 13.67 | |||
| Range | (30–54) | (4–36) | |||
| SLE7 | F | 36 | 4 | MMF | ChIP |
| SLE8 | F | 27 | 6 | GC | ChIP |
| SLE9 | F | 39 | 5 | HCQ | ChIP |
| SLE10 | F | 32 | 8 | Aza | ChIP |
| Average | 33.5 | 5.75 | |||
| Range | (27–39) | (4–8) | |||
Naïve CD4+ T cells from healthy controls were purified from total T cell suspension using the Human Naïve CD4+ T Cell Isolation kit II (Miltenyi Biotec). For activation assays, naïve CD4+ T cells were incubated in the absence or presence of plate-bound anti-CD3 and anti-CD28 antibodies (BioXCell; both at 1 μg/ml) for 72 h.
mRNA Analysis
Total RNA was isolated, using the RNeasy Mini kit (Qiagen). cDNA was generated using a first strand cDNA synthesis kit (Invitrogen). For gene expression analyses, real-time PCR was performed using Taqman site-specific primers and probes (Roche Diagnostics) on an ABI OneStepPlus Real-time PCR System. Results were normalized to GAPDH.
ChIP
Anti-HDAC1, anti-histone 3 lysine 18 acetylation (H3K18ac), and anti-histone 3 lysine 27 trimethylation (H3K27me3), normal rabbit IgG, and normal mouse IgG were from Upstate (Millipore). Anti-CREMα antibodies were generated as described previously (19), and anti-DNMT3a was from Abcam. ChIP Grade protein A/G plus agarose was purchased from Pierce (ThermoScientific). ChIP assays were carried out essentially according to the manufacturer's instructions (Upstate Biotechnology/Millipore). Briefly, 1–2 × 106 cells were cross-linked with 1% formaldehyde, washed with cold PBS, and lysed in buffer containing protease inhibitors (Roche Applied Science). Lysates were sonicated to shear DNA and sedimented, and diluted supernatants were immunoprecipitated with antibodies. A proportion (20%) of the diluted supernatants was kept as “input” (input represents PCR amplification of the total sample). The bead-bound protein-DNA complexes were eluted in 1% SDS, 0.1 m NaHCO3, and cross-linking was reversed at 65 °C. DNA was recovered with Qiaprep DNA Miniprep kits (Qiagen) and subjected to PCR analysis on an ABI OneStepPlus Real-time PCR System. The sequences of the primer pairs used for quantitative PCR are listed in Table 2.
TABLE 2.
Primer sequences for H3K18ac, H3K27me3, and CREMα ChIP
| Region | Forward primer | Reverse primer | Product length | Annealing temperature for PCR |
|---|---|---|---|---|
| bp | º C | |||
| IL2CNS1 | GGGCTTTTCCAATGAGGT | CTACAGGGTCCCAGAAGTAT | 111 | 55 |
| IL2CNS2 | GTGATAGGGAACTCTTGAAC | TCAGACAGGTAAAGTCTTTG | 118 | 55 |
| IL2-180CRE | ACCCCCTTTCTGACTGAGTTACCA | CCCCACCCCCTTAAAGAAAGGAGG | 139 | 55 |
| IL2CNS3 | AGCACCTATAATGCCTTCAG | AATAGGAGCCATCACTTCAC | 110 | 55 |
| IL2CNS4 | ATGCTTACTCTGTGCTGTTTC | GGGCTCTAAAATGGTTTCACT | 163 | 55 |
Methylated CpG-DNA Immunoprecipitation
The methylated CpG-DNA immunoprecipitation assay was carried out according to the manufacturer's instructions (Zymo Research). Briefly, genomic DNA from T cells obtained from SLE patients and healthy control individuals was purified using the AllPrep RNA/DNA/protein Mini kit (Qiagen), sheared to fragments of ∼200 bp using DNA shearase (Zymo Research). Subsequently, 100 ng of sheared genomic DNA was used for methylated CpG-DNA immunoprecipitation. Equal amounts (100 ng) of completely (100%) methylated human DNA, and unmethylated human DNA (Zymo Research) were included as input and negative control respectively. Methylated DNA was recovered and subjected to PCR analysis on an ABI OneStepPlus real-time PCR system. Sequences of PCR primers are listed in Table 1.
Gene Expression Plasmids
An expression plasmid for human CREMα (in pcDNA3.1/V5-His-TOPO plasmid; Invitrogen) was provided by G. N. Europe-Finner (Faculty of Medical Sciences, Newcastle upon Tyne, UK) (20). DNMT3a expression plasmids have been described before (21). Three million Jurkat T cells were transfected with a total amount of 3 μg of expression plasmid by the Amaxa transfection system (Lonza).
