Abstract
The β-N-acetylhexosaminidase (EC 3.2.1.52) from glycoside hydrolase family 20 (GH20) catalyzes the hydrolysis of the β-N-acetylglucosamine (NAG) group from the nonreducing end of various glycoconjugates. The putative surface-exposed N-acetylhexosaminidase StrH/Spr0057 from Streptococcus pneumoniae R6 was proved to contribute to the virulence by removal of β(1,2)-linked NAG on host defense molecules following the cleavage of sialic acid and galactose by neuraminidase and β-galactosidase, respectively. StrH is the only reported GH20 enzyme that contains a tandem repeat of two 53% sequence-identical catalytic domains (designated as GH20-1 and GH20-2, respectively). Here, we present the 2.1 Å crystal structure of the N-terminal domain of StrH (residues Glu-175 to Lys-642) complexed with NAG. It adopts an overall structure similar to other GH20 enzymes: a (β/α)8 TIM barrel with the active site residing at the center of the β-barrel convex side. The kinetic investigation using 4-nitrophenyl N-acetyl-β-d-glucosaminide as the substrate demonstrated that GH20-1 had an enzymatic activity (kcat/Km) of one-fourth compared with GH20-2. The lower activity of GH20-1 could be attributed to the substitution of active site Cys-469 of GH20-1 to the counterpart Tyr-903 of GH20-2. A complex model of NAGβ(1,2)Man at the active site of GH20-1 combined with activity assays of the corresponding site-directed mutants characterized two key residues Trp-443 and Tyr-482 at subsite +1 of GH20-1 (Trp-876 and Tyr-914 of GH20-2) that might determine the β(1,2) substrate specificity. Taken together, these findings shed light on the mechanism of catalytic specificity toward the β(1,2)-linked β-N-acetylglucosides.
Keywords: Cell Surface Enzymes, Enzyme Kinetics, Hydrolases, Protein Structure, X-ray Crystallography, β-N-Acetylhexosaminidase, Substrate Specificity
Introduction
Streptococcus pneumoniae is a commensal Gram-positive, encapsulated pathogen that is widely distributed in nature. It causes acute pneumonia, otitis media, meningitis, and several other serious diseases that lead to the death of millions of people worldwide annually. During colonization and infection to the human host, S. pneumoniae encounters a variety of glycoconjugates, including mucin, host defense molecules, and glycans associated with the epithelial surface. To deglycosylate these host glycoconjugates, a number of S. pneumoniae glycosidases have evolved (1). Genome sequencing studies suggested that a large and diverse array of glycosidases is necessary for full virulence of the pneumococci (2, 3). Among them, three surface-exposed exoglycosidases, neuraminidase, β-galactosidase, and StrH, have been previously demonstrated to sequentially remove sialic acid, galactose, and NAG4 to expose the mannose on glycoconjugates of the host defense molecules such as human secretory component, lactoferrin, and immunoglobulin A1 (4). This cleavage was proposed to alter the clearance function of these molecules, facilitating the persistence of Streptococci in the airway. Moreover, the monosaccharides liberated from these glycoconjugates could be utilized for the growth of bacteria (5).
The N-acetylhexosaminidase StrH from S. pneumoniae is a pneumococcal surface protein that was proved to be a virulence factor (4, 6). It belongs to glycoside hydrolase family 20 (GH20) (7–9), the members of which catalyze the hydrolysis of the β1-linked NAG group from the nonreducing end of various glycoconjugates, such as glycans, glycoproteins, and glycolipids (10). Members of this family, such as N-acetylhexosaminidase (EC 3.2.1.52) and lacto-N-biosidase (EC 3.2.1.140), have activities toward different substrates. They have been postulated to have specialized physiological functions, including posttranslational modification of N-glycans, degradation of glycoconjugates, as well as egg-sperm recognition (10). StrH is a 1312-residue protein that contains a tandem repeat of two GH20 domains. The recombinant protein had β-N-acetylhexosaminidase activity and cleaved the β(1,2)-linked NAG group from human defense molecules (11). The tandemly connected GH20 domains are indispensable for StrH to hydrolyze versatile physiological glycans or glycoconjugates. The primary sequences of both GH20 domains are homologous to the following GH20 enzymes of known structure: Serratia marcescens CHB (12), human HexA and HexB (13, 14), Actinobacillus actinomycetemcomitans DspB (15), Streptomyces plicatus Hex (16), Streptococcus gordonii GcnA (17), Paenibacillus sp. TS12 Hex (18), and Ostrinia furnacalis Hex1 (19). Despite the catalytic domain having a common (β/α)8 TIM barrel fold harboring the active site, these GH20 enzymes demonstrate versatile physiological functions and specificity toward diverse substrates.
