Background: Many signaling molecules coordinately play roles in cell movement.
Results: ADIP (afadin dilute domain-interacting protein) mediates Rac activation by interacting with Vav2 for the formation of the leading edge.
Conclusion: ADIP integrates the intracellular signaling to promote cell movement.
Significance: This provides a new insight into the molecular mechanism of cell movement.
Keywords: Cell Migration, Growth Factors, Guanine Nucleotide Exchange Factor (GEF), Rac, Signal Transduction
Abstract
Cell movement is an important cellular function not only in physiological but also in pathological conditions. Although numerous studies have been conducted to reveal the mechanism of cell movement, the full picture has yet to be depicted, likely due to the complex features of cell movement. We show here that the scaffold protein afadin dilute domain-interacting protein (ADIP), an afadin-binding protein, is involved in the regulation of cell movement. ADIP localized at the leading edge of moving cells in response to platelet-derived growth factor (PDGF) and was required for the formation of the leading edge and the promotion of cell movement. Impaired cell movement observed in ADIP knockdown cells was not rescued by expression of an ADIP mutant that is incapable of binding to afadin, leading to the notion that the function of ADIP in moving cells depends on its interaction with afadin. Knockdown of ADIP as well as knockdown of afadin inhibited the activation of the small G protein Rac, which is important for the formation of the leading edge and the promotion of cell movement. Furthermore, ADIP interacted with Vav2, a GDP/GTP exchange factor for Rac, in a Src phosphorylation-dependent manner, suggesting that ADIP mediates the activation of Rac through Vav2. These results indicate that ADIP plays an essential role in PDGF-induced cell movement by interacting with afadin and Vav2 and regulating the activation of Rac.
Introduction
Cell movement is essential for the development of embryos as well as for wound healing and tissue remodeling of adults in multicellular organisms (1, 2). Cell movement is also associated with the progression of diseases. Diverse patterns of tumor cell movement into neighboring tissues have been observed in the process of invasion and metastasis of cancers (3). Thus, the understanding of the mechanism of cell movement is important not only for cell biology and physiology but also for pathology. However, the mechanism of cell movement implies complex features of cell shape changes and signal transduction, making it difficult to reveal the complete picture of the mechanism of cell movement.
One of the important morphogenetic events for cell movement is the formation of the leading edge at the front of moving cells (4). The leading edge consists of several characteristic structures: membrane ruffles, lamellipodia, and focal complexes (5–7). Growth factor receptors, such as platelet-derived growth factor (PDGF) receptor, and integrins, such as integrin αvβ3, preferentially accumulate at the leading edge and play key roles in its formation by regulating the intracellular signaling (8–10). However, the molecular basis for the functional and physical association of PDGF receptor with integrin αvβ3 had been poorly understood for a long time. In a previous study, we demonstrated that nectin-like molecule 5 (Necl-5),2 an immunoglobulin-like cell adhesion molecule, forms a ternary complex with PDGF receptor and integrin αvβ3 at the leading edge and enhances the PDGF-induced cell movement (11, 12). More recently, we also clarified that the accumulation and stabilization of the ternary complex at the leading edge are controlled by afadin (13). Afadin was identified as an actin filament (F-actin)-binding protein that localizes at cell-cell adherens junctions (AJs; Ref. 14) and functions in cell polarity, survival, and movement as well as cell-cell adhesion (15). Our recent studies also revealed that afadin plays central roles in the PDGF-induced dynamic activation and inactivation of the Rho family of small G proteins that are essential for cell movement at the leading edge (16–18). Although vigorous investigations into the molecular mechanism of cell movement are being conducted worldwide, the mechanism has yet to be fully elucidated.
In this study, we discovered that the scaffold protein afadin dilute domain-interacting protein (ADIP) localizes at the leading edge of individually moving cells and regulates the formation of the leading edge and cell movement. ADIP was originally identified as an afadin-binding protein by the yeast two-hybrid system and is concentrated at AJs in epithelial cells (19). Similar to the role of ADIP at AJs, the interaction of ADIP with afadin is necessary for the formation of the leading edge and for cell movement. For these functions, ADIP mediates the activation of Rac, a small G protein, by interacting with Vav2, a GDP/GTP exchange factor for Rac.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Expression vectors for HA-tagged mouse ADIP (pCMV-HA-ADIP) and Myc- and FLAG-tagged Vav2 (pCIneo-myc-Vav2 and pEFBOS-FLAG-Vav2, respectively) were prepared as described previously (19, 20). An expression vector expressing the T7-tagged dilute (DIL) domain of rat afadin (amino acids 606–983; pCMV-T7-afadin-DIL) was also prepared as described previously (19). Expression vectors for enhanced green fluorescent protein (EGFP)-tagged mouse ADIP (amino acids 1–615) and ADIPΔABR (amino acids 1–226 and 501–615), which lacks the afadin-binding region, were constructed with pEGFP-C1 (Clontech). For rescue experiments, RNAi-resistant vectors for EGFP-ADIP and EGFP-ADIPΔABR were created by alteration of several nucleotides in the RNAi target sequence by mutagenesis.
Cell Culture and Knockdown Experiments
NIH3T3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum. NIH3T3 cells stably expressing EGFP and afadin knockdown NIH3T3 cells stably expressing GFP-afadin were generated as described previously (21). Following the construction of an expression vector for RNAi-resistant EGFP-tagged rat afadin from which the DIL domain was deleted (amino acids 1–646 and 893–1829; pMSCVpuro-EGFP-afadinΔDIL), afadin knockdown NIH3T3 cells stably expressing EGFP-afadinΔDIL were generated by retrovirus-mediated introduction of the vector and selection with both 500 μg/ml G418 and 10 μg/ml puromycin. HEK293, HeLa, and SYF cells were cultured in DMEM supplemented with 10% fetal bovine serum. Lipofectamine 2000 reagent (Invitrogen) was used to transfect expression vectors into cells.
The small interfering RNA (siRNA) method was applied to knock down the expression of ADIP. Stealth RNAi duplexes against mouse ADIP and a Stealth RNAi negative control duplex were purchased from Invitrogen. The sequences of RNAi duplexes against mouse ADIP were as follows: 5′-CAGGAGAGAACAUUGAACAAAGUAU-3′ (siRNA 1), 5′-GCAAGAACAGGAGUUUGCAUCAGCU-3′ (siRNA 2), and 5′-CAGCUCGUUAUGAACAAGAAGGAUA-3′ (siRNA 3). siRNA transfection was performed using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's protocol.