Co-immunoprecipitation of DNMT3a with CREMα
One million HEK293T cells were transfected with expression plasmids for (i) pCDNA3 and CREMα, (ii) pCDNA3 and DNMT3a, or (iii) CREMα and DNMT3 (2 μg of each plasmid/transfection) using Lipofectamine (Invitrogen) according to the manufacturer's instructions. 48 h after transfection cells were harvested and lysed in 400 μl of RIPA buffer including protease inhibitors (Roche). Cell lysates were precleared for 30 min at 4 °C with 40 μl of Pansorbin (Calbiochem) and 4 μl of nonspecific rabbit IgG1 (Santa Cruz Biotechnology). After centrifugation (14,000 rpm, 10 min, 4 °C) 200 μl of supernatant was incubated with either anti-CREMα (19) or anti-DNMT3a (Abcam) antibodies at 4 °C overnight. 20 μl of Pansorbin was added and incubated for 1 h at 4 °C and Pansorbin-bound antibody-protein complexes were pelleted by centrifugation at 14,000 rpm. After several washes with RIPA buffer (including protease inhibitors) pellets were resuspended in 50 μl of RIPA buffer, reducing sample buffer, boiled for 5 min at 95 °C and pelleted. Supernatants were subjected to SDS-PAGE as described before (19). Proteins were transferred to PVDF membranes and detected by anti-DNMT3a antibody, suitable secondary peroxidase-linked anti-rabbit antibody (Santa Cruz Biotechnology), and ECL (Amersham Biosciences) as chemiluminescent. Input controls to confirm overexpression of the respective proteins was performed by immunoblotting the nonimmunoprecipitated cell lysates.
Data Analysis
A paired two-tailed Student's t test was used for statistical analysis.
RESULTS
Bioinformatic Analysis of the IL2 Gene
To investigate epigenetic patterns across the human IL2 gene, we defined regions of interest, based on bioinformatic approaches. We aligned the mouse and human IL2 genes (VISTA Genome Browser) and searched for conserved noncoding sequences (CNS), exons and conserved untranslated regions (UTRs) (Fig. 1). CNS sites were defined as regions with sequence homology of >75% between human and mouse over at least 200 bp.
Four regions of interest (CNS1–CNS4) were defined, based on the degree of sequence conservation and the presence of reported regulatory regions. CNS1–CNS3 are located within the proximal promoter of the IL2 gene; CNS4 localizes to the highly conserved 3′-UTR.
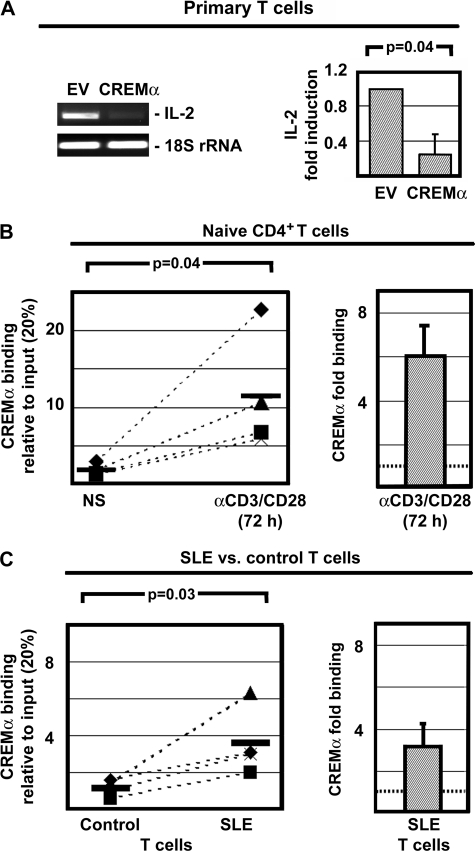
CREMα Binds to −180 CRE and Reduces IL-2 Expression in SLE T Lymphocytes
In line with previous reports from our group, CREMα overexpression in primary human T lymphocytes resulted in reduced IL-2 expression (Fig. 2A). To investigate the involvement of CREMα binding in autoregulatory limitation of IL-2 expression, we investigated CREMα binding to the −180 CRE site in naïve and activated CD4+ T lymphocytes. We detected weak CREMα binding to the −180 CRE site in naïve CD4+ T lymphocytes and a 6-fold increase in response to T lymphocyte activation with anti-CD3/CD28 antibodies (p = 0.04) (Fig. 2B). Because CREMα expression is increased in T lymphocytes from SLE patients and it mediates transcriptional silencing of IL2, we investigated CREMα recruitment to the −180 CRE site in T lymphocytes from SLE patients and compared our findings with controls. We detected weak binding of CREMα to the −180 CRE element in unstimulated control T lymphocytes. In SLE T lymphocytes CREMα binding to the −180 CRE site was significantly stronger (3-fold; p = 0.03) (Fig. 2C). This confirms in vivo CREMα binding to the −180 CRE site of the IL2 promoter and subsequent transcriptional silencing.
FIGURE 2.