All GH20 enzymes of known structure have only one catalytic GH20 domain and cleave either the β(1,4)- or β(1,6)-linked NAG group from diverse glycoconjugates. In contrast, StrH possesses a tandem repeat of two GH20 domains (designated as GH20-1 and GH20-2, respectively) and cleaves the substrate of a β(1,2)-linkage. Here we report the 2.1 Å crystal structure of the first GH20 domain of StrH in complex with NAG. Despite it sharing a typical (β/α)8 TIM barrel core structure with other GH20 members, two loops at the entrance of the substrate binding pocket are distinct. Structural comparison in combination with enzymatic analyses revealed that residues Trp-443 and Tyr-482, which reside on these two loops, respectively, are crucial for the stringent specificity toward the β(1,2) substrate. This is the first report of the structural basis for the cleavage of β(1,2)-linkage catalyzed by N-acetylhexosaminidase.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of StrH and Its Mutants
The coding region of the strH gene was amplified from the genomic DNA of S. pneumoniae R6. Genes encoding StrH and its mutants were individually cloned into a pET28a-derived expression vector with an N-terminal His6 tag and overexpressed in Escherichia coli strain BL21-RIL (DE3) (Novagen) using LB culture medium (10 g of NaCl, 10 g of Bacto-Tryptone, and 5 g of yeast extract/liter). The transformed cells were grown at 37 °C in LB medium containing 30 μg/ml kanamycin and 34 μg/ml chloramphenicol until the A600 nm reached ∼0.6. Expression of the recombinant proteins was then induced with 0.2 mm isopropyl β-d-1-thiogalactopyranoside for another 20 h at 16 °C before harvesting. The cells were collected and resuspended in 40 ml of lysis buffer (50 mm HEPES, pH 7.5, 150 mm NaCl). After 2.5 min of sonication and centrifugation at 12,000 × g for 25 min, the supernatant containing the target protein was collected and loaded onto a nickel-nitrilotriacetic acid column (GE Healthcare) equilibrated with the binding buffer (50 mm HEPES, pH 7.5, 150 mm NaCl). The target protein was eluted with 300 mm imidazole, and further loaded onto a Superdex 200 column (GE Healthcare; 50 mm HEPES, pH 7.5, 150 mm NaCl). Fractions containing the target protein were combined and concentrated to 10 mg/ml for crystallization. Samples for enzymatic activity assays were collected at the highest peak fractions without concentration. The purity of protein was assessed by electrophoresis, and the protein sample was stored at −80 °C.
The selenium-Met (Se-Met)-labeled StrH protein was expressed in E. coli strain B834 (DE3) (Novagen). Transformed cells were grown at 37 °C in Se-Met medium (M9 medium with 25 μg/ml Se-Met and the other essential amino acids at 50 μg/ml) containing 30 μg/ml kanamycin until the A600 nm reached ∼0.6 and were then induced with 0.2 mm isopropyl β-d-1-thiogalactopyranoside for 20 h at 16 °C. Se-Met substituted His6-StrH was purified in the same manner as native His6-StrH.
Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with the plasmid encoding the wild-type StrH as the template. The mutant proteins were expressed, purified, and stored in the same manner as the wild-type protein.
Crystallization, Data Collection, and Processing
Both native and Se-Met substituted StrH (amino acids 175–642) were concentrated to 10 mg/ml by ultrafiltration (Millipore Amicon) for crystallization. Crystals were grown at 16 °C using the hanging drop vapor diffusion method, with the initial condition of mixing 1 μl of protein solution with an equal volume of the reservoir solution (30% PEG 4000, 0.2 m ammonium acetate, 0.1 m sodium citrate tribasic dehydrate, pH 5.6). The crystals were transferred to cryoprotectant (reservoir solution supplemented with 25% glycerol) and flash-cooled with liquid nitrogen. The Se-Met derivative data for a single crystal were collected at 100 K in a liquid nitrogen stream using beamline 17U with an MX225 CCD (MARresearch) at the Shanghai Synchrotron Radiation Facility. All of the diffraction data were integrated and scaled with the program HKL2000 (20).
Structure Determination and Refinement
The crystal structure of StrH was determined using the single-wavelength anomalous dispersion phasing (21) method from a single Se-Met-substituted protein crystal to a maximum resolution of 2.1 Å. The Autosol program from PHENIX (22) was used to locate the heavy atoms, and the phase was calculated and further improved with the program SOLVE/RESOLVE (23, 24). Electron density maps showed clear features of secondary structural elements. Automatic model building was carried out using Autobuild in PHENIX. The initial model was refined using the maximum likelihood method implemented in REFMAC5 (25) as part of CCP4i (26) program suite and rebuild interactively using the program COOT (27). The final model was evaluated with the programs MOLPROBITY (28) and PROCHECK (29). Crystallographic parameters are listed in Table 1. All of the structure figures were prepared with PyMOL (30).
TABLE 1.
Crystal parameters, data collection, and structure refinement
| StrH-N | |
|---|---|
| Data collection | |
| Space group | P3121 |
| Unit cell | |
| a, b, c (Å) | 173.98, 173.98, 108.20 |
| α, β, γ (°) | 90.00, 90.00, 120.00 |
| Resolution range (Å) | 50.00–2.10 (2.14–2.10)f |
| Unique reflections | 209,284 (9,191) |
| Completeness (%) | 98.2 (86.7) |
| <I/σ(I)> | 21.3 (3.7) |
| Rmerge (%)a | 10.9 (47.5) |
| Average redundancy | 10.9 (8.2) |
| Structure refinement | |
| Resolution range (Å) | 45.56–2.10 |
| Rfactorb/Rfreec (%) | 19.2/22.1 |
| Number of protein atoms | 6,825 |
| Number of water atoms | 636 |
| RMSD bond lengths (Å)d | 0.012 |
| RMSD bond angles (°) | 1.192 |
| Mean B factors (Å2) | 36.9 |
| Ramachandran plot (residues, %)e | |
| Most favored (%) | 97.46 |
| Additional allowed (%) | 2.54 |
| Outliers (%) | 0 |
| Protein Data Bank entry | 3RPM |
a Rmerge = ΣhklΣi|Ii(hkl) − <I(hkl)>|/ΣhklΣiIi(hkl), where Ii(hkl) is the intensity of an observation, and <I(hkl)> is the mean value for its unique reflection. Summations are over all reflections.
b Rfactor = Σh|Fo(h) − Fc(h)|/ΣhFo(h), where Fo and Fc are the observed and calculated structure-factor amplitudes, respectively.
c Rfree was calculated with 5% of the data excluded from the refinement.
d RMSD, root mean square deviation from ideal values.
e The categories were defined by Molprobity.
f The values in parentheses refer to statistics in the highest bin.