Antibodies and Reagents
A rabbit anti-afadin polyclonal antibody (pAb) and a mouse anti-afadin monoclonal Ab (mAb) were prepared as described (14, 22). A rabbit anti-ADIP pAb and a rat anti-Necl-5 mAb (1A8-8) were also prepared as described (19, 23). Abs listed below were purchased from commercial sources and used for immunofluorescence microscopy and Western blotting: a goat anti-ADIP pAb (Abcam), a rabbit anti-PDGF receptor β (Tyr-92) mAb (Abcam), a mouse anti-Rac1 (102) mAb (BD Transduction), a rabbit anti-Rap1 pAb (Santa Cruz Biotechnology), a mouse anti-v-Src (327) mAb (Sigma-Aldrich), a rabbit anti-Src (Tyr(P)-418) phosphospecific pAb (BIOSOURCE), a rabbit anti-Vav2 pAb (Santa Cruz Biotechnology), a rabbit anti-GAPDH (14C10) mAb (Cell Signaling Technology), a mouse anti-phosphotyrosine (PY20) mAb (Santa Cruz Biotechnology), a mouse anti-phosphotyrosine (4G10) mAb (Upstate Biotechnology), a mouse anti-T7 mAb (Novagen), a mouse anti-FLAG (M2) mAb (Sigma-Aldrich), and a rabbit anti-GFP pAb (MBL International Corp.). F-actin was labeled with Alexa Flour 568-phalloidin (Invitrogen). Horseradish peroxidase-conjugated and fluorophore-conjugated secondary Abs were purchased from GE Healthcare and Invitrogen, respectively. Vitronectin was purified from human plasma (Kohjinbio) as described previously (24).
Directional Stimulation by PDGF
To generate a concentration gradient of PDGF, a μ-Slide VI flow dishes (uncoated; Ibidi) was used as described previously (12). Briefly, cells were seeded at a density of 5 × 103 cells/cm2 on the vitronectin-coated μ-Slide VI flow dishes, cultured for 18 h, and serum-starved with DMEM containing 0.5% BSA for 4 h. A concentration gradient of 30 ng/ml PDGF was prepared according to the manufacturer's protocol. After a 30-min PDGF stimulation, cells were fixed with acetone/methanol (1:1) or with 3.7% PFA followed by 0.2% Triton X-100 penetration, incubated with 1% BSA in PBS, and then incubated with 20% BlockAce in PBS followed by immunofluorescence microscopy.
Wound Healing Assay
The wound healing assay was performed as described previously (25). The width of the wound space at 0, 2, and 8 h after scratching the confluent cell monolayer was measured at least at five different points in each experiment to quantify the extent of wound closure (26). Data from at least three independent experiments are expressed as mean ± S.D.
Boyden Chamber Assay
The Boyden chamber assay was performed as described previously (27). FalconTM cell culture inserts (8.0-μm pores; BD Biosciences) were coated with 3 μg/ml vitronectin at 37 °C for 1 h. The inserts were then blocked with 1% BSA at 37 °C for 30 min. Cells were serum-starved in DMEM supplemented with 0.5% fatty acid-free BSA for 1 h and then detached with 0.05% trypsin and 0.53 mm EDTA followed by treatment with a trypsin inhibitor. Cells were resuspended in DMEM supplemented with 0.5% fatty acid-free BSA and seeded at a density of 2.5 × 104 cells/insert. Cells were incubated at 37 °C for 16 h in the presence or absence of 30 ng/ml PDGF. Following incubation, cells were fixed with 3.7% formaldehyde and permeabilized with 0.2% Triton X-100. The culture inserts were then washed with PBS three times and blocked with a 1% BSA and PBS solution and 20% BlockAce followed by DAPI staining (Sigma-Aldrich). After removing the cells from the upper part of the filter by cotton sticks, the number of stained cells that crossed the filter was counted in five randomly chosen fields per filter using a microscope. Data from at least three independent experiments are expressed as mean ± S.D.
Reverse Transcription (RT)-PCR
Total mRNA extraction from NIH3T3 cells and first strand cDNA synthesis were performed using the SuperScript III CellsDirect cDNA Synthesis System (Invitrogen) according to the manufacturer's protocol. Second strand cDNA synthesis by PCR was carried out with the following primers: ADIP, GGAGATTGGATGACTGTGACAGAT (forward) and GGTTATCGAGTTTTTCTACATGAC (reverse); and GAPDH, GTGAAGGTCGGTGTGAACGGATTT (forward) and CTCCTTGGAGGCCATGTAGGCCAT (reverse).
Western Blotting and Immunoprecipitation
Cells were washed with ice-cold PBS and harvested using Lysis Buffer A (25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm CaCl2, 1 mm MgCl2, 1% Nonidet P-40, 10 μg/ml leupeptin, 10 mm 4-amidinophenylmethylsulfonyl fluoride hydrochloride, 0.5 mm EDTA, and a phosphatase inhibitor mixture (Sigma-Aldrich)), boiled for 5 min, and rotated at 4 °C for 15 min. Cell lysates were centrifuged at 20,000 × g for 15 min, and the supernatant was collected. Protein concentrations were determined using the RC DC protein assay kit (Bio-Rad) with BSA as a reference protein. Samples were separated by SDS-PAGE followed by Western blotting with the indicated Abs.
For the immunoprecipitation assay using HEK293 cells expressing FLAG-Vav2 and/or EGFP-ADIP, cells were lysed with Lysis Buffer A at 4 °C for 15 min. Cell lysates were centrifuged at 20,000 × g for 15 min, and the supernatant was incubated with anti-FLAG M2-agarose beads (Sigma-Aldrich) or anti-GFP-agarose beads (MBL International Corp.) at 4 °C for 3 h. Then the beads were washed twice with Washing Buffer A (10 mm Tris-HCl, pH 8, 500 mm NaCl, 0.5% Nonidet P-40, and 0.05% SDS), once with Washing Buffer B (10 mm Tris-HCl, pH 8, 150 mm NaCl, 0.5% Nonidet P-40, 0.05% SDS, and 0.5% dideoxycholate), and finally once with Washing Buffer C (10 mm Tris-HCl, pH 8 and 0.05% SDS). For the immunoprecipitation assay for endogenous proteins in HeLa cells, cell lysates were prepared as described above, and the supernatant was precleared by incubation with protein A-Sepharose beads at 4 °C for 15 min. The precleared supernatant was then incubated with the anti-Vav2 pAb at 4 °C for 3 h followed by incubation with protein A-Sepharose beads at 4 °C for 1 h. The beads were washed as described above. Proteins bound to agarose or Sepharose beads were eluted from the beads by boiling in SDS sample buffer for 5 min and subjected to SDS-PAGE followed by Western blotting with the indicated Abs.
Pulldown Assay for Small G Proteins
The pulldown assay was performed as described (16, 28). In brief, after a 4-h serum starvation, cells were incubated in medium with or without 15 ng/ml PDGF for 1 min. Cells were then washed with ice-cold PBS containing 1 mm sodium vanadate and lysed in Lysis Buffer B (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm phenylmethylsulfonyl fluoride, and 1 mm sodium vanadate) containing GST fusion proteins (GST-RalGDS-RBD for Rap1 or GST-PAK-CRIB for Rac) at 2 °C for 30 min. Cell lysates were centrifuged at 20,000 × g at 0 °C for 5 min, and the supernatant was incubated with glutathione-agarose beads (GE Healthcare) at 2 °C for 1 h. After the beads were washed with Lysis Buffer B, proteins bound to the beads were eluted with SDS sample buffer and subjected to SDS-PAGE followed by Western blotting with the indicated Abs.