CREMα and its effects on IL-2 expression. A, overexpression of CREMα in primary human T lymphocytes (for 5 h) results in reduced IL-2 mRNA expression (n = 4). B, CREMα weakly binds to the IL2 −180 CRE in naïve CD4+ T lymphocytes. CREMα binding is significantly (p = 0.04) increased in response to T cell activation with anti-CD3/CD28 antibodies (for 72 h). Pairs of naïve CD4+ T lymphocytes and activated CD4+ T cells from the same individual are connected by dashed lines. C, CREMα binds to the IL2 −180 CRE of T lymphocytes from healthy control individuals. Corresponding results from age-, gender-, and ethnicity-matched controls are connected to data from SLE T cells by dashed lines. CREMα binding to the IL2 −180 CRE site is significantly (p = 0.03) increased in T lymphocytes from SLE patients. Error bars, S.D.
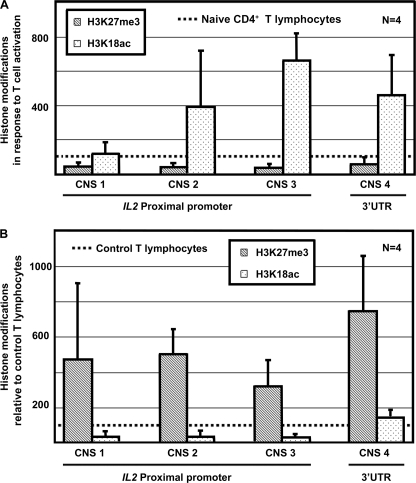
IL-2 Expression Is Associated with Histone H3K18 Acetylation and H3K27 Demethylation
It has been reported previously that activation of murine T lymphocytes with anti-CD3/CD28 antibodies results in hyperacetylation of histones H3 and H4 within the IL2 gene (22). Furthermore, CREMα binding to the IL2 promoter in Jurkat T cells has been shown to result in HDAC1 recruitment and transcriptional silencing of IL2 (23). To understand further the regulation of IL-2 expression in SLE T lymphocytes, we investigated dynamic modifications during the activation of primary naïve human CD4+ T lymphocytes (with anti-CD3 and anti-CD28 antibodies). In naïve CD4+ T lymphocytes histone H3 was hypomethylated at lysine 18 and hypermethylated at lysine 27. In response to T cell activation, we observed increased acetylation of histone H3K18 and decreased methylation of H3K27 (Fig. 3A). This reflects chromatin remodeling and results in a transcriptionally active “euchromatic” state. Because T lymphocytes from SLE patients fail to express IL-2 (8), we investigated histone modifications in T cells from four SLE patients and corresponding age-, gender-, and ethnicity-matched healthy controls (Table 1). All patients included in the study were female. The average SLE disease activity index (SLEDAI) score of the four patients included in this part of the study was 5.75, representing an overall mild to moderate disease activity (24). Compared with healthy controls, SLE T cells were hypermethylated at histone H3K27 and hypoacetylated at H3K18, reflecting a transcriptionally inactive or “heterochromatic” state (Fig. 3B).
FIGURE 3.
Histone modifications of the IL2 gene in response to T lymphocyte activation and in SLE T cells. A, activation of naïve CD4+ T lymphocytes with anti-CD3 and anti-CD28 antibodies results in increased histone H3K18 acetylation and decreased histone H3K27 trimethylation. Results are displayed as relative increase in histone H3K18 acetylation and H3K27 trimethylation based on the situation in naïve CD4+ T lymphocytes (indicated by dashed line = 100%). B, T lymphocytes from SLE patients display decreased histone H3K18 acetylation and increased histone H3K27 trimethylation compared with T lymphocytes from healthy controls. Results are displayed as relative histone H3K18 acetylation and H3K27 trimethylation based on the situation in control T lymphocytes (indicated by dashed line = 100%). Error bars, S.D.
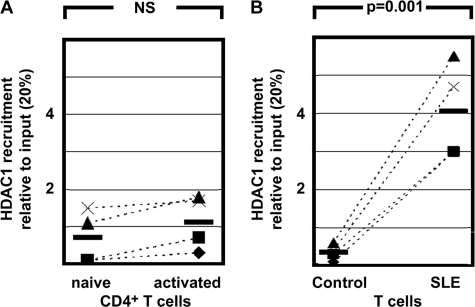
CREMα Binding to the IL2 −180 CRE Site Mediates HDAC1 Recruitment and Reduced IL-2 Expression
It has been reported that CREMα overexpression in Jurkat T cells results in transcriptional silencing of IL2. It has further been reported that CREMα recruits HDAC1 to the IL2 promoter, resulting in hypoacetylation of the IL2 gene (12, 23). Physical interaction between HDAC1 and CREMα has been demonstrated by co-immunoprecipitation experiments (23). To investigate further the effects of CREMα on IL-2 expression in primary human T cells, and the involvement of CREMα in IL-2 regulation under physiological conditions, we investigated HDAC1 recruitment to the −180 CRE site in naïve CD4+ T lymphocytes and in response to activation with anti-CD3/CD28 antibodies. We detected weak binding of HDAC1 to the −180 CRE site and no significant induction of HDAC1 recruitment after T lymphocyte activation (Fig. 4A). In SLE T lymphocytes, we detected significantly more HDAC1 recruitment to this region compared with control T cells (Fig. 4B). This is in agreement with the aforementioned differences in histone H3K18 acetylation between naïve CD4+ T lymphocytes in response to activation and SLE T cells. Thus, our findings suggest that CREMα mediated HDAC1 recruitment to IL2 −180 CRE that results in gene-wide deacetylation of histone H3K18 and transcriptional silencing. This may be due to increased CREMα levels in SLE T lymphocytes compared with controls.