Enzymatic Activity Assay
The enzyme kinetic parameters of native StrH and its mutants were measured using 4-nitrophenyl N-acetyl-β-d-glucosaminide (pNp-NAG; Sigma) as substrate to a standard curve of 4-nitrophenol, which was described by Prag et al. (31) with minor changes. All of the assays were performed at 37 °C in the buffer containing 50 mm NaH2PO4, pH 7.5. The reactions were initiated by the addition of StrH. Using a DU800 spectrophotometer (Beckman Coulter, Fullerton, CA), the increase in absorption at 405 nm was monitored continuously. Michaelis-Menten parameters (Vmax and Km) were extracted from these data by nonlinear fitting to the Michaelis-Menten equation using the program Origin 7.5.
Preparation of 1-Phenyl-3-methyl-5-pyrazolone (PMP) Derivatives of Saccharides
PMP derivatization of saccharides was carried out as described previously (32, 33) with minor changes. Briefly, the 10-μl reaction mixture was terminated by mixing with an equal volume of 0.3 m aqueous NaOH, and a 10-μl 0.5 m methanol solution of PMP was added to each. Each mixture was allowed to react for 30 min at 70 °C, then cooled to room temperature, and neutralized with 10 μl of 0.3 m HCl. The resulting solution was dissolved in 100 μl of chloroform. After vigorous shaking and centrifuging, the organic phase was carefully discarded to remove the excess reagents. The extraction process was repeated three times; then the aqueous layer was diluted with 40 μl of water before HPLC analysis.
HPLC Analysis
The assays were performed at 37 °C in a 10-μl system containing the buffer of 50 mm citric acid/sodium phosphate, pH 5.0, and the disaccharide NAGβ(1,2)Man (Dextra, UK) with a series of concentrations. The reactions were triggered by adding the purified protein solution and terminated by mixing with an equal volume of 0.3 m NaOH. After PMP derivatization as mentioned above, the mixture was centrifuged at 12,000 × g for 10 min, and the supernatant was analyzed by HPLC system (Agilent 1200 Series). Mannose and NAG standards were quantified by HPLC analysis using a series concentrations ranging from 0.1 to 5 mm. Acetonitrile and 100 mm K2HPO4/KH2PO4, pH 7.0, were mixed to give a final mixture of 20% acetonitrile, which was used for equilibration of the column (Eclipse XDS-C18 column, 4.6 × 150 mm; Agilent) and separation of the components at a flow rate of 1 ml/min. The samples were injected in volumes of 10 μl. Retention times of monosaccharides were determined by separation of standard monosaccharide solutions in 10-μl injections, individually as well as in mixture. Three independent kinetic determinations were made to calculate the means and standard deviations for the reported Km and kcat values.
RESULTS AND DISCUSSION
Domain Organization of StrH
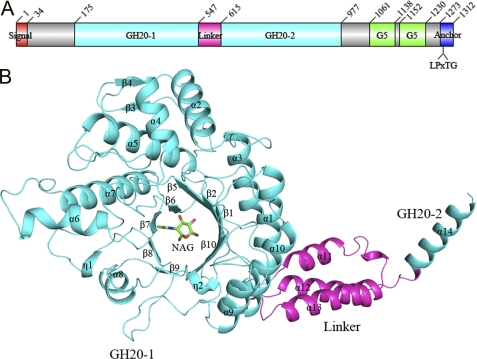
The 1312-residue StrH is composed of six distinct domains: a putative N-terminal signal peptide of 34 residues, a tandem repeat of two GH20 domains, followed by two G5 domains and a C-terminal domain to anchor to the cell wall (Fig. 1A). The two 53% sequence-identical GH20 domains of β-N-acetylhexosaminidase are bridged by a linker of 67 residues. Each G5 domain is a module of ∼80 residues that is found in a variety of enzymes such as Streptococcal IgA peptidases and various glycoside hydrolases in bacteria. These enzymes are usually involved in metabolism of bacterial cell walls and related to the adhesive function to the host (34, 35). The C-terminal cell wall anchor domain contains a canonical LPXTG motif, which is covalently anchored to the cell wall upon cleavage of the LPXTG sequence by a transpeptidase (a sortase) (6).
FIGURE 1.
Domain organization of StrH and overall structure of GH20-1. A, five distinct domains of StrH were drawn by Domain Graph, version 2.0 (40). B, overall structure of GH20-1 (cyan) together with the linker (magenta) and the N-terminal α-helix of GH20-2 (cyan). The secondary structural elements were labeled sequentially. The NAG molecule at the active site of GH20-1 is shown as green sticks.
Overall Structure of StrH-N
In an initial effort to obtain the full-length structure of StrH, we found that the protein was degraded during purification. Limited proteolysis combined with liquid chromatography-mass spectrometry enabled us to define a relatively stable fragment comprising residues Glu-175–Lys-642 (termed StrH-N). We obtained the Se-Met derivative crystals of StrH-N in the presence of NAG and determined the complex structure at 2.1 Å by the single-wavelength anomalous diffraction phasing method. The structure was refined to R/Rfree factor of 19.2%/22.1% and showed good geometry as determined by the programs MOLPROBITY (28) and PROCHECK (29). Each asymmetric unit contains two identical subunits (A and B), with an overall root mean square deviation of 0.7 Å over 422 Cα atoms. Residues Asn-181–Ala-630 in subunit A and residues Asn-181–Val-559 and Thr-567–Asn-609 in subunit B are well fitted in the final model. In addition, a molecule of NAG could be well defined at the active sites of each subunit. An interface of ∼500 Å2 between the two subunits indicated that StrH-N exists as a monomer in solution, which was proved by size exclusion chromatography (data not shown).