RESULTS
Localization of ADIP at Leading Edge Formed by PDGF Stimulation
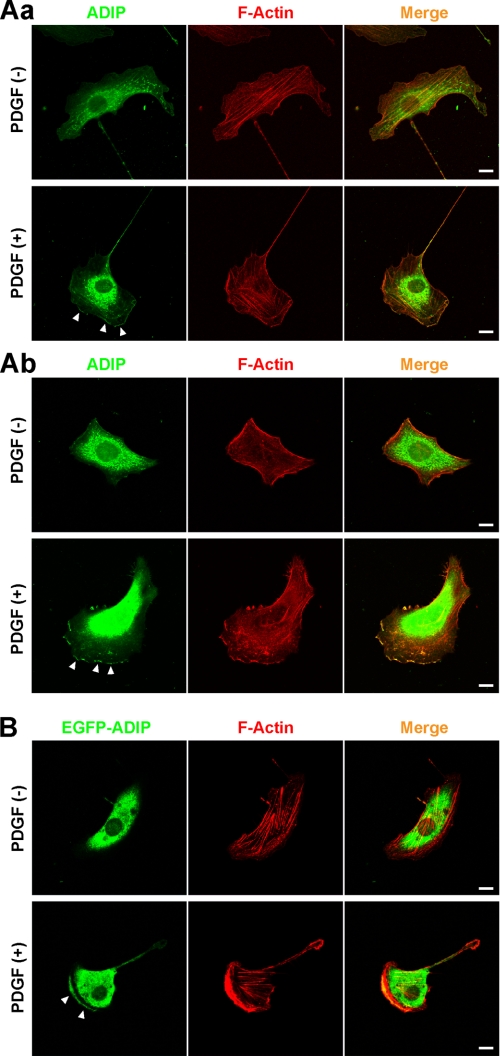
Previously, we reported that afadin is important for the formation of the leading edge in moving cells and that afadin interacts with ADIP at AJs (13, 16, 19, 29). Based on these results, we first examined whether ADIP is concentrated at the leading edge of moving cells. NIH3T3 cells were sparsely plated on μ-Slide VI flow dishes to directionally stimulate them with or without PDGF. In the absence of PDGF stimulation, no obvious leading edge was observed in NIH3T3 cells, and the immunofluorescence signal for ADIP was broadly distributed in the cytosol (Fig. 1A, panel a, upper panels). When NIH3T3 cells were treated with PDGF, they became polarized with a well spreading leading edge toward the higher concentration of PDGF, and the signal for ADIP was observed at the leading edge (Fig. 1A, panel a, lower panels). The accumulation of ADIP at the leading edge was confirmed using another cell line. When HeLa cells were directionally stimulated with PDGF, they formed the leading edge where the signal for ADIP accumulated similarly to NIH3T3 cells (Fig. 1A, panel b). Furthermore, when EGFP-ADIP was ectopically expressed in NIH3T3 cells, which were then directionally stimulated with PDGF, EGFP-ADIP accumulated at the leading edge (Fig. 1B). Collectively, these results indicate that in response to PDGF ADIP localizes at the leading edge in individually moving cells.
FIGURE 1.
Localization of ADIP in moving cells. A, localization of ADIP at the leading edge in response to PDGF. After serum starvation, NIH3T3 cells (A, panel a) and HeLa cells (A, panel b) seeded on the μ-Slide VI flow dishes were directionally stimulated with or without 30 ng/ml PDGF for 30 min from the bottom, then fixed, and stained with the anti-ADIP pAb. F-actin was labeled with fluorophore-conjugated phalloidin. B, localization of EGFP-ADIP at the leading edge in response to PDGF. NIH3T3 cells expressing EGFP-ADIP were treated as described in A. Arrowheads indicate the accumulation of ADIP. Scale bars, 10 μm. The results shown are representative of three independent experiments.
Necessity of ADIP for Formation of Leading Edge and Cell Movement
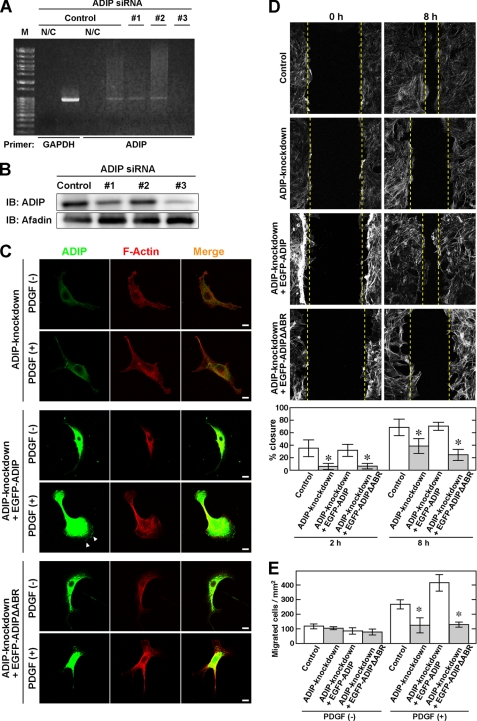
To explore the role of ADIP in the formation of the leading edge, ADIP was knocked down in NIH3T3 cells. In this experiment, we used three siRNAs, but only siRNA 3 effectively knocked down ADIP in these cells (Fig. 2, A and B). Thus, siRNA 3 was used in the following experiments. In the absence of PDGF stimulation, cell shape and F-actin organization in ADIP knockdown NIH3T3 cells did not significantly differ from those in wild-type NIH3T3 cells (Fig. 2C, upper panels; see also Fig. 1A, panel a). However, the formation of the leading edge in response to PDGF was clearly inhibited in ADIP knockdown NIH3T3 cells (Fig. 2C, lower panels; see also Fig. 1A, panel a). Next, we performed the wound healing and Boyden chamber assays to examine the involvement of ADIP in the regulation of cell movement. The closure of the wound, which was made by scratching the confluent cell monolayer, was significantly delayed in ADIP knockdown NIH3T3 cells (Fig. 2D). Similar to this result, the Boyden chamber assay showed that ADIP knockdown NIH3T3 cells were less responsive to PDGF than control cells because the number of ADIP knockdown cells crossing the membrane (migrated cells) was lower than that of control cells (Fig. 2E). These results indicate that ADIP plays a role in the formation of the leading edge and crucially regulates cell movement.
FIGURE 2.
ADIP-mediated formation of leading edge and cell movement. A and B, knockdown of ADIP in NIH3T3 cells. NIH3T3 cells were transfected with siRNA against ADIP (siRNAs 1, 2, and 3) or control siRNA (Control). Knockdown of ADIP was evaluated by RT-PCR (A) and Western blotting with the anti-ADIP pAb and the anti-afadin mAb (B). In RT-PCR experiments, GAPDH was used as an internal control. N/C, RT-PCR was conducted without the reverse transcriptase; M, molecular weight markers; IB, immunoblot. C, inhibition of the formation of the leading edge by knockdown of ADIP. After serum starvation, NIH3T3 cells transfected with siRNA against ADIP (ADIP knockdown), ADIP knockdown NIH3T3 cells expressing siRNA-resistant EGFP-ADIP, and ADIP knockdown NIH3T3 cells expressing siRNA-resistant EGFP-ADIPΔABR were seeded on the μ-Slide VI flow dishes. ΔABR indicates the lack of the afadin-binding region. These cells were directionally stimulated with 30 ng/ml PDGF for 30 min from the bottom, then fixed, and stained with the anti-ADIP pAb. F-actin was labeled with fluorophore-conjugated phalloidin. D, cell movement estimated by the wound healing assay. Confluent cell monolayers of control NIH3T3 cells, ADIP knockdown NIH3T3 cells, ADIP knockdown NIH3T3 cells expressing EGFP-ADIP, or ADIP knockdown NIH3T3 cells expressing EGFP-ADIPΔABR were manually scratched and cultured for 8 h in the presence of 30 ng/ml PDGF. F-actin was labeled with fluorophore-conjugated phalloidin to determine the cell front. The dotted lines indicate the front of moving cells. The percent wound closure at 2 and 8 h after scratching was calculated as described under “Experimental Procedures.” E, cell movement estimated by the Boyden chamber assay. The four types of cells indicated in D were incubated on cell culture inserts coated with vitronectin in the presence of 30 ng/ml PDGF in the bottom well for 12 h. The number of cells that migrated into the bottom well was counted after the cells were stained with DAPI. The error bars in D and E indicate ± S.D. *, p < 0.05 versus control. The results shown are representative of three independent experiments.