FIGURE 4.
HDAC1 recruitment to the IL2 −180 CRE site as detected by ChIP. A, Naïve and activated (anti-CD3/CD28) CD4+ T lymphocytes display no relevant HDAC1 binding. Pairs of naïve CD4+ T lymphocytes and activated CD4+ T cells from the same individual are connected by dashed lines. NS, not significant. B, HDAC1 only weakly binds to the IL2 −180 CRE site in control T lymphocytes. In T lymphocytes from SLE patients, we detected significantly more CREMα recruitment to the −180 CRE site (p = 0.001). Corresponding results from age-, gender-, and ethnicity-matched controls are connected to data from SLE T cells by dashed lines.
The IL2 Gene Is Hypermethylated in SLE T Lymphocytes
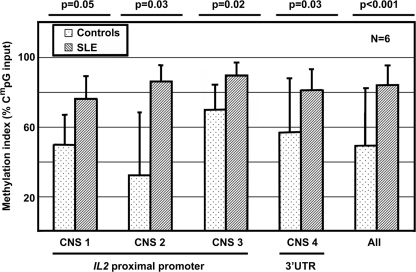
It has been reported that epigenetic alterations play a role in the pathophysiology of SLE. To define epigenetic mechanisms that are responsible for transcriptional repression of IL-2 in T lymphocytes from SLE patients, we investigated the degree of IL2 CpG-DNA methylation in T lymphocytes of six SLE patients and age-, gender-, and ethnicity-matched controls. All patients included in the study were female, and presented with mildly to highly active disease as assessed, using SLEDAI scores (Table 1). The average SLEDAI score of the six patients included in this part of the study was 13.67, representing an overall high disease activity (24). Using immunoprecipitating antibodies against methylated CpG-nucleotides, we detected significantly increased CpG-DNA methylation across the IL2 gene in SLE patients compared with controls. Across all investigated CNS regions, we detected significantly higher degrees of DNA methylation in T lymphocytes from SLE patients: methylation indexes of 84% in SLE T cells versus 49% in healthy control T cells (Fig. 5).
FIGURE 5.
CpG-DNA methylation of the IL2 gene as detected by immunoprecipitation of methylated CpG sequences. CNS1–CNS3 are located in the 2 kb spanning proximal promoter, CNS4 is located in the 3′-UTR. Values are normalized to 100% methylated (input) DNA and unmethylated (control) DNA. The IL2 gene T lymphocytes from SLE patients is methylated to a significantly lower degree compared with control T cells (p < 0.001). Error bars, S.D.
CpG-DNA Methylation through DNMT3a Results in Reduced IL-2 Expression
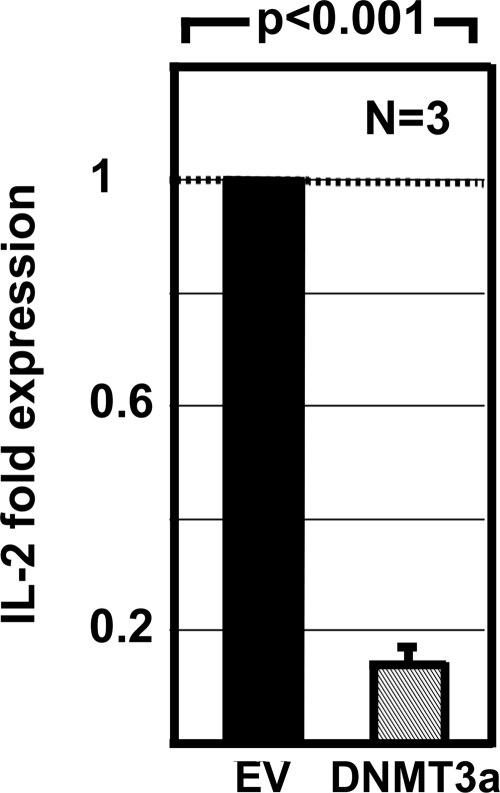
To investigate a possible role of CpG-DNA methylation in the epigenetic regulation of IL2, we overexpressed DNA methyltransferase DNMT3a in Jurkat T cells. DNMT3a overexpression resulted in significantly reduced IL-2 mRNA expression (Fig. 6).
FIGURE 6.