The subunit A of StrH-N is composed of GH20-1 (Glu-175–Ala-547), the linker (Asp-548–Pro-614), and an α-helix of GH20-2 (Glu-615–Ala-630). In addition, a molecule of NAG presents at the active site (Fig. 1B). In contrast, the α-helix of GH20-2 is not visible in subunit B. Like all GH20 enzymes of known structure, the core structure of GH20-1 folds into a canonical (β/α)8 TIM barrel: a central eight-stranded β-barrel (β1, β2, and β5–β10) surrounded by eight helices (α1, α3, α5, α7, η1, and α8–α10), including a 310 helix η1. In addition, three α-helices (α2, α4, and α6) and two β-strands (β3 and β4) are located at the lateral side of the TIM barrel. The 67-residue linker forms a three-helix bundle (α11–α13), which bridges GH20-1 and GH20-2 (Fig. 1B).
Overall structure comparison of GH20-1 using the Dali server (http://ekhidna.biocenter.helsinki.fi/dali_server/) (36) gave 53 hits for eight unique proteins with a Z-score higher than 17.0. All of these proteins were members of the GH20 family. Although GH20-1 of StrH shares a sequence identity of less than 17% with these GH20 enzymes, they all share a highly conserved central (β/α)8-barrel. The major differences are from the segments beyond the core structure, including the loops, helices α2, α4, and α6 and β-strands (β3 and β4), because of frequent sequence variation, deletion, or insertion in these regions of all glycosidases.
The Active Site
Previous studies suggested a substrate-assisted catalytic mechanism for GH20 enzymes (12, 16, 37). First, a catalytic glutamate residue provides a general acid to catalyze the hydrolysis of the glycosidic linkage. As the glycosidic linkage is breaking, the anomeric carbon migrates toward the oxygen atom of the 2-acetamido group of the substrate to form a cyclic NAG-oxazolinium ion intermediate. Then a water molecule activated by the deprotonated glutamate attacks the anomeric center to form the final product. Another catalytic residue asparate is responsible for stabilizing the positively charged nitrogen of the 2-acetamide group (12, 16, 31, 38). At the center of the (β/α)8 TIM barrel convex side of StrH-N, there is a molecule of NAG, the pyranose ring of which adopts a chair-like conformation (Fig. 2A). Three residues, Phe-415, Trp-439, and Trp-517, form a hydrophobic pocket to accommodate the hydrophobic pyranose ring. In addition, several polar residues at the active site form a hydrogen-bond network with NAG. The Nη2 and Nη1 of Arg-196 form two hydrogen bonds with O3 and O4 of NAG, respectively, whereas the carboxyl group of Asp-519 also makes two hydrogen bonds with O4 and O6 of NAG, respectively. Asn-225 interacts with O3 of NAG through a water molecule Wat-1. The catalytic residue Asp-360 forms a hydrogen bond with NAG via Wat-1, whereas another catalytic residue Glu-361 is stabilized by two hydrogen bonds with Nϵ and Nη2 of Arg-324.
FIGURE 2.
The active site of GH20-1. A, NAG binding site. The residues are shown as sticks, and the water molecule Wat-1 is shown as a sphere. The polar interactions are indicated by dashed lines. B, active site comparison of GH20-1 (cyan) and SpHex (pink).
Compared with the structure of N-acetylhexosaminidase SpHex in complex with NAG (Protein Data Bank 1M01, 2.1 Å), most residues at the active site of GH20-1 are structurally conserved. The catalytic residue Asp-360 adopts a similar pose to Asp-313 of SpHex. Three aromatic residues, Phe-415, Trp-439, and Trp-517, of StrH-N could be well superimposed with Trp-344, Trp-361, and Trp-442 in SpHex, respectively. In addition, residues Arg-196, Glu-519, and Asn-225 in StrH-N adopt a similar conformation to their counterparts in SpHex (Fig. 2B). However, there are significant differences in the active site between these two enzymes. In SpHex, the hydroxyl group of Tyr-393 donates a hydrogen bond to the oxygen atom of the 2-acetamido group. This hydrogen bond is important for catalysis, because it can fix the carbonyl oxygen of the NAG 2-acetamido group in the position for nucleophilic attack on the anomeric carbon C1 (16). However, SpHex-Tyr-393 is substituted by StrH-Cys-469, which has no interaction with NAG. Moreover, Tyr-393 in SpHex is conserved in other GH20 enzymes of known structure, such as Tyr-669 of SmCHB and Tyr-278 of AaDspB. This substitution of Tyr by Cys-469 in GH20-1 results in NAG adopting a distinct conformation. In SpHex, the 2-acetamido group of NAG turns back to its pyranose ring, and consequently the carbonyl oxygen lies under the mean plane of the pyranose ring and shows a distance of 2.5 Å to the anomeric carbon C1 for nucleophilic attack. In contrast, the 2-acetamido group in StrH-N points outwards against the pyranose ring, which results in a larger distance between the carbonyl oxygen of the 2-acetamido group and the anomeric carbon C1 (4.1 Å) (Fig. 2B). This conformation of NAG in GH20-1 is much more similar to that in the structure of SpHex D313N mutant (Protein Data Bank 1M04) (38), which has been proved to prevent the 2-acetamido group from providing efficient anchimeric assistance, resulting in the large reduction in enzymatic activity. Furthermore, another remarkable difference is that the catalytic residue Glu-361 adopts a distinct orientation compared with its counterpart, Glu-314 in SpHex (Fig. 2B). In SpHex, Glu-314 points to the substrate-binding site, and its carboxyl group makes a hydrogen bond (2.5 Å) with the anomeric hydroxyl of NAG. This conformation of Glu-314 favors its general acid-base catalysis to the glycosidic linkage. However, the carboxyl group of Glu-361 of StrH-N points 10.6 Å away from NAG and is stabilized by two hydrogen bonds formed with Nϵ and Nη2 of Arg-324 (Fig. 2A). The E361A mutant of GH20-1 was found to have a comparable Km, whereas a kcat of one-fifth compared with the wild-type protein suggests that Glu-361 functions in catalysis (Table 2). Taken together, NAG at the active site of StrH-N adopts a conformation that is not suitable for catalysis. This is mainly due to the effect of Cys-469.