Cooperative Roles of ADIP and Afadin in Formation of Leading Edge and Cell Movement
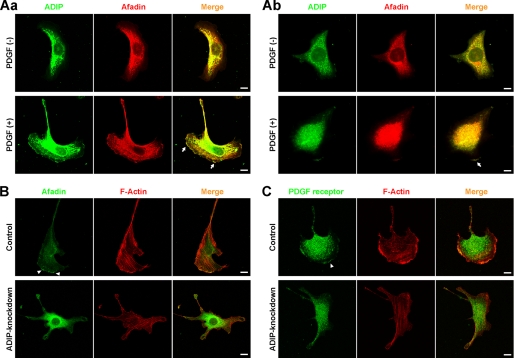
We next investigated whether the interaction of ADIP with afadin is necessary for the formation of the leading edge and for cell movement. ADIP and afadin co-localized at the leading edge of both NIH3T3 and HeLa cells, which were directionally stimulated with PDGF (Fig. 3A). We also confirmed that HeLa cells expressed PDGF receptor, integrin αvβ3, and Necl-5 and that these molecules accumulated at the leading edge in response to PDGF (supplemental Fig. S1, A and B). In control NIH3T3 cells, the leading edge formed in the direction of the PDGF stimulation, and the signals for afadin and PDGF receptor were observed at the leading edge (Fig. 3, B and C, upper panels), whereas in ADIP knockdown NIH3T3 cells, the formation of the leading edge was remarkably perturbed (Fig. 3, B and C, lower panels). The signals for afadin and PDGF receptor still remained at the tip of residual small cell protrusions, but the levels of the signal intensity were suppressed in comparison with control cells. Essentially the same results were observed for the signal for phospho-PDGF receptor (supplemental Fig. S2). The knockdown of ADIP did not alter the expression level of afadin (Fig. 2B). These results indicate that ADIP is necessary for the PDGF-induced formation of the leading edge and that there seems to be feedback regulation in the recruitment of afadin and PDGF receptor to the leading edge by ADIP.
FIGURE 3.
Localization of afadin and PDGF receptor in moving cells. A, co-localization of ADIP with afadin at the leading edge in response to PDGF. NIH3T3 cells (A, panel a) and HeLa cells (A, panel b) were directionally stimulated with 30 ng/ml PDGF for 30 min, then fixed, and stained with the anti-ADIP pAb and the anti-afadin mAb. Arrows indicate the co-localization of ADIP with afadin. B and C, impaired signals for afadin and PDGF receptor at the tip of residual small cell protrusions in ADIP knockdown NIH3T3 cells in response to PDGF. Control and ADIP knockdown NIH3T3 cells were treated as described in A. Arrowheads indicate the accumulation of afadin (B) or PDGF receptor (C). Scale bars, 10 μm. The results shown are representative of three independent experiments.
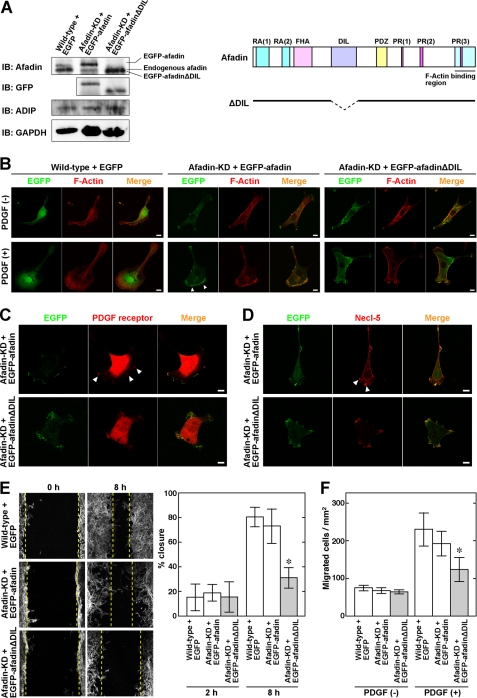
Because ADIP binds to afadin at the DIL domain, we generated afadin knockdown NIH3T3 cells expressing EGFP-afadinΔDIL, which lacks the DIL domain (Fig. 4A), to analyze the effect of the interaction of ADIP with afadin on the formation of the leading edge. In the absence of PDGF stimulation, there was no obvious difference in cell shape and F-actin organization among NIH3T3 cells expressing EGFP, afadin knockdown NIH3T3 cells expressing EGFP-afadin, and afadin knockdown NIH3T3 cells expressing EGFP-afadinΔDIL (Fig. 4B, upper panels). The expression level of ADIP was similar in these cells (Fig. 4A). After stimulation with PDGF, both NIH3T3 cells expressing EGFP and afadin knockdown NIH3T3 cells expressing EGFP-afadin formed the leading edge (Fig. 4B, lower panels). In the latter cells, EGFP-afadin accumulated at the leading edge. In contrast, afadin knockdown NIH3T3 cells expressing EGFP-afadinΔDIL formed only small cell protrusions that were independent of the direction of PDGF stimulation. Although EGFP-afadinΔDIL was detected at the tip of those protrusions, the signal intensity was low. Similarly, the signals for PDGF receptor and Necl-5 were also observed at the leading edge in afadin knockdown NIH3T3 cells expressing EGFP-afadin, whereas those signals were visible but subtle at the tip of the small protrusions in afadin knockdown NIH3T3 cells expressing EGFP-afadinΔDIL (Fig. 4, C and D). Essentially the same results were observed for the signal for phospho-PDGF receptor (supplemental Fig. S2). We further examined the cell motility of these cells and found that the wound closure in the wound healing assay and the number of cells that migrated into the bottom chamber in the Boyden chamber assay were significantly suppressed in afadin knockdown NIH3T3 cells expressing EGFP-afadinΔDIL (Fig. 4, E and F). Collectively, these results indicate that the interaction of ADIP with afadin is important for the formation of the leading edge and consequent cell movement.
FIGURE 4.