Overexpression of DNMT3a in Jurkat T cells suppresses the expression of IL-2 mRNA. Empty vector (EV; 3 μg), or DNMT3a (3 μg) expression plasmids were transfected into Jurkat T cells. Cells were cultured overnight, stimulated with anti-CD3 and anti-CD28 antibodies for 5 h, and harvested. RNA was purified from and IL-2 transcripts were measured and normalized to GAPDH by real-time RT-PCR. The expression levels of the transcripts were significantly decreased in Jurkat T cells transfected with DNMT3a compared with cells transfected with empty control plasmids (p < 0.001). The results represent the mean ± S.D. (error bar) from three independent experiments.
CREMα Interacts with DNMT3a, Resulting in DNMT3a Recruitment to the −180 CRE Site in SLE T Lymphocytes
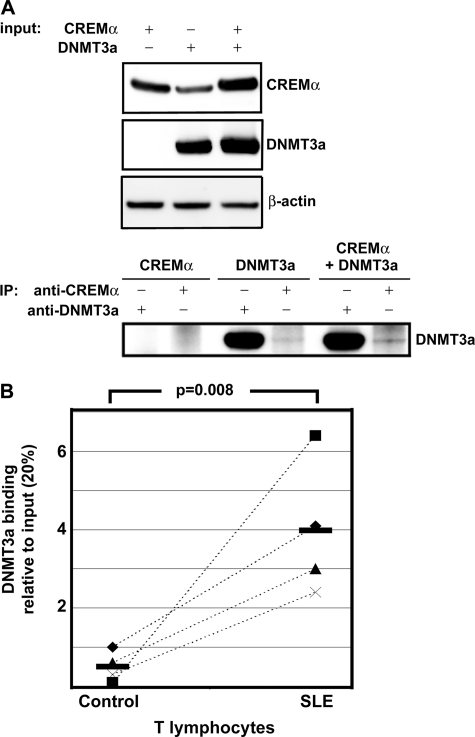
Because we found increased CpG-DNA methylation of the IL2 gene in SLE T cells compared with controls, we aimed to investigate whether CREMα can interact directly with DNMT3a and possibly recruit DNMT3a to the IL2 promoter. Therefore, we overexpressed DNMT3a and CREMα in HEK293T cells and determined protein expression in nuclear extracts (Fig. 7A, upper). When we overexpressed both proteins, DNMT3a could be co-immunoprecipitated by the anti-CREMα antibody, suggesting a direct interaction between CREMα and DNMT3a (Fig. 7A, lower, sixth lane). This band was markedly weaker when only DNMT3a was overexpressed (fourth lane). This seems to reflect DNMT3a interaction with endogenous CREMα.
FIGURE 7.
DNMT3a interacts with CREMα and gets recruited to IL2 −180 CRE. A, upper, CREMα and DNMT3a were transfected into HEK293T cells as indicated, and overexpression is shown by immunoblotting of cell lysates. Lower, co-immunoprecipitated (IP) DNMT3a is shown. Both endogenous CREMα (fourth lane 4) and overexpressed CREMα protein (sixth lane) co-immunoprecipitate overexpressed DNMT3a protein. B, SLE T cells that produce significantly less IL-2 compared with controls show increased DNMT3a recruitment to the IL2 −180 CRE site compared with control T lymphocytes. Corresponding results from age-, gender-, and ethnicity-matched controls are connected to data from SLE T cells by dashed lines.
Subsequently, we investigated DNMT3a recruitment to the IL2 −180 CRE element and detected significantly more DNMT3a recruitment to the IL2 promoter in SLE T lymphocytes when compared with healthy controls (Fig. 7B). These findings suggest a direct interaction of CREMα with DNMT3a that may result in DNMT3a recruitment to the −180 CRE site, resulting in CpG-DNA methylation of the IL2 gene and transcriptional silencing.
DISCUSSION
Increased CREMα binding to the IL2 promoter has been reported to result in transcriptional silencing of IL2 that is mediated by several mechanisms: (i) trans-repression of the IL2 promoter (8), (ii) the failure to activate the histone acetyltransferase p300 by CREMα (12), and (iii) HDAC1 recruitment through CREMα (23). These events suggest histone hypoacetylation and subsequently reduced IL-2 expression. Still, the involvement of these mechanisms in IL-2 silencing in SLE T lymphocytes has not been shown yet.
In the present study, we demonstrate for the first time that the IL2 gene in T lymphocytes from SLE patients undergoes epigenetic remodeling at different levels. We present evidence that CREMα mediates gene-wide histone deacetylation in primary SLE T lymphocytes. Furthermore, we demonstrate increased histone H3K27 trimethylation and CREMα-mediated CpG-DNA methylation through DNMT3a. This suggests an important role of these mechanisms in transcriptional silencing of IL2 in SLE T cells.
Modifications to histone tails, such as acetylation, methylation, and phosphorylation, are responsible for changes in the organization of nucleosomes, thus regulating genomic accessibility for transcription factors and RNA polymerases (25). Generally, histone acetylation is associated with transcriptional activation whereas histone trimethylation reduces transcriptional activity. Epigenetic patterns in SLE are complex (3, 14–16). Recent evidence suggests that region- and tissue-specific histone acetylation is associated with high disease activity, whereas histone acetylation in other regions seems to have protective effects (3, 14, 26–30).