TABLE 2.
Kinetic constants of different constructs of StrH towards pNp-NAG
| Enzyme | Residues | Km | kcat | kcat/Km |
|---|---|---|---|---|
| μm | s−1 | ×10−3s−1 μm−1 | ||
| GH20-1&2a | 175–977 | 500 ± 5.0 | 11.0 ± 0.2 | 22 ± 1.0 |
| GH20-1 | 175–547 | 850 ± 5.9 | 1.3 ± 0.8 | 1.5 ± 0.5 |
| GH20-2 | 615–977 | 837 ± 6.4 | 4.8 ± 0.2 | 5.8 ± 0.9 |
| GH20-1&Lb | 175–642 | 700 ± 5.8 | 1.4 ± 0.1 | 2.0 ± 0.6 |
| GH20-2&Lc | 547–977 | 787 ± 5.4 | 4.9 ± 0.3 | 6.3 ± 0.9 |
| E361A | 175–642 | 900 ± 4.9 | 0.4 ± 0.3 | 0.4 ± 0.1 |
| C469Y | 175–547 | 650 ± 6.2 | 5.1 ± 0.5 | 7.8 ± 0.5 |
a GH20-1&2 is a construct of StrH covering GH20-1, the linker, and GH20-2.
b GH20-1&L is a construct of StrH covering GH20-1 and the linker.
c GH20-2&L is a construct of StrH covering the linker and GH20-2.
Activity Analyses of the Two GH20 Domains
The kinetic parameters of the wild-type, truncated, and mutant proteins were determined using pNp-NAG as the substrate (Table 2). GH20-1 had a comparable activity with the fragment covering GH20-1 and the linker. This is also the case for GH20-2, indicating that the linker has no effect on the activity of either individual GH20-1 or GH20-2 (Table 2). In addition, GH20-1 had an enzymatic activity (kcat/Km) one-fourth of GH20-2 and approximately one-fifteenth of GH20-1&2 (residues 175–977, covering both GH20-1 and GH20-2). Such a significantly lower activity of GH20-1 might be due to the substitution of Cys-469 of GH20-1 by Tyr-903 of GH20-2, because the counterpart residue Tyr in other GH20 enzymes was reported to favor the catalysis by critically stabilizing the intermediate state of the substrate (16). In fact, the C469Y mutant of GH20-1 had an activity comparable with that of GH20-2 (Table 2).
Substrate Specificity of StrH
To date, all GH20 enzymes of known structure are reported to catalyze the hydrolysis of NAG with a β(1,4)- or β(1,6)-glycosidic linkage. The substrate-binding site could be divided into two subsites, termed subsites −1 and +1, respectively (39). Aromatic residues at subsite +1 were thought to play a crucial role in determining substrate specificity. For instance, Trp-685 at subsite +1 of SmCHB is responsible for stabilizing the +1 sugar in a proper pose, which is twisted 90° relative to the −1 NAG to facilitate the cleavage of the β(1,4)-glycosidic linkage (12). Similar conformations in other β(1,4)-N-acetylhexosaminidases were also observed, such as Trp-408 in SpHex (16), Trp-410 in PsHex (18), and Trp-490 in OfHex1 (19). However, in the structure of β(1,6)-N-acetylhexosaminidase AaDspB, the open binding pocket contains no conserved aromatic residue for stacking the +1 sugar, which was proposed to favor the conformation of the β(1,6)-linked polymer (15).
Superposition of GH20-1 against SpHex yields an overall root mean square deviation of 2.9 Å over 293 Cα atoms, and 1.9 Å over 72 Cα atoms of the central (β/α)8-barrels. The main differences concern the loops connecting the central (β/α)8-barrel. It has been demonstrated that StrH specifically hydrolyzes the N-linked sugars with a glycosidic linkage of NAGβ(1,2)Man (11). For a better understanding of the structural basis of this substrate specificity, we attempted to obtain crystals in the presence of the disaccharide NAGβ(1,2)Man by either soaking or co-crystallization but were unsuccessful. Alternatively, we first calculated a putative substrate entrance path with the CAVER program (http://loschmidt.sci.muni.cz/caver/index.php). It revealed a dumbbell-shaped tunnel that was gated by two unique loops, LA (Trp-439–Ser-450) and LB (Cys-469–Asn-483) (Fig. 3A). LA between β8 and α8 exhibits an extended conformation, whereas LB connecting β9 and α9 shows a twisted conformation. Two aromatic residues, Trp-443 on LA and Tyr-482 on LB, were found to guard the entrance of the tunnel (Fig. 3A). Furthermore, we manually constructed a model with NAGβ(1,2)Man in the active site of our present structure by fixing the NAG moiety to subsite −1. The mannose at subsite +1 was fitted into the position with the best steric geometry (Fig. 3B). The results showed that the +1 mannose moiety of NAGβ(1,2)Man was twisted ∼90° relative to the −1 NAG, packing against Trp-443 and Tyr-482. The distance from the +1 mannose moiety to Trp-443 and Tyr-482 is ∼4.0 and 3.3 Å, respectively (Fig. 3B). Compared with SmCHB, subsite +1 of GH20-1 is structurally distinct, which could be attributed to the different substrate specificity. Interestingly, sequence analysis revealed that these two aromatic residues are conserved in the GH20-1 and GH20-2 domains of StrH. The counterparts in GH20-2 are Trp-876 and Tyr-914, respectively.
FIGURE 3.