Necessity of interaction of ADIP with afadin for formation of leading edge and cell movement. A, expression of EGFP-tagged afadin mutants in afadin knockdown NIH3T3 cells. To examine the expression level of afadin mutants and ADIP, cell lysates were subjected to Western blotting. The expression of GAPDH was detected as a loading control. KD, afadin knockdown; IB, immunoblot. A schematic representation of the afadin mutant that lacks the dilute domain (ΔDIL) is shown on the right. FHA, forkhead-associated domain; PDZ, PDZ domain; RA, Ras association domain; PR, proline-rich domain. B, impaired formation of the leading edge in afadin knockdown NIH3T3 cells expressing EGFP-afadinΔDIL. After serum starvation, each NIH3T3 cell type seeded on the μ-Slide VI flow dishes was directionally stimulated with or without 30 ng/ml PDGF for 30 min from the bottom. Cells were fixed, and F-actin was labeled with fluorophore-conjugated phalloidin. Arrowheads indicate the accumulation of EGFP-afadin or EGFP-afadinΔDIL. C and D, impaired signals for PDGF receptor and Necl-5 at the tip of residual small cell protrusions in afadin knockdown NIH3T3 cells expressing EGFP-afadinΔDIL. After serum starvation, afadin knockdown NIH3T3 cells expressing EGFP-afadin or EGFP-afadinΔDIL were seeded on the μ-Slide VI flow dishes and then directionally stimulated with 30 ng/ml PDGF for 30 min from the bottom. Cells were fixed and stained with the anti-PDGF receptor mAb or the anti-Necl-5 mAb. Arrowheads indicate the accumulation of PDGF receptor (C) or Necl-5 (D). Scale bars, 10 μm. E, cell movement estimated by the wound healing assay. Confluent cell monolayers of each NIH3T3 cell type were manually scratched and cultured for 8 h in the presence of 30 ng/ml PDGF. F-actin was labeled with fluorophore-conjugated phalloidin to determine the cell front. The dotted lines indicate the front of moving cells. The percent wound closure at 2 and 8 h after scratching was calculated as described under “Experimental Procedures.” F, cell movement estimated by the Boyden chamber assay. Each NIH3T3 cell type was incubated on cell culture inserts coated with vitronectin in the presence of 30 ng/ml PDGF in the bottom well for 12 h. The number of cells that migrated into the bottom well was counted after the cells were stained with DAPI. The error bars in E and F indicate ± S.D. *, p < 0.05 versus wild type + GFP. The results shown are representative of three independent experiments.
This importance was additionally confirmed by use of the ADIP mutant (ADIPΔABR) that does not contain the afadin-binding region (supplemental Fig. S3, A and B). In the absence of PDGF stimulation, the cell morphology in ADIP knockdown NIH3T3 cells expressing EGFP-ADIP and ADIP knockdown NIH3T3 cells expressing EGFP-ADIPΔABR resembled that in ADIP knockdown cells (Fig. 2C). When these cells were stimulated with PDGF, ADIP knockdown NIH3T3 cells expressing EGFP-ADIP formed the leading edge in the direction of the PDGF stimulation, and EGFP-ADIP accumulated at the leading edge. In contrast, ADIP knockdown NIH3T3 cells expressing EGFP-ADIPΔABR did not form the obvious leading edge, and the accumulation of EGFP-ADIPΔABR at the tip of cell protrusions was poor. The wound healing and Boyden chamber assays also showed that expression of EGFP-ADIP in ADIP knockdown NIH3T3 cells restored the cell motility but that expression of EGFP-ADIPΔABR did not (Fig. 2, D and E).
Involvement of Rac and Vav2 in ADIP-mediated Formation of Leading Edge
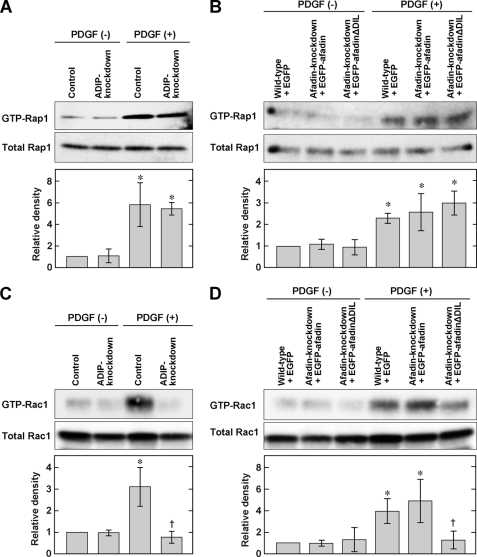
Recently, we demonstrated that the activation of Rap1, a small G protein, is important for the formation of the leading edge and is regulated by afadin (16, 18). In this study, we investigated whether ADIP itself or the interaction of ADIP with afadin is involved in the activation of Rap1. Neither knockdown of ADIP nor expression of EGFP-afadinΔDIL in afadin knockdown cells significantly altered the level of GTP-bound activated Rap1 in response to PDGF (Fig. 5, A and B), indicating that ADIP has no effect on the regulation of Rap1 activity. Next, we examined whether ADIP regulates the activation of another small G protein, Rac, which is also essential for the formation of the leading edge by producing lamellipodial cell protrusions. We found that knockdown of ADIP significantly decreased the level of GTP-bound Rac following PDGF stimulation (Fig. 5C) and that the effect of ADIP on the activation of Rac was similar to that of afadin (supplemental Fig. S3C). Expression of full-length afadin in afadin knockdown NIH3T3 cells restored the activation of Rac, whereas expression of afadinΔDIL in such cells did not (Fig. 5D). Although we showed previously that the PDGF-induced inhibition of RhoA was perturbed by knockdown of afadin (16), ADIP is not likely to be involved in this perturbation because neither knockdown of ADIP nor expression of EGFP-afadinΔDIL in afadin knockdown cells increased the activation of RhoA in response to PDGF (supplemental Fig. S3, D and E). These results indicate that among the small G proteins Rap1, Rac, and RhoA, ADIP specifically regulates the activation of Rac by interacting with afadin in NIH3T3 cells directionally stimulated with PDGF.
FIGURE 5.
ADIP-mediated Rac activation. A, no effect of ADIP on the activation of Rap1 in NIH3T3 cells stimulated with PDGF. At 4 h after serum starvation, control or ADIP knockdown NIH3T3 cells were stimulated with or without 15 ng/ml PDGF for 1 min. Cell lysates were used for the pulldown assay and subjected to Western blotting using the anti-Rap1 pAb. B, no effect of the ADIP-afadin interaction on the activation of Rap1 in NIH3T3 cells stimulated with PDGF. The assay was performed as described in A. C, necessity of ADIP for the activation of Rac in NIH3T3 cells stimulated with PDGF. Cells were treated as described in A. Cell lysates from each NIH3T3 cell type were used for the pulldown assay and subjected to Western blotting using the anti-Rac1 mAb. D, necessity of the ADIP-afadin interaction for the activation of Rac1 in NIH3T3 cells stimulated with PDGF. The assay was performed as described in C. The bar graphs represent the relative density of GTP-bound Rap1 or GTP-bound Rac1 normalized to the total amount of Rap1 or Rac1, respectively. These values were compared with the value of the control NIH3T3 cells or wild-type NIH3T3 cells expressing EGFP without PDGF stimulation, which is expressed as 1. The error bars indicate ±S.D. *, p < 0.05 versus control without PDGF stimulation in A and C or wild type + GFP without PDGF stimulation in B and D. †, p < 0.05 versus wild type + GFP with PDGF stimulation. The results shown are representative of three independent experiments.