The IL2 gene in naïve CD4+ T lymphocytes undergoes histone H3K18 hyperacetylation, and histone H3K27 demethylation in response to T lymphocyte activation. We observed reverse effects in T lymphocytes from SLE patients. We detected higher degrees of H3K18 acetylation and lower degrees of H3K27 methylation in control T lymphocytes compared with SLE T cells. In agreement with this, we detected increased HDAC1 recruitment to the −180 CRE site within the IL2 promoter in SLE T lymphocytes, but not in control T cells, or in response to activation of naïve CD4+ T lymphocytes. This could reflect an important mechanism in the transcriptional silencing of IL2 in SLE T lymphocytes and is in agreement with previous findings of our group (23). We demonstrated HDAC1 recruitment to the −180 CRE site of the IL2 promoter in response to overexpression of CREMα and stimulation with anti-CD3 and anti-CD28 antibodies (23). Because T lymphocytes from SLE patients were reported to produce increased amounts of CREMα compared with healthy controls, HDAC1 recruitment could likely be “dose-dependent” (13) (Fig. 8). Our observation that HDAC1-mediated deacetylation of IL2 results in transcriptional silencing is in line with reports that the application of HDAC inhibitors resulted in down-regulation proinflammatory cytokines in MRL/lpr splenocytes in vitro and in ameliorated proteinuria, glomerulonephritis, and splenomegaly in vivo (28).
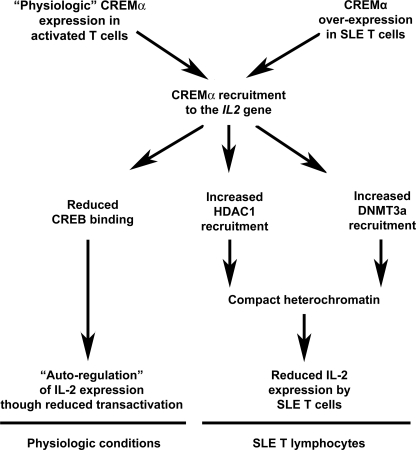
FIGURE 8.
Model for the involvement of CREMα in the regulation of IL2 gene expression under physiologic conditions and in SLE. Under physiologic conditions, CREMα gets expressed in response to T lymphocyte activation. Increased CREMα expression results in binding to the −180 CRE site and competition with the IL2 trans-activator pCREB, an activator of IL-2 expression. This results in an autoregulatory reduction of IL-2 expression. SLE T lymphocytes overexpress CREMα. This results in competition with pCREB and reduced trans-activation of IL2. CREMα binding to the IL2 −180 CRE site results in recruitment of HDAC1 and DNMT3a to the IL2 promoter. This results in epigenetic remodeling and transcriptional repression through histone deacetylation and CpG-DNA methylation.
CpG-DNA methylation plays an important role in the epigenetic control of gene expression (31, 32). Transcription factors need to bind cis-regulatory regions to exert physiologic functions which can be prevented by CpG-DNA methylation and are mediated through DNMTs. In SLE and other autoimmune diseases, “global” CpG-DNA methylation has been reported to be reduced, resulting in overexpression of primarily proinflammatory cytokines (3, 18, 21).
The murine Il2 gene has been shown to be under epigenetic control through CpG-DNA methylation (33). It has been reported that following activation of murine naïve CD4+ T lymphocytes, the Il2 promoter undergoes rapid and sustained demethylation (34). In this study, we detected similar patterns, comparing the human IL2 promoter of SLE T cells with controls. The IL2 promoter in SLE T lymphocytes that fail to produce IL-2 is highly methylated compared with control samples. Additionally, we detected DNMT3a recruitment to the IL2 promoter that co-localized to a CREMα binding site (−180 CRE), and overexpression of DNMT3a resulted in reduced IL-2 transcription that was most likely caused by increased CpG-DNA methylation. For the first time, we detected a direct interaction between CREMα and DNMT3a. DNMT3a is responsible for DNA de novo methylation and is independent of cell division. This suggests that DNMT3a recruitment to the IL2 promoter is mediated by CREMα and results in increased CpG-DNA methylation of the IL2 gene. Because activated CD4+ T lymphocytes and lymphocytes from healthy controls did not exhibit DNMT3a recruitment to the −180 CRE site, we propose that DNMT3a recruitment to the IL2 promoter, such as the aforementioned HDAC1 recruitment, could be a “dose-dependent” effect of CREMα. Increased histone H3K27 trimethylation of the IL2 gene is increased in SLE T lymphocytes and could be a reflection of CpG-DNA methylation in the same region. Several reports indicate that CpG-DNA methylation and histone modifications coincide and that they are interconnected by several mechanisms (35). Methyl-CpG-binding domain proteins for instance selectively bind methylated CpG-DNA and recruit HDACs and histone methyltransferases (36).