The substrate specificity of StrH. A, the putative substrate entrance tunnel calculated by CAVER program, denoted in orange mesh. Trp-443 and Tyr-482 in the vicinity were shown as sticks and labeled. B, a manually built model of NAGβ(1,2)Man in complex with GH20-1.
To determine the special roles of Trp-443 and Tyr-482 in GH20-1 (Trp-876 and Tyr-914 in GH20-2), enzymatic activity assays were performed using the wild-type proteins and the corresponding mutants. All of the protein samples were quality-controlled by circular dichroism spectroscopy, the results of which showed that the mutations did not introduce significant changes to the protein structures (supplemental Fig. S1). The disaccharide NAGβ(1,2)Man and pNp-NAG were used as the substrates of StrH. Compared with the wild-type enzyme, the W443A mutant of GH20-1 had comparable Km but much lower kcat values, resulting in enzymatic activities (kcat/Km) ∼1% toward pNp-NAG and one-fourth toward NAGβ(1,2)Man (Table 3). It suggested that Trp-443 at the subsite +1 had no considerable effect on substrate binding but played important roles in enzymatic activity. The Y482A mutant of GH20-1 had 2-fold increase in Km and one-third of kcat value, leading to a much lower activity toward pNp-NAG. Noticeably, the mutation of Y482A completely abolished the enzymatic activity toward NAGβ(1,2)Man. It indicated that Tyr-482 at subsite +1 is crucial for both substrate binding and catalysis, and more importantly, indispensable for the activity toward NAGβ(1,2)Man (Table 3). In addition, enzymatic activities toward pNp-NAG of W443F and Y482F mutants of GH20-1, and W876F and Y914F mutants of GH20-2 were measured. Compared with the wild-type enzymes, the kcat/Km values for W443F and W876F mutants were approximately one-sixteenth and one-eighth, whereas they were approximately one-third for Y482F and comparable for Y914F, respectively (Table 3). On the other hand, all mutants to Phe showed somewhat higher enzymatic activities toward pNp-NAG compared with the corresponding mutants to Ala. Moreover, double mutant W443A/Y482A of GH20-1 had no detectable activity toward either pNp-NAG or NAGβ(1,2)Man. Similar results of the enzymatic activity were also observed for the Trp-876 and/or Tyr-914 mutants of GH20-2, suggesting the same roles of these residues at the subsite +1 of GH20-2 (Table 3). Furthermore, double mutant W443A/W876A of GH20-1&2 had significant lower activities toward pNp-NAG and NAGβ(1,2)Man, whereas Y482A/Y914A completely abolished the activities. These results further confirmed the essential roles of these residues in substrate binding and catalysis for the full-length enzyme. To further investigate the substrate specificity of StrH, we also performed the enzymatic assays toward chitobiose of a β(1,4)-linkage. The results showed that neither the wild-type StrH nor the Y482A or W443A mutants of GH20-1 (mutants Y914A or W876A of GH20-2) could hydrolyze the β(1,4)-linked chitobiose (data not shown). Multiple-sequence alignment revealed that the key residues at the subsite +1 (Trp-443 and Tyr-482 in GH20-1 and Trp-876 and Tyr-914 in GH20-2) of StrH are highly conserved among Lactobacillales of Gram-positive bacteria (Fig. 4). We suggest that these proteins could also cleave the substrates with a β(1,2)-linkage in a way similar to StrH.
TABLE 3.
Kinetic constants of different constructs of StrH and mutants towards pNp-NAG and NAGβ(1,2)Man
| Enzyme | pNp-NAG |
NAGβ(1,2)Man |
||||
|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| μm | s−1 | ×10−3s−1μm−1 | mm | min−1 | min−1mm−1 | |
| GH20-1 | 850 ± 5.9 | 1.3 ± 0.8 | 1.5 ± 0.5 | 1.03 ± 0.06 | 1.50 ± 0.07 | 1.46 ± 0.11 |
| W443A | 857 ± 4.5 | 0.015 ± 0.003 | 0.018 ± 0.004 | 1.34 ± 0.05 | 0.52 ± 0.03 | 0.39 ± 0.03 |
| W443F | 1120 ± 4.8 | 0.11 ± 0.03 | 0.09 ± 0.004 | |||
| Y482A | 1800 ± 4.7 | 0.46 ± 0.05 | 0.25 ± 0.01 | ND | ND | ND |
| Y482F | 1500 ± 5.2 | 0.66 ± 0.09 | 0.44 ± 0.03 | |||
| W443A/Y482A | NDa | ND | ND | ND | ND | ND |
| GH20-2 | 837 ± 6.4 | 4.8 ± 0.2 | 5.8 ± 0.9 | 1.31 ± 0.03 | 6.10 ± 0.22 | 4.66 ± 0.13 |
| W876A | 1000 ± 5.2 | 0.51 ± 0.04 | 0.51 ± 0.03 | 1.51 ± 0.04 | 0.54 ± 0.23 | 0.36 ± 0.15 |
| W876F | 1292 ± 4.4 | 0.98 ± 0.09 | 0.75 ± 0.05 | |||
| Y914A | 1560 ± 5.8 | 0.033 ± 0.002 | 0.022 ± 0.005 | ND | ND | ND |
| Y914F | 950 ± 5.4 | 4.7 ± 0.2 | 5.0 ± 0.5 | |||
| W876A/Y914A | ND | ND | ND | ND | ND | ND |
| GH20-1&2 | 500 ± 5.0 | 11.0 ± 0.2 | 22.0 ± 1.0 | 0.98 ± 0.02 | 27.50 ± 0.22 | 38.70 ± 0.13 |
| W443A/W876A | 900 ± 4.9 | 0.04 ± 0.005 | 0.044 ± 0.0002 | 1.30 ± 0.03 | 0.58 ± 0.22 | 0.44 ± 0.16 |
| Y482A/Y914A | ND | ND | ND | ND | ND | ND |
| W443A/W876A/Y482A/Y914A | ND | ND | ND | ND | ND | ND |
a ND, not detectable.