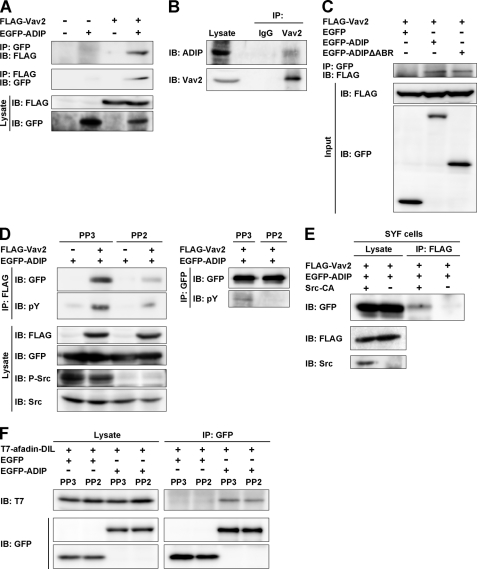
We further examined the mechanism by which ADIP regulates the activation of Rac. The activation of Rac is directly regulated by GDP/GTP exchange factors, such as Vav2 (30). To test our assumption that ADIP regulates the activation of Rac through Vav2, we performed a co-immunoprecipitation assay by transfecting FLAG-Vav2 and EGFP-ADIP expression vectors into HEK293 cells and found that these molecules were co-immunoprecipitated (Fig. 6A). This co-immunoprecipitation was confirmed endogenously using the lysates of HeLa cells (Fig. 6B). The level of Vav2 co-immunoprecipitated with ADIP or ADIPΔABR was similar (Fig. 6C), suggesting no involvement of the afadin-ADIP interaction in this co-immunoprecipitation. On the other hand, the interaction of ADIP with Vav2 was dependent on the Src-mediated tyrosine phosphorylation of these molecules because in the presence of PP2, a specific Src inhibitor, the phosphorylation of these two molecules as well as Src was impaired, and the level of co-immunoprecipitation was markedly reduced (Fig. 6D). The importance of Src for the ADIP-Vav2 interaction was confirmed using SYF cells (Fig. 6E). These cells are deficient for the Src family kinases, including Src, Yes, and Fyn (31). In addition, interaction of ADIP with afadin was independent of the activation state of Src (Fig. 6F). Taken together, these results indicate that ADIP promotes the activation of Rac by interacting with both afadin and Vav2 for the formation of the leading edge in moving cells.
FIGURE 6.
Interaction of ADIP with Vav2 in Src phosphorylation-dependent manner. A, co-immunoprecipitation of ADIP with Vav2. Lysates of HEK293 cells transiently expressing FLAG-Vav2 and/or EGFP-ADIP were immunoprecipitated with anti-FLAG M2-agarose or anti-GFP-agarose beads followed by Western blotting with the anti-FLAG mAb or the anti-GFP pAb. B, co-immunoprecipitation of endogenous ADIP with Vav2. Lysates of HeLa cells were immunoprecipitated with the anti-Vav2 pAb or preimmune IgG as a control followed by Western blotting with the anti-Vav2 and the anti-ADIP pAbs. C, no requirement of the afadin-binding region of ADIP for the interaction of ADIP with Vav2. Lysates of HEK293 cells transiently expressing FLAG-Vav2 with EGFP, EGFP-ADIP, or EGFP-ADIPΔABR were immunoprecipitated with anti-GFP-agarose beads followed by Western blotting with the anti-FLAG mAb or the anti-GFP pAb. D, inhibition of the co-immunoprecipitation of ADIP with Vav2 by a Src inhibitor. HEK293 cells transiently expressing FLAG-Vav2 and/or EGFP-ADIP were pretreated with the Src inhibitor PP2 or the inactive analog PP3 for 1 h. Cells were lysed followed by immunoprecipitation with anti-FLAG M2-agarose or anti-GFP-agarose beads. The immunoprecipitates were subjected to Western blotting with the indicated Abs. E, no co-immunoprecipitation of ADIP with Vav2 in SYF cells. Lysates of SYF cells transiently expressing FLAG-Vav2 and EGFP-ADIP with or without the constitutively active form of Src (Src-CA) were immunoprecipitated with anti-FLAG M2-agarose beads. The immunoprecipitates were subjected to Western blotting with the indicated Abs. F, no requirement of the Src-mediated phosphorylation of ADIP for the interaction of ADIP with afadin. Lysates of HEK293 cells transiently expressing T7-afadin-DIL with EGFP or EGFP-ADIP in the presence of PP2 or PP3 were immunoprecipitated with anti-GFP-agarose beads followed by Western blotting with the anti-T7 mAb or the anti-GFP pAb. The results shown are representative of three independent experiments. IP, immunoprecipitation; IB, immunoblot; P-Src, phosphorylated Src; pY, phosphotyrosine.
DISCUSSION
It is well established that the function of Rho family small G proteins is crucial for cell movement by rearranging the actin cytoskeleton and integrating various signaling networks (4, 32). Several scaffold proteins, kinases, and phosphatases as well as F-actin-binding proteins are involved in such networks. We show here for the first time that the scaffold protein ADIP regulates cell movement in cooperation with afadin. We also found the molecular relationship among ADIP; afadin; and small G proteins Rap1, Rac, and RhoA in the formation of the leading edge and cell movement in response to PDGF. Afadin enhances the activation of Rac through Vav2 and inhibits the activation of RhoA through ARAP1, a GTPase-activating protein for RhoA (16), whereas ADIP only mediates the activation of Rac. Afadin regulates the PDGF-induced, Crk- and C3G-mediated activation of Rap1 and directly interacts with activated Rap1 to stabilize the localization of the ternary complex containing PDGF receptor, integrin αvβ3, and Necl-5 at the leading edge (13, 16), whereas ADIP has no effect on the activation of Rap1. Thus, afadin and ADIP play distinct roles in the intracellular signaling for cell movement, although these proteins co-localize at the leading edge and are reported to directly interact with each other (19). In contrast, the interaction of ADIP with afadin is necessary for the activation of Rac and the subsequent enhancement of cell movement because cells expressing the afadin or ADIP mutant that lacks the region necessary for binding to the other partner showed impaired activation of Rac, delayed wound closure, and a decreased number of cells detected in the lower chamber in the Boyden chamber assay. Besides PDGF, other growth factors, such as fibroblast growth factor (FGF), also promote cell movement (33). However, ADIP does not seem to mediate the FGF-induced signaling for cell movement because knockdown of ADIP did not significantly affect the formation of the leading edge in response to FGF (data not shown). These data suggest that FGF activates signaling pathways in which ADIP is not involved for cell movement.