Our findings could also help to interpret conflicting data on DNMT expression in SLE patients. Authors have reported normal (37, 38), up-, and down-regulated (39, 40) DNMT expression in SLE patients. Conflicting results may be caused by differences in disease activity, but also by discrepancies between DNMT expression levels, protein activity, and the recruitment to specific sites that depend on further modulators, such as CREMα.
Whether our observations can be applied as biomarkers for disease activity remains to be elucidated. The patients included in our studies had moderate to high disease activity (24), which documents the involvement of epigenetic alterations in these cases. In this context it also needs to be mentioned that shifts in relative T lymphocyte numbers may influence our observations. To achieve sufficient cell numbers, whole/bulk T lymphocytes from SLE patients and controls were used in our studies rather than purified T cell subsets. Furthermore, environmental factors including medication have been reported to impact epigenetic patterns (3, 32). In the present study, treatment was rather heterogeneous. However, epigenetic patterns of SLE patients did not vary significantly. Further studies in larger cohorts are warranted to investigate the influence of disease activity and immunosuppressive treatment on epigenetic marks in SLE.
In contrast to IL-2, the proinflammatory cytokine IL-17A is overexpressed in T lymphocytes from SLE patients (41). In an accompanying manuscript (42), we report activation (through CD3/CD28 stimulation)-mediated CREMα recruitment to a CRE site within the IL17A promoter, resulting in trans-activation and epigenetic remodeling of the IL17A gene. Chromatin remodeling within and around the IL17A gene is associated with increased expression of IL-17A in naïve CD4+ T lymphocytes in response to activation, and in T lymphocytes from SLE patients. This could reflect basal T lymphocyte activation in SLE patients through increased T cell receptor signaling. In contrast to the IL2 gene, CREMα does not recruit HDAC1 and DNMT3a to the IL17A promoter in control and SLE T (42). These findings are contrary to the effects of CREMα on the IL2 promoter and its effects on gene expression, indicating activating and repressing features of the transcription factor CREMα. Thus, our findings add further evidence to the discussion of whether mediators of epigenetic modifications (such as CREMα) and resulting epigenetic marks display region- and tissue-specific patterns in health and disease.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI42269, R01 AI49954, and R01 AI85567 (to G. C. T.). This work was also supported by Deutsche Forschungsgemeinschaft Research Fellowship RA1927-1/1 (to T. R.).
- SLE
- systemic lupus erythematosus
- CNS
- conserved noncoding sequence
- CpG
- cytosine phosphate guanosine
- CRE
- cAMP-responsive element
- CREB
- CRE-binding protein
- CREM
- CRE modulator
- DNMT
- DNA methyltransferase
- HDAC1
- histone deacetylase 1
- RIPA
- radioimmune precipitation assay
- SLEDAI
- SLE disease activity index.
REFERENCES
- 1. Ma A., Koka R., Burkett P. (2006) Annu. Rev. Immunol. 24, 657–679 [DOI] [PubMed] [Google Scholar]
- 2. Crispín J. C., Tsokos G. C. (2009) Autoimmun. Rev. 8, 190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hedrich C. M., Tsokos G. C. (2011) Trends Mol. Med., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alcocer-Varela J., Alarcón-Segovia D. (1982) J. Clin. Invest. 69, 1388–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Faucal P., Godard A., Peyrat M. A., Moreau J. F., Soulillou J. P. (1984) Ann. Immunol. 135D, 161–172 [DOI] [PubMed] [Google Scholar]
- 6. Wigfall D. R., Sakai R. S., Wallace D. J., Jordan S. C. (1988) Clin. Immunol. Immunopathol. 47, 354–362 [DOI] [PubMed] [Google Scholar]
- 7. Ishida H., Kumagai S., Umehara H., Sano H., Tagaya Y., Yodoi J., Imura H. (1987) J. Immunol. 139, 1070–1074 [PubMed] [Google Scholar]
- 8. Lieberman L. A., Tsokos G. C. (2010) J. Biomed. Biotechnol. 2010, 740619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Linker-Israeli M., Bakke A. C., Kitridou R. C., Gendler S., Gillis S., Horwitz D. A. (1983) J. Immunol. 130, 2651–2655 [PubMed] [Google Scholar]