FIGURE 4.
Conservation of Trp-443 and Tyr-482 in StrH and its homologs. Multiple-sequence alignment of putative GH20 enzymes with β(1,2) substrate specificity. The active site residues at subsite −1 are labeled with green triangles. Residues Trp-443 and Tyr-482 at subsite +1 of GH20-1 were marked with red triangles.
During the second round revision of our manuscript, we found the recently released structures of GH20-1 from S. pneumoniae TIGR4 complexed with NAGβ(1,2)Man (Protein Data Bank code 2YL8) and GH20-2 complexed with pentasaccharide Nβ2Mα3M(Nβ4)β4N (Protein Data Bank code 2YLA), deposited by the group of Boraston et al. These two structures in combination with our structure made it possible for us to elucidate the structural insights into the unique substrate specificity of StrH (11). The active site pockets of both GH20-1 and GH20-2, which are specific for the β(1,2)-linked NAG, could accommodate either of the branches of these oligosaccharides. In the structure of pentasaccharide-complexed GH20-2, the substrate Nβ2Mα3M(Nβ4)β4N is finely buried in the active site, with O4 of its +1 mannose directed outwards from the active site pocket (supplemental Fig. S2). Thus O4 of the +1 mannose could be further decorated by an NAG molecule. This explains why only the double branches of β(1,2)- and β(1,4)-linked NAG to the +1 mannose of the tetraantennary oligosaccharide could enter the active site for hydrolysis (11). In addition, only the full-length StrH of S. pneumoniae TIGR4, but not the GH20-1, could hydrolyze the bisected biantennary oligosaccharide. Structural comparison of S. pneumoniae TIGR4 GH20-1 (Protein Data Bank 2YL8) and GH20-2 (Protein Data Bank 2YLA) revealed that the active site pockets are similar, both of which have two extended loops (LA and LB) and adopt similar conformations. The most significant difference is the variable sequences of LB loops from GH20-1 and GH20-2. Especially Tyr-482 in GH20-1 is substituted by Gly-914 in GH20-2. This substitution alters the active site accessibility and thus broadens the substrate specificity. Tyr-482 of GH20-1 partially occupies the space for the bisected NAG molecule in GH20-2 (supplemental Fig. S2), making it impossible for GH20-1 to accommodate the bisected biantennary oligosaccharide. This explains why only the full-length StrH, but not the GH20-1 alone, could hydrolyze the bisected biantennary oligosaccharide. More notably, multiple-sequence alignment revealed that some S. pneumoniae strains have a Tyr instead of Gly at LB loop of StrH GH20-2, such as R6 in our study. Thus we propose that StrH from these strains including R6 could not hydrolyze the bisected biantennary oligosaccharide, possibly because of the steric hindrance of Tyr residue to the active site.
Distinct from other GH20 enzymes, Streptococcal StrH possesses a tandem repeat of two GH20 domains. The enzymatic activity of GH20-1&2 is significantly higher than the sum of two individual GH20 domains (∼15- and 4-fold higher than GH20-1 and GH20-2, respectively). These results suggest that GH20-1 and GH20-2 may have a synergistic effect during hydrolysis of the glycoconjugates of the host defense molecules. To further verify this effect, we simply mixed GH20-1 and GH20-2 at a 1:1 molar ratio and compared its activity toward pNp-NAG with the tandem repeat (GH20-1&2) and individual domains. The mixed GH20 domains had a Km value of 840 ± 7.3 μm, similar to that of either individual GH20 domain, and a kcat value of 6.6 ± 0.9 s−1, which is comparable to the sum of that for two domains. However, the mixed GH20 domains had an enzymatic activity (kcat/Km) of only approximately one-third that of GH20-1&2. The results indicated that the tandemly repeated two GH20 domains of StrH had a synergistic effect. This effect could probably be attributed to the flexible interdomain linker that enables the two GH20 domains to easily approach each other during hydrolysis. Because of the substitution of the critical Tyr to Cys at the active site, GH20-1 only showed an enzymatic activity of approximately one-fourth compared with GH20-2. A sequence homology search revealed that all orthologs in other Streptococci have a conserved Tyr at the corresponding site. The Tyr → Cys mutation was only found in GH20-1 from the avirulent strain S. pneumoniae R6. The proposed attenuated activity of StrH toward the glycoconjugates of human host is in agreement with the low virulence of the R6 strain.
Supplementary Material
Acknowledgments
We thank the staff at the Shanghai Synchrotron Radiation Facility. We are grateful to all of the developers of the CCP4 Suite, ESPript, TreeView, and PyMOL.
Note Added in Proof
During the revision of this manuscript, an article was published by Pluvinage et al. (41) describing the relevant structures.
This work was supported by Ministry of Science and Technology of China Grant 2009CB918804 and National Natural Science Foundation of China 30870488.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- NAG
- β-N-acetylglucosamine
- GH20
- glycoside hydrolase family 20
- pNp-NAG
- 4-nitrophenyl N-acetyl-β-d-glucosaminide
- Se-Met
- selenium-methionine
- PMP
- 1-phenyl-3-methyl-5-pyrazolone.