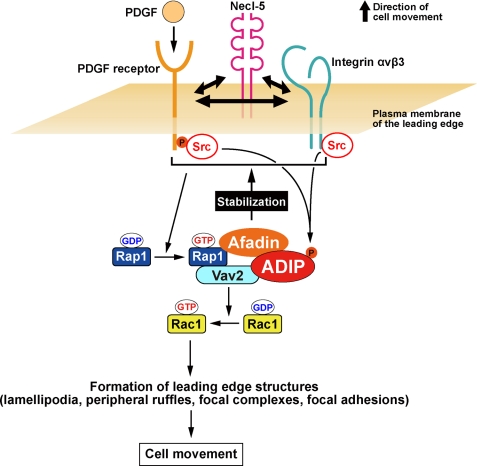
The molecular mechanism by which ADIP mediates the activation of Rac through Vav2 is also elucidated in this study. Vav2 performs its exchange activity of GDP to GTP on Rac by its tyrosine phosphorylation (34). The structural analysis of Vav proteins has revealed that at the resting state the GDP/GTP exchange factor activity of Vav is inhibited by its N-terminal arm, which occludes the kinase domain, called the Dbl homology domain. When the tyrosine residue (Tyr-174) in the N-terminal arm of Vav is phosphorylated by activated Src, the N-terminal arm is released from the Dbl homology domain, resulting in the exposure of the GDP/GTP exchange factor activity site and the activation of Rac. Thus, the tyrosine phosphorylation of Vav2 is required for its activation. However, the tyrosine phosphorylation alone is not likely to be sufficient for maximal activation of Vav2 on Rac, and other Vav2-interacting proteins appear to be necessary (35). It has been reported that Rap1 interacts with Vav2 and is involved in Vav2-mediated activation of Rac at the edge of spreading cells (36). We found that ADIP is a novel Vav2-interacting protein necessary for the activation of Rac and that this interaction is dependent on the Src-mediated phosphorylation of Vav2 and/or ADIP because the association of ADIP with Vav2 was markedly reduced in the presence of the Src inhibitor PP2 and in Src-deficient cells. Src has been shown to be activated by PDGF receptor and integrin αvβ3 (37, 38). Taken together with previous and our present results, ADIP, afadin, and Rap1 play important roles in the enhancement of the GDP/GTP exchange activity of Vav2 on Rac by forming a complex with Vav2 at the leading edge of moving cells. The relationship of these molecules for the regulation of cell movement is depicted in Fig. 7.
FIGURE 7.
Schematic model for ADIP-mediated regulation of cell movement. For details, see the “Discussion.”
Individually moving and proliferating cells gradually form cell-cell adhesions by contacting with neighboring cells to establish the epithelial tissue. This phenomenon is known as mesenchymal-epithelial transition. After the formation of mature cell-cell junctions, cells cease movement and proliferation. In contrast, under certain physiological and pathological conditions, such as embryonic development and cancer progression, disruption of cell-cell junctions preferentially occurs. Thus, cells under those conditions lose their connection with neighboring cells and restart migration and proliferation. This phenomenon is called epithelial-mesenchymal transition. The dynamic regulation of mesenchymal-epithelial transition and epithelial-mesenchymal transition is crucial for the maintenance of tissue functions, leading to the survival of multicellular organisms. Interestingly, the same molecules are important for and often seamlessly participate in both cell movement and intercellular junction formation. For example, integrin αvβ3 accumulates at the leading edge of moving cells and enhances cell movement together with PDGF receptor and Necl-5 as described above. Integrin αvβ3 also plays a key role in the formation of nectin-initiated cell-cell junctions by supporting the activation of intracellular molecules that reorganize the actin cytoskeleton, promoting the cadherin-based formation of AJs (39). In addition to integrin αvβ3, the cell-cell adhesion molecule occludin and the scaffold proteins IQGAP and vinculin have been reported to regulate both cell movement and cell-cell junctions (40–44). Our previous and present data also demonstrate that afadin and ADIP are involved in cell movement and cell-cell junctions (13, 19, 45). For the physiological regulation of mesenchymal-epithelial transition and epithelial-mesenchymal transition, it may be advantageous that some molecules mutually function in cell movement and cell-cell junctions.
Another line of evidence indicates that the human orthologue of ADIP (human ADIP) is the synovial sarcoma X breakpoint 2-interacting protein based on its interaction with SSX2 (46). The expression of SSX2 is usually very low in normal tissues except for the testis but is up-regulated in various types of cancer, including melanoma (47). In contrast to SSX2, human ADIP (synovial sarcoma X breakpoint 2-interacting protein) as well as mouse ADIP is expressed in a wide variety of normal tissues (19, 46). Human ADIP was also identified as a leukemia-associated antigen and was detected in 33% of patients with acute myeloid leukemia but not in normal donors (48). However, the function of ADIP and its molecular mechanism in acute myeloid leukemia have yet to be investigated in detail. Although we demonstrated the role of ADIP in cell-cell junction and cell movement, the pathological significance of ADIP is largely unknown. Thus, further studies are required to reveal the clinical implication of ADIP, especially in malignancies, including acute myeloid leukemia, which would contribute to the development of novel diagnosis and therapy against these diseases.
Supplementary Material
This work was supported by grants-in-aid for scientific research and by the Targeted Proteins Research Program from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (2010 and 2011); The Uehara Memorial Foundation; and the Japan Foundation for Applied Enzymology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- Necl-5
- nectin-like molecule 5
- F-actin
- actin filament
- AJ
- adherens junction
- Ab
- antibody
- pAb
- polyclonal antibody
- ADIP
- afadin dilute domain-interacting protein
- DIL
- dilute
- EGFP
- enhanced green fluorescent protein
- ABR
- afadin-binding region.
REFERENCES
- 1. Affolter M., Weijer C. J. (2005) Dev. Cell 9, 19–34 [DOI] [PubMed] [Google Scholar]
- 2. Aman A., Piotrowski T. (2010) Dev. Biol. 341, 20–33 [DOI] [PubMed] [Google Scholar]
- 3. Friedl P., Wolf K. (2003) Nat. Rev. Cancer 3, 362–374 [DOI] [PubMed] [Google Scholar]
- 4. Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003) Science 302, 1704–1709 [DOI] [PubMed] [Google Scholar]
- 5. Abercrombie M., Heaysman J. E., Pegrum S. M. (1970) Exp. Cell Res. 60, 437–444 [DOI] [PubMed] [Google Scholar]
- 6. Condeelis J. (1993) Annu. Rev. Cell Biol. 9, 411–444 [DOI] [PubMed] [Google Scholar]
- 7. Zaidel-Bar R., Cohen M., Addadi L., Geiger B. (2004) Biochem. Soc. Trans. 32, 416–420 [DOI] [PubMed] [Google Scholar]