- 10. Kovacs B., Thomas D. E., Tsokos G. C. (1996) Clin. Exp. Rheumatol. 14, 695–697 [PubMed] [Google Scholar]
- 11. Xu L., Zhang L., Yi Y., Kang H. K., Datta S. K. (2004) Nat. Med. 10, 411–415 [DOI] [PubMed] [Google Scholar]
- 12. Tenbrock K., Juang Y. T., Tolnay M., Tsokos G. C. (2003) J. Immunol. 170, 2971–2976 [DOI] [PubMed] [Google Scholar]
- 13. Solomou E. E., Juang Y. T., Tsokos G. C. (2001) J. Immunol. 166, 5665–5674 [DOI] [PubMed] [Google Scholar]
- 14. Zhao M., Sun Y., Gao F., Wu X., Tang J., Yin H., Luo Y., Richardson B., Lu Q. (2010) J. Autoimmun. 35, 58–69 [DOI] [PubMed] [Google Scholar]
- 15. Hewagama A., Richardson B. (2009) J. Autoimmun. 33, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Strickland F. M., Richardson B. C. (2008) Autoimmunity 41, 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hochberg M. C. (1997) Arthritis Rheum. 40, 1725. [DOI] [PubMed] [Google Scholar]
- 18. Sunahori K., Juang Y. T., Tsokos G. C. (2009) J. Immunol. 182, 1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Juang Y. T., Wang Y., Solomou E. E., Li Y., Mawrin C., Tenbrock K., Kyttaris V. C., Tsokos G. C. (2005) J. Clin. Invest. 115, 996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bailey J., Tyson-Capper A. J., Gilmore K., Robson S. C., Europe-Finner G. N. (2005) J. Mol. Endocrinol. 34, 1–17 [DOI] [PubMed] [Google Scholar]
- 21. Sunahori K., Juang Y. T., Kyttaris V. C., Tsokos G. C. (2011) J. Immunol. 186, 4508–4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adachi S., Rothenberg E. V. (2005) Nucleic Acids Res. 33, 3200–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tenbrock K., Juang Y. T., Leukert N., Roth J., Tsokos G. C. (2006) J. Immunol. 177, 6159–6164 [DOI] [PubMed] [Google Scholar]
- 24. Mosca M., Bombardieri S. (2006) Clin. Exp. Rheumatol. 24, S-99–104 [PubMed] [Google Scholar]
- 25. Renaudineau Y., Youinou P. (2011) Keio J. Med. 60, 10–16 [DOI] [PubMed] [Google Scholar]
- 26. Sullivan K. E., Suriano A., Dietzmann K., Lin J., Goldman D., Petri M. A. (2007) Clin. Immunol. 123, 74–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nambiar M. P., Warke V. G., Fisher C. U., Tsokos G. C. (2002) J. Cell. Biochem. 85, 459–469 [DOI] [PubMed] [Google Scholar]
- 28. Mishra N., Reilly C. M., Brown D. R., Ruiz P., Gilkeson G. S. (2003) J. Clin. Invest. 111, 539–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao M., Wu X., Zhang Q., Luo S., Liang G., Su Y., Tan Y., Lu Q. (2010) Arthritis Res. Ther. 12, R227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou Y., Qiu X., Luo Y., Yuan J., Li Y., Zhong Q., Zhao M., Lu Q. (2011) Lupus, in press [DOI] [PubMed] [Google Scholar]
- 31. Hedrich C. M., Ramakrishnan A., Dabitao D., Wang F., Ranatunga D., Bream J. H. (2010) Mol. Immunol. 48, 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hedrich C. M., Bream J. H. (2010) Immunol. Res. 47, 185–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Frommer M., McDonald L. E., Millar D. S., Collis C. M., Watt F., Grigg G. W., Molloy P. L., Paul C. L. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bruniquel D., Schwartz R. H. (2003) Nat. Immunol. 4, 235–240 [DOI] [PubMed] [Google Scholar]
- 35. Brenner C., Fuks F. (2007) Dev. Cell 12, 843–844 [DOI] [PubMed] [Google Scholar]
- 36. Ng H. H., Zhang Y., Hendrich B., Johnson C. A., Turner B. M., Erdjument-Bromage H., Tempst P., Reinberg D., Bird A. (1999) Nat. Genet. 23, 58–61 [DOI] [PubMed] [Google Scholar]
- 37. Wang G. S., Zhang M., Li X. P., Zhang H., Chen W., Kan M., Wang Y. M. (2009) Lupus 18, 1037–1044 [DOI] [PubMed] [Google Scholar]
- 38. Balada E., Ordi-Ros J., Serrano-Acedo S., Martinez-Lostao L., Rosa-Leyva M., Vilardell-Tarrés M. (2008) Immunology 124, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lei W., Luo Y., Yan K., Zhao S., Li Y., Qiu X., Zhou Y., Long H., Zhao M., Liang Y., Su Y., Lu Q. (2009) Scand. J. Rheumatol. 38, 369–374 [DOI] [PubMed] [Google Scholar]
- 40. Januchowski R., Wudarski M., Chwalińska-Sadowska H., Jagodzinski P. P. (2008) Clin. Rheumatol. 27, 21–27 [DOI] [PubMed] [Google Scholar]
- 41. Crispín J. C., Tsokos G. C. (2010) Curr. Opin. Rheumatol. 22, 499–503 [DOI] [PubMed] [Google Scholar]
- 42. Rauen T., Hedrich C. M., Juang Y. T., Tenbrock K., Tsokos G. C. (2011) J. Biol. Chem. 286, 43437–43446 [DOI] [PMC free article] [PubMed] [Google Scholar]