REFERENCES
- 1. García-Suárez Mdel M., Vázquez F., Méndez F. J. (2006) Enferm. Infecc. Microbiol. Clin. 24, 512–517 [DOI] [PubMed] [Google Scholar]
- 2. Tettelin H., Nelson K. E., Paulsen I. T., Eisen J. A., Read T. D., Peterson S., Heidelberg J., DeBoy R. T., Haft D. H., Dodson R. J., Durkin A. S., Gwinn M., Kolonay J. F., Nelson W. C., Peterson J. D., Umayam L. A., White O., Salzberg S. L., Lewis M. R., Radune D., Holtzapple E., Khouri H., Wolf A. M., Utterback T. R., Hansen C. L., McDonald L. A., Feldblyum T. V., Angiuoli S., Dickinson T., Hickey E. K., Holt I. E., Loftus B. J., Yang F., Smith H. O., Venter J. C., Dougherty B. A., Morrison D. A., Hollingshead S. K., Fraser C. M. (2001) Science 293, 498–506 [DOI] [PubMed] [Google Scholar]
- 3. Hava D. L., Camilli A. (2002) Mol. Microbiol. 45, 1389–1406 [PMC free article] [PubMed] [Google Scholar]
- 4. King S. J., Hippe K. R., Weiser J. N. (2006) Mol. Microbiol. 59, 961–974 [DOI] [PubMed] [Google Scholar]
- 5. Burnaugh A. M., Frantz L. J., King S. J. (2008) J. Bacteriol. 190, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bergmann S., Hammerschmidt S. (2006) Microbiology 152, 295–303 [DOI] [PubMed] [Google Scholar]
- 7. Henrissat B. (1991) Biochem. J. 280, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henrissat B., Bairoch A. (1993) Biochem. J. 293, 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Henrissat B., Bairoch A. (1996) Biochem. J. 316, 695–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Intra J., Pavesi G., Horner D. S. (2008) BMC Evol. Biol. 8, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clarke V. A., Platt N., Butters T. D. (1995) J. Biol. Chem. 270, 8805–8814 [DOI] [PubMed] [Google Scholar]
- 12. Tews I., Perrakis A., Oppenheim A., Dauter Z., Wilson K. S., Vorgias C. E. (1996) Nat. Struct. Biol. 3, 638–648 [DOI] [PubMed] [Google Scholar]
- 13. Maier T., Strater N., Schuette C. G., Klingenstein R., Sandhoff K., Saenger W. (2003) J. Mol. Biol. 328, 669–681 [DOI] [PubMed] [Google Scholar]
- 14. Lemieux M. J., Mark B. L., Cherney M. M., Withers S. G., Mahuran D. J., James M. N. (2006) J. Mol. Biol. 359, 913–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ramasubbu N., Thomas L. M., Ragunath C., Kaplan J. B. (2005) J. Mol. Biol. 349, 475–486 [DOI] [PubMed] [Google Scholar]
- 16. Mark B. L., Vocadlo D. J., Knapp S., Triggs-Raine B. L., Withers S. G., James M. N. (2001) J. Biol. Chem. 276, 10330–10337 [DOI] [PubMed] [Google Scholar]
- 17. Langley D. B., Harty D. W., Jacques N. A., Hunter N., Guss J. M., Collyer C. A. (2008) J. Mol. Biol. 377, 104–116 [DOI] [PubMed] [Google Scholar]
- 18. Sumida T., Ishii R., Yanagisawa T., Yokoyama S., Ito M. (2009) J. Mol. Biol. 392, 87–99 [DOI] [PubMed] [Google Scholar]
- 19. Liu T., Zhang H., Liu F., Wu Q., Shen X., Yang Q. (2011) J. Biol. Chem. 286, 4049–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Otwinowski Z., Minor W. (1997) Macromol. Crystallogr. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 21. Brodersen D. E., de La Fortelle E., Vonrhein C., Bricogne G., Nyborg J., Kjeldgaard M. (2000) Acta Crystallogr. D. Biol. Crystallogr. 56, 431–441 [DOI] [PubMed] [Google Scholar]
- 22. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D. Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Terwilliger T. C. (2003) Acta Crystallogr. D. Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murshudov. G. N., Vagin. A. A., Dodson. E. J. (1997) Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 26. Collaborative Computational Project, No. 4. (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 27. Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laskowski R., Macarthur M., Moss D., Thornton J. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 30. DeLano W. (2002) The PyMOL Molecular Graphics System, De Lano Scientific, San Carlos, CA [Google Scholar]
- 31. Prag G., Papanikolau Y., Tavlas G., Vorgias C. E., Petratos K., Oppenheim A. B. (2000) J. Mol. Biol. 300, 611–617 [DOI] [PubMed] [Google Scholar]
- 32. Yang X., Zhao Y., Wang Q., Wang H., Mei Q. (2005) Anal. Sci. 21, 1177–1180 [DOI] [PubMed] [Google Scholar]
- 33. Honda S., Akao E., Suzuki S., Okuda M., Kakehi K., Nakamura J. (1989) Anal. Biochem. 180, 351–357 [DOI] [PubMed] [Google Scholar]
- 34. Bateman A., Holden M. T., Yeats C. (2005) Bioinformatics 21, 1301–1303 [DOI] [PubMed] [Google Scholar]
- 35. Ruggiero A., Tizzano B., Pedone E., Pedone C., Wilmanns M., Berisio R. (2009) J. Mol. Biol. 385, 153–162 [DOI] [PubMed] [Google Scholar]
- 36. Holm L., Sander C. (1993) J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 37. Drouillard S., Armand S., Davies G. J., Vorgias C. E., Henrissat B. (1997) Biochem. J. 328, 945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Williams S. J., Mark B. L., Vocadlo D. J., James M. N., Withers S. G. (2002) J. Biol. Chem. 277, 40055–40065 [DOI] [PubMed] [Google Scholar]
- 39. Davies G. J., Wilson K. S., Henrissat B. (1997) Biochem. J. 321, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. (2009) Cell Res. 19, 271–273 [DOI] [PubMed] [Google Scholar]
- 41. Pluvinage B., Higgins M. A., Abbott D. W., Robb C., Dalia A. B., Deng L., Weiser J. N., Parsons T. B., Fairbanks A. J., Vocadlo D. J., Boraston A. B. (2011) Structure 19, 1603–1614 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.