- 8. Kiosses W. B., Shattil S. J., Pampori N., Schwartz M. A. (2001) Nat. Cell Biol. 3, 316–320 [DOI] [PubMed] [Google Scholar]
- 9. Rönnstrand L., Heldin C. H. (2001) Int. J. Cancer 91, 757–762 [DOI] [PubMed] [Google Scholar]
- 10. Woodard A. S., García-Cardeña G., Leong M., Madri J. A., Sessa W. C., Languino L. R. (1998) J. Cell Sci. 111, 469–478 [DOI] [PubMed] [Google Scholar]
- 11. Amano H., Ikeda W., Kawano S., Kajita M., Tamaru Y., Inoue N., Minami Y., Yamada A., Takai Y. (2008) Genes Cells 13, 269–284 [DOI] [PubMed] [Google Scholar]
- 12. Minami Y., Ikeda W., Kajita M., Fujito T., Amano H., Tamaru Y., Kuramitsu K., Sakamoto Y., Monden M., Takai Y. (2007) J. Biol. Chem. 282, 18481–18496 [DOI] [PubMed] [Google Scholar]
- 13. Miyata M., Ogita H., Komura H., Nakata S., Okamoto R., Ozaki M., Majima T., Matsuzawa N., Kawano S., Minami A., Waseda M., Fujita N., Mizutani K., Rikitake Y., Takai Y. (2009) J. Cell Sci. 122, 4319–4329 [DOI] [PubMed] [Google Scholar]
- 14. Mandai K., Nakanishi H., Satoh A., Obaishi H., Wada M., Nishioka H., Itoh M., Mizoguchi A., Aoki T., Fujimoto T., Matsuda Y., Tsukita S., Takai Y. (1997) J. Cell Biol. 139, 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takai Y., Miyoshi J., Ikeda W., Ogita H. (2008) Nat. Rev. Mol. Cell Biol. 9, 603–615 [DOI] [PubMed] [Google Scholar]
- 16. Miyata M., Rikitake Y., Takahashi M., Nagamatsu Y., Yamauchi Y., Ogita H., Hirata K., Takai Y. (2009) J. Biol. Chem. 284, 24595–24609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nagamatsu Y., Rikitake Y., Takahashi M., Deki Y., Ikeda W., Hirata K., Takai Y. (2008) J. Biol. Chem. 283, 14532–14541 [DOI] [PubMed] [Google Scholar]
- 18. Takahashi M., Rikitake Y., Nagamatsu Y., Hara T., Ikeda W., Hirata K., Takai Y. (2008) Genes Cells 13, 549–569 [DOI] [PubMed] [Google Scholar]
- 19. Asada M., Irie K., Morimoto K., Yamada A., Ikeda W., Takeuchi M., Takai Y. (2003) J. Biol. Chem. 278, 4103–4111 [DOI] [PubMed] [Google Scholar]
- 20. Kawakatsu T., Ogita H., Fukuhara T., Fukuyama T., Minami Y., Shimizu K., Takai Y. (2005) J. Biol. Chem. 280, 4940–4947 [DOI] [PubMed] [Google Scholar]
- 21. Kanzaki N., Ogita H., Komura H., Ozaki M., Sakamoto Y., Majima T., Ijuin T., Takenawa T., Takai Y. (2008) J. Cell Sci. 121, 2008–2017 [DOI] [PubMed] [Google Scholar]
- 22. Sakisaka T., Nakanishi H., Takahashi K., Mandai K., Miyahara M., Satoh A., Takaishi K., Takai Y. (1999) Oncogene 18, 1609–1617 [DOI] [PubMed] [Google Scholar]
- 23. Ikeda W., Kakunaga S., Itoh S., Shingai T., Takekuni K., Satoh K., Inoue Y., Hamaguchi A., Morimoto K., Takeuchi M., Imai T., Takai Y. (2003) J. Biol. Chem. 278, 28167–28172 [DOI] [PubMed] [Google Scholar]
- 24. Yatohgo T., Izumi M., Kashiwagi H., Hayashi M. (1988) Cell Struct. Funct. 13, 281–292 [DOI] [PubMed] [Google Scholar]
- 25. Ikeda W., Kakunaga S., Takekuni K., Shingai T., Satoh K., Morimoto K., Takeuchi M., Imai T., Takai Y. (2004) J. Biol. Chem. 279, 18015–18025 [DOI] [PubMed] [Google Scholar]
- 26. Goldfinger L. E., Hopkinson S. B., deHart G. W., Collawn S., Couchman J. R., Jones J. C. (1999) J. Cell Sci. 112, 2615–2629 [DOI] [PubMed] [Google Scholar]
- 27. Fujito T., Ikeda W., Kakunaga S., Minami Y., Kajita M., Sakamoto Y., Monden M., Takai Y. (2005) J. Cell Biol. 171, 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fukuyama T., Ogita H., Kawakatsu T., Fukuhara T., Yamada T., Sato T., Shimizu K., Nakamura T., Matsuda M., Takai Y. (2005) J. Biol. Chem. 280, 815–825 [DOI] [PubMed] [Google Scholar]
- 29. Nakata S., Fujita N., Kitagawa Y., Okamoto R., Ogita H., Takai Y. (2007) J. Biol. Chem. 282, 37815–37825 [DOI] [PubMed] [Google Scholar]
- 30. Hornstein I., Alcover A., Katzav S. (2004) Cell. Signal. 16, 1–11 [DOI] [PubMed] [Google Scholar]
- 31. Klinghoffer R. A., Sachsenmaier C., Cooper J. A., Soriano P. (1999) EMBO J. 18, 2459–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jaffe A. B., Hall A. (2005) Annu. Rev. Cell Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 33. Nugent M. A., Iozzo R. V. (2000) Int. J. Biochem. Cell Biol. 32, 115–120 [DOI] [PubMed] [Google Scholar]
- 34. Aghazadeh B., Lowry W. E., Huang X. Y., Rosen M. K. (2000) Cell 102, 625–633 [DOI] [PubMed] [Google Scholar]
- 35. Bustelo X. R. (2001) Oncogene 20, 6372–6381 [DOI] [PubMed] [Google Scholar]
- 36. Arthur W. T., Quilliam L. A., Cooper J. A. (2004) J. Cell Biol. 167, 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arias-Salgado E. G., Lizano S., Sarkar S., Brugge J. S., Ginsberg M. H., Shattil S. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13298–13302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kypta R. M., Goldberg Y., Ulug E. T., Courtneidge S. A. (1990) Cell 62, 481–492 [DOI] [PubMed] [Google Scholar]
- 39. Sakamoto Y., Ogita H., Hirota T., Kawakatsu T., Fukuyama T., Yasumi M., Kanzaki N., Ozaki M., Takai Y. (2006) J. Biol. Chem. 281, 19631–19644 [DOI] [PubMed] [Google Scholar]
- 40. Du D., Xu F., Yu L., Zhang C., Lu X., Yuan H., Huang Q., Zhang F., Bao H., Jia L., Wu X., Zhu X., Zhang X., Zhang Z., Chen Z. (2010) Dev. Cell 18, 52–63 [DOI] [PubMed] [Google Scholar]
- 41. le Duc Q., Shi Q., Blonk I., Sonnenberg A., Wang N., Leckband D., de Rooij J. (2010) J. Cell Biol. 189, 1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. White C. D., Brown M. D., Sacks D. B. (2009) FEBS Lett. 583, 1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ziegler W. H., Liddington R. C., Critchley D. R. (2006) Trends Cell Biol. 16, 453–460 [DOI] [PubMed] [Google Scholar]
- 44. Tsukita S., Furuse M. (1999) Trends Cell Biol. 9, 268–273 [DOI] [PubMed] [Google Scholar]
- 45. Sato T., Fujita N., Yamada A., Ooshio T., Okamoto R., Irie K., Takai Y. (2006) J. Biol. Chem. 281, 5288–5299 [DOI] [PubMed] [Google Scholar]
- 46. de Bruijn D. R., dos Santos N. R., Kater-Baats E., Thijssen J., van den Berk L., Stap J., Balemans M., Schepens M., Merkx G., van Kessel A. G. (2002) Genes Chromosomes Cancer 34, 285–298 [DOI] [PubMed] [Google Scholar]
- 47. Türeci O., Sahin U., Schobert I., Koslowski M., Scmitt H., Schild H. J., Stenner F., Seitz G., Rammensee H. G., Pfreundschuh M. (1996) Cancer Res. 56, 4766–4772 [PubMed] [Google Scholar]
- 48. Guinn B. A., Bland E. A., Lodi U., Liggins A. P., Tobal K., Petters S., Wells J. W., Banham A. H., Mufti G. J. (2005) Biochem. Biophys. Res. Commun. 335, 1293–1304 